Abstract
Muscleblind-like 1 (MBNL1) is a splicing regulator that controls developmentally regulated alternative splicing of a large number of exons including exon 11 of the Insulin Receptor (IR) gene and exon 5 of the cardiac Troponin T (cTNT) gene. There are three paralogs of MBNL in humans, all of which promote IR exon 11 inclusion and cTNT exon 5 skipping. Here, we identify a cluster of three binding sequences located downstream of IR exon 11 that constitute the MBNL1 response element and a weaker response element in the upstream intron. In addition, we used sequential deletions to define the functional domains of MBNL1 and MBNL3. We demonstrate that the regions required for splicing regulation are separate from the two pairs of zinc-finger RNA-binding domains. MBNL1 and MBNL3 contain core regulatory regions for both activation and repression located within an 80-amino-acid segment located downstream of the N-terminal zinc-finger pair. Deletions of these regions abolished regulation without preventing RNA binding. These domains have common features with the CUG-BP and ETR3-like Factor (CELF) family of splicing regulators. These results have identified protein domains required for splicing repression and activation and provide insight into the mechanism of splicing regulation by MBNL proteins.
INTRODUCTION
Ninety-four percent of human genes contain introns such that following transcription into a pre-mRNA, exons are joined and introns are removed to produce mature mRNA. This process takes place in the nucleus and is catalyzed by the spliceosome (1). Alternative splicing produces multiple mRNAs from individual genes most often resulting in expression of different protein isoforms (2). More than 90% of human genes express pre-mRNAs that undergo alternative splicing and more than 50% of the mRNA species generated differ between tissues (3,4). Thus, alternative splicing is a major mechanism for generating and regulating the expression of protein isoform diversity.
Inclusion of an exon into the mature mRNA depends on the efficiency of spliceosome recruitment to the flanking splice sites. In general, splicing is suboptimal for most alternative splice sites due to at least one weak feature such as non-consensus splice site, suboptimal exon length, strong competing splice sites or RNA secondary structure (5). Inefficient exon recognition allows for modulated exon use by trans-acting factors, primarily RNA binding proteins, which bind to cis-acting elements within the exon or flanking introns, usually within 300 nt of the regulated splice site(s) (6). Alternative splicing is often regulated through combinatorial control by a splicing code made up of multiple cis-acting elements. These elements are bound by proteins that can have either positive or negative effects to ultimately control exon identification and regulated splicing (7,8). In addition, splicing can be regulated without specific auxiliary splicing factors, indicating a likely role for components of the basal splicing machinery as modulators of alternative splicing (9).
Muscleblind-like 1 (MBNL1) is an RNA-binding protein that functions as both a positive and negative splicing regulator (10). All three human paralogs (MBNL1, MBNL2 and MBNL3) contain four CCCH zinc-finger domains which are structured in pairs and function as RNA-binding domains (11,12). We previously identified a MBNL-binding motif [YGCU(U/G)Y] required for negative regulation of cardiac troponin T (cTNT) exon 5 (10). Recently, other groups have refined this to the YGCY motif with a preference for UGCU (13,14). Except for the RNA-binding domains, MBNL proteins do not contain recognizable domains that could provide information about the mechanism of splicing regulation. A critical question toward understanding the mechanisms by which RNA-binding proteins regulate splicing is how the proteins communicate with the splicing machinery to promote exon inclusion or skipping. To understand this mechanism, it is critical to identify the regions of the protein that are required for splicing repression and/or activation.
Splicing misregulation by MBNL is a key component in the pathogenesis of the neuromuscular disorder myotonic dystrophy (DM). DM is an autosomal dominant disease caused by expansion of CTG or CCTG microsatellite repeats in the DMPK or ZNF9 gene, respectively (15–17). Pathogenesis results from a toxic RNA gain-of-function mechanism in which CUG- or CCUG-repeat-containing RNA transcribed from the expanded allele form RNA foci that sequester and reduce MBNL activity without affecting the protein or mRNA levels (11,18). Loss of MBNL activity results in abnormal regulation of at least 200 alternative splicing events (13,14), including cardiac Troponin T (cTNT) exon 5, insulin receptor (IR) exon 11 and exon 7a of the muscle-specific chloride channel (ClC1) resulting in specific features of the disease (19). MBNL1 knockout mice reproduced the phenotype of the disease (20) as do MBNL2 knockout mice, although to a lesser degree (21), highlighting the relevance of this family of splicing regulators to DM pathogenesis.
The IR isoform that contains exon 11 (IR-B) is expressed in adult tissues, whereas the embryonic isoform (IR-A) lacks exon 11 (22,23). The expression of the two isoforms is also tissue specific (24). IR-A and IR-B function differently such that insulin binding to IR-B efficiently transduces the signal while the kinase activity of the IR-A receptor isoform is reduced and the signal is less efficiently transduced (25,26). Sequestration of MBNL1 in DM tissues results in exon 11 skipping and reduced insulin signaling, contributing to an insulin resistance phenotype (22,23).
In this study, we defined three MBNL1-binding sites located 93 nt downstream from IR exon 11. We demonstrated that the binding sites serve as the major response element for splicing regulation by MBNL1 and MBNL3. We also demonstrate that a segment of intron 10 that is proximal to the exon responds to MBNL expression, although direct binding to this region was minimal. We also used sequential deletions of MBNL1 and MBNL3 to identify protein domains required for both splicing activation (IR exon 11) and repression (cTNT exon 5). We found that the domains required for splicing activity are physically and functionally distinguished from the RNA-binding domains. It is likely that regions required for splicing regulation mediate protein–protein interactions between MBNL and spliceosomal components or co-regulators involved in MBNL-mediated responses.
MATERIALS AND METHODS
In vitro transcription and gel-shift
RNA probes used in gel-shift assays were synthesized from polymerase chain reaction (PCR) products using primers with the T7 promoter sequence upstream of the region of interest. 32P-UTP-labeled RNA was incubated with reaction mix (yeast tRNA 0.5 mg/ml, heparin 2 mg/ml, BSA 50 μg/ml, rATP 0.5 mM, KCl 75 mM) to a final concentration of 10−7 M with or without His6-MBNL1 at 30°C for 30 min and immediately analyzed by electrophoresis at 200 V for 2 h on a 5% (acrylamide:bis-acrylamide, 37.5:1) native gel in 1x Tris-boric acid-EDTA at room temperature. Gels were pre-run at 250 V for 30 min prior to loading. Gels were dried down and the bands were visualized by autoradiography. The ratio of bound RNA/total RNA was determined using Typhoon Trio phosphorimager (GE Healthcare, UK). Apparent Kd values were calculated as the amount of MBNL1 where 50% of the RNA was shifted.
Recombinant protein
Human MBNL1 protein (41 kDa isoform, NP_066368) was cloned into the pET30a (+) vector at the KpnI and BamHI sites, which contains a His6-tag and induced in BL21 bacteria cells by 0.4 mM IPTG for 4 h at 30°C. Cells were harvested by centrifugation at 6000 g for 10 min and resuspended in extraction buffer [25 mM HEPES (pH 7.5), 20% glycerol, 1 mM DTT, 20 mM NaCl, 0.2 mM EDTA, 0.05% SDS, 1% TritonX-100 and proteinase inhibitor cocktail buffer]. Cells were lysed by sonicating three times for 10 s each. Supernatant was collected after centrifugation at 14 000 g for 10 min. The protein was purified using the Novagen kit according to the manufacturer’s protocol.
