Abstract
EndA is a sequence non-specific endonuclease that serves as a virulence factor during Streptococcus pneumoniae infection. Expression of EndA provides a strategy for evasion of the host's neutrophil extracellular traps, digesting the DNA scaffold structure and allowing further invasion by S. pneumoniae. To define mechanisms of catalysis and substrate binding, we solved the structure of EndA at 1.75 Å resolution. The EndA structure reveals a DRGH (Asp-Arg-Gly-His) motif-containing ββα-metal finger catalytic core augmented by an interesting ‘finger-loop’ interruption of the active site α-helix. Subsequently, we delineated DNA binding versus catalytic functionality using structure-based alanine substitution mutagenesis. Three mutants, H154A, Q186A and Q192A, exhibited decreased nuclease activity that appears to be independent of substrate binding. Glu205 was found to be crucial for catalysis, while residues Arg127/Lys128 and Arg209/Lys210 contribute to substrate binding. The results presented here provide the molecular foundation for development of specific antibiotic inhibitors for EndA.
INTRODUCTION
Streptococcus pneumoniae commonly colonizes the human upper respiratory tract (1). However, when the delicate balance between benign harboring of bacterial flora and deleterious microbial outgrowth is disrupted, invasive pneumococcal disease (IPD) can result (2). IPD is one of the leading causes of opportunistic infections in young children, the elderly and in individuals immunocompromised due to cancer, diabetes or human immunodeficiency virus infection (3–6). Though favorable outcomes are primarily expected as a result of early detection and prompt antibacterial therapy, severe complications of IPD include bacteremia, meningitis and death (2,6,7,8).
Bacterial colonization in the lungs of infected individuals triggers neutrophil recruitment to the site of infection (9), resulting in extrusion of DNA/protein scaffolds known as neutrophil extracellular traps [NETs (10)]. These filamentous NETs physically trap bacteria, leaving them vulnerable to phagocytic attack and/or destruction by NET-bound antimicrobial proteins located in the NETs (9–12). To combat the innate immune response, constitutive expression of membrane-bound DNA-entry nuclease EndA (13) is instrumental for respiratory track invasion and increased virulence of S. pneumoniae. EndA localization to the outer face of the bacterial membrane (14,15) enables its sequence non-specific nuclease activity to destroy the NETs. EndA, a DNA/RNA endonuclease (16), is normally associated with DNA degradation and uptake during the biochemical state of competence, further promoting S. pneumoniae pathogenicity through gene transfer (13,17). By helping streptococci to establish genetic diversity, EndA may influence the ability of this organism to adapt to changing environmental conditions, providing a significant advantage during infection.
Nuclease digestion of the NETs’ DNA scaffold is associated with increased bacterial migration from the upper airway to the lungs, and in 20–30% of cases, invasion into the bloodstream (8,18,19). Streptococcus pneumoniae strains expressing EndA have been shown to evade and destroy NETs, and cause more virulent, invasive forms of pneumonia (18). The role of EndA as a virulence factor in pneumococcal infection makes it an attractive target for antimicrobial therapeutics. However, despite the central importance of EndA in promoting virulence, the mechanism by which EndA degrades DNA remains enigmatic, due in part to the lack of EndA protein structural information. Furthermore, the toxicity of recombinantly expressed enzymes (15,20) and the subsequent inability to purify active nuclease have hampered biochemical and mechanistic studies of EndA.
To define mechanisms by which DNA-entry nucleases bind and degrade DNA substrates, we report the structure and detailed mutagenesis of S. pneumoniae EndA. We engineered an active site glycine substitution mutant, EndA(H160G), which yielded quantifiable imidazole rescue nuclease activity. Structural information, coupled with imidazole rescue, allowed for biochemical characterization of EndA variants surrounding the active site, delineating catalytic activity versus DNA substrate binding. Based on these results, histidine-to-glycine substitution paired with imidazole rescue could be utilized as an all-purpose strategy for expression, purification and biochemical characterization of other ββα-metal finger nucleases.
MATERIALS AND METHODS
Cloning, expression and purification of EndA(H160A)
The sequence encoding Ala31-Gln274 of EndA(H160A) from S. pneumoniae was cloned into the pET30M expression vector as described previously (20). EndA(H160A) was expressed as a His-GST fusion protein in Rosetta2(DE3) cells, overnight at 18°C. The cells were pelleted by centrifugation, resuspended in 25 mM Tris pH 7.5, 500–750 mM NaCl and lysed by sonication. The soluble fraction was bound in-batch to glutathione sepharose 4B resin and EndA(H160A) was cleaved from the resin by TEV protease, overnight at 4°C. Uncleaved fusion protein contaminants were removed by a brief incubation with Ni-NTA agarose. Unbound protein was concentrated and loaded onto a Superdex 200 16/60 column equilibrated with 25 mM Tris pH 7.5, 75 mM NaCl. Peak fractions were collected and concentrated to ∼50 mg/ml.
Crystallization and data collection
EndA(H160A) was crystallized by mixing 1 μl of protein (51 mg/ml) with 1 μl of 0.1 M Tris pH 7, 17% PEG8K, 0.2 M MgCl2. Crystals were grown by sitting drop vapor diffusion (21) at room temperature. The highest resolution data set was obtained from a glutaraldehyde cross-linked crystal. This crystal was transferred from the original drop into a 3.5 μl hanging drop containing 0.1 M Tris pH 7, 75 mM NaCl, 0.2 M MgCl2 and 20% PEG8K (mother liquor). The crystal was placed in a drop over a vapor diffusion chamber containing a sitting drop bridge with 3.5 μl of 25% glutaraldehyde and 500 μl mother liquor for 8 min at room temperature. The crystal was then soaked in a solution containing 0.1 M Tris pH 7, 75 mM NaCl, 0.2 M MgCl2, 20% PEG8K, 11.6 mM trimethyl lead acetate and 9.63 mM triethyl lead acetate for 43 h at room temperature for potential phasing opportunities. The soaked crystal was transferred to a cryoprotectant (0.1 M Tris pH 7, 75 mM NaCl, 0.2 M MgCl2, 20% PEG8K, 15% ethylene glycol, 11.9 mM trimethyl lead acetate and 13.75 mM triethyl lead acetate) in four steps, then flash frozen in liquid nitrogen and placed in a stream of nitrogen gas cooled to −180°C. The data were collected at Southeast Regional Collaborative Access Team (SER-CAT) BM-22 beamline at the Advanced Photon Source (Argonne National Laboratory), at a wavelength of 0.94 Å, with the MAR225 area detector. The data were processed at 1.75 Å, in space group P212121 using HKL2000 (22) (Table 1). All attempts at solving the structure of EndA(H160A) using molecular replacement with other DRGH nucleases as search models, heavy metal soaking or multi-wavelength anomalous dispersion with engineered selenomethionine-substituted protein failed to provide useful phase information. The phase problem was solved by crystallizing EndA as a MBP(wt)-EndA(H160A) fusion protein (23) and using a partial model of EndA(H160A) from this data to solve the 1.75 Å structure by molecular replacement (Supplementary Data). The quality of the electron density maps was improved by density modification and non-crystallographic averaging. An initial round of refinement was performed in crystallographic refinement package (CNS) (24), and the resulting structure was used as a starting model for iterative building using AutoBuild (25) in PHENIX (26). About 420 amino acids for two molecules of EndA(H160A) (86.1%) were built, with an Rfree of 23%. The final model for EndA(H160A) was generated by iterative cycles of manual model building and refinement in O (27) and CNS (24) (Rfree 21.6%). The refined model for EndA(H160A) contains amino acid residues Ser46-Tyr124 and Ser136-Gln274 in molecule A and residues Lys43-Lys125 and Ser134-Gln274 in molecule B (Table 1). No glutaraldehyde cross-linkages or lead derivative molecules were found in the density. Ramachandran statistics were determined by MolProbity (28), with 98.7% of residues in the favored regions, and 100% in allowed regions.
