Abstract
Multiple regulatory modules contribute to the complex expression programs realized by many loci. Although long thought of as isolated components, recent studies demonstrate that such regulatory sequences can physically associate with promoters and with each other and may localize to specific sub-nuclear transcription factories. These associations provide a substrate for putative interactions and have led to the suggested existence of a transcriptional interactome. Here, using a controlled strategy of transgenesis, we analyzed the functional consequences of regulatory sequence interaction within the myelin basic protein (mbp) locus. Interactions were revealed through comparisons of the qualitative and quantitative expression programs conferred by an allelic series of 11 different enhancer/inter-enhancer combinations ligated to a common promoter/reporter gene. In a developmentally contextual manner, the regulatory output of all modules changed markedly in the presence of other sequences. Predicted by transgene expression programs, deletion of one such module from the endogenous locus reduced oligodendrocyte expression levels but unexpectedly, also attenuated expression of the overlapping golli transcriptional unit. These observations support a regulatory architecture that extends beyond a combinatorial model to include frequent interactions capable of significantly modulating the functions conferred through regulatory modules in isolation.
INTRODUCTION
Gene transcription is controlled at multiple levels with interactions between cis-acting regulatory modules and the proximal promoter figuring prominently in the control of both developmental and lineage restricted programming. Notable features of many regulatory modules include clustering of transcription factor-binding sites (TFBS) (1–3) and inter-species sequence conservation (4–6). Regulatory function is typically accompanied by a myriad of local changes including DNA methylation patterns and the accumulation of specific histone isoforms with activity specific modifications including acetylation, methylation and phosphorylation (7,8).
A number of model loci have been investigated in sufficient depth to expose at least part of the complexity intrinsic to transcriptional control mechanisms. Notable among these is the β-globin locus where expanding models incorporate multiple levels of regulation including promoter-proximal, enhancer-mediated and domain-wide components of the transcriptional control mechanism. These regulatory components include locus control regions (LCRs) (9,10), insulators and boundary elements such as CCTC (11,12), DNA methylation; chromatin modifiers and physical interactions mediated among additional distal regulatory elements [reviewed in (13); 14–16]. Illuminating further levels of regulatory sequence integration, in sea urchin, the output of transgenes bearing different regulatory sequence domains revealed an extensive network of functionally significant positive and negative interactions largely accommodated by a logic circuit model (17–19).
Recent investigations, using chromatin conformation capture assays and 3D FISH, demonstrate that physical associations among regulatory sequences are common (20,21). For the immunoglobulin heavy chain locus, achieving a close proximity of distant regulatory sequences appears to involve the folding of intervening chromatin into bundles of loops (22). Similarly, for the TNF (tumor necrosis factor) locus, intra-chromosomal looping, inferred from chromatin conformation capture assays, appears to support simultaneous interactions among dispersed regulatory modules and the proximal promoter (15). Predicting their functional significance, associations among regulatory sequences are observed to change through development in association with evolving gene expression programs (13,23). Providing a further link between gene expression and cell type-specific chromatin structure, cohesin and mediator localize to and participate in DNA loop formation at active genes (24). Additionally, active genes may associate at sub-nuclear transcription factories (25,26) providing a potential substrate for functionally important cis and trans interactions. This may be of particular significance for co-regulated genes where their dynamic co-localization at such sites may provide access to environments enriched in lineage specific transcription factors.
Despite the accumulating evidence demonstrating extensive physical associations and functional integration among diverse regulatory sequences, isolated regulatory modules frequently confer expression programs seemingly accommodated faithfully within the developmental and lineage-restricted program realized by the endogenous locus. This is the case for four previously identified regulatory modules situated upstream of the myelin basic protein (mbp) locus, three of which showed autonomous targeting to mbp expressing glia (27–31). However, in earlier investigations we also obtained results suggesting that interactions with functional consequences occurred among some of these modules and here we sought to expose the extent and consequences of interactions among these four modules in a more comprehensive manner. We designed reporter constructs such that each module could be investigated either alone or in partnership with multiple combinations of the other modules. The in vivo qualitative and quantitative expression programs conferred at three developmentally significant time points by all constructs were then compared in mice. If a simple combination of their autonomous outputs were to be reflected in their combined expression programs, functionally significant interactions would not be indicated. Conversely, if unique expression programming were encountered, functionally significant interactions would be implicated.
