Abstract
In conventionally-expressed eukaryotic genes, transcription start sites (TSSs) can be identified by mapping the mature mRNA 5′-terminal sequence onto the genome. However, this approach is not applicable to genes that undergo pre-mRNA 5′-leader trans-splicing (SL trans-splicing) because the original 5′-segment of the primary transcript is replaced by the spliced leader sequence during the trans-splicing reaction and is discarded. Thus TSS mapping for trans-spliced genes requires different approaches. We describe two such approaches and show that they generate precisely agreeing results for an SL trans-spliced gene encoding the muscle protein troponin I in the ascidian tunicate chordate Ciona intestinalis. One method is based on experimental deletion of trans-splice acceptor sites and the other is based on high-throughput mRNA 5′-RACE sequence analysis of natural RNA populations in order to detect minor transcripts containing the pre-mRNA’s original 5′-end. Both methods identified a single major troponin I TSS located ∼460 nt upstream of the trans-splice acceptor site. Further experimental analysis identified a functionally important TATA element 31 nt upstream of the start site. The two methods employed have complementary strengths and are broadly applicable to mapping promoters/TSSs for trans-spliced genes in tunicates and in trans-splicing organisms from other phyla.
INTRODUCTION
Identification and mapping of promoters is a key aspect of gene regulatory studies. Precise localization of promoters requires determination of transcription start sites (TSSs). In conventionally-expressed eukaryotic genes TSSs can be precisely mapped by 5′-RACE analysis or other methods to determine the nucleotide sequence at the 5′-end of the mature mRNA (1). Mapping the mRNA 5′-sequence onto the genome precisely localizes the TSS because the first nucleotide at the 5′-end of the nascent pre-mRNA transcript is capped with m7G early during gene transcription and is maintained as the 5′-end of the mature mRNA. However, this simple and direct approach of mature mRNA 5′ sequence analysis is not applicable to TSS mapping in genes that undergo pre-mRNA spliced-leader (SL) trans-splicing.
SL trans-splicing, a gene expression mechanism found in several but not all animal and protist phyla, consists of the spliceosomal transfer of the 5′-segment of a specialized donor RNA, the SL RNA, to unpaired splice acceptor sites in the 5′-region of target pre-mRNAs (2–4). In those species in which SL trans-splicing occurs, a significant fraction, in some cases the majority, of the total gene number produce trans-spliced mRNAs. The net result of SL trans-splicing is the loss of the outron, i.e. the 5′-capped pre-mRNA’s original 5′-segment upstream of the trans-splice acceptor site (5), and its replacement by the 5′-capped spliced leader (SL) sequence. Because the outron is discarded, the 5′-end of a mature trans-spliced mRNA has lost the sequence information that would otherwise permit TSS mapping. Thus TSS mapping for trans-spliced genes requires alternative approaches.
A plausible general approach to determining TSSs for trans-spliced genes is 5′-end sequence analysis, not of the mature mRNA, but of the discarded outron, or of transcripts in which the outron is retained. Outron-retaining transcripts could be generated by experimental genetics approaches that avoid or block SL trans-splicing or, alternatively, they may exist at low levels in natural RNA preparations as occasional failures of trans-splicing or as pre-mRNAs that have not yet undergone trans-splicing. Experimental genetic approaches and natural minor transcript analyses have both been applied to TSS mapping in previous studies of several nematode trans-spliced genes (detailed below). We report here the development and application of new and general methods based on both experimental genetics, and natural minor transcript analyses, and show that these distinct approaches give precisely concordant results on the trans-spliced troponin I gene (CiTnI) of the ascidian tunicate chordate Ciona intestinalis.
Previous studies have established candidate TSSs for several trans-spliced genes, all in nematodes. TSSs for the Onchocerca volvulus superoxide dismutase genes Ov-sod-1, -2, and -3, and glutathione-S-transferase gene Ov-GST1a, were mapped in studies in which trans-splicing was experimentally avoided by transcription of recombinant gene constructs in transfected heterologous cell cultures (6), or in vitro in heterologous nuclear extracts (7), derived from species in which SL trans-splicing does not occur, i.e. Homo sapiens or Drosophila melanogaster. The 5′-ends of the non-trans-spliced transcripts thus produced were mapped by primer-extension reverse transcription, thus identifying TSSs (6,7), albeit with significant caveats relating to in vitro methodology and to uncertain cross-phylum transcriptional fidelity. An alternative experimental genetic approach that could block trans-splicing without resorting to heterologous or in vitro transcription systems is suggested by a study of the rol-6 collagen gene of the nematode Caenorhabditis elegans. Experimental insertion of a splice donor site into the outron of that gene blocked trans-splicing of the modified gene in transgenic nematodes by preferentially driving cis-splicing from the inserted donor site to the acceptor site normally used for trans-splicing (8). This led to the production of rol-6 transcripts that retained the 5′-segment of the wild-type gene’s outron, including the pre-mRNA’s original 5′-end, which was already known from minor transcript analysis (see below). This approach of inserting donor sites into trans-spliced gene outrons has not been used to map unknown TSSs, but it, or other experimental genetic modifications that might similarly block trans-splicing of the modified gene, could in principle be used for that purpose, and we have developed and applied such a method.
Analysis of low-abundance naturally occurring outron-retaining transcripts has been used to determine TSSs for several trans-spliced Caenorhabditis genes: ubiquitin UbiA (9), and collagens col-13 (10) and rol-6 (11). These minor transcripts that presumably represent either trans-splicing failures or not-yet-trans-spliced molecules were identified and characterized through primer-extension reverse-transcription analysis of RNA preparations from normal animals. Given the recent development of high-throughput sequencing methods for mRNA 5′-ends (12), the direct detection of rare non-trans-spliced RNA molecules, or of discarded outrons, in the natural RNA population could represent a convenient approach for the high-throughput definition of TSSs for trans-spliced genes. We report here the use of high-throughput 5′-RACE sequence data for mapping the TSS of a trans-spliced gene.
We describe here studies of the trans-spliced C. intestinalis CiTnI gene in which both experimental molecular genetic studies and analysis of naturally occurring minor transcripts were used to identify the TSS. We eliminated trans-splicing of CiTnI experimental constructs in a Ciona gene expression system using a novel and simple method based on deletion of the trans-splice acceptor site and associated branchpoint. Analysis of the resulting non-trans-spliced transcripts by 5′-RACE identified a TSS ∼460 bp upstream of the trans-splice acceptor site. We also searched a data set of C. intestinalis mRNA oligocapping 5′-RACE 5′-sequence tags generated by high-throughput sequencing of normal tailbud embryos. Although the vast majority of CiTnI transcripts revealed in this analysis were, as expected, trans-spliced, a minority ∼2–3% were non-trans-spliced and the 5′-ends of most of these mapped to exactly the same site we had previously determined as the TSS by experimental molecular genetics, confirming the use of that TSS in the endogenous CiTnI gene.