Cell culture
QT35 quail fibrosarcoma cells were grown in F-10 media supplemented with 5% fetal bovine serum, 1% chicken serum, L-glutamine, penicillin–streptomycin (Gibco; Invitrogen, Carlsbad, CA, USA) and 2% Tryptose phosphate (Sigma-Aldrich, St. Louis, MO, USA). COSM6 cells were grown in Dulbecco’s modified Eagle medium supplemented with 10% fetal bovine serum, 1% L-glutamine and penicillin–streptomycin.
Transient transfections
QT35 and COSM6 cells were plated in six-well plates at 200 000 or 80 000 cells/well, respectively. Transfection was performed 24 h later after fresh medium was added. Minigene plasmid (500 ng) and 500 ng of plasmid encoding MBNL1, MBNL3, or empty vector were transfected using Fugene6 (Roche, Indianapolis, IN, USA) according to the manufacturer’s directions. RNA and protein were extracted 48 h post-transfection.
Minigenes and MBNL plasmids
For the minigenes used in Figure 4, the region under study was cloned into SalI and XbaI sites of the RHCglo construct that has been described previously (27). The IRN minigene was used to assay splicing activity of the deletion mutants of MBNL1 and MBNL3 and it has been described previously (28). The MBNL1 and MBNL3 deletion mutants were cloned into the p-EGFP-C1 vector (Clontech Laboratories, Inc, Mountain View, CA, USA) at SalI and EcoRI sites downstream of the GFP protein. The endpoints of each of deletion are presented in Supplementary Figure 2.
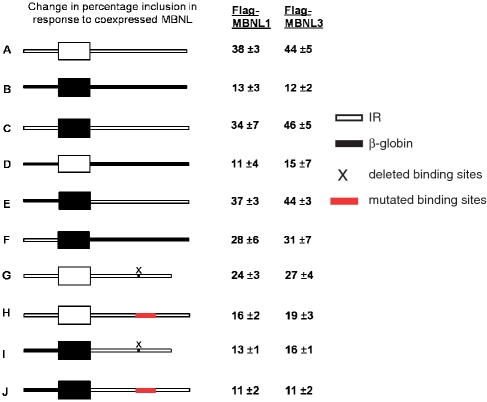
Figure 4.
The region downstream of IR exon 11 bound by MBNL1 is required for regulation by MBNL1. (A–J) Diagram of the genomic segments tested in the RHCglo minigene to identify which segment of the 273-nt region contains the MBNL response element. White indicates IR wild-type sequences and black indicates heterologous sequences from the human β-globin gene. X indicates deletion of the 30-nt sequence containing the binding site and the red box indicates the binding site with mutated sequences, which abolish MBNL1 direct interaction (substitutions from probe 8, Figure 3). The minigenes were co-transfected into QT35 quail fibroblasts with plasmids expressing Flag-MBNL1, Flag-MBNL3 or empty vector and splicing was determined by RT–PCR. The numbers indicate the change in percentage inclusion compared to basal inclusion levels (no MBNL). Results are from three independent experiments.
Reverse transcription–PCR and western blot analysis
RNA isolation and reverse transcription–PCR (RT–PCR) for all minigenes were performed as described previously (29). Primers RSV5U and TNIE4 (27) were used for all minigenes except IRN for which IR-D and IR-U were used as described (30). PCR product bands were quantified by using the Kodak Gel Logic 2200 with Molecular Imaging Software. Protein expression was confirmed by western blotting as described previously (27). Protein extracts from cells were resolved with SDS/PAGE, followed by transfer to PVDF membrane and antibody incubation. Anti-FLAG M2 peroxidase conjugate monoclonal antibody (Sigma-Aldrich, St. Louis, MO, USA) was used at 1:5000 for 1 h at room temperature and anti-GFP HRP-conjugate (Santa Cruz Biotechnology, Santa Cruz, CA, USA) at 100 ng/ml for 2 h at room temperature. Anti-MBNL1 polyclonal antibody has been described previously (31). Anti-MBNL3 monoclonal antibody was provided by G. Morris (32). Both antibodies were used at 1:1000 at 4°C overnight. Sheep anti-mouse (Jackson ImmunoResearch, West Grove, PA, USA) and goat anti-rabbit (Calbiochem, Gibbstown, NJ, USA) secondary antibodies were used at 1:10 000 for 1 h at room temperature.
In situ hybridization
COSM6 cells were plated on lysine-coated coverslips in six-well plates and transfected with DT960 (which expresses RNA containing 960 interrupted CUG repeats) (10) and GFP-tagged MBNL. Forty-eight hours post-transfection, coverslips were washed with phosphate-buffered saline (PBS) and fixed with 4% paraformaldehyde. Permeabilization took place with 0.02% Triton X-100 followed by incubation with 40% formamide and Cy3-(CAG)5 probe. The coverslips were mounted and DAPI stained using Vectashield hard-set mounting media.
RESULTS
MBNL1 binds to a 30-nt segment located 93 nt downstream of IR exon 11
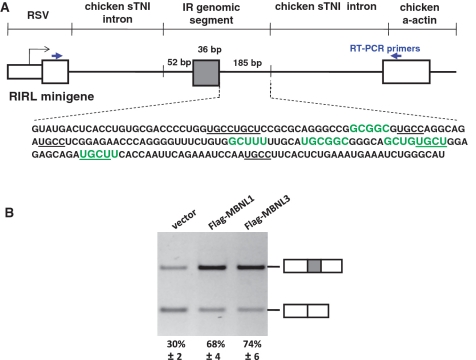
We have previously shown that all three MBNL paralogs strongly promoted inclusion of IR exon 11 (10). The minigenes used in the previous studies contained large segments of introns 10 and 11 as well as the flanking exons (28). To identify the cis-acting elements required for the splicing response to MBNL, we inserted an IR genomic segment containing exon 11, 52 nt of the upstream intron and 185 nt of the downstream intron into the RHCglo minigene reporter (27) to generate the RIRL minigene (Figure 1A). RIRL was co-expressed with Flag-tagged MBNL1 or MBNL3 or the empty expression vector. The experiments throughout used the 41-kDa isoform of human MBNL1 (NP_066368) and the isoform of human MBNL3 represented by CAI43107. Primers complementary to the flanking exons were used for RT–PCR to determine the percent of mRNAs that include the alternative exon (Figure 1A and ‘Materials and Methods’ section). The results demonstrate that the 273-nt IR genomic fragment was sufficient for a response to MBNL that was comparable to the minigene containing the larger genomic segment (Figure 1B and ref. 10). To determine whether MBNL1 and MBNL3 differed in their intrinsic activity, we transfected increasing amounts of Flag-tagged MBNL1 or MBNL3 plasmids with the RIRL minigene to express different levels of protein and compared the response of exon 11 inclusion. The results showed that when the two paralogs were expressed at comparable levels, MBNL3 induced a stronger response of exon 11 splicing. Western blots for MBNL1 and MBNL3 were performed to assay for increase in MBNL protein expression (Supplementary Figure 1).
Figure 1.