Mbp is expressed at highest levels in oligodendrocytes in the central nervous system (CNS) and to lower levels by Schwann cells in the peripheral nervous system (PNS). In both cell types, expression begins perinatally, rises to maximal levels while myelin is elaborated during the pre-weaning period declining thereafter to reach stable levels maintained in mature animals (32,33). Four modules (M1–M4) demonstrating extensive inter-species conservation are recognized in the first 10 kb of mouse mbp 5′-flanking sequence (29). M4, the module furthest upstream at −9.5 kb, is a Schwann cell enhancer (27,29,31). M3, at −5 kb, is an oligodendrocyte enhancer that also demonstrates cryptic, albeit transient, Schwann cell targeting activity (28). M2, at −700 bp has no autonomous targeting activity while M1, extending to −377 bp, serves as the proximal promoter and demonstrates autonomous targeting activity in oligodendrocytes (27–30). To support inter-construct comparisons, constructs were inserted at a common site in single copy and common orientation using the HPRT based method of controlled transgenesis (34). Furthermore, we deleted M3 from the endogenous mbp locus and compared the expression program realized in mice by the mutant allele to that of both the intact mbp locus and relevant reporter genes. Beyond their previously characterized autonomous regulatory activities, all mbp modules revealed developmentally contextual interactions that significantly altered their own regulatory capacities and/or those conferred by other regulatory sequences. These observations lead to a model in which the autonomous activity revealed by isolated enhancers is but a limited predictor of their role within endogenous loci where functionally significant interactions with other regulatory sequences appear to be common.
MATERIALS AND METHODS
Generation of constructs for HPRT transgenesis
9.5 kb is a construct previously described (29) consisting of 9.5 kb of the 5′-flanking sequence of the mouse mbp gene fused to the Escherichia coli lacZ gene. The 9.5ΔM3 construct corresponds to 9.5 kb with a deletion of a PmlI-DraI (687 bp) fragment containing M3. The 9.5ΔM4 construct corresponds to 9.5 kb with a deletion of a SacI-AfeI (769 bp) fragment containing M4. The construct in which the intervening sequence was deleted (9.5ΔI) was generated by cutting out a BtrI-PmlI (3.8 kb) fragment from 9.5 kb.
Modules with linkers were generated by PCR and cloned into pENTR1A MCSLacZ a modification of pENTR1A (Invitrogen) containing a multiple cloning site and a lacZ reporter gene. The primers used to amplify the modules were the following: (all coordinates are from the UCSC Mouse browser, July 2007 Assembly).
XM1F: GGACTCGAGGCGTAACTGTGCGTTTTATAGGAGA
(chr18:82 723 599–82 723 623)
KM1R: TTTGGTACCCCGGAAGCTGCTGTGGGGTC
(chr18:82 723 939–82 723 958)
NM2F: AGTGCGGCCGCTGCAGAAAGATGTGGGAAGTCCT
(chr18:82 723 267–82 723 291)
XM2R: ATACTCGAGGTTTAAAAGGCCACCAGTGCACA
(chr18:82 723 352–82 723 375)
HM3F: ATAAAGCTTGTGGCAGATTTAGACTCCTTACCA
(chr18:82 718 977–82 719 000)
NM3R: ATAGCGGCCGCAGCCTGGTTCTGGAGTTGCG
(chr18:82 719 215–82 719 234)
BM4F: ATAAGATCTTGTGACCCCTTTCTCGATGTGG
(chr18:82 714 888–82 714 910)
HM4R: TATAAGCTTAGGCAGCCTAGGCCTGCAGA
(chr18:82 715 238–82 715 257)
M2(chr18:82 722 949–82 723 392) was cloned into an hsp-lacZ ‘Entry’ vector (27) based on pENTR1A (Invitrogen, San Diego, CA, USA) to generate M2hsplacZ. M2 was ligated upstream of the hsp promoter in reverse orientation relative to endogenous mbp.
Entry vectors were recombined in vitro with the HPRT Gateway destination vector using LR clonase (Invitrogen). Hprt-targeting vectors were sequenced and 40 µg was linearized for transfection of embryonic stem cells.