Table 1.
Data collection and refinement statistics
| EndA(H160A)a | MBP(wt)- EndA(H160A)a,b | |
|---|---|---|
| Data collection | ||
| Space group | P212121 | P21 |
| Cell dimensions | ||
| a, b, c (Å) | 67.96, 75.93, 90.78 | 53.63, 64.23, 89.88 |
| α, β, γ (°) | 90, 90, 90 | 90, 96.32, 90 |
| Resolution (Å) | 50–1.75 (1.78–1.75)c | 50–2.6 (2.69–2.60) |
| Unique reflections | 46 402 | 18 270 |
| Rsym (%) | 9.7 (37.1) | 12.9(39.2) |
| I / σI | 26.51 (3.56) | 9.8 (2.4) |
| Completeness (%) | 96.8 (68.7) | 97.2 (97.0) |
| Redundancy | 13.6 (7.2) | 4.3 (2.9) |
| Model refinement | ||
| Resolution (Å) | 50–1.75 | |
| No. reflections | 45 460 | |
| Rwork / Rfreed | 19.6/21.6 | |
| No. atoms | ||
| Protein | 3285 | |
| Ions | 7 | |
| Water | 495 | |
| B-factors | ||
| Protein (MolA/MolB) | 22.74/21.63 | |
| Ions (MolA/MolB) | 16.71/18.89 | |
| Water | 31.99 | |
| RMSD | ||
| Bond lengths (Å) | 0.005 | |
| Bond angles (°) | 1.2 | |
| Ramachandran plot | ||
| Most favorable (%) | 98.7 | |
| Allowed (%) | 100 |
aA single crystal was used for data collection.
bData quality for MBP(wt)-H160A was insufficient for complete structure solution and refinement. Therefore, only the statistics for data collection are included.
cValues in parentheses refer to the highest resolution shell.
dRwork = Σ | |Fobs| − |Fcalc| | / Σ |Fobs|, where Rfree is calculated for a randomly chosen 5% of reflections, which were not used for structure refinement and Rwork is calculated for the remaining reflections.
Generation of EndA(H160G) mutants
The H160G mutation in EndA was obtained by site-directed mutagenesis, using pET30M-EndA(H160A) as a template for the reaction. Additional mutants were generated on the H160G background using the same method. All EndA proteins were expressed and purified as for EndA(H160A).
Nuclease activity assay
EndA proteins were diluted to 20 nM in activity buffer (20 mM Tris, 10 mM sodium acetate, 10 mM MES, 25 mM NaCl, 5 mM MgCl2, pH 8) and incubated with 15 ng/μl of supercoiled pBluescript SK(+) plasmid for 30–35 min, in the presence or absence of 0.2 M imidazole. This concentration of imidazole was chosen for the reactions, as all phases (supercoiled, nicked open circle, linearized and degraded) of digestion are visible.
The reactions were quenched by addition of loading dye containing EDTA. Samples were run on a 0.8% (w/v) agarose gel, dissolved in 1× Tris–Acetate–EDTA buffer. DNA species in the gel were visualized by ethidum bromide staining, scanned with a Typhoon fluorescence imager and analyzed using ImageQuant. Nuclease activity assays were performed in triplicate.
Single radial enzyme diffusion (SRED) assay
Agarose (1% w/v) was dissolved in activity buffer (20 mM Tris, 10 mM sodium acetate, 10 mM MES, 25 mM NaCl, 5 mM MgCl2, ±0.2 M imidazole, pH 8) and high molecular weight salmon sperm DNA (30 μg/ml) and ethidium bromide (1 μg/ml) were added immediately prior to casting. EndA proteins were diluted to 5 mg/mL in activity buffer (20 mM Tris, 10 mM sodium acetate, 10 mM MES, 25 mM NaCl, 5 mM MgCl2, ±0.2 M imidazole, pH 8) and 1 μl of each protein solution was placed in a small well on the SRED plates. About 0.5 μl of DNase I (1 unit/μl) was added to each plate as a positive control. The plates were incubated overnight at 37°C. Nuclease activity is identified by visualization of a dark ‘halo’ around the well site, as seen under UV254nm illumination. The relative extent of nuclease activity for each protein was determined by measurement of the halo radius. This experiment was performed in triplicate.
DNA-binding assays
A 24mer oligonucleotide (5′-GCAAGGTGCGTGCAGCCTTGGACC-3′) was annealed to a complementary oligonucleotide by denaturation and slow cooling. The annealed DNA was mixed with 10 μM EndA protein in a 4:1 molar ratio (11.5 mM Tris pH 8, 10 mM MgCl2, 7.5 mM NaCl) and incubated on ice at 4°C for 2 h. About 2 μl of High Density Tris Borate-EDTA (TBE) Loading Buffer was added to each sample, and 5μl of this mixture was loaded onto a 1% agarose/1% acrylamide gel in 0.25× TBE. DNA and protein species were serially stained on the same gel with ethidium bromide and SYPRO Orange, respectively and scanned using a Typhoon fluorescence imager. About 610 nm band-pass and 555 nm band-pass emission filters were used to detect the ethidium bromide and SYPRO Orange signals, respectively.
RESULTS
Structural organization of EndA(H160A)
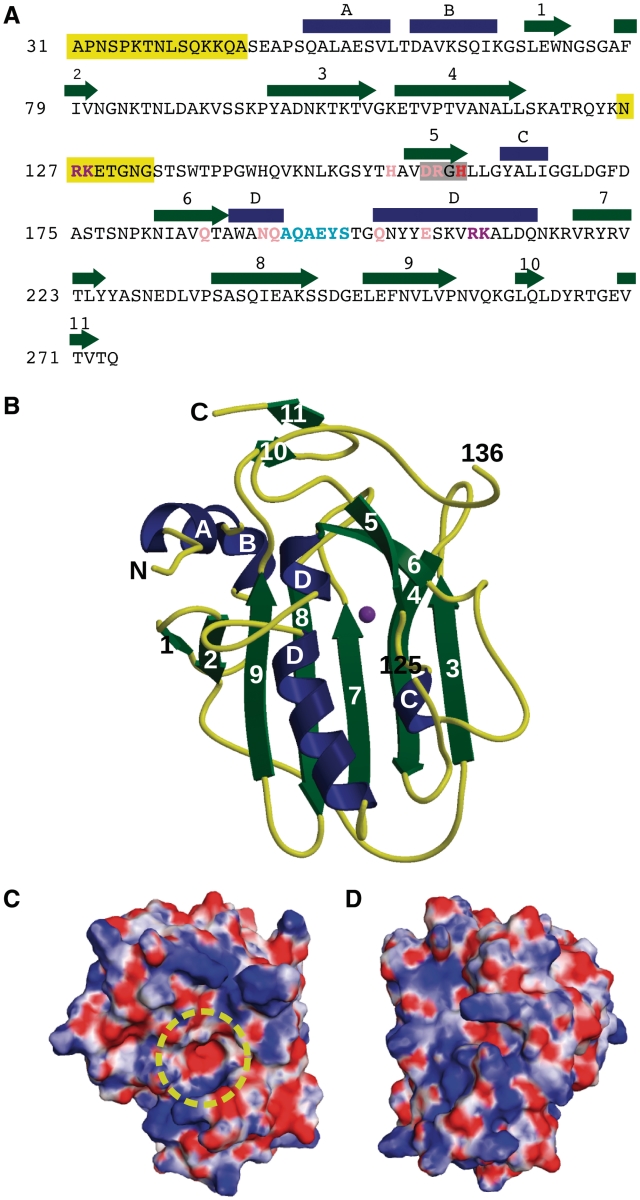
The nuclease domain (Ala31-Gln274) of EndA from S. pneumoniae, that contained the inactivating H160A mutation and also lacking the N-terminal membrane localization signal sequence, was crystallized and refined as summarized in Table 1 (PDB ID code 3OWV). A model for molecular replacement of EndA was obtained by crystallizing an N-terminal MBP fixed-arm fusion with the EndA nuclease domain and building a partial model to the EndA density which contained the core structural elements (see ‘Materials and Methods’ section, and Supplementary Data). The final structure for the EndA nuclease domain was refined through a combination of automated and manual model building. The protein core is comprised of a central β-sheet consisting of five antiparallel β-strands (β-strands 3,4,7,8 and 9, Figure 1 and Supplementary Figure S1). The ‘front’ face of the molecule contains the active site and putative DNA-binding surface. β-strands 5/6 and α-helix D form the ββα-metal finger motif that creates the catalytic center (Figures 1B and 2A). The ‘back’ face of the molecule contains little secondary structure. Aside from a single ordered loop between β-strands 2 and 3 traversing this face, most of the sheet is exposed to solvent (Supplementary Figure S2). Analysis of the electrostatic surface of EndA underscores the small size of the active site (Figure 1C and D), and reveals a fairly even distribution of positively and negatively charged residues on both faces of the protein. Residues Asn126-Gly135 connecting β-strands 4 and 5 are disordered in this structure, as in other crystal forms we were able to obtain, and were not included in the calculation of the electrostatic surface potential. Mass spectrometric analysis of dissolved crystals yielded a molecular weight consistent with the expected protein construct size rather than proteolyzed fragments (data not shown). Due to the exclusion of the putative transmembrane domain (Leu9–Leu18), and the lack of overt hydrophobic patches on the protein surface, the current structural information reveals little information on the mechanism by which EndA associates with the bacterial outer membrane.