The precise agreement of the two approaches is an important cross-validation of both methods for TSS determination. Thus this study defines a powerful set of techniques, each with distinct strengths, to be exploited in future studies of TSSs in Ciona or in other organisms that carry out SL trans-splicing. Cross-methodology agreement also strengthens confidence in the CiTnI TSS determination that is the first for a trans-spliced chordate gene. Identification of the TSS allowed us to perform further experiments to show that the CiTnI promoter includes a functionally important TATA element 31 nt upstream, and that no essential transcriptional control elements reside within the transcribed part of the gene. In addition it permitted a comparison with the previously determined TSSs of two non-trans-spliced ascidian actin genes, which revealed a number of similarities.
MATERIALS AND METHODS
DNA constructs
Experimental gene constructs were based on the 5′-part of an allele of the CiTnI troponin I gene of C. intestinalis [GenBank AF237978 (13–15)] isolated by PCR amplification from an Atlantic coast (Massachusetts, USA) animal. This differs slightly from the allele represented in the assembled genome sequence [version 1,(16)] derived from a Pacific coast (California, USA) animal. All coordinates reported in this paper refer to the ‘Atlantic’ allele except where indicated by inclusion of the term ‘(P)’ to identify ‘Pacific’ allele coordinates. Table 1 summarizes the location of key gene features in the two alleles with reference to the ATG translation start codon.
Table 1.
ATG-based coordinates of CiTnI gene features in ‘Atlantic’ (AF237978.2) and ‘Pacific’ [version 1 genome assembly (16)] alleles
| Atlantic | Pacific | |
|---|---|---|
| Trans-splice acceptor nucleotide | −64 | −58(P) major site, with satellite at −55(P) |
| Transcription start site (determined in this study) | −523 | −521(P) |
| Length of outron (determined in this study) | 459 nt | 463 nt |
Vector and LacZ reporter backbone
All constructs were based on the plasmid vector pBluescript II SK (+) (Stratagene) into whose SmaI and BamHI sites was cloned a ∼3.6 kb SmaI/BglII fragment of pSp72-1.27 (17), which contained a promoterless gene encoding a nuclear-localized derivative of Escherichia coli LacZ β-galactosidase (18). This vector/reporter backbone is termed pBnZ. CiTnI DNA segments were introduced in their natural orientations at the SmaI site in the 5′-untranslated region of the pBnZ LacZ reporter gene. All CiTnI DNA segments were derived from regions upstream of the protein-coding region. Construct names identify the first and last nucleotides of CiTnI DNA present, verified by DNA sequencing and numbered with respect to the ATG translation start codon.
Large-scale CiTnI deletions
Large-scale deletions were produced by restriction enzyme cleavage. The parent construct for this series was CiTnI(–1437/–24)nZ, previously termed 1.5 kb TnI/β-gal (15) (note revised nucleotide numbering). CiTnI restriction sites and corresponding vector ends are summarized in Table 2.
Table 2.
Restriction sites used for large-scale CiTnI deletion constructs
| Construct | Upstream CiTnI end (vector end) | Downstream CiTnI end (vector end) |
|---|---|---|
| CiTnI(−1437/−24)nZ | KpnI (KpnI) | Blunted Bpu1102I (SmaI) |
| CiTnI(−1437/−383)nZ | KpnI (KpnI) | Blunted Bpu1102I (SmaI) |
| CiTnI(−819/−383)nZ | Blunted AatII (blunted KpnI) | Blunted Bpu1102I (SmaI) |
| CiTnI(−1437/−826)nZ | KpnI (KpnI) | Blunted AatII (SmaI) |
| CiTnI(−384/−24)nZ | Blunted Bpu1102I (blunted KpnI) | Blunted Bpu1102I (SmaI) |
| CiTnI(−644/−383)nZ | Blunted XbaI (blunted KpnI) | Blunted Bpu1102I (SmaI) |
| CiTnI(−819/−641)nZ | Blunted AatII (blunted KpnI) | Blunted XbaI (SmaI) |
Smaller-scale nested end-deletion constructs
The CiTnI DNA segments in smaller-scale nested-deletion constructs were generated by PCR amplification with Pfu DNA polymerase using rightward and leftward primers containing added KpnI and Eco147I sites, respectively. Amplified fragments were cut with KpnI and cloned by insertion into KpnI/SmaI-cut pBluescript II SK (+) after which the usual ∼3.6 kb LacZ reporter gene-containing SmaI/BglII fragment was introduced into Eco147I and BamHI sites. The ‘full-length’ construct for this series was CiTnI(−836/−335)nZ and the series included 5′ deletion construct CiTnI(−639/−335)nZ and 3′ deletion constructs CiTnI(−836/−623)nZ, CiTnI(−836/−571)nZ, and CiTnI(−836/−422)nZ.
CiTnI constructs lacking the transcribed region
CiTnI(−836/−523)nZ was prepared by subcloning into pBluescript II SK (+) a 334-bp KpnI/HincII Ciona DNA fragment (−836 to −503) from CiTnI(−836/−335)nZ, cutting this plasmid intermediate at CiTnI position −523 with BccI, blunting with Klenow, cutting with KpnI and recovering the 314-bp BccI/KpnI CiTnI gene fragment. This was used in a three-way ligation with the 3.6-kb SmaI/BglII fragment of pSp72-1.27 LacZ and pBluescript II SK (+) cut with KpnI and BamHI. The TATA-element mutation CiTnI(−836/−523)mutTATAnZ was produced in parallel by the same procedure applied to a starting plasmid identical to CiTnI(−836/−335)nZ but in which the TATA element had been mutated by overlap extension PCR (19) as follows: wt sequence (-555)CTATTTAAGG(-546); mutant CTAggcAAGG.
Promoter test constucts
This series included a 151-bp segment of the H. roretzi HrMA4a actin gene [−216 to −66, numbered with respect to the TSS identified by Hikosaka et al. (20)] amplified by PCR from a cloned genomic DNA fragment kindly provided by Dr Yutaka Satou, Kyoto University. The rightward and leftward amplification primers included added KpnI and HindIII sites, respectively, and the fragment was cloned into pBnZ vector HindIII and KpnI sites. The resulting plasmid, HrnZ, contains the HrMA4a enhancer, and the LacZ reporter gene, but no eukaryotic promoter. Fragments of CiTnI DNA to be tested for promoter activity were inserted between the HrMA4a enhancer and LacZ gene, in place of the HindIII-to-SmaI region of the vector polylinker. CiTnI DNA fragments were produced by PCR amplification using rightward and leftward primers that contained added HindIII and Eco147I sites, respectively, and were joined to the HrMA4a enhancer by a HindIII/HindIII fusion and to the LacZ reporter gene by an Eco147I/SmaI fusion.