A 273-bp IR genomic segment is sufficient for MBNL response and contains MBNL consensus binding sites in the downstream intron. (A) The RIRL minigene was generated by inserting the indicated 273-bp segment into the RHCglo minigene (27). Five putative MBNL-binding sites were found using the consensus sequences originally identified (highlighted in green) (10) or the minimal motif YGCY (underlined) (14). (B) Exon 11 and the proximal flanking intronic sequences are sufficient for alternative splicing regulation by MBNL1 and MBNL3. QT35 quail fibroblasts were transfected with vector only, or plasmids expressing Flag-MBNL1 or Flag-MBNL3. The averaged RT–PCR results from three independent transfections are shown as percentages of mRNA including the alternative exon.
We previously identified the motif YGCU(U/G)Y as a potential MBNL-binding site (10) and a recent study showed that YGCY is sufficient for MBNL1 binding (14). Five motifs that were identical or similar to the predicted binding sites were identified in the 273 nt IR genomic fragment and all were in the downstream intron (highlighted in Figure 1A). The core YGCY motif was present only on the last two identified motifs.
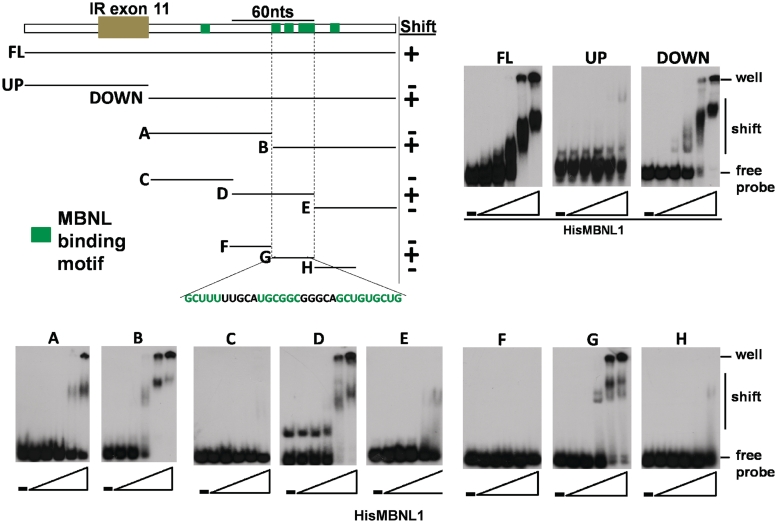
To identify where MBNL1 binds within the MBNL1-responsive 273 nt IR RNA segment, we performed a systematic analysis using a gel-shift binding assay. Increasing amounts of purified recombinant His6-tagged MBNL1 protein were incubated with in vitro transcribed and radiolabeled RNA. Bound and unbound RNA were separated on non-denaturing gels. MBNL1 bound to the RNA transcribed from the whole 273 nt IR genomic fragment (FL) with high affinity (Figure 2 and Supplementary Table 1). The region was separated into the downstream intronic segment (DOWN) and upstream with the exon (UP). The affinity of MBNL1 to the downstream segment was nearly as strong as to the FL while binding to the upstream fragment was barely detectable (Figure 2 and Supplementary Table 1). The 185-nt-long downstream segment was divided into smaller fragments of either 92 nt (A and B) or 61 nt (C, D and E), and MBNL1 bound with highest affinity to fragments B and D. These two fragments have an overlap of 30 nt from 90 to 120 nt downstream of the regulated exon that contains three of the five predicted binding sites. A gel-shift assay showed that MBNL1 binds to this region (G) with affinity approaching the larger FL and DOWN segments but not to the immediately adjacent upstream or downstream neighboring fragments (F and H) (Figure 2 and Supplementary Table 1). From these results, we conclude that MBNL1 binds predominantly to a 30-nt RNA segment downstream of exon 11 that contains three potential binding motifs and only one of them contains the core YGCY motif.
Figure 2.
Systematic analysis demonstrated that MBNL1 binds to a region with three binding motifs on intron 11 of IR pre-mRNA. (Upper left panel) Diagram of the RNA probes used to assay direct interactions with MBNL1 in a gel-shift assay. Green boxes indicate potential binding sites. The minimal region for direct interaction is probe G and the sequence is indicated. Gel-shift images are shown in the upper right panel and lower panel. RNA probes and increasing amounts of His6-tagged MBNL1 were incubated and complexes were separated on a native acrylamide gel. The RNA/MBNL1 molar ratios were 1/0, 1/0.33, 1/1, 1/3, 1/10 and 1/33. All gel-shift assays in Figures 2 and 3 were performed with the same preparation of recombinant MBNL1 to allow for direct comparisons of relative affinities. Apparent Kd values are listed in Supplementary Table 1.
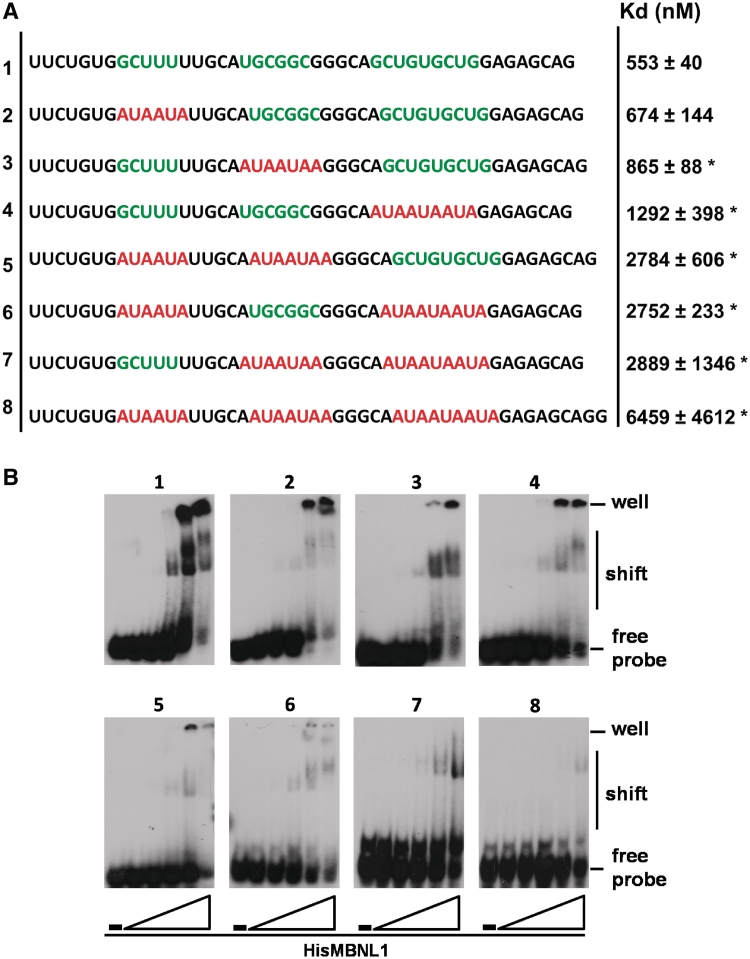
To directly test whether the putative binding motifs shown in Figure 1A were required for MBNL1 binding, we used a 45-nt RNA probe containing combinations of wild type and mutated putative MBNL-binding sequences. Each binding motif was mutated with AUAAUA, which was previously shown to abolish binding of MBNL1 to RNA (Figure 3A) (10). First, RNA containing the WT sequence (Probe 1) was compared to RNAs in which each motif was individually mutated (Probes 2, 3 and 4). The mutations in two of the three motifs demonstrated that each motif contributed to binding (Probes 5, 6 and 7). Importantly, when all three sites were mutated, binding was nearly abolished (Probe 8) (Figure 3A and B, and Supplementary Table 1), demonstrating that these three motifs are primarily responsible for binding of MBNL1 to the genomic segment sufficient for MBNL-mediated regulation of IR exon 11.