Figure 1.
Sequence and structure of EndA. (A) Sequence and secondary structural elements of EndA from S. pneumoniae. α-Helices are indicated as blue rectangles and β-strands as green arrows. The DRGH motif is boxed in gray. The position of the general base (His160) is shown in red. Mutated residues involved in catalysis are shown in pink. Residues likely involved in DNA substrate binding and/or catalysis are shown in purple. The ‘finger-loop’ is shown in cyan. Disordered regions are highlighted in yellow. (B) Ribbon diagram of EndA, showing the ‘front face’ of the molecule that contains the active site. Secondary structural elements are color-coded as in (A) and the magnesium ion is colored purple. The positions of the termini and the ends of the disordered loop are labeled. (C) Electrostatic surface potential of the ‘front face’ of EndA, in the same orientation as in (B). The surface was calculated using the Adaptive Poisson–Boltzmann Solver tool in PyMOL (47). The potential ranges from −8 kTe−1 (red) to 8 kTe−1 (blue). The location of the active site is highlighted by a yellow circle. (D) Electrostatic surface potential of the ‘back face’ of EndA [Disordered side chains lacking density were modeled onto the EndA surface for panels (C) and (D).].
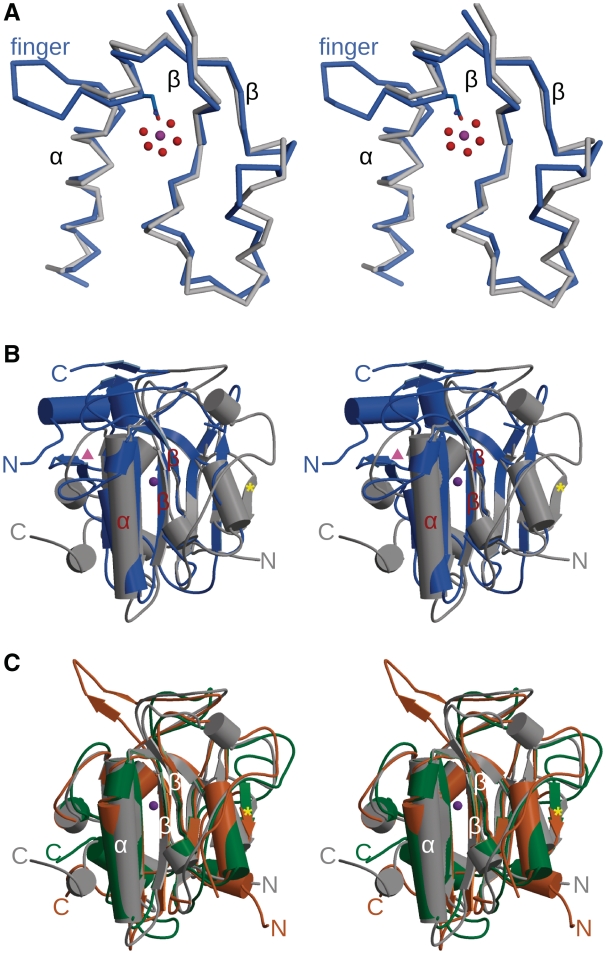
Figure 2.
Comparison of ββα-metal finger nuclease structures. (A) Stereo superposition of the ββα-metal finger motif from EndA (blue) and Serratia nuclease [gray, PDB ID code 1G8T (34)]. Asn191 from EndA is drawn in ball-and-stick in blue, along with the hydrated magnesium (Mg2+ as a purple sphere, and water molecules in red). (B) Stereo superposition of the structures of EndA from S. pneumoniae (blue) and SM endonuclease from S. marcescens [gray, PDB ID code 1G8T (34)]. The ββα-metal finger motif is labeled in red. The β-strand found in Serratia, NucA and EndoG that is lacking in EndA is labeled with a yellow star. The strand-loop-strand protecting the opposite side of the β-sheet in EndA is labeled with a pink triangle. (C) Stereo superposition of the structures of SM endonuclease from S. marcescens [gray, PDB ID code 1G8T (34)], NucA from Anabaena sp. [green, PDB ID code 1ZM8 (31)] and EndoG from Drosophila melanogaster [orange, PDB ID code 3ISM (35)]. The β-strand found in Serratia, NucA and EndoG that is lacking in EndA is labeled with a yellow star. For the dimeric EndoG and Serratia nucleases, only the monomer is shown.
Sequence comparison of EndA with other DRGH nucleases using ClustalW (29) shows, at best 16.3% sequence identity over all core residues with the SR nuclease from Syncephalastrum racemosum (30) although this alignment failed to align the DRGH motif properly. Structural comparisons by the Dali server (31) demonstrate some core features are conserved with the DRGH nucleases: NucA (32,33), Serratia nuclease (34,35), and EndoG (36) (Figure 2). The SM endonuclease from Serratia marcescens showed the greatest similarity to EndA, with an RMSD of 2.9 Å (over 137 out of 235 Cα atoms). In this structure-based alignment, only 18.2% of the aligned backbone atoms exhibited corresponding side chain conservation (Supplementary Table S1). There is minimal structural similarity with EndA beyond the conserved structure of the central β-sheet and the ββα-Me motif which forms the active site (Figure 2A). For example, EndA shows a distinguishable difference in the location of its N-terminus, compared to that of the other DRGH nucleases. For NucA, Serratia nuclease, and EndoG, the N-terminal residues lie along the ‘back’ face of the molecule, covering hydrophobic residues from the core β-sheet. However, in EndA, the N-terminus lies on the ‘front’ face of the sheet, traveling via two α-helices (A and B) to the ‘back’ face through a hydrophobic channel along the top of the sheet (Figure 1B and Supplementary Figure S2). N-terminal α-helices A and B appear to be crucial for proper protein folding, since constructs lacking these residues exhibited decreased protein expression and stability. Another difference characterizing the central β-sheet in EndA is the absence of a small β-strand found in NucA (Tyr58-Val61) and EndoG (Val87-His90) (Figure 2B and C, yellow star). In NucA and EndoG, this β-strand lies slightly out of the plane of the rest of the β-sheet, possibly protecting the sheet and preventing unwanted edge-on aggregation (37). For Serratia nuclease, this β-strand becomes an ordered loop (Val21–Val24) that could serve a similar purpose. The opposite edge of the sheet in EndA is protected by an antiparallel strand-loop-strand motif (β-strands 1 and 2) not seen in the other nucleases (Figure 2B, pink triangle). Instead, there is a short helix protecting this side of the sheet in Serratia nuclease (Phe200-Phe203), NucA (Trp238–Tyr241), and EndoG (Ile275–Phe278).