Generation of targeting vector for M3 deletion in situ
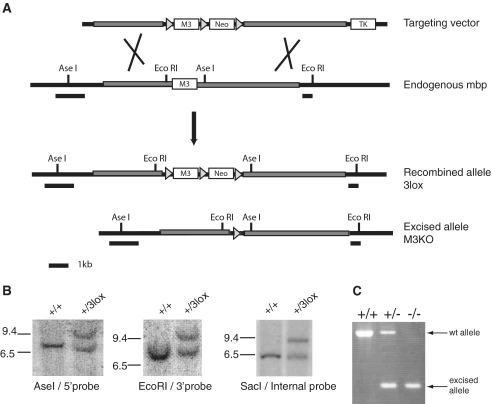
To generate classical and conditional M3 knockout ES cells, we used a gene targeting vector that contained a loxP site upstream of M3, a neo cassette for positive selection flanked by two other loxP sites, and a thymidine kinase cassette outside of the homology arms for negative selection (Figure 5). The pPNT vector containing pgk-neo and thymidine kinase (kindly given by Dr Brigitte Boldyreff) was modified by cloning a PCR product of floxed M3 (759 bp). M3 was amplified with primers that contained loxP sites: CACCGCTCGAGCCGCACGTGATAACTTCGTATAATGTATGCTATACGAAGTTATTTTAAATGGTAAAAACCCC and CGGCACCTGCAGGGTTTAAACATAACTTCGTATAGCATACATTATACGAAGTTATACACGTGGCACTCTGAG. Right (4.6 kb) and left (3.7 kb) homology arms were cut from a vector containing 9.5 kb of mbp promoter sequence with PmlI-KpnI and AfeI-DraI respectively, and then cloned into pPNT. Upon homologous recombination in ES cells, clones with floxed M3 that lost the thymidine kinase cassette were selected, screened, and used to generate transgenic mice.
Figure 5.
M3 knockout from the golli-mbp locus. (A) Gene targeting strategy for knocking out M3 in the endogenous mbp gene. Diagrams of the targeting vector, endogenous mbp gene, recombined allele bearing 3 lox P sites, and the excised allele or M3KO are shown. Homology arms are shown as gray rectangles and LoxP sites as gray triangles. M3, the neomycin resistance gene and the thymidine kinase gene are shown as white, labeled boxes. Relevant restriction sites for Southern blots are indicated in the diagrams. Probe sequences are shown as black rectangles underneath each diagram. (B) Identification of ES clones with the recombined +/3Lox allele using external and internal probes by Southern blot analysis. The correct targeting event was confirmed by the appearance of an 8.7-kb band after digestion with AseI and hybridization with the 5′-probe and a 9.5-kb band after digestion with EcoRI and hybridization with the 3′-probe. With the same probes, the wild-type allele revealed 6.8- and 7.6-kb bands, respectively. After SacI digestion, an internal probe (not shown) detected 6.3- and 2.3-kb bands with the wild-type allele and 8.2- and 2.3-kb bands with the +/3lox allele. (2.3-kb bands are not shown). (C) Strategy to discriminate between wt and M3KO alleles by PCR.
Derivation of transgenic mice
Reporter gene constructs in destination vectors were transfected into BPES-11 embryonic stem cells generated in the lab as previously described (35), which bear the hprt deletion originally described by Hooper et al. (36). Upon homologous recombination the transgenes insert in single copy upstream of the hprt gene and only cells with the insertion are able to grow in hypoxantin–aminopterin–thymine selection media. Selected and genotyped embryonic stem cell clones were injected into C57BL/6 blastocysts, which were then transplanted into pseudo-pregnant females. Male chimeras were mated to C57BL/6 females for germline passage, which was confirmed by transgene specific PCR genotyping, and BC2 mice were derived by a subsequent cross between germ-line females and C57Bl/6 males.
Derivation of M3 knockout mice
The targeting vector was linearized with NotI and transfected by electroporation into BPES-6 mouse embryonic stem cells (derived together with BPES-11, but without the hprt deletion). Clones were selected with G418 (200 µg/ml) and ganciclovir (2 µM). Clones with the correct recombination event, as determined by Southern blot hybridization, were used to obtain germline-transmitting chimeras after injection of C57BL/6 blastocysts. Chimeras were crossed with C57BL/6 wild-type mice. Heterozygous offspring were crossed to Prm-Cre transgenic mice (Jackson Laboratory), which express Cre recombinase from the protamine promoter in sperm (37), and further backcrossed to C57BL/6.
Genotyping of M3KO mice by Southern blot and PCR: Genomic DNA was extracted from targeted ES cells and mouse tails. Southern blot hybridization was done with AseI, EcoRI and SacI-digested genomic DNA using internal and external probes (Figure 5). Digested gDNA was transferred to Hybond-N+ membrane (Amersham Biosciences) and hybridized with probes labeled with HexaLabelTM DNA-Labeling Kit (Fermentas). For PCR genotyping the following primers were used:
M3Hom1F (CAAGGACACATAGAAGATGGGCTAC) and
M3Hom1R (AGGAAAGGGCAAGACTGAGTGG).