Figure 3.
All three MBNL recognition motifs contribute to MBNL1 binding. (A) Sequences of the RNA probes used for the gel-shift assay. Wild-type sequences of the binding sites (contained in the region encompassed by probe G, Figure 2) are indicated in green and mutated sequences are indicated in red. The column on the right indicates the average of the apparent Kd values obtained from three independent experiments along with the standard deviation values (*P < 0.05). (B) Images of gel shifts. RNA probes alone and increasing amounts of His6-tagged MBNL1 were incubated and run on a native acrylamide gel. The RNA/MBNL1 molar ratios were 1/0, 1/0.33, 1/1, 1/3, 1/10 and 1/33.
The MBNL-binding sites comprise the primary response element for splicing regulation
To determine whether the binding sites can function as response elements for MBNL1 and MBNL3, we performed the in vivo splicing assay using modified RIRL minigenes transfected into QT35 quail fibroblast cells. Co-expression of the RIRL minigene with MBNL1 or MBNL3 produced a 38- and 44-percentage-point increase in IR exon 11 inclusion, respectively (Figures 1B and 4). To determine whether the effect is specific to the IR genomic segment, we used the RHCglo minigene that was the parental construct for RIRL and contains an artificial alternatively spliced exon and flanking intronic segments derived from the human β-globin gene (27) (Figure 4, construct B). While the artificial exon is not expected to be responsive to MBNL1, it showed 13- and 12-point increase in percentage inclusion for MBNL1 and MBNL3, respectively, indicating the background level of response. For a systematic analysis of which segment of the IR genomic segment contains the MBNL response element, we separately tested the exon and each flanking intron for MBNL-dependent regulation. The heterologous exon flanked by both IR introns (construct C) showed a change in percentage inclusion that was the same as the RIRL wild-type construct (construct A), whereas the exon alone (construct D) gave a response similar to background (Figure 4), indicating that the response element lies outside of the alternative exon. When the intronic segments were tested individually, the downstream intron (construct E) showed as high a response to MBNL1 and MBNL3 as the RIRL minigene. The upstream intron (construct F) also showed a mild response (Figure 4). This result was unexpected because direct binding of MBNL1 to RNA containing this region was minimal (Figure 2, upper panel). The results suggest the presence of a response element upstream of exon 11 and MBNL is recruited to this site through an indirect mechanism. Since the response was not as strong as the downstream intron, these results indicate that the predominant response element for MBNL lies downstream of exon 11.
The results from the transfection analysis strongly suggested that the binding sites identified in the gel-shift assay serve as the predominant response elements for MBNL proteins. To test this hypothesis, we either deleted the region containing all three binding sites or introduced the same substitutions that were used to abolish binding of MBNL1 (Probe 8, Figure 3A and B). Deleting the sequences produced a reduced response for MBNL1 and MBNL3 (24 and 27 percentage points, respectively), while the mutations of these sites reduced the response to background levels (constructs G and H, respectively, Figure 4). The reason for the residual activity upon deletion of the MBNL-binding site is unclear, but could be related to the residual activity in the upstream intronic segment observed in construct F. For this reason, we used the minigene containing only the IR downstream intron segment containing the same deletion or substitution of the MBNL-binding site. In this architecture, both the deletion and the substitution reduced the response to the background levels (constructs I and J, Figure 4). These results show that the MBNL-binding sites defined in Figures 2 and 3 serve as the MBNL splicing response elements.
MBNL contains regions required for splicing activity that are separate from the RNA binding domains
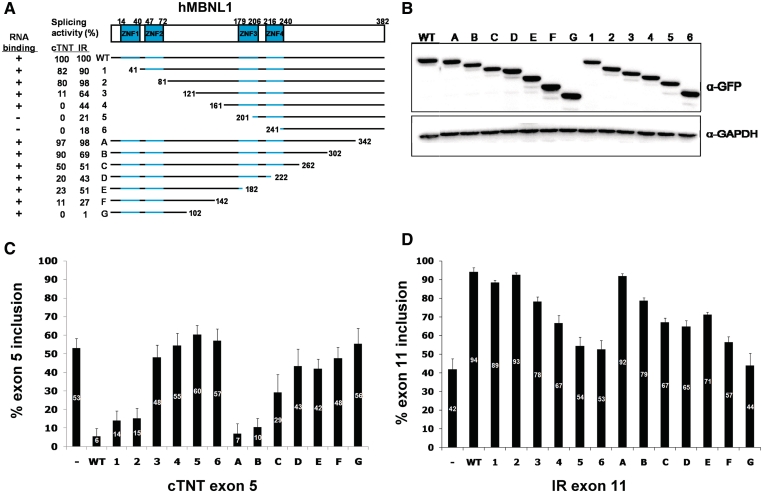
The human MBNL paralogs contain four zinc-finger RNA-binding domains which are paired near the N-terminus (ZNF1 and ZNF2) and in the center of the protein (ZNF3 and ZNF4) (Figure 5A). The homology between the two pairs is very high (33,34); however, the remainder of the protein does not contain recognizable functional domains. To identify region(s) of MBNL1 that are required for splicing activity, we generated sequential 40 amino acid deletions of MBNL1 from either the N-terminus or the C-terminus (Figure 5A, deletion endpoints are indicated on the protein sequence in Supplementary Figure 2). These deletions were made in proteins in which GFP was fused to the N-terminus and expression of the proteins was assayed with a GFP antibody (Figure 5B). We tested the effects of the deletions on splicing activation using the IRN insulin receptor minigene (28) and on splicing repression of human cTNT exon 5 using the previously described minigene (29). The splicing results are shown in Figure 5C and D, and in Figure 5A the activity of each deletion was normalized to full-length MBNL1 which is considered 100%.
Figure 5.
The regions of MBNL1 required for splicing activation and repression are located predominantly between the two zinc-finger pairs. (A) Diagram of MBNL1 deletion mutants. The upper panel shows the protein structure. There are two zinc-finger pairs (ZNF1–4). The N-terminal deletion mutants are named 1–6 and C-terminal mutants A-G. All mutants contain an N-terminal GFP tag. The columns on the left show the percentage splicing activity on cTNT exon 5 (repression) and IR exon 11 (activation) relative to full-length protein (WT). The first column shows whether each mutant can bind to CUG-repeat RNA (Figure 7). (B) Western-blot results of the transfected GFP-tagged deletion mutants. (Upper panel) Blot probed with HRP-tagged GFP antibody. (Lower panel) Blot probed with GAPDH antibody. (C, D) RT–PCR results of inclusion levels of cTNT exon 5 (C) and IR exon 11 (D) when COSM6 cells were transfected with MBNL1 WT, deletion mutants or empty vector. The results are from three independent experiments.