One of the most notable differences between EndA and other ββα-Me nucleases is the ‘finger-loop’ interruption of α-helix D (Figure 2A). Interestingly, superposition of α-helix D from EndA with the same helix in the Serratia nuclease [PDB ID 1G8T (35)] clearly shows that the presence of the ‘finger-loop’ does not cause a true break in the canonical hydrogen bonding network indicative of α-helices, allowing a smooth transition across the site of the extrusion. Conservation of the architecture of α-helix D is likely critical as the catalytic residue which coordinates the metal ion, immediately precedes the loop. This residue from EndA, Asn191, superimposes precisely with structurally homologous residues from NucA (Asn155), Serratia nuclease (Asn119) and EndoG (Asn185), despite the presence of the ‘finger-loop’.
Improved imidazole chemical rescue of nuclease activity by structure-based mutagenesis
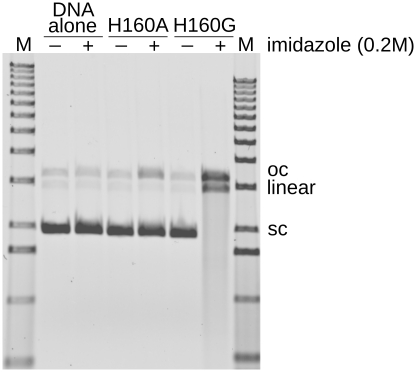
Historically, active wild-type EndA is purified directly from S. pneumoniae or expressed in cell-free systems (15,20). In contrast, bacterial expression of the inactive EndA(H160A) variant yielded large amounts of extremely soluble (>75 mg/ml) and stable protein. Nuclease activity for EndA(H160A) could be rescued by addition of imidazole (38,39) using a plasmid conversion nuclease activity assay (20). This assay measures conversion of supercoiled substrate to nicked open circle, linear duplex or degraded fragments. Nuclease activity was recovered in the presence of imidazole, however, the extent of rescue was relatively low (Figure 3). One goal of our crystallographic studies was to use structural information from EndA(H160A) to develop a platform from which more robust imidazole rescue experiments could be performed, using easily expressed, catalytically inactive nuclease variants.
Figure 3.
Plasmid conversion nuclease activity assay for EndA(H160A) and EndA(H160G). The assay measures conversion of the supercoiled plasmid substrate to nicked open circle or linear (double strand breaks). The assay was performed in the presence of 200 mM imidazole and 10 mM MgCl2).
Based on examination of the EndA(H160A) structure, it appeared that an imidazole molecule substituting for histidine in the catalytic center would create steric clashes with the Cβ of H160A when properly positioned for catalysis. The modeled imidazole ring would lie within 1.5 Å of the Cβ, whereas, if the alanine were substituted with glycine, the distance from its Cα to the imidazole (2.58 Å) would significantly reduce the steric clash. We therefore hypothesized that the extent of imidazole rescue could be improved by substituting the alanine with a glycine residue. The resulting H160G mutant behaved similarly to the H160A mutant during expression and purification, and displayed excellent solubility. Nuclease assays measuring conversion of a supercoiled plasmid substrate (15) to open circle (nicked) or linear (containing double-strand DNA breaks) showed that the H160G mutant exhibited substantially increased imidazole-dependent nuclease activity, compared to the H160A mutant (Figure 3, Table 2). This observation is consistent with results seen for imidazole rescue in other types of enzymes (40,41).
Table 2.
Comparison of imidazole-rescued nuclease activity between EndA H160A and H160G
| Mutant | Conversion to oc | Conversion to linear | Degraded |
|---|---|---|---|
| H160A | 5.4 ± 0.18%a | ND | ND |
| H160G | 22.29 ± 1.25% | 12.39 ± 2.24% | 58.92 ± 2.51% |
oc: open circle (nicked) plasmid; ND: not detected.
aThese nuclease activity assays were performed in triplicate, scanned using a Typhoon fluorescence imager (GE Healthcare), and analyzed using ImageQuant (Molecular Dynamics, Sunnyvale, CA, USA).
In order to determine the optimum concentration of imidazole, the plasmid conversion assay was repeated for EndA(H160G), titrating the imidazole concentration from 10 nM to 500 mM. Conversion of supercoiled plasmid to nicked open circle surpassed background levels upon addition of 1 μM imidazole and reached maximum levels at ∼10 mM (Supplementary Figure S3A). At this concentration, the plasmid substrate was completely digested to nearly undetectable small fragments. Recovery of nuclease activity at imidazole concentrations higher than 100 mM appeared slightly less robust, likely due to the high-ionic strength of the reaction buffer. Though 10 mM imidazole yielded the most potent EndA(H160G) nuclease activity, the 200 mM concentration was chosen as the optimal concentration for the plasmid conversion assay, since all phases of the digestion (nicked open circle, linearized and high-molecular weight degraded oligonucleotides) are visible and readily quantifiable.
Divalent cation preference displayed by EndA(H160G)
Many ββα-metal finger nucleases exhibit an interesting evolutionary active site design, which is optimized to bind a hydrated divalent cation (32,34–36). The structure of EndA contains a bound magnesium ion coordinated by Asn191, and five water molecules (Figure 4). This ion was refined as magnesium because of the high concentration of MgCl2 (200 mM) present in the crystallization buffer, the octahedral coordination geometry and because the bond distances (2.07–2.21 Å) between the ion and the water molecules were consistent with those expected from magnesium. In order to determine whether EndA displays a preference for certain divalent metal cations, the plasmid conversion assay was performed in the presence of 200 mM imidazole and 10 mM of various cations. Imidazole-rescued EndA(H160G) activity appears to have little preference between Mg2+ and Mn2+, but shows little detectable activity in the presence of Ca2+ or Zn2+ (Supplementary Figure S3B).
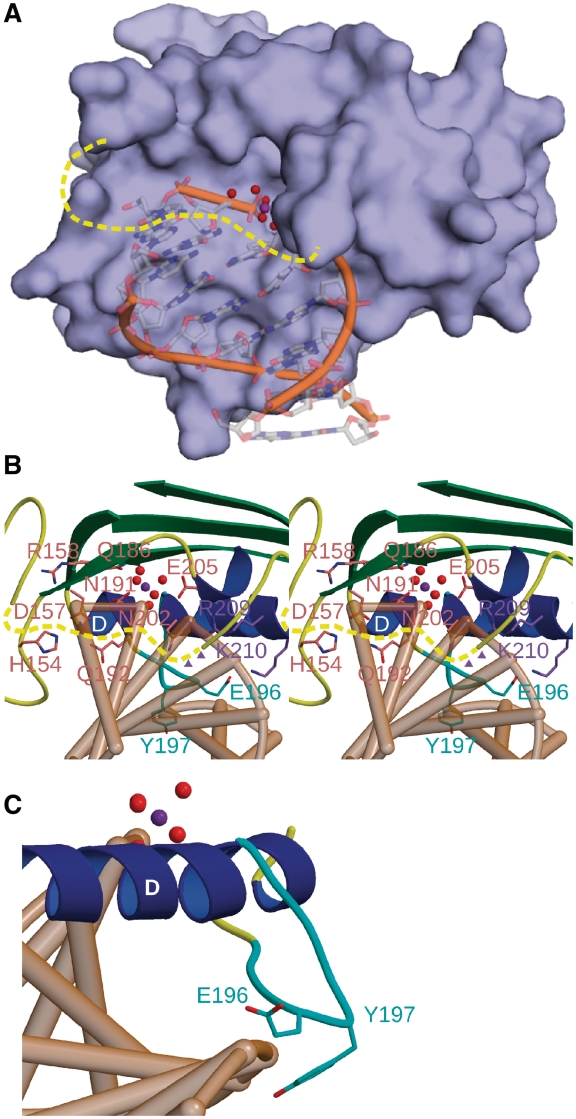
Figure 4.