Gene expression assay
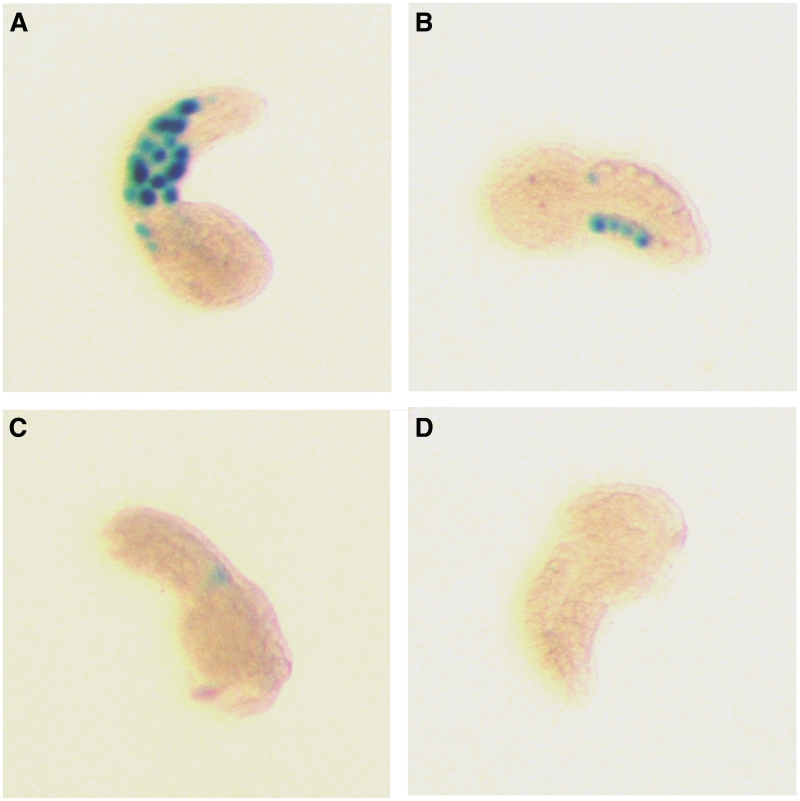
Plasmid DNA constructs (25 μg) purified using the Qiagen Plasmid Maxikit were introduced by electroporation as described by Corbo et al. (17) into dechorionated zygotes produced from adult animals obtained in Rhode Island and Massachusetts, USA. Following ∼12 h development at 18°C, normal-appearing tailbud embryos were sorted and were either fixed for histochemical detection of reporter nuclear-localized β-galactosidase by X-Gal staining (17) or RNA was extracted by lysis with sodium dodecyl sulfate, phenol and chloroform extraction and ethanol precipitation. Construct expression levels were scored as the percentage of normal embryos showing detectable X-Gal staining. This population parameter correlated very well with expression levels in individual embryos. In each X-Gal-treated embryo, β-galactosidase expression was semiquantitatively assessed as either negative, or positive in three increasing grades, i.e. + (1 or 2 faintly-stained cells), ++ (1–5 well-stained cells) and +++ (6 or more well-stained cells) (see Figure 1). We noted that with every construct showing expression in >60% of embryos, the most numerous positive staining category was +++, in every construct showing expression in 6–60% of embryos the most numerous positive staining category was ++ and in every construct showing expression in <5% of embryos the most numerous positive staining category was + (data not shown). Each construct was tested in two or more independent transformations and results were concordant and were pooled. For most constructs, including all constructs showing weak or no activity, two independent DNA preparations were analyzed. Several constructs were tested in each experimental session, at least one of which showed strong activity, thus providing a positive control in parallel for each inactive or weakly expressed construct. In general, LacZ expression was in tail muscle cells, with a very low level in some tail-adjacent cells in the trunk. None of the mutant constructs tested showed marked ectopic expression.
Figure 1.
Gene expression assay. Range of X-Gal staining intensities observed in individual tailbud embryos following zygote electroporation (17) of CiTnI/nuclear β-galactosidase reporter constructs: (A) +++, (B) ++, (C) +, (D) undetectable. DNA constructs used were CiTnI(−836/−335)nZ (A, B) and CiTnI(−836/−571)nZ (C, D). Construct gene expression levels are reported in Figures 2 and 4 as the percentage of normal-appearing embryos showing detectable staining, a population parameter that correlated well with individual embryo X-Gal staining intensities (see ‘Materials and Methods’ section).
TnI/LacZ mRNA 5′-RACE
5′-RACE PCR was performed with LacZ-specific primers for reverse transcription (5′ CGCTGATTTGTGTAGTC 3′) and leftward PCR synthesis (5′ TCACTCCAACGCAGCACCATCA 3′, and for nested reamplification 5′ ATCGCACTCCAGCCAGCTTTCC 3′) using the ‘5′ RACE System for Rapid Amplification of cDNA Ends Version 2.0′ (Invitrogen) based on oligo(dC)-tailing of first-strand cDNA with terminal deoxynucleotidyl transferase and second-strand synthesis primed by an anchor sequence linked to oligo(dI/dG).
High-throughput mRNA 5′-RACE analysis
Total RNA from mid-tailbud stage embryos produced from adults collected in Murotsu harbor, Hyogo, Japan was subjected to oligo-capping (ligation of an arbitrary 5′-anchor sequence to initially capped 5′ ends) before recovery of poly(A)+ RNA and reverse-transcription using random hexamer primers linked 3′ to an arbitrary 3′-anchor sequence (21). RNA 5′-segments were PCR-amplified with 5′ and 3′ anchor primers and the products were sequenced using the Illumina Genome Analyzer (12). Reads were aligned, at a 90% match criterion including indels, with the KH genome assembly (22) using SeqMap (23) for 34-nt reads and BLAT (24) for 46-nt reads. This data set included 5 216 665 uniquely-mapped reads (38% non-trans-spliced and the remainder SL trans-spliced; 45% 34-nt reads and the remainder 46-nt reads). A full description of this data and its application to mapping TSSs for non-trans-spliced genes will be presented elsewhere (K. Okamura et al., manuscript in preparation). Here we analyze in detail the ∼8 × 103 reads that uniquely mapped to the trans-spliced CiTnI gene.