For the N-terminal deletions, splicing repression of cTNT exon 5 was essentially lost by deletion mutant 3, particularly between mutants 2 and 3 (Figure 5A and C), while activation of IR exon 11 was lost incrementally with each subsequent deletion (Figure 5A and D). The loss of repression activity of mutants 3 and 4 is not due to general protein misfolding, as these deletion mutations exhibit 64% and 44% of full-length activity for activation of IR exon 11. For the C-terminal deletions, while activity was decreased by deletions downstream of the central zinc-finger domains, neither repression nor activation was lost until the majority of the segment between the two pairs of RNA-binding domains was deleted. The conclusions from the MBNL1 deletion analysis are: (i) regions separate from the RNA-binding domains are required for splicing activation (mutants 2 and E). For splicing repression, the second pair of zinc fingers is important for splicing activity (mutants D, E and F); (ii) either the N-terminal or central pair of zinc-finger RNA-binding domains is sufficient for MBNL1 to function as a splicing activator of IR (mutants 2 and E). This does not seem to be the case for splicing repressor activity of MBNL1 in which mutant E has only 23% of the activity of full-length protein; (iii) MBNL1 has more than one activation domain since mutants 4 and F retain the ability to activate IR exon 11 inclusion but do not contain overlapping regions; (iv) two regions are required simultaneously for ≥20% activity of full-length protein for repression of cTNT exon 5 (mutants 3 and D).
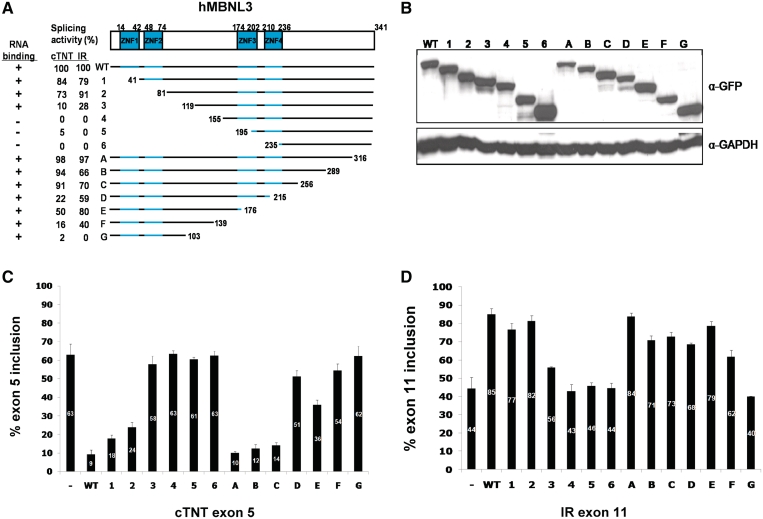
MBNL3 is nearly identical to MBNL1 in the zinc-finger domains but substantially differs within the regions found to contain activation and repression domains (Supplementary Figure 2). To gain further insight into the regions and possibly specific amino acids required for splicing activation and repression, we performed a similar functional analysis of sequential N-terminal and the C-terminal deletions of MBNL3 (Figure 6A). Deletion endpoints were at the comparable positions determined by alignment of MBNL1 and MBNL3 (Supplementary Figure 2). As for MBNL1, both splicing repression and splicing activation were assayed (Figure 6C and D, respectively) and the splicing activity was normalized to the full-length protein (Figure 6A). The full-length and deletion mutants were N-terminal GFP-tagged and expression of the proteins was assayed by western blots using a GFP antibody (Figure 6B).
Figure 6.
Splicing activation and repression regions of MBNL3 reside between the two zinc-finger pairs. Panels are the same as for MBNL1 in Figure 5. The MBNL3 isoform used contains 342 amino acids; however, the initiating Met was deleted from the GFP-MBNL3 fusion protein to prevent internal translation initiation.
As for MBNL1, either pair of zinc-finger domains in MBNL3 was sufficient for positive regulation and MBNL3 retains at least 50% activity containing one pair of zinc fingers (mutants 2 and E). Unlike MBNL1, the region from residues 80 to 155 is required for both activation and repression (mutants 2 and 4). This region is active whether it is associated with the N-terminal or central zinc-finger RNA-binding domain (mutants 2 and E). Strikingly, MBNL3 mutant E has 80% of the activity of full-length protein, although it contains only the N-terminal zinc-finger domains and the downstream 102 residues. Deletion of an additional 37 amino acids produced a protein that retained 40% of full-length activity (mutant F). To further narrow down the region of splicing activation, we used smaller deletions between mutants F and G. This analysis demonstrated that deletion of an additional 8 amino acids from mutant F (positions 131–139, mutant G1), lost all splicing activity (Supplementary Figure 3). Replacing the deleted MBNL3 segment with the heterologous C-terminal 42 amino acids of luciferase did not restore activity, suggesting that the specific residues removed are required for splicing activity (data not shown).
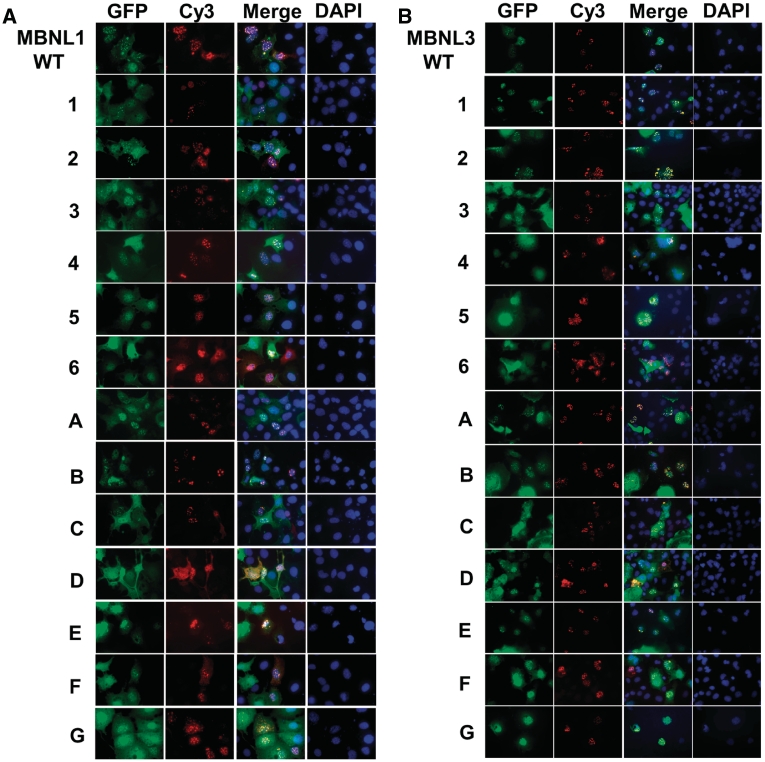
Deletion mutants that lack splicing activity retain RNA-binding activity in vivo
The regions required for splicing activity were found to be separate from the RNA-binding domains; however, it was possible that the deletions that defined these regions disrupted RNA binding and that loss of splicing activity was secondary to loss of RNA binding. To determine whether or not loss of splicing activity correlated with a loss of RNA binding in vivo, we took advantage of the observation that MBNL1 and MBNL3 bind to and colocalize with nuclear foci of CUG-repeat RNA (11,12). The same MBNL1 and MBNL3 deletion mutants tested for splicing activity in Figures 5 and 6 were co-expressed with a plasmid that expresses RNA containing 960 CUG repeats. We have previously shown that co-expressed GFP-MBNL1 colocalizes with nuclear foci of CUG repeat RNA that accumulate from this plasmid (10). Colocalization was visualized using fluorescence of GFP fusion proteins and in situ hybridization using a Cy3-labeled CAG locked nucleic acid (LNA) probe to detect the CUG repeat RNA foci (Figure 7). For MBNL1, mutants 5 and 6 were the only mutants that did not colocalize with RNA foci and these are also the only deletions that do not contain one intact pair of zinc fingers (Figures 5A and 7A). From this result, we conclude that at least one pair of zinc fingers is required for MBNL1 to bind to RNA in vivo. Of particular interest, mutants 4 and G, which lost splicing repression and activation activities, respectively, retained in vivo RNA-binding activity. From these results we conclude that the loss of splicing activity is not due to the loss of RNA-binding activity.