Mutagenesis of catalytic and DNA-binding residues in the active site of EndA. (A) Surface rendering of EndA, with the modeled DNA substrate from the VVN nuclease [PDB ID code 1OUP (42)] superimposed in the active site. (B) Stereo ribbon diagram of the active site of EndA, displaying the ‘finger-loop’ (cyan), catalytic residues (pink) and DNA-binding residues (purple) targeted for mutagenesis. The disordered loop is drawn in dashed yellow. Putative positions of Arg127 and Lys128 on the disordered loop are shown as purple triangles. (C) Ribbon diagram of the ‘finger-loop’ position (cyan), relative to the putative position of a bound DNA substrate (tan).
Effects of alanine substitution mutagenesis on EndA nuclease activity
Since the EndA(H160G) exhibited greatly increased nuclease activity in imidazole rescue experiments, relative to EndA(H160A), the greater dynamic range available with the glycine substitution allowed for a more precise measurement of small changes in catalytic activity. Therefore, we used this mutant as a ‘background’ for evaluating the effects of additional mutations. These mutants were then tested for levels of nuclease activity in the plasmid conversion assay and the single-radial enzyme diffusion (SRED) assay (42).
To date, attempts to crystallize EndA in the presence of various DNA substrates have been unsuccessful. To determine the residues involved in catalysis and DNA binding outside of the canonical DRGH motif, DNA from another ββα-Metal finger nuclease—VVN from Vibrio vulnificus [PDB ID code 1OUP (43)]—was superimposed into the active site of EndA (Figure 4) based on the ββα-Me motif. The superimposed DNA fits snugly into a shallow cleft on the surface of EndA, immediately outside the active site (Figure 4A). It should be noted that the disordered loop between β-strands 4 and 5 would likely lie in close proximity to the DNA, over the small cavity containing the catalytic center (Figure 4A and B). Upon detailed examination of the superimposed DNA into the EndA(H160A) structure, we mutated several residues in the immediate vicinity of the active site (Arg127/Lys128, His154, Gln186, Gln192, Asn202, Glu205 and Arg209/Lys210) to alanine and evaluated their effects on DNA substrate binding and/or catalysis. We included N191A in our study as a negative control, since that mutation was nearly as deactivating as alanine substitution for His160 (20), which is proposed to act as the general base (44). To elucidate the role of the ‘finger-loop’ in α-helix D (Figure 4C), we created alanine substitution mutants for Glu196/Tyr197 and a deletion mutant where residues Gln194-Thr199 were removed from the construct (ΔLoop). Given the ‘finger-loop’ position proximal to the putative DNA binding site, it is tempting to speculate that this feature could play a role in DNA substrate binding or recognition.
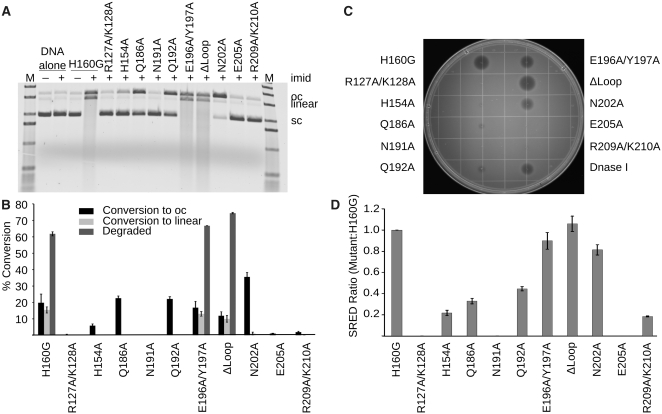
All alanine substitution mutants appeared indistinguishable from the original EndA(H160G) with regards to solubility, stability and behavior during size exclusion chromatography. Plasmid conversion nuclease activity assays with imidazole rescue were performed for each mutant, and evaluated with respect to EndA(H160G). These assays showed that, as expected, the H160G/N191A mutant displayed no detectable nuclease activity assay (Figure 5A and B). Therefore, mutations made on the H160G background, with the aid of imidazole rescue, behave similarly to the same mutations on wildtype background. The H160G/E205A and H160G/R127A/K128A mutations also had no detectable activity. H160G/H154A and H160G/R209A/K210A had minimal activity levels, while H160G/Q186A and H160G/Q192A were less affected. H160G/N202A showed activity levels nearly comparable to the original H160G alone. Surprisingly, the ‘finger-loop’ mutants showed no significant differences from the H160G mutant, though the ΔLoop variant appeared slightly more active.
Figure 5.
Nuclease activity assays for EndA variants generated on the background of H160G. (A) Plasmid conversion nuclease activity assay for EndA(H160G) variants generated on the background of H160G. Supercoiled plasmid substrate was incubated with EndA(H160G) variants in the presence or absence of 200 mM imidazole. The supercoiled substrate (sc) is first nicked to create the open circle (oc) species, further nicking activity eventually creates double-strand breaks, linearizing the plasmid. A high level of nuclease activity degrades the linearized substrate to small oligonucleotide fragments, which can be visualized as smears. (B) Bar graph illustrating the relative levels of nuclease activity for the EndA(H160G) variants. The error bars reflect the standard deviation for n = 3 replicates of this experiment. (C) Single radial enzyme diffusion assay for EndA(H160G) variants. A SRED agarose plate containing high molecular weight salmon sperm DNA, MgCl2 and ethidium bromide. EndA(H160G) variants were spotted onto this plate in equal amounts. Nuclease digestion is visualized by a dark ‘halo’ around each injection site. (D) Halo radii for each EndA(H160G) variant were measured, and are displayed on the bar graph as a ratio of the H160G background. The error bars reflect the standard deviation for n = 3 replications of this experiment.
As independent confirmation of these results, the nuclease activity was tested using the SRED assay (42). Equal amounts of the EndA(H160G) variants were spotted onto agarose plates containing MgCl2, high-molecular weight salmon sperm DNA, imidazole and ethidum bromide. Since ethidum bromide fluoresces only when bound to undigested DNA (45,46), nuclease digestion by the EndA variants results in decreased signal, which is visualized by a dark ‘halo’ surrounding the injection site. Enzymes with higher levels of nuclease activity are easily differentiated by a larger ‘halo’ (Figure 5C and 5D). Results from the SRED assay are consistent with those obtained from the plasmid conversion assay.