RESULTS
Experimental molecular genetic analysis
To identify genetic elements important in transcription of the muscle-specific CiTnI gene encoding the contractile regulatory protein troponin I, we studied expression of gene constructs in which segments of genomic DNA including the CiTnI 5′-region were linked to an otherwise promoterless reporter gene encoding a nuclear-localized derivative of Escherichia coli β-galactosidase (LacZ) (18). Plasmid DNA was introduced into C. intestinalis zygotes by electroporation (17) and reporter gene expression was assessed histochemically in tailbud embryos by X-Gal staining (Figure 1). Expression levels were scored as the percentage of normally developed embryos showing detectable expression, which also correlated with expression levels within individual embryos estimated from the number of stained cells and their staining intensities (see ‘Materials and Methods’ section). In the parent construct CiTnI(−1437/−24)nZ a 1414-bp segment of Ciona CiTnI DNA (‘Atlantic’ allele, see ‘Materials and Methods’ section) from −1437 to −24 was used (-1 is the nucleotide preceding the ATG translation start codon). As we have previously reported (15), this construct (there termed CiTnI 1.5 nucLacZ) drives high-level LacZ expression and β-galactosidase activity in a large proportion (100% in Figure 2) of tailbud stage embryos, specifically in the tail muscle cells, indicating that regulatory elements sufficient to drive effective tissue-specific expression reside within this 1414-bp segment. This presumably includes both core promoter/TSS, and any necessary accessory elements such as enhancers. Because the natural CiTnI pre-mRNA undergoes SL trans-splicing at position −64 (14) we know the location of the core promoter and TSS must be upstream of that point, but the loss of the CiTnI outron that occurs during trans-splicing has to date precluded precise TSS localization.
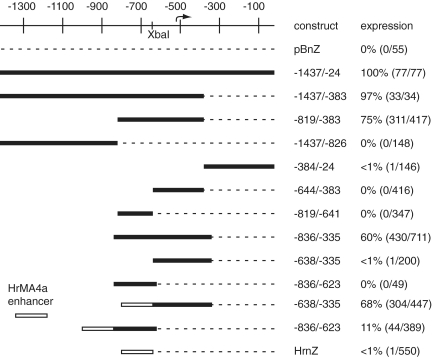
Figure 2.
Localization of CiTnI transcriptional elements through large-scale end-deletion analysis. The figure schematically depicts CiTnI gene DNA segments. ATG start codon-based coordinates are shown at the top, with the TSS mapped in the present study indicated by a curved arrow. The XbaI site at −645 to −640 is indicated. Each DNA region tested (black bars) was linked at the right end to a promoterless nuclear β-galactosidase reporter gene (dashed lines indicate the deleted CiTnI DNA downstream of the region being tested). Recombinant reporter constructs were assayed as in Figure 1. The column ‘construct’ lists the construct name or the precise range of the CiTnI DNA segment being tested and the column ‘expression’ gives the percentage of normally-developed embryos that showed detectable X-Gal staining (absolute numbers in brackets). CiTnI DNA segments −638/−335 and −836/−623 were tested with and without the HrMA4 enhancer (white bar), as indicated.
Identification of an upstream regulatory region including a promoter
From the parent construct CiTnI(−1437/−24)nZ we found that sequential deletion first of downstream region −382 to −24 (construct CiTnI(−1437/−383)nZ) and then of upstream region −1437 to −820 (construct CiTnI(−819/−383)nZ) had little effect on gene expression (Figure 2), indicating that these regions did not contain essential regulatory elements. These non-essential regions were also found to be incapable of driving reporter gene expression on their own (constructs CiTnI(−1437/−826)nZ and CiTnI(−384/−24)nZ, Figure 2). These experiments localized all essential elements, including enhancer and promoter elements, to a 437 bp regulatory region, −819 to −383. Johnson et al. (25) identified a homologous region in the orthologous C. savignyi troponin I gene (CsTnI), which they termed the minimal sufficient regulatory region (see also below).
Upon cleavage of the 437 bp CiTnI regulatory region at an XbaI restriction site we found that neither the upstream (−819 to −641) nor the downstream (−644 to −383) segments were capable of driving expression on their own (Figure 2). It seemed possible that a core promoter on one subfragment had been separated from an essential enhancer element on the other. To assess this we devised an enhancer-complementation assay for core promoter function. This assay was based on a promoter-dependent enhancer from the HrMA4a muscle actin gene from the distantly-related ascidian H. roretzi. (The HrMA4a gene, like C. intestinalis muscle actin genes (26) does not undergo SL trans-splicing.) The HrMA4a gene contains a 38-bp muscle-specific enhancer located 66 bp upstream of the TSS (27). We recovered a 151-bp HrMA4a gene segment containing this enhancer, but not the promoter, and placed it upstream of the LacZ reporter gene. As expected, this promoterless construct, HrnZ, showed no significant expression in tailbud embryos (Figure 2). Thus, it could now be used as a vehicle to test for core promoter function in segments of the CiTnI gene regulatory region. Insertion, between the enhancer and reporter gene in HrnZ, of a PCR-amplified segment of the CiTnI regulatory region including the XbaI site and downstream DNA led to strong muscle-specific expression (construct HrMA-CiTnI(−638/−335)nZ). In contrast, insertion of a segment including the XbaI site and upstream DNA (construct HrMA-CiTnI(−836/−623)nZ) resulted in much weaker activity (Figure 2). These results indicated the presence of a functional core promoter in the downstream part of the regulatory region, −638 to −335, and indicated that the upstream part had at best weak core promoter function.
Precise TSS mapping by experimentally blocking trans-splicing
We noted that CiTnI constructs lacking the natural trans-splice acceptor site at −64 and its probable branchpoint at −82 (14), e.g. construct CiTnI(−819/−383)nZ, nonetheless showed reporter gene expression levels similar to the parent construct CiTnI(−1437/−24)nZ (Figure 2). The absence of the normal trans-splice acceptor site raised the possibility that the LacZ-encoding mRNAs produced from these constructs would not be trans-spliced and would therefore retain the outron including the original pre-mRNA 5′-end, thus permitting mapping of the TSS. 5′-RACE analysis showed that LacZ mRNAs produced from CiTnI(−819/−383)nZ in tailbud embryos were in fact not trans-spliced, but were entirely colinear with CiTnI genomic DNA extending to a unique 5′-end that mapped to genomic position −523, thus identifying that as a TSS (Figure 3A). Similar 5′-RACE analysis of LacZ mRNAs produced by a different 3′ deletion construct that also removed the normal trans-splice acceptor site and branchpoint, CiTnI(−836/−422)nZ (see below) identified the same start site (data not shown). This TSS, at −523, was located within the region −638 to −335 that we had identified in the HrMA4a enhancer-complementation assay as containing a functional enhancer-dependent promoter (Figure 2). Interestingly, we also found that the promoter prediction program NNPP (28) predicted site −523 as the most likely TSS within the −819 to −383 sequence of the CiTnI regulatory region.
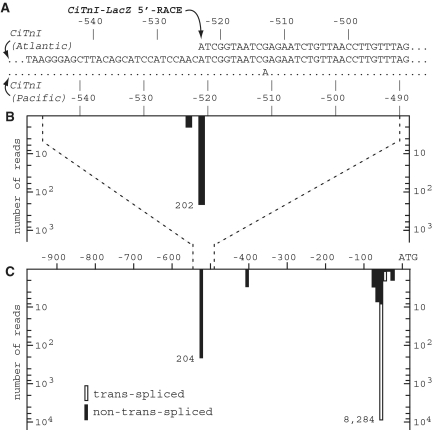
Figure 3.