Figure 7.
CUG-repeat RNA-binding assay with deletion mutants. MBNL1 (A) and MBNL3 (B) mutants were co-transfected with a plasmid expressing 960 CTG repeats into COSM6 cells. Deletion mutant proteins are visualized using green fluorescence of the GFP tag. CUG RNA foci were detected by in situ hybridization with a Cy3-CAG probe and DAPI was used to stain the nucleus.
For MBNL3, we obtained results similar to those obtained for MBNL1 (Figures 6A and 7B) in that at least one pair of zinc-finger domains was required for colocalization (mutants 5 and 6) and loss of splicing activity is not due to loss of RNA-binding activity (mutant G). One difference was that mutant 4 did not colocalize with RNA foci, despite the fact that the deletion was 56 residues upstream from the RNA-binding domains. We conclude that there are regions required for splicing activity that are separate from regions required for RNA binding (Figure 8A). It is likely that the regions required for activation/repression mediate protein–protein interactions between MBNL and co-regulators.
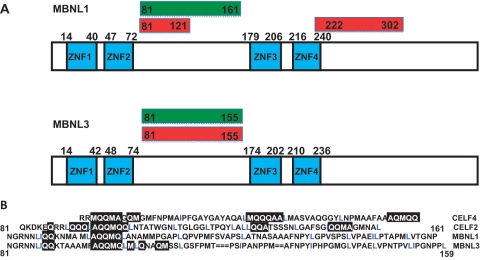
Figure 8.
Summary of core regions of MBNL1 and MBNL3 required for repression and activation of splicing. (A) Diagram of the MBNL1 (upper panel) and MBNL3 (lower panels) protein domains and associated regions required for activation and repression. Green boxes indicate domains required for splicing activation of IR exon 11 and red boxes indicate domains required for splicing repression of cTNT exon 5. (B) Alignment of splicing activity domains among CELF2, CELF4 and the domains between the two zinc-finger pairs of MBNL1 and MBNL3. The common motifs that were identified include regions rich in A, Q, M (black boxes) and L and I (blue).
We have used similar deletion analyses to define regions that are required for splicing regulation by CELF2 and CELF4 (30,35). A comparison of the sequences of activation domains mapped within CELF2 and CELF4 to those regions mapped between the two pairs of zinc fingers of MBNL1 and MBNL3 revealed two common features (Figure 8B). First, the regulatory regions of all four proteins contain regions rich in grouped M, L, Q and A residues including a match of ‘LAQQMQ’ between the regulatory region of CELF2 and MBNL1 and MBNL3. A similar motif was identified between mutants A and B of MBNL1 and MBNL3 where there is a drop in splicing activation (Figures 5A, 6A and Supplementary Figure 2). Second is the presence of similarly spaced hydrophobic residues, primarily L but also I and V. These are unlikely to be NES motifs, which are typically less spread out (consensus = LX1-3LX2-3LXL). These similarities suggest a common mode of interactions between these two families of splicing regulators with either co-regulators or spliceosomal components.
DISCUSSION
In this study, we investigated several aspects of alternative splicing regulation by MBNL proteins. We identified a 273-nt-long segment in the IR pre-mRNA containing alternatively spliced exon 11 that is necessary and sufficient for regulation of exon 11 by MBNL1 and MBNL3. We demonstrated that MBNL1 directly interacts with a 30-nt region downstream of exon 11 that contains three separate consensus-binding sequences. Similar clusters of binding sites have been identified in other MBNL targets (14). Binding of MBNL1 to each of the motifs contributes to binding of MBNL1 to the downstream intron. These results support the possibility of cooperative binding correlating with previous data demonstrating potential dimerization of MBNL1 (36). Importantly, the 30-nt region was required for responsiveness of exon 11 splicing to MBNL1 or MBNL3 expression demonstrating that the 30-nt segment mediates splicing regulation via direct interactions with MBNL1. These results are consistent with recently published results (37). While there was no direct interaction of MBNL1 on the upstream intronic sequence, the response to MBNL expression was higher than the background levels (construct F, Figure 4). This implies that MBNL proteins can still respond to splicing through an indirect mechanism potentially through protein–protein interactions of another RNA-binding factor. However, results from construct H (Figure 4) are inconsistent with a response element located within intron 10. Therefore, the nature of an MBNL response element in intron 10 remains to be delineated.
The consensus motif that we used in this study was YGCY(U/G)Y (10). It was recently found that the simplified form of YGCY motif is enough for MBNL1 binding (13,14). The binding sites that were identified in the present study contained one YGCY motif and several YGC or GCY motifs. Secondary structure of RNA can also play a role in the interaction of RNA-binding proteins (38) and MBNL1 binds to GC base-pairs interrupted by pyrimidine mismatches (36,39,40). We were unable to identify a secondary structure within the functionally defined 30 nt MBNL response element in IR (data not shown), suggesting that the probes form single-stranded RNA structures as previously described (14).
A second goal of this investigation was to identify regions of MBNL1 and MBNL3 that are required for splicing activation or repression and are separate from the RNA-binding domains. We performed parallel analyses on MBNL1 and MBNL3 because both strongly induce IR exon 11 inclusion and cTNT exon 5 skipping and yet differ outside of the RNA-binding domains. To test the ability of MBNL deletion mutants to bind RNA in vivo, we took advantage of the ability of MBNL proteins to bind and colocalize with CUG-repeat RNA nuclear foci (41).
The analyses of MBNL1 and MBNL3 protein domains provided several conclusions. First, for both MBNL1 and MBNL3, regions required for positive splicing activity were localized in regions that were separate from the RNA-binding domains (Figure 8A). Most deletions that lost the majority of splicing activity retained the ability to bind RNA (MBNL1 mutations 3, 4, G and MBNL3 mutations 3, G), indicating that a domain required for intrinsic splicing activity rather than RNA binding was affected by the deletion. We cannot rule out that deletions resulted in general disruption of protein structure resulting in a loss of activity, although each truncated protein was expressed at similar levels as the FL proteins. N-terminal deletions were essentially replaced by the N-terminal fusion with GFP rather than being simply truncated and for one deletion, MBNL3 mutation G, replacing the deleted region by a luciferase segment did not restore activity. Our results also indicate that efficient splicing repression of cTNT exon 5 requires the second pair of zinc fingers. In addition, while MBNL1 mutation 4 was inactive on cTNT exon 5, it retained nearly half of its activity on IR exon 11, indicating that the effect on cTNT splicing regulation was likely to be specific to repressor activity and not a general loss of activity. MBNL3 mutant 4 lost RNA-binding activity even though the deletion was 56 residues from the zinc-finger domains. It is unclear whether this deletion disrupted general protein structure having a secondary effect on RNA binding or this region of the protein could provide a more specific role in promoting or stabilizing RNA binding.