Delineation of EndA(H160G) nuclease activity and DNA substrate binding
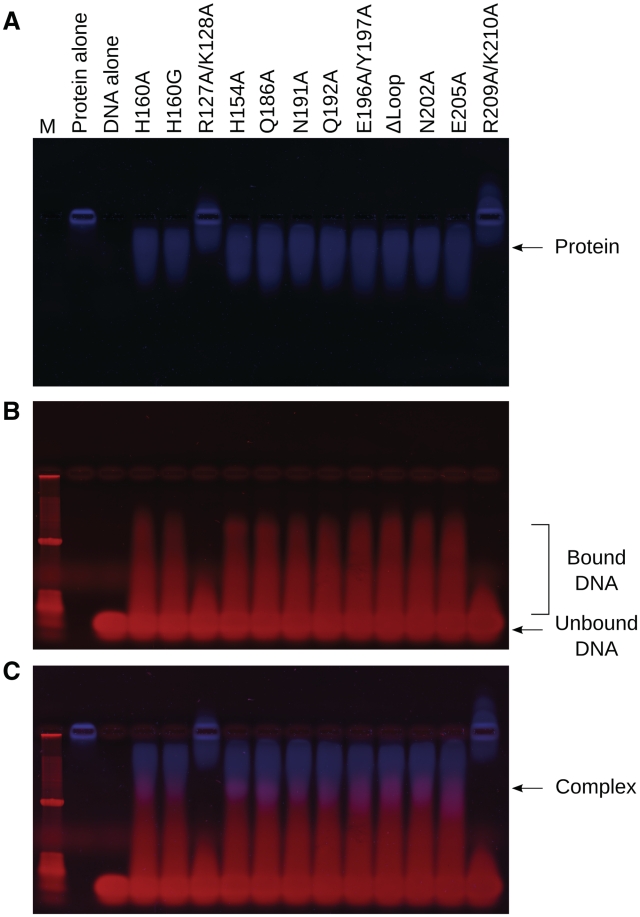
In order to discover whether loss of nuclease activity could be correlated with DNA substrate binding, an electrophoretic mobility shift assay was performed using all EndA(H160G) variants (Figure 6). Due to its high predicted pI (9.51), native gel electrophoresis of EndA(H160G) alone results in negligible migration of the protein (Figure 6A, lane 2). A complex formed by non-specific binding of the 24 base pair duplex DNA substrate to the protein (Figure 6B) alters the pI, enabling its migration toward the cathode. The DNA/protein complexes formed in this assay appear to be relatively unstable and of low affinity, presenting as a low-molecular weight smear migrating above the unbound DNA (Figure 6B and C). This behavior is consistent with the non-sequence specific nature of EndA interactions with DNA. Mutations which are thought to undermine the catalytic mechanism (H160G/N191A) would be expected to exhibit ‘wildtype’ substrate binding, but no nuclease activity. Interestingly, all variants except for the H160G/R127A/K128A and H160G/R209A/K210A mutants were capable of binding the DNA substrate to an extent similar to that of the ‘background’ H160G mutant (Figure 6C). These results suggest that residues Arg127/Lys128 and Arg209/Lys210 are likely involved in substrate binding. Conversely, the E205A mutation displays no alteration in substrate binding, yet has no detectable nuclease activity. Therefore, Glu205 appears to be crucial for catalysis.
Figure 6.
DNA-binding assays for EndA(H160G) variants. Equal amounts of EndA(H160G) proteins were mixed with a 24 base pair duplex DNA substrate under low salt conditions, then separated using an electrophoretic mobility shift assay. (A) Protein-containing species were stained with SYPRO Orange (blue, Sigma-Aldrich). (B) Molecular species containing DNA were stained with ethidium bromide (red) on the same gel. (C) Images showing DNA (red) and protein (blue) were merged using ImageJ (48), highlighting the DNA/protein complexes in purple.
Due to the markedly deleterious effects of the H160G/R127A/K128A and the H160G/R209A/K210A multiple mutants, individual mutations for each residue were created and analyzed for both DNA binding and nuclease activity. All four individual mutations (H160G/R127A, H160G/K128A, H160G/R209A and H160G/K210A) exhibited greatly decreased nuclease activity (Supplementary Figure S4A and B), though not to the extent of the multiple mutations. Interestingly, the individual mutants did not show as drastic of a reduction in DNA binding (Supplementary Figure S4C–E), compared to the two double mutants.
DISCUSSION
Previous biochemical studies with EndA from S. pneumoniae have been guided primarily by sequence alignments with other members of the nuclease superfamily. EndA contains a DRGH motif specific to some related nucleases. Residues of the DRGH motif (Asp157, Arg158 and His160) have been substituted with alanine, resulting in severely deleterious effects on catalytic activity (20). Beyond alignments of select active site motifs, however, low sequence conservation between EndA and other DRGH nucleases has hindered efforts to understand the function and behavior of this sequence non-specific nuclease. Therefore, in this work, we present the X-ray crystal structure of the ββα-metal finger EndA nuclease from S. pneumoniae, at 1.75 Å. Comparison of this structure with other nucleases within the same family reveals a conserved core β-sheet and ββα-metal finger architecture for the catalytic center. Aside from these conserved secondary structural elements, there is minimal similarity to other nuclease structures within this superfamily.
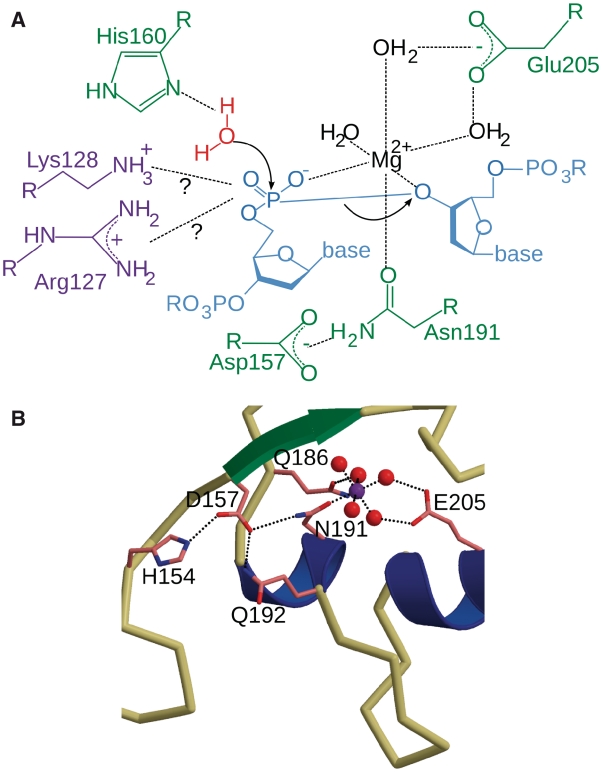
The catalytic mechanism for EndA is likely to be similar to that proposed for Serratia nuclease (34,35) and NucA (32), whereby His160 positions and activates a catalytic water molecule for nucleophilic attack on the scissile phosphate. Asn191 directly coordinates the catalytic metal ion believed to be involved in stabilization of the charge buildup at the transition state (Figure 7A). Based on their proximity to the nuclease active site, several residues were substituted with alanine. Despite its putative location within the DNA binding surface, Asn202 had no apparent effect on either DNA binding or catalysis. N202A displayed nuclease activity indistinguishable from the H160G background mutant. Residues His154, Gln186, Gln192 and Glu205 were identified as being catalytically relevant, the most crucial of these being Glu205. Based on the structure, Glu205 is homologous to Glu163 from NucA, Glu127 from Serratia nuclease and Glu195 from EndoG. Glu205 forms hydrogen bonds with two of the waters coordinating the magnesium ion, possibly aiding in active site assembly for catalysis (Figure 7B). The structure revealed that Gln186 forms a hydrogen bond with another of the coordinating water molecules. The Q186A substitution was not as catalytically deleterious as E205A, presumably related to the fact that Gln186 interacts with only one of the inner sphere water molecules coordinating the divalent ion. Residues His154 and Gln192 are positioned farther from the active site, in what could be described as ‘second shell’ active site residues. These residues form a hydrogen bond network, which likely helps stabilize the position of Asn191, using Asp157 of the DRGH motif as an intermediary (Figure 7B).
Figure 7.
EndA active site residues involved in catalysis. (A) Diagram of the putative catalytic mechanism employed by EndA. (B) Diagram of the residues in the active site that effect catalysis based on this study (ball-and-stick in pink). The magnesium ion is shown as a purple sphere, and its coordinating water molecules are shown in red.
In addition to catalytically important residues, we also identified several residues that appear to be important for binding of the DNA substrate. Double mutants R127A/K128A and R209A/K210A exhibited severely diminished nuclease activity that correlates with abolished DNA substrate binding. Therefore, some of these residues appear to be involved in binding of the DNA substrate. It is also possible that either Arg127 or Lys128 could play a catalytic role in stabilizing the transition state during phosphodiester bond hydrolysis (Figure 7), similar to the putative role of Arg93 in NucA (32). At present, determining which of these residues is more likely to assume this role is difficult, since Arg127 and Lys128 lie on a disordered loop and their exact positions are not defined. Furthermore, individual alanine substitution of Arg127, Lys128, Arg209 and Lys210 showed a more marked decrease in catalytic activity, in contrast to DNA binding capability. It appears likely that Arg127/Lys128 and Arg209/Lys210 act synergistically to bind the DNA for catalysis. This study presents the first reported incidence of structure-driven mutagenesis resulting in severely abrogated substrate binding by a nuclease in the ββα-metal finger superfamily. That substrate binding can be so greatly influenced by so few residues is intriguing, given the relatively non-specific interactions of EndA with DNA.