CiTnI TSS identification by experimental molecular genetics and by high-throughput 5′-RACE sequence analysis of minor transcripts present in normal tailbud embryo RNA. Panel (A) TSS mapping by experimental molecular genetics. The panel shows, on the second sequence line, CiTnI (Atlantic allele) DNA in the region −550 to −491, and beneath it the very similar sequence of the Pacific allele (dots indicate identitity, and note that the ATG-based coordinates are shifted by 2 nt). The topmost sequence line shows the 5′-terminal sequence of a ∼500 bp CiTnI-LacZ mRNA 5′-RACE product generated from embryos expressing construct CiTnI(−819/−383)nZ. Four independent 5′-RACE clones all gave the same sequence, as shown (arbitrary anchor/oligo(dG) sequences introduced during the 5′-RACE procedure have been trimmed). The 5′-end of all four products mapped precisely to CiTnI nucleotide −523 (Atlantic allele, corresponding to −521 Pacific allele). Panels (B and C) CiTnI TSS mapping by high-throughput sequencing of random-primed oligocapping 5′-RACE products from naturally occurring transcripts present in normal tailbud embryo RNA. The panels show the distribution of 5′-reads mapping uniquely to CiTnI (Pacific allele) region −550 to −490 (B) or region −900 to −1 (C). Panel B corresponds spatially to Panel A, with each histogram bar representing a single nucleotide position. The spatial relationships of Panels B and C are indicated by dashed lines. In Panel C each histogram bar represents a 10-nt bin. The genomic position mapped for each read corresponds to the first nucleotide of the read for non-trans-spliced reads (black bars), and the first nt following the 16-nt SL sequence for trans-spliced reads (white bars). Note the logarithmic scale of the read count axes; all positions/bins in the region with no bars shown had zero reads mapped, and the fact that several bins in Panel C contained a single mapped read is shown, arbitrarily, by the shortest black bars visible. All other bars shown represent two or more mapped reads. Exact numbers for peak counts of non-trans-spliced and trans-spliced reads are indicated.
High-throughput 5′-RACE analysis of naturally occurring minor CiTnI transcripts
Because outron-retaining CiTnI transcripts could be useful for TSS mapping and might exist at low levels in normal cells/embryos, we searched for novel minor CiTnI transcripts in RNA extracted from experimentally unperturbed normal tailbud embryos. In a high-throughput random-primed oligocapping 5′-RACE analysis of tailbud embryo RNA, we found that 8523 reads (of a genome-wide total of ∼5 × 106 5′-end reads) uniquely mapped to the 5′-region of the CiTnI gene (‘Pacific’ allele present in the version 1 (16) and KH (22) genome assemblies). Of these, 97.2 % (8284) were SL trans-spliced 5′-end reads whose CiTnI-derived sequences 5′-mapped to the known major trans-splice acceptor site at −58(P) and a satellite site at −55(P) (note Pacific allele numbering indicated by P in parenthesis; see ‘Materials and Methods’ section). In addition, as shown in Figure 3B and C, a set of 237 non-trans-spliced 5′-end reads also uniquely mapped to the CiTnI region, including 202 reads that 5′-mapped to a single site at position −521(P). Because such non-trans-spliced transcripts retain the original pre-mRNA’s 5′-end, these reads identify a TSS. Moreover, the site identified corresponded to the same nucleotide that we had previously identified as the TnI TSS in the experimental genetic analysis described above (−523, Atlantic allele numbering, see Figure 3A). The perfect correspondence of TSS localization through both experimental genetics and minor transcript analysis adds confidence to the identification of the site as a major TSS and also indicates the utility of high-throughput analysis of natural RNA populations in for mapping TSSs for trans-spliced genes.
Identification of a functionally important TATA element
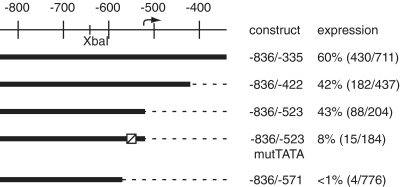
Having confidently identified a TSS by independent concordant methods, we looked for associated core promoter elements. To assess the possible presence of downstream core promoter elements we made a 3′ deletion up to but not including −523, i.e. complete deletion of the transcribed portion of the gene, retaining only the first nucleotide of the natural CiTnI pre-mRNA. This construct, CiTnI(−836/−523)nZ, was actively expressed (Figure 4), indicating that no elements within the transcribed gene region, except perhaps the first nucleotide, were essential for promoter activity. However, larger 3′ deletions, to −571 or to −623, that removed the RNA start site at −523 and nearby upstream DNA, were inactive [constructs CiTnI(−836/−571)nZ (Figure 4) and CiTnI(−836/−623)nZ (Figure 2)]. Thus DNA upstream of, and within 48 bp of, the TSS (i.e. −523 to −571) includes information required for transcription, including core promoter function.
Figure 4.
Smaller-scale 3′ deletion analysis of the transcriptionally-active region of CiTnI and TATA element mutation. Conventions as in Figure 2. Site-directed mutation of the TATA element located at −555 to −546 is symbolized by a diagonally-lined square.
This 48-bp region includes the 28-bp segment CH3, one of four blocks of high C. savignyi/C. intestinalis sequence conservation noted by Johnson et al. (25) in their study of the CsTnI gene. The most highly-conserved part of CH3 includes an 8-nt A/T-rich motif, TATTTAAG, beginning 31 nt upstream of the TSS we identifed at −523. This sequence conforms to the TATA element consensus of Patikoglou et al. (29) T >> c > a ∼ g/A >> t/T >> a∼ c/A >> t/T >> a/A >> g > c∼ t/A∼ T > g > c/G∼ A > c∼ t, and the extended TATA consensus of Bucher (30), G∼C/T/A>t/T/A>t/A∼T/A/A∼T/G∼A/G∼C/C∼G/G∼C/G∼C/G∼C/G∼C (bolded letters show agreement). Moreover, the motif’s location is within the known range of functional TATA elements in vertebrate promoters, where the first T is 22–38 nt upstream of the TSS (30), with the most frequently-observed distances being 30 and 31 nt (31). We found that targeted mutation of this TATA-like element from TATTTAAG to TAggcAAG in the setting of the CiTnI(−836/−523)nZ construct greatly attenuated expression (Figure 4). Thus this upstream TATA-like element appears to play an important role in CiTnI promoter function.
DISCUSSION
We report the first precise mapping of a TSS for a trans-spliced gene in the ascidian tunicate chordate C. intestinalis, an important genetic model organism. Of more general significance, our work establishes two approaches for TSS mapping in SL trans-spliced genes that are broadly applicable to tunicates and trans-splicing organisms in other phyla.
Nature of the CiTnI promoter and outron
Diverse core promoter elements have been identified in various eukaryotic genes, including the TATA element located upstream of the TSS and the Inr element which, when present, generally includes the TSS (32,33).