Second, for both MBNL1 and MBNL3, either the N-terminal or central pair of zinc-finger RNA-binding domains was sufficient to function as splicing activators of IR. MBNL1 and MBNL3 mutants 2 and E contain either the N-terminal or central zinc-finger domains, respectively, and retain substantial activation and repression activity. The observation that different RNA-binding domains within the same protein target the protein appropriately has also been observed for the CELF proteins which contain RRM-type RNA-binding domains (35).
Third, MBNL1 and MBNL3 contain more than one activation domain. For MBNL1, this is based on results from mutants 4 and E in which N- and C-terminal regions of the protein each have nearly 50% activity of full-length and overlap by only 21 residues. As mutant F retains more than a quarter of full-length activity with no residues overlapping with mutant 4, separate halves of MBNL1 can activate IR exon 11. The MBNL1 domains required for repression are between residues 80 and 160 (we consider the 11% activity of MBNL1 mutant F for cTNT repression to be negligible) and between residues 222 and 302. For MBNL3, the majority of the activity for both repression and activation is located between residues 81 and 176 based on results from mutants 2 and E. The most telling is deletion of 73 residues from mutant E to mutant G, which completely eliminates activity. Importantly, the inactive deletion mutant (G) retains RNA-binding activity, indicating that this region is required for an activity other than RNA binding.
Interestingly, the regions within MBNL1 and MBNL3 that are required for splicing regulation have sequence similarities and even identity with regions within CELF proteins that are required for splicing regulation (Figure 8B). Regions rich in alanine, glutamine and methionine as well as spaced leucines or isoleucines are suggestive of protein–protein interaction domains. Overall, our results support a model in which separate domains of MBNL proteins function to bind specific motifs within the RNA and interact directly with components of the spliceosome or with co-regulators to mediate splicing regulation. It will be of particular interest to identify these interacting proteins and to compare the proteins that are required for activation and repression.
Splicing repression of cTNT exon 5 by MBNL1 results from its antagonism of U2AF65 binding upstream of the regulated exon (39). The mechanism by which MBNL activates splicing remains elusive. However, the results from this study have identified the sites on IR pre-mRNA where MBNL binds and the regions of the MBNL1 and MBNL3 proteins that are required for splicing regulation, which likely contribute to its mechanism of splicing activation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institutes of Health (HL045565 to T.C.); and an American Heart Association postdoctoral fellowship (Texas Affiliate) (0625119Y to Y.-H.G.). Funding for open access charge: NIH HL045565.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Dr Nuno Andre Faustino for generating the MBNL1 deletion mutants and Donnie Bundman for generating MBNL3 deletion mutants. We thank Rachel Rawle for help in generating data for part of Figure 7 and part of Supplementary Figure 3. We also thank Dr Nicholas Webster for providing the IRN minigene and Dr Glenn Morris for providing the antibody for MBNL3.
REFERENCES
- 1.Wahl MC, Will CL, Luhrmann R. The spliceosome: design principles of a dynamic RNP machine. Cell. 2009;136:701–718. doi: 10.1016/j.cell.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 2.Nilsen TW, Graveley BR. Expansion of the eukaryotic proteome by alternative splicing. Nature. 2010;463:457–463. doi: 10.1038/nature08909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 4.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cooper TA. Use of minigene systems to dissect alternative splicing elements. Methods. 2005;37:331–340. doi: 10.1016/j.ymeth.2005.07.015. [DOI] [PubMed] [Google Scholar]
- 6.Barash Y, Calarco JA, Gao W, Pan Q, Wang X, Shai O, Blencowe BJ, Frey BJ. Deciphering the splicing code. Nature. 2010;465:53–59. doi: 10.1038/nature09000. [DOI] [PubMed] [Google Scholar]
- 7.Blencowe BJ. Alternative splicing: new insights from global analyses. Cell. 2006;126:37–47. doi: 10.1016/j.cell.2006.06.023. [DOI] [PubMed] [Google Scholar]
- 8.Licatalosi DD, Darnell RB. RNA processing and its regulation: global insights into biological networks. Nat. Rev. Genet. 2010;11:75–87. doi: 10.1038/nrg2673. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yu Y, Maroney PA, Denker JA, Zhang XH, Dybkov O, Luhrmann R, Jankowsky E, Chasin LA, Nilsen TW. Dynamic regulation of alternative splicing by silencers that modulate 5′ splice site competition. Cell. 2008;135:1224–1236. doi: 10.1016/j.cell.2008.10.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ho TH, Charlet BN, Poulos MG, Singh G, Swanson MS, Cooper TA. Muscleblind proteins regulate alternative splicing. EMBO J. 2004;23:3103–3112. doi: 10.1038/sj.emboj.7600300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miller JW, Urbinati CR, Teng-Umnuay P, Stenberg MG, Byrne BJ, Thornton CA, Swanson MS. Recruitment of human muscleblind proteins to (CUG)(n) expansions associated with myotonic dystrophy. EMBO J. 2000;19:4439–4448. doi: 10.1093/emboj/19.17.4439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fardaei M, Rogers MT, Thorpe HM, Larkin K, Hamshere MG, Harper PS, Brook JD. Three proteins, MBNL, MBLL and MBXL, co-localize in vivo with nuclear foci of expanded-repeat transcripts in DM1 and DM2 cells. Hum. Mol. Genet. 2002;11:805–814. doi: 10.1093/hmg/11.7.805. [DOI] [PubMed] [Google Scholar]
- 13.Du H, Cline MS, Osborne RJ, Tuttle DL, Clark TA, Donohue JP, Hall MP, Shiue L, Swanson MS, Thornton CA, et al. Aberrant alternative splicing and extracellular matrix gene expression in mouse models of myotonic dystrophy. Nat. Struct. Mol. Biol. 2010;17:187–193. doi: 10.1038/nsmb.1720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Goers ES, Purcell J, Voelker RB, Gates DP, Berglund JA. MBNL1 binds GC motifs embedded in pyrimidines to regulate alternative splicing. Nucleic Acids Res. 2010;38:2467–2484. doi: 10.1093/nar/gkp1209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mankodi A, Logigian E, Callahan L, McClain C, White R, Henderson D, Krym M, Thornton CA. Myotonic dystrophy in transgenic mice expressing an expanded CUG repeat. Science. 2000;289:1769–1773. doi: 10.1126/science.289.5485.1769. [DOI] [PubMed] [Google Scholar]