Interestingly, we could not determine a specific role for the unusual ‘finger-loop’ interruption of α-helix D. The E196A/Y197A and the ΔLoop deletion mutants were competent in both DNA substrate binding and in catalysis. It is tempting to speculate that this loop could be involved in substrate specificity—by sensing differences in substrate structure—in delineation of endonucleolytic versus exonucleolytic activity, or in protein–protein interactions with other components of the DNA uptake complex during competence.
To the best of our knowledge, structure-driven mutagenesis of EndA has yielded the first reported incidence of catalytic versus DNA binding separation of function for a ββα-metal finger nuclease sequence non-specific nuclease. This technique could also be applied to other members of the ββα-metal finger nuclease superfamily, which could prove invaluable due to the lack of sequence conservation between family members. Further characterization of the differences between EndA and other sequence non-specific nucleases will be critical for structure-based design of specific small-molecule inhibitors as therapeutics for S. pneumoniae infection.
ACCESSION NUMBER
PDB ID code 3OWV.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research was funded by the Division of Intramural Research of the National Institute of Environmental Health Sciences, US National Institutes of Health. Intramural Research Program of the National Institutes of Health; National Institute of Environmental Health Sciences; US Department of Energy, Office of Science, Office of Basic Energy Sciences, Advanced Photon (contract no. W-31-109-Eng-38). Funding for open access charge: The Intramural Research Program of the National Institute of Environmental Health Sciences; National Institutes of Health.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank D. Stumpo for generous donation of the pBluescript SK(+) and J. Krahn for insightful aid during manual model building. We thank J. Williams in the Protein Microcharacterization Core Facility for his expertise in mass spectrometry. We also thank G. Mueller and R. Williams for critical reading of the manuscript. A.F.M., crystallization of EndA(H160A), structural refinement and biochemical assays. M.M., G.M. and A.P., initial characterization of the EndA(H160A) mutant. R.E.L., critical review of experimental design. L.C.P., structural refinement, experimental design and oversight. All authors participated in manuscript preparation.
REFERENCES
- 1.Murphy TF, Bakaletz LO, Smeesters PR. Microbial interactions in the respiratory tract. Pediatr. Infect. Dis. J. 2009;28:S121–S126. doi: 10.1097/INF.0b013e3181b6d7ec. [DOI] [PubMed] [Google Scholar]
- 2.Lynch JP, 3rd, Zhanel GG. Streptococcus pneumoniae: epidemiology and risk factors, evolution of antimicrobial resistance, and impact of vaccines. Curr. Opin. Pulm. Med. 2010;16:217–225. doi: 10.1097/MCP.0b013e3283385653. [DOI] [PubMed] [Google Scholar]
- 3.Janoff EN, Breiman RF, Daley CL, Hopewell PC. Pneumococcal disease during HIV infection. Epidemiologic, clinical, and immunologic perspectives. Ann. Intern. Med. 1992;117:314–324. doi: 10.7326/0003-4819-117-4-314. [DOI] [PubMed] [Google Scholar]
- 4.Janoff EN, Rubins JB. Invasive pneumococcal disease in the immunocompromised host. Microb. Drug Resist. 1997;3:215–232. doi: 10.1089/mdr.1997.3.215. [DOI] [PubMed] [Google Scholar]
- 5.WHO. Pneumococcal vaccines. Weekly Epidemiological Record. 2003;78:110–119. [Google Scholar]
- 6.Mattei SM, Falleiros-Carvalho LH, Cavalcante NJ. Invasive pneumococcal disease in HIV seropositive children and adolescents. J. Pediatr. (Rio J.) 2008;84:276–280. doi: 10.2223/JPED.1762. [DOI] [PubMed] [Google Scholar]
- 7.Kim KS. Acute bacterial meningitis in infants and children. Lancet Infect. Dis. 2010;10:32–42. doi: 10.1016/S1473-3099(09)70306-8. [DOI] [PubMed] [Google Scholar]
- 8.Ortqvist A, Kalin M, Julander I, Mufson MA. Deaths in bacteremic pneumococcal pneumonia. A comparison of two populations–Huntington, WVa, and Stockholm, Sweden. Chest. 1993;103:710–716. doi: 10.1378/chest.103.3.710. [DOI] [PubMed] [Google Scholar]
- 9.Kadioglu A, Andrew PW. The innate immune response to pneumococcal lung infection: the untold story. Trends Immunol. 2004;25:143–149. doi: 10.1016/j.it.2003.12.006. [DOI] [PubMed] [Google Scholar]
- 10.Brinkmann V, Reichard U, Goosmann C, Fauler B, Uhlemann Y, Weiss DS, Weinrauch Y, Zychlinsky A. Neutrophil extracellular traps kill bacteria. Science. 2004;303:1532–1535. doi: 10.1126/science.1092385. [DOI] [PubMed] [Google Scholar]
- 11.Hirsch JG. Bactericidal action of histone. J. Exp. Med. 1958;108:925–944. doi: 10.1084/jem.108.6.925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weinrauch Y, Drujan D, Shapiro SD, Weiss J, Zychlinsky A. Neutrophil elastase targets virulence factors of enterobacteria. Nature. 2002;417:91–94. doi: 10.1038/417091a. [DOI] [PubMed] [Google Scholar]
- 13.Lacks S, Greenberg B, Neuberger M. Identification of a deoxyribonuclease implicated in genetic transformation of Diplococcus pneumoniae. J. Bacteriol. 1975;123:222–232. doi: 10.1128/jb.123.1.222-232.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lacks S, Neuberger M. Membrane location of a deoxyribonuclease implicated in the genetic transformation of Diplococcus pneumoniae. J. Bacteriol. 1975;124:1321–1329. doi: 10.1128/jb.124.3.1321-1329.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Puyet A, Greenberg B, Lacks SA. Genetic and structural characterization of endA. A membrane-bound nuclease required for transformation of Streptococcus pneumoniae. J. Mol. Biol. 1990;213:727–738. doi: 10.1016/S0022-2836(05)80259-1. [DOI] [PubMed] [Google Scholar]
- 16.Rosenthal AL, Lacks SA. Nuclease detection in SDS-polyacrylamide gel electrophoresis. Anal. Biochem. 1977;80:76–90. doi: 10.1016/0003-2697(77)90627-3. [DOI] [PubMed] [Google Scholar]
- 17.Lacks S, Greenberg B, Neuberger M. Role of a deoxyribonuclease in the genetic transformation of Diplococcus pneumoniae. Proc. Natl Acad. Sci. USA. 1974;71:2305–2309. doi: 10.1073/pnas.71.6.2305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Beiter K, Wartha F, Albiger B, Normark S, Zychlinsky A, Henriques-Normark B. An endonuclease allows Streptococcus pneumoniae to escape from neutrophil extracellular traps. Curr. Biol. 2006;16:401–407. doi: 10.1016/j.cub.2006.01.056. [DOI] [PubMed] [Google Scholar]
- 19.Buchanan JT, Simpson AJ, Aziz RK, Liu GY, Kristian SA, Kotb M, Feramisco J, Nizet V. DNase expression allows the pathogen group A Streptococcus to escape killing in neutrophil extracellular traps. Curr. Biol. 2006;16:396–400. doi: 10.1016/j.cub.2005.12.039. [DOI] [PubMed] [Google Scholar]