Upstream of the experimentally-determined CiTnI TSS our study revealed a functionally important TATA element. Although TATA-like elements have been observed in tunicate genes (25,34) ours is the first demonstration of a functional TATA element appropriately positioned vis-a-vis an experimentally-determined TSS. This TATA element was essential for high-level expression in a construct lacking all CiTnI sequences downstream of the TSS. Interestingly, the data of Johnson et al. (25) showed that mutation of a 20-bp sequence block (scrambled window 19), including the corresponding TATA element, inactivated a C. savignyi CsTnI reporter gene construct, consistent with the important role we document here. However, further data of Johnson et al. showed that in a CsTnI construct extending farther downstream, the entire 27-bp CH3 region including the TATA element could be deleted without compromising expression. This could perhaps reflect compensation by a downstream element, or by an upstream element brought closer to the TSS by the CH3 deletion.
The CiTnI TSS conforms to the general consensus initiation sequence for mammalian promoters, YR (31) however there is only limited agreement with the mammalian Inr element functional consensus YYANWYY (35) (IUPAC nomenclature, matches in bold, and where R or A is the start nucleotide). We also found no appropriately-positioned sequences resembling BREu, XCPE1, MTE, or DCE, (32,33) and only a weak resemblance of the TATA-flanking downstream sequence to BREd (36), GGAGCTT versus RTDKKKK (matches in bold). An AGTCA sequence at +32/+36 matches the DPE consensus RGWYV (32) though it is not positioned precisely at the +28/+32 location expected for a functional DPE, and we have shown that no elements downstream of the first transcribed nucleotide are essential for transcription. Thus the only clearly recognizable and experimentally-validated CiTnI core promoter element we identify is the TATA element.
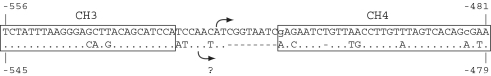
Our high-throughput 5′-RACE sequence data suggest a single major TSS in the region, at −523, indicating that the CiTnI promoter is of the ‘focused’ variety (33). This site was also precisely predicted by NNPP. The success of NNPP in predicting an actual TSS for the CiTnI gene implies conformity of this ascidian promoter to the NNPP training regime, which is focused on TATA and Inr elements in vertebrate promoters (28). NNPP’s second choice was at −576, corresponding to its topscore prediction for the C. savignyi CsTnI gene (25). However, there is no direct evidence that this upstream site is used as a TSS in C. savignyi and, because we found that no 5′-RACE reads aligned with that apparently single-copy region of the CiTnI gene, it does not appear to be used as a significant alternative TSS in C. intestinalis tailbud embryos. We note, as shown in Figure 5, that NNPP’s second-choice TSS prediction for CsTnI is located very close to the established CiTnI TSS, suggesting this as a candidate TSS for CsTnI. Figure 5 also shows that the established CiTnI TSS is not located within a highly conserved sequence block but between two such blocks, CH3 and CH4, whereas the upstream TATA element is perfectly conserved within CH3. Stronger conservation of the TATA element region than of the TSS per se is a general rule for mammalian TATA-containing promoters (31).
Figure 5.
The CiTnI TSS is located between highly conserved sequence blocks. The upper sequence shows the C. intestinalis CiTnI gene (Atlantic allele) in the −556 to −481 regions including the TSS at −523 (arrow). Atlantic and Pacific CiTnI alleles are identical throughout the region, except for the two bases shown in lower case, where the Pacific allele matches the CsTnI sequence. The lower sequence is that of the C. savignyi orthologue CsTnI, with identical nucleotides indicated by dots, and gaps by dashes. Highly-conserved sequence blocks CH3 and CH4, identified by Johnson et al (25), are boxed. The lower arrow marks the NNPP second-choice TSS prediction for the CsTnI gene region −821 to −354, i.e., position −515, with the question mark denoting the absence of experimental data to confirm or refute the use of this predicted TSS in the CsTnI gene.
The only other tunicate TSSs determined to date are those of the non-trans-spliced muscle actin genes of the ascidian H. roretzi, HrMA4a (20,37) and HrMA1a/b (34). Like the CiTnI TSS, these TSSs have appropriately placed TATA-like elements (20,34), although their function has not been investigated by mutation analysis. Apart from the TATA similarity we also note that the TSSs are located within a common CATC motif (Figure 6).
Figure 6.
Similar features in the core promoter regions of CiTnI and the H. roretzi actin genes HrMA4a and HrMA1a. Sequences (from GenBank AF237978.2, S76735.1 and D29014.1) are aligned on the common CATC sequence that includes the experimentally-identified TSSs (bolded nucleotide) (20,34). TATA elements and TSS CATC motifs are shown in upper case.
Identification of the CiTnI TSS also delineates the outron, i.e. the primary transcript’s 5′-segment upstream of the trans-splice acceptor site. The CiTnI outron is 459 nt long (463 nt in the Pacific allele). Putative outrons established by heterologous transcription of nematode Onchocerca volvulus genes range in length from 12 to 68 nt for Ov-sod genes (7) and <79 nt for Ov-GST1a, (6). Outrons in Caenorhabditis, more definitively delineated on the basis of natural minor transcript analysis, were 65 nt for collagen col-13 (10), 173 nt for collagen rol-6(8), and 450 nt for ubiquitin Ubi A (9). Experimental studies in Caenorhabditis indicate that functional outrons must be A+U-rich (∼70%) and >∼50 nt in length (8,38). The 459-nt length of the CiTnI outron, and its 68% A+U content (versus the 55% A+U content of the adjacent 76-nt first-exon sequence downstream of the trans-splice acceptor site) are consistent with these prior findings in nematodes.
One of the functions proposed for SL trans-splicing is 5′-untranslated region sanitization, i.e. the removal of sequence elements present in the outron that may be deleterious to mRNA maturation, stability, or function (3). In this connection, we note that the CiTnI outron contains 3 upstream in-frames, and an additional 3 out-of-frame, ATG codons, which could perhaps interfere with proper translation.
General applicability of the TSS mapping methods
Both approaches we employed to map the TSS for the CiTnI gene—experimental molecular genetics, and analytical high-throughput 5′-RACE sequencing—have broad potential application to TSS mapping in organisms that carry out SL trans-splicing.
The experimental molecular genetic approach consisted of two phases, (i) approximate promoter mapping (using large-scale deletions coupled with an assay for enhancer-dependent core promoter function) and (ii) precise TSS localization by mutational elimination of trans-splicing leading to outron retention in the mature mRNA. Both phases could be carried out in any organism for which gene introduction techniques exist. In the case of ascidian species (and perhaps other tunicates) for which gene introduction techniques are not available, it is likely that gene introduction into Ciona zygotes would be workable. In addition, it is likely that the Halocynthia HrMA4a muscle actin enhancer would be able to drive embryonic muscle-specific expression from the core promoters of many ascidian/tunicate genes, as this enhancer drives muscle-specific expression in ascidian embryos even from the minimal promoter of a mammalian virus, SV40 (27). Application of the full experimental genetic approach to transfectable species from other trans-splicing phyla may require the identification of comparable promoter-dependent enhancers.