Histochemical detection and quantitation of β-galactosidase activity
Histochemical staining was performed as described previously (31). Mice were anesthetized and perfused with 0.5% paraformaldehyde and 2.5% glutaraldehyde in 0.1 M phosphate buffer pH 7.4. Whole mounts were fixed for 1 h and incubated in staining solution with 0.4 mg/ml of Bluo-Gal.
Quantitation of β-galactosidase activity was done in tissues from mice backcross two to C57BL/6, at P14, P21 and P90. Mice were anesthetized and tissues (cervical spinal cords and sciatic nerves) were dissected and snap-frozen in liquid nitrogen. Four to 10 samples per construct were homogenized in a mixer mill (Qiagen) in 1 ml (cervical cords) or 0.25 ml (sciatic nerves) of lysis buffer. β-galactosidase activity was detected using the Galacto-StarTM System (Applied Biosystems). Total protein concentrations were measured in triplicate by Bradford (Bio-Rad) using BSA in a standard curve. Analysis of variance between groups was performed using GraphPad Software (GraphPad Software Inc. San Diego, CA USA) and Statistics tools from Saint John’s University, Department of physics (www.physics.csbsju.edu/stats/anova.html). Significant differences between constructs were determined by ANOVA and the criterion was P < 0.01.
Extraction and quantitation of mRNA by real-time PCR
Tissues (cervical spinal cord and optic nerves) from P12, P21 and P60 anesthetized mice were dissected and snap-frozen in liquid nitrogen. RNA was prepared from single tissues of at least five mice per genotype with the Qiagen RNAeasy Lipid mini Kit according to the protocol. For the optic nerves, RNA was prepared from pools of five mice. RNA was reverse transcribed using Superscript II (Invitrogen) and random hexamers according to the protocol of the manufacturer. Quantitative PCR was performed using the LightCycler® FastStart DNA MasterPLUSSYBR Green I Kit (Roche). The cDNA was diluted 1/10 and 1/20 and measured using 2 µl in a 20-µl reaction. Golli transcripts were measured using primers specific for golli exons 2 and 3 that are present in the BG21 and J37 isoforms (TTATCTGCTGAGAAGGCCAGT and CCACGCTTCTCTTCTTTCCA) while mbp transcripts were measured using primers for mbp exons 3 and 4 (CGAGAACTACCCATTATGGCTCCC and TGGAGGTGGTGTTCGAGGTGTC) that are present in the 14-, 17-, 18.5- and 21.5-kDa isoforms as well as the J37 golli isoform. The much greater abundance of mbp transcripts in the tissues examined ensures that the mbp RT–PCR measures mbp transcripts and not the overlapping golli transcripts. Glyceradehyde-3-phosphate dehydrogenase (gapdh) cDNA was amplified with primers: ACCACAGTCCATGCCATCAC and TCCACCACCCTGTTGCTGTA. hprt cDNA was amplified with primers: GCTGGTGAAAAGGACCTCT and CACAGGACTAGAACACCTGC. Gapdh and hprt cDNA quantification were used to normalize golli and mbp quantification.
RESULTS
Reporter constructs containing most combinations of the known mbp enhancer and inter-enhancer sequences were transfected into ES cells where homologous recombination led to their insertion, in single copy and common orientation, upstream of HPRT (34). Chimeras were generated and the single copy transgenes were derived into lines of mice where their targeting and quantitative expression phenotypes were evaluated at three developmentally significant stages (Figure 1). Throughout the PNS, Schwann cells initiate myelination perinatally and by post-natal (P) Day 14, myelin elaboration and myelin gene expression approach peak levels. Oligodendrocytes initiate myelinogenesis in different regions of the CNS during the first three post-natal weeks but in the cervical spinal cord analyzed here, the program is similar to the PNS, initiating at birth, approaching peak levels by P14 and nearing completion by P21. In mature mice, myelin protein turnover is thought to be low (38) and myelin gene expression continues at stable, but reduced from peak, levels (39–41). To evaluate relative levels of reporter gene expression in the context of myelin maturation, β-galactosidase accumulation was measured in cervical spinal cord (CNS) and sciatic nerve (PNS) samples obtained at P14, P21 and P90.
Figure 1.