- 20.Midon M, Schafer P, Pingoud A, Ghosh M, Moon AF, Cuneo MJ, London RE, Meiss G. Mutational and biochemical analysis of the DNA-entry nuclease EndA from Streptococcus pneumoniae. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq802. doi:10.1093/nar/gkq802 [Epub ahead of print, 15 September] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Chayen NE. Comparative studies of protein crystallization by vapour-diffusion and microbatch techniques. Acta Crystallogr. D Biol. Crystallogr. 1998;54:8–15. doi: 10.1107/s0907444997005374. [DOI] [PubMed] [Google Scholar]
- 22.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 23.Moon AF, Mueller GA, Zhong X, Pedersen LC. A synergistic approach to protein crystallization: combination of a fixed-arm carrier with surface entropy reduction. Protein Sci. 2010;19:901–913. doi: 10.1002/pro.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Brunger AT, Adams PD, Clore GM, DeLano WL, Gros P, Grosse-Kunstleve RW, Jiang JS, Kuszewski J, Nilges M, Pannu NS, et al. Crystallography & NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 1998;54:905–921. doi: 10.1107/s0907444998003254. [DOI] [PubMed] [Google Scholar]
- 25.Terwilliger TC, Grosse-Kunstleve RW, Afonine PV, Moriarty NW, Zwart PH, Hung LW, Read RJ, Adams PD. Iterative model building, structure refinement and density modification with the PHENIX AutoBuild Wizard. Acta Crystallogr. D Biol. Crystallogr. 2008;64:61–69. doi: 10.1107/S090744490705024X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Adams PD, Afonine PV, Bunkoczi G, Chen VB, Davis IW, Echols N, Headd JJ, Hung LW, Kapral GJ, Grosse-Kunstleve RW, et al. PHENIX: a comprehensive Python-based system for macromolecular structure solution. Acta Crystallogr. D Biol. Crystallogr. 2010;66:213–221. doi: 10.1107/S0907444909052925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jones TA, Zou JY, Cowan SW, Kjeldgaard M. Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Crystallogr. A. 1991;47(Pt 2):110–119. doi: 10.1107/s0108767390010224. [DOI] [PubMed] [Google Scholar]
- 28.Lovell SC, Davis IW, Arendall WB, 3rd, de Bakker PI, Word JM, Prisant MG, Richardson JS, Richardson DC. Structure validation by Calpha geometry: phi,psi and Cbeta deviation. Proteins. 2003;50:437–450. doi: 10.1002/prot.10286. [DOI] [PubMed] [Google Scholar]
- 29.Thompson JD, Higgins DG, Gibson TJ. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 1994;22:4673–4680. doi: 10.1093/nar/22.22.4673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ho HC, Liao TH. Protein structure and gene cloning of Syncephalastrum racemosum nuclease. Biochem. J. 1999;339(Pt 2):261–267. [PMC free article] [PubMed] [Google Scholar]
- 31.Holm L, Rosenstrom P. Dali server: conservation mapping in 3D. Nucleic Acids Res. 2010;38(Suppl.):W545–W549. doi: 10.1093/nar/gkq366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh M, Meiss G, Pingoud A, London RE, Pedersen LC. Structural insights into the mechanism of nuclease A, a betabeta alpha metal nuclease from Anabaena. J. Biol. Chem. 2005;280:27990–27997. doi: 10.1074/jbc.M501798200. [DOI] [PubMed] [Google Scholar]
- 33.Ghosh M, Meiss G, Pingoud AM, London RE, Pedersen LC. The nuclease A-inhibitor complex is characterized by a novel metal ion bridge. J. Biol. Chem. 2007;282:5682–5690. doi: 10.1074/jbc.M605986200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Miller MD, Cai J, Krause KL. The active site of Serratia endonuclease contains a conserved magnesium-water cluster. J. Mol. Biol. 1999;288:975–987. doi: 10.1006/jmbi.1999.2729. [DOI] [PubMed] [Google Scholar]
- 35.Shlyapnikov SV, Lunin VV, Perbandt M, Polyakov KM, Lunin VY, Levdikov VM, Betzel C, Mikhailov AM. Atomic structure of the Serratia marcescens endonuclease at 1.1 A resolution and the enzyme reaction mechanism. Acta Crystallogr. D Biol. Crystallogr. 2000;56:567–572. doi: 10.1107/s090744490000322x. [DOI] [PubMed] [Google Scholar]
- 36.Loll B, Gebhardt M, Wahle E, Meinhart A. Crystal structure of the EndoG/EndoGI complex: mechanism of EndoG inhibition. Nucleic Acids Res. 2009;37:7312–7320. doi: 10.1093/nar/gkp770. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Richardson JS, Richardson DC. Natural beta-sheet proteins use negative design to avoid edge-to-edge aggregation. Proc. Natl Acad. Sci. USA. 2002;99:2754–2759. doi: 10.1073/pnas.052706099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Huang S, Tu SC. Identification and characterization of a catalytic base in bacterial luciferase by chemical rescue of a dark mutant. Biochemistry. 1997;36:14609–14615. doi: 10.1021/bi9722554. [DOI] [PubMed] [Google Scholar]
- 39.Newmyer SL, de Montellano PR. Rescue of the catalytic activity of an H42A mutant of horseradish peroxidase by exogenous imidazoles. J. Biol. Chem. 1996;271:14891–14896. doi: 10.1074/jbc.271.25.14891. [DOI] [PubMed] [Google Scholar]
- 40.Lehoux IE, Mitra B. (S)-Mandelate dehydrogenase from Pseudomonas putida: mutations of the catalytic base histidine-274 and chemical rescue of activity. Biochemistry. 1999;38:9948–9955. doi: 10.1021/bi9907532. [DOI] [PubMed] [Google Scholar]
- 41.McCartney SA, Brignole EJ, Kolegraff KN, Loveland AN, Ussin LM, Gibson W. Chemical rescue of I-site cleavage in living cells and in vitro discriminates between the cytomegalovirus protease, assemblin, and its precursor, pUL80a. J. Biol. Chem. 2005;280:33206–33212. doi: 10.1074/jbc.M506876200. [DOI] [PubMed] [Google Scholar]
- 42.Nadano D, Yasuda T, Kishi K. Measurement of deoxyribonuclease I activity in human tissues and body fluids by a single radial enzyme-diffusion method. Clin. Chem. 1993;39:448–452. [PubMed] [Google Scholar]
- 43.Li CL, Hor LI, Chang ZF, Tsai LC, Yang WZ, Yuan HS. DNA binding and cleavage by the periplasmic nuclease Vvn: a novel structure with a known active site. EMBO J. 2003;22:4014–4025. doi: 10.1093/emboj/cdg377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Huang H, Yuan HS. The conserved asparagine in the HNH motif serves an important structural role in metal finger endonucleases. J. Mol. Biol. 2007;368:812–821. doi: 10.1016/j.jmb.2007.02.044. [DOI] [PubMed] [Google Scholar]
- 45.Kim HS, Liao TH. Isoelectric focusing of multiple forms of DNase in thin layers of polyacrylamide gel and detection of enzymatic activity with a zymogram method following separation. Anal. Biochem. 1982;119:96–101. doi: 10.1016/0003-2697(82)90671-6. [DOI] [PubMed] [Google Scholar]
- 46.Yasuda T, Mizuta K, Ikehara Y, Kishi K. Genetic analysis of human deoxyribonuclease I by immunoblotting and the zymogram method following isoelectric focusing. Anal. Biochem. 1989;183:84–88. doi: 10.1016/0003-2697(89)90175-9. [DOI] [PubMed] [Google Scholar]
- 47.DeLano WL. The PyMOL Molecular Graphics System. San Carlos, CA: DeLano Scientific; 2002. [Google Scholar]
- 48.Collins TJ. ImageJ for microscopy. Biotechniques. 2007;43:25–30. doi: 10.2144/000112517. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.