A potential complication of the experimental genetic approach is the possible unmasking or activation of upstream cryptic trans-splice acceptor sites upon deletion of the natural acceptor site and putative branch point. This would preclude precise mapping of the TSS because the extreme 5′-end of the initial pre-mRNA transcript would still be discarded during the resultant trans-splicing reaction (albeit on a shorter than wild-type outron). Activation of cryptic upstream acceptor sites following deletion/mutation of the natural trans-splice acceptor site has been observed in several Caenorhabditis genes (38) and in trypanosomes (39), and we also observed this in some CiTnI experimental constructs (data not shown). However, cryptic acceptors do not appear to be densely packed within the CiTnI outron sequence, and we readily found constructs in which no cryptic trans-splicing occurred and in which the entire construct outron was maintained in the mature mRNA.
The approach to TSS mapping through high-throughput 5′-RACE sequence analysis of naturally occurring minor transcripts is independent of the experimental molecular genetic approach. Our analysis identified a minority of non-trans-spliced CiTnI transcripts in normal embryos, most of which 5′-mapped precisely to the experimentally determined CiTnI TSS. The existence of such molecules, presumably trans-splicing failures or not-yet-trans-spliced mRNAs/pre-mRNAs, makes high-throughput 5′-RACE a potentially powerful approach for TSS determination for large numbers of trans-spliced genes. High-throughput validation methods would be useful for assessing candidate TSSs mapped through natural minor transcript analysis. Although the experimental molecular genetic approach is too labor-intensive for high-throughput application, an alternative validation approach, suggested by the fact that NNPP predicted the CiTnI TSS precisely, is the use of NNPP or other promoter prediction programs to give a rank of confidence to candidate TSSs. An additional potential validation parameter is genomic location an appropriate distance upstream of a known trans-splice acceptor site. The latter may themselves be mapped from the same high-throughput 5′-RACE sequence data (see e.g. ref. 40) if the readlength is longer than the SL sequence.
The high-throughput 5′-RACE method for trans-spliced gene TSS mapping could be applied to any organism for which genomic DNA sequence information is available, including organisms for which no gene transfer methodology has yet been developed. It could therefore have a broad and immediate applicability.
FUNDING
Natural Sciences and Engineering Research Council of Canada (to K.E.M.H); U.S. National Institutes of Health (2R15 HD47357-02, to T.H.M.), Japan Society for the Promotion of Science, Grants-in-Aid for Scientific Research (17310114, 20310115, and 22310120, to T.K. and K.N.) Funding for open access charge: Publication charges will be paid from K. Hastings NSERC Canada research grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Yutaka Satou for providing the HrMA4a DNA.
REFERENCES
- 1.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW. Trans-splicing of nematode premessenger RNA. Annu. Rev. Microbiol. 1993;47:413–440. doi: 10.1146/annurev.mi.47.100193.002213. [DOI] [PubMed] [Google Scholar]
- 3.Hastings KEM. SL trans-splicing: easy come or easy go? Trends Genet. 2005;21:240–247. doi: 10.1016/j.tig.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 4.Blumenthal T. Trans-splicing and polycistronic transcription in Caenorhabditis elegans. Trends Genet. 1995;11:132–136. doi: 10.1016/s0168-9525(00)89026-5. [DOI] [PubMed] [Google Scholar]
- 5.Conrad R, Thomas J, Spieth J, Blumenthal T. Insertion of part of an intron into the 5′ untranslated region of a Caenorhabditis elegans gene converts it into a trans-spliced gene. Mol. Cell Biol. 1991;11:1921–1926. doi: 10.1128/mcb.11.4.1921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Krause S, Sommer A, Fischer P, Brophy PM, Walter RD, Liebau E. Gene structure of the extracellular glutathione S-transferase from Onchocerca volvulus and its overexpression and promoter analysis in transgenic Caenorhabditis elegans. Mol. Biochem. Parasitol. 2001;117:145–154. doi: 10.1016/s0166-6851(01)00342-5. [DOI] [PubMed] [Google Scholar]
- 7.Tawe W, Walter RD, Henkle-Duhrsen K. Onchocerca volvulus superoxide dismutase genes: identification of functional promoters for pre-mRNA transcripts which undergo trans-splicing. Exp. Parasitol. 2000;94:172–179. doi: 10.1006/expr.2000.4488. [DOI] [PubMed] [Google Scholar]
- 8.Conrad R, Liou RF, Blumenthal T. Conversion of a trans-spliced C. elegans gene into a conventional gene by introduction of a splice donor site. Embo J. 1993;12:1249–1255. doi: 10.1002/j.1460-2075.1993.tb05766.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Graham RW, Van Doren K, Bektesh S, Candido EP. Maturation of the major ubiquitin gene transcript in Caenorhabditis elegans involves the acquisition of a trans-spliced leader. J. Biol. Chem. 1988;263:10415–10419. [PubMed] [Google Scholar]
- 10.Park YS, Kramer JM. Tandemly duplicated Caenorhabditis elegans collagen genes differ in their modes of splicing. J. Mol. Biol. 1990;211:395–406. doi: 10.1016/0022-2836(90)90360-X. [DOI] [PubMed] [Google Scholar]
- 11.Park YS, Kramer JM. The C. elegans sqt-1 and rol-6 collagen genes are coordinately expressed during development, but not at all stages that display mutant phenotypes. Dev. Biol. 1994;163:112–124. doi: 10.1006/dbio.1994.1127. [DOI] [PubMed] [Google Scholar]
- 12.Tsuchihara K, Suzuki Y, Wakaguri H, Irie T, Tanimoto K, Hashimoto S, Matsushima K, Mizushima-Sugano J, Yamashita R, Nakai K, et al. Massive transcriptional start site analysis of human genes in hypoxia cells. Nucleic Acids Res. 2009;37:2249–2263. doi: 10.1093/nar/gkp066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.MacLean DW, Meedel TH, Hastings KEM. Tissue-specific alternative splicing of ascidian troponin I isoforms. Redesign of a protein isoform-generating mechanism during chordate evolution. J. Biol. Chem. 1997;272:32115–32120. doi: 10.1074/jbc.272.51.32115. [DOI] [PubMed] [Google Scholar]