- 16.Liquori CL, Ricker K, Moseley ML, Jacobsen JF, Kress W, Naylor SL, Day JW, Ranum LP. Myotonic dystrophy type 2 caused by a CCTG expansion in intron 1 of ZNF9. Science. 2001;293:864–867. doi: 10.1126/science.1062125. [DOI] [PubMed] [Google Scholar]
- 17.Orengo JP, Chambon P, Metzger D, Mosier DR, Snipes GJ, Cooper TA. Expanded CTG repeats within the DMPK 3′ UTR causes severe skeletal muscle wasting in an inducible mouse model for myotonic dystrophy. Proc. Natl Acad. Sci. USA. 2008;105:2646–2651. doi: 10.1073/pnas.0708519105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mankodi A, Urbinati CR, Yuan QP, Moxley RT, Sansone V, Krym M, Henderson D, Schalling M, Swanson MS, Thornton CA. Muscleblind localizes to nuclear foci of aberrant RNA in myotonic dystrophy types 1 and 2. Hum. Mol. Genet. 2001;10:2165–2170. doi: 10.1093/hmg/10.19.2165. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler TM, Thornton CA. Myotonic dystrophy: RNA-mediated muscle disease. Curr. Opin. Neurol. 2007;20:572–576. doi: 10.1097/WCO.0b013e3282ef6064. [DOI] [PubMed] [Google Scholar]
- 20.Kanadia RN, Johnstone KA, Mankodi A, Lungu C, Thornton CA, Esson D, Timmers AM, Hauswirth WW, Swanson MS. A muscleblind knockout model for myotonic dystrophy. Science. 2003;302:1978–1980. doi: 10.1126/science.1088583. [DOI] [PubMed] [Google Scholar]
- 21.Hao M, Akrami K, Wei K, De Diego C, Che N, Ku JH, Tidball J, Graves MC, Shieh PB, Chen F. Muscleblind-like 2 (Mbnl2) -deficient mice as a model for myotonic dystrophy. Dev. Dyn. 2008;237:403–410. doi: 10.1002/dvdy.21428. [DOI] [PubMed] [Google Scholar]
- 22.Savkur RS, Philips AV, Cooper TA. Aberrant regulation of insulin receptor alternative splicing is associated with insulin resistance in myotonic dystrophy. Nat. Genet. 2001;29:40–47. doi: 10.1038/ng704. [DOI] [PubMed] [Google Scholar]
- 23.Denley A, Wallace JC, Cosgrove LJ, Forbes BE. The insulin receptor isoform exon 11- (IR-A) in cancer and other diseases: a review. Horm. Metab. Res. 2003;35:778–785. doi: 10.1055/s-2004-814157. [DOI] [PubMed] [Google Scholar]
- 24.Moller DE, Yokota A, Caro JF, Flier JS. Tissue-specific expression of two alternatively spliced insulin receptor mRNAs in man. Mol. Endocrinol. 1989;3:1263–1269. doi: 10.1210/mend-3-8-1263. [DOI] [PubMed] [Google Scholar]
- 25.McClain DA. Different ligand affinities of the two human insulin receptor splice variants are reflected in parallel changes in sensitivity for insulin action. Mol. Endocrinol. 1991;5:734–739. doi: 10.1210/mend-5-5-734. [DOI] [PubMed] [Google Scholar]
- 26.Kellerer M, Lammers R, Ermel B, Tippmer S, Vogt B, Obermaier-Kusser B, Ullrich A, Haring HU. Distinct alpha-subunit structures of human insulin receptor A and B variants determine differences in tyrosine kinase activities. Biochemistry. 1992;31:4588–4596. doi: 10.1021/bi00134a008. [DOI] [PubMed] [Google Scholar]
- 27.Singh G, Cooper TA. Minigene reporter for identification and analysis of cis elements and trans factors affecting pre-mRNA splicing. Biotechniques. 2006;41:177–181. doi: 10.2144/000112208. [DOI] [PubMed] [Google Scholar]
- 28.Kosaki A, Nelson J, Webster NJ. Identification of intron and exon sequences involved in alternative splicing of insulin receptor pre-mRNA. J. Biol. Chem. 1998;273:10331–10337. doi: 10.1074/jbc.273.17.10331. [DOI] [PubMed] [Google Scholar]
- 29.Philips AV, Timchenko LT, Cooper TA. Disruption of splicing regulated by a CUG-binding protein in myotonic dystrophy. Science. 1998;280:737–741. doi: 10.1126/science.280.5364.737. [DOI] [PubMed] [Google Scholar]
- 30.Han J, Cooper TA. Identification of CELF splicing activation and repression domains in vivo. Nucleic Acids Res. 2005;33:2769–2780. doi: 10.1093/nar/gki561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kalsotra A, Xiao X, Ward AJ, Castle JC, Johnson JM, Burge CB, Cooper TA. A postnatal switch of CELF and MBNL proteins reprograms alternative splicing in the developing heart. Proc. Natl Acad. Sci. USA. 2008;105:20333–20338. doi: 10.1073/pnas.0809045105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Holt I, Jacquemin V, Fardaei M, Sewry CA, Butler-Browne GS, Furling D, Brook JD, Morris GE. Muscleblind-like proteins: similarities and differences in normal and myotonic dystrophy muscle. Am. J. Pathol. 2009;174:216–227. doi: 10.2353/ajpath.2009.080520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.He F, Dang W, Abe C, Tsuda K, Inoue M, Watanabe S, Kobayashi N, Kigawa T, Matsuda T, Yabuki T, et al. Solution structure of the RNA binding domain of the human muscleblind-like protein 2. Protein Sci. 2009;18:80–91. doi: 10.1002/pro.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Liu YF, Liu HY, Tu LC, Lin CW, Hsiao KM, Pan H. Zebrafish muscleblind-like genes: identification, structural features and expression. Comp. Biochem. Physiol. B Biochem. Mol. Biol. 2008;151:118–124. doi: 10.1016/j.cbpb.2008.06.005. [DOI] [PubMed] [Google Scholar]
- 35.Singh G, Charlet BN, Han J, Cooper TA. ETR-3 and CELF4 protein domains required for RNA binding and splicing activity in vivo. Nucleic Acids Res. 2004;32:1232–1241. doi: 10.1093/nar/gkh275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuan Y, Compton SA, Sobczak K, Stenberg MG, Thornton CA, Griffith JD, Swanson MS. Muscleblind-like 1 interacts with RNA hairpins in splicing target and pathogenic RNAs. Nucleic Acids Res. 2007;35:5474–5486. doi: 10.1093/nar/gkm601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Sen S, Talukdar I, Liu Y, Tam J, Reddy S, Webster NJ. Muscleblind-like 1 (Mbnl1) promotes insulin receptor exon 11 inclusion via binding to a downstream evolutionarily conserved intronic enhancer. J. Biol. Chem. 2010;285:25426–25437. doi: 10.1074/jbc.M109.095224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Warf MB, Berglund JA. Role of RNA structure in regulating pre-mRNA splicing. Trends Biochem. Sci. 2010;35:169–178. doi: 10.1016/j.tibs.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Warf MB, Diegel JV, von Hippel PH, Berglund JA. The protein factors MBNL1 and U2AF65 bind alternative RNA structures to regulate splicing. Proc. Natl Acad. Sci. USA. 2009;106:9203–9208. doi: 10.1073/pnas.0900342106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Warf MB, Berglund JA. MBNL binds similar RNA structures in the CUG repeats of myotonic dystrophy and its pre-mRNA substrate cardiac troponin T. RNA. 2007;13:2238–2251. doi: 10.1261/rna.610607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Dansithong W, Paul S, Comai L, Reddy S. MBNL1 is the primary determinant of focus formation and aberrant insulin receptor splicing in DM1. J. Biol. Chem. 2005;280:5773–5780. doi: 10.1074/jbc.M410781200. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.