- 14.Vandenberghe AE, Meedel TH, Hastings KEM. mRNA 5′-leader trans-splicing in the chordates. Genes Dev. 2001;15:294–303. doi: 10.1101/gad.865401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cleto CL, Vandenberghe AE, MacLean DW, Pannunzio P, Tortorelli C, Meedel TH, Satou Y, Satoh N, Hastings KEM. Ascidian larva reveals ancient origin of vertebrate-skeletal-muscle troponin I characteristics in chordate locomotory muscle. Mol. Biol. Evol. 2003;20:2113–2122. doi: 10.1093/molbev/msg227. [DOI] [PubMed] [Google Scholar]
- 16.Dehal P, Satou Y, Campbell RK, Chapman J, Degnan B, De Tomaso A, Davidson B, Di Gregorio A, Gelpke M, Goodstein DM, et al. The draft genome of Ciona intestinalis: insights into chordate and vertebrate origins. Science. 2002;298:2157–2167. doi: 10.1126/science.1080049. [DOI] [PubMed] [Google Scholar]
- 17.Corbo JC, Levine M, Zeller RW. Characterization of a notochord-specific enhancer from the Brachyury promoter region of the ascidian, Ciona intestinalis. Development. 1997;124:589–602. doi: 10.1242/dev.124.3.589. [DOI] [PubMed] [Google Scholar]
- 18.Fire A, Harrison SW, Dixon D. A modular set of lacZ fusion vectors for studying gene expression in Caenorhabditis elegans. Gene. 1990;93:189–198. doi: 10.1016/0378-1119(90)90224-f. [DOI] [PubMed] [Google Scholar]
- 19.Horton RM. In: PCR Cloning Protocols From Molecular Cloning to Genetic Enginerring. White BA, editor. Vol. 67. Totowa, New Jersey: Humana Press; 1997. pp. 141–150. [Google Scholar]
- 20.Hikosaka A, Kusakabe T, Satoh N. Short upstream sequences associated with the muscle-specific expression of an actin gene in ascidian embryos. Dev. Biol. 1994;166:763–769. doi: 10.1006/dbio.1994.1354. [DOI] [PubMed] [Google Scholar]
- 21.Suzuki Y, Yoshitomo-Nakagawa K, Maruyama K, Suyama A, Sugano S. Construction and characterization of a full length-enriched and a 5′-end-enriched cDNA library. Gene. 1997;200:149–156. doi: 10.1016/s0378-1119(97)00411-3. [DOI] [PubMed] [Google Scholar]
- 22.Satou Y, Mineta K, Ogasawara M, Sasakura Y, Shoguchi E, Ueno K, Yamada L, Matsumoto J, Wasserscheid J, Dewar K, et al. Improved genome assembly and evidence-based global gene model set for the chordate Ciona intestinalis: new insight into intron and operon populations. Genome Biol. 2008;9:R152. doi: 10.1186/gb-2008-9-10-r152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Jiang H, Wong WH. SeqMap: mapping massive amount of oligonucleotides to the genome. Bioinformatics. 2008;24:2395–2396. doi: 10.1093/bioinformatics/btn429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johnson DS, Davidson B, Brown CD, Smith WC, Sidow A. Noncoding regulatory sequences of Ciona exhibit strong correspondence between evolutionary constraint and functional importance. Genome Res. 2004;14:2448–2456. doi: 10.1101/gr.2964504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Satou Y, Hamaguchi M, Takeuchi K, Hastings KEM, Satoh N. Genomic overview of mRNA 5′-leader trans-splicing in the ascidian Ciona intestinalis. Nucleic Acids Res. 2006;34:3378–3388. doi: 10.1093/nar/gkl418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Satou Y, Satoh N. Two cis-regulatory elements are essential for the muscle-specific expression of an actin gene in the ascidian embryo. Dev. Growth Differ. 1996;38:565–573. doi: 10.1046/j.1440-169X.1996.t01-1-00013.x. [DOI] [PubMed] [Google Scholar]
- 28.Reese MG. Application of a time-delay neural network to promoter annotation in the Drosophila melanogaster genome. Comput. Chem. 2001;26:51–56. doi: 10.1016/s0097-8485(01)00099-7. [DOI] [PubMed] [Google Scholar]
- 29.Patikoglou GA, Kim JL, Sun L, Yang SH, Kodadek T, Burley SK. TATA element recognition by the TATA box-binding protein has been conserved throughout evolution. Genes Dev. 1999;13:3217–3230. doi: 10.1101/gad.13.24.3217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bucher P. Weight matrix descriptions of four eukaryotic RNA polymerase II promoter elements derived from 502 unrelated promoter sequences. J. Mol. Biol. 1990;212:563–578. doi: 10.1016/0022-2836(90)90223-9. [DOI] [PubMed] [Google Scholar]
- 31.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CA, Taylor MS, Engstrom PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 32.Smale ST, Kadonaga JT. The RNA polymerase II core promoter. Annu. Rev. Biochem. 2003;72:449–479. doi: 10.1146/annurev.biochem.72.121801.161520. [DOI] [PubMed] [Google Scholar]
- 33.Juven-Gershon T, Kadonaga JT. Regulation of gene expression via the core promoter and the basal transcriptional machinery. Dev. Biol. 2010;339:225–229. doi: 10.1016/j.ydbio.2009.08.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kusakabe T, Hikosaka A, Satoh N. Coexpression and promoter function in two muscle actin gene complexes of different structural organization in the ascidian Halocynthia roretzi. Dev. Biol. 1995;169:461–472. doi: 10.1006/dbio.1995.1161. [DOI] [PubMed] [Google Scholar]
- 35.Lo K, Smale ST. Generality of a functional initiator consensus sequence. Gene. 1996;182:13–22. doi: 10.1016/s0378-1119(96)00438-6. [DOI] [PubMed] [Google Scholar]
- 36.Deng W, Roberts SG. A core promoter element downstream of the TATA box that is recognized by TFIIB. Genes Dev. 2005;19:2418–2423. doi: 10.1101/gad.342405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kusakabe T, Makabe KW, Satoh N. Tunicate muscle actin genes. Structure and organization as a gene cluster. J. Mol. Biol. 1992;227:955–960. doi: 10.1016/0022-2836(92)90237-e. [DOI] [PubMed] [Google Scholar]
- 38.Conrad R, Liou RF, Blumenthal T. Functional analysis of a C. elegans trans-splice acceptor. Nucleic Acids Res. 1993;21:913–919. doi: 10.1093/nar/21.4.913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Matthews KR, Tschudi C, Ullu E. A common pyrimidine-rich motif governs trans-splicing and polyadenylation of tubulin polycistronic pre-mRNA in trypanosomes. Genes Dev. 1994;8:491–501. doi: 10.1101/gad.8.4.491. [DOI] [PubMed] [Google Scholar]
- 40.Matsumoto J, Dewar K, Wasserscheid J, Wiley GB, Macmil SL, Roe BA, Zeller RW, Satou Y, Hastings KEM. High-throughput sequence analysis of Ciona intestinalis SL trans-spliced mRNAs: alternative expression modes and gene function correlates. Genome Res. 2010;20:636–645. doi: 10.1101/gr.100271.109. [DOI] [PMC free article] [PubMed] [Google Scholar]