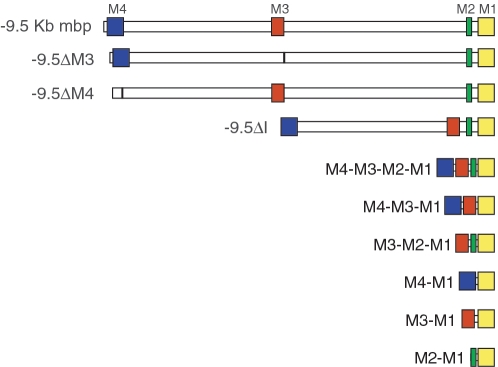
Constructs docked as single copy at the hprt locus in transgenic mice. The constructs generated in this study contain different deletions and combinations of mbp regulatory sequences, ligated to a lacZ reporter gene. Mbp regulatory modules (M1–M4) are represented as colored boxes whereas non-conserved inter-modular sequences are shown in white. The first construct contains 9.5 kb of mbp 5′-flanking sequence while the others bear deletions of modules and/or intermodular sequences.
Murine mbp 5′-flanking sequence extending to −9.5 kb contains the mbp regulatory sequence so far characterized for function using reporter genes and it confers cell specificity and quantitative programming similar to that realized by the endogenous mbp locus (29,31). Each conserved mbp module (M1, M2, M3 and M4) has been extensively analyzed by controlled transgenesis in order to characterize its function and composite elements (27–30). Consequently, the targeting and quantitative expression programs conferred by 9.5 kb of mbp 5′-flanking sequence and each of the modules studied independently, served as internal references for comparison of the constructs analyzed in this investigation.
M3 enhances, but is not required for, mbp expression in oligodendrocytes
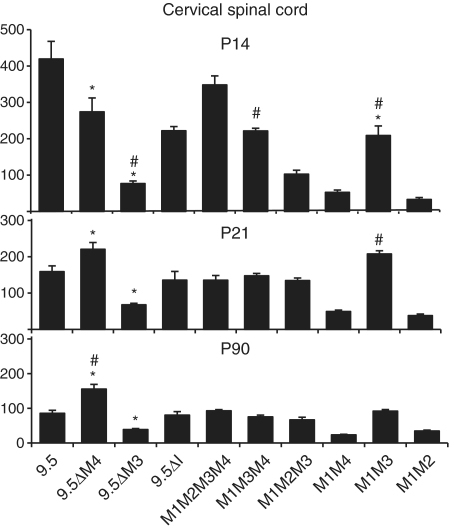
The most proximal module, M1, contains the mbp promoter and is capable of targeting expression to oligodendrocytes, but not Schwann cells (29,30). However, earlier observations, using both random and controlled transgenesis, suggested that activity from M1 alone was transient and that sustained expression in the oligodendrocytes of mature mice required M3 (29,39). Subsequent investigations using controlled transgenesis demonstrated that constructs lacking M3 were capable of continued expression in mature mice, but only when inserted in one orientation relative to HPRT (30). Here we show that the 9.5-kb 5′-sequence deleted of M3 (9.5ΔM3) expresses in oligodendrocytes of both young and mature mice, but at a level only 45% (P90) of that realized from the intact 9.5-kb construct (Figure 2).
Figure 2.
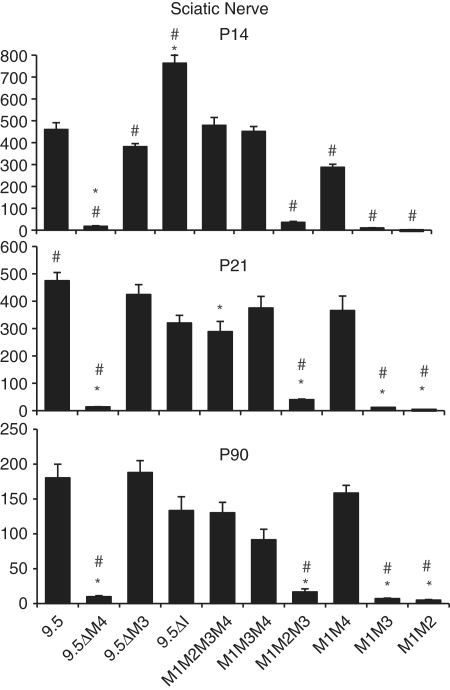
Expression phenotypes of mbp reporter constructs in oligodendrocytes. The function of mbp regulatory sequences in oligodendrocytes was analyzed by measuring the β-galactosidase activity at post-natal Days 14, 21 and 90 in cervical spinal cords. Significant differences (when P < 0.01) are represented by asterisk or hash compared to 9.5 kb or M1M2M3M4, respectively. Comparisons between all constructs are shown in Supplementary Table S1.
M1M3 drives maximal expression in adult oligodendrocytes while maximal expression during myelin biogenesis requires additional input from M2 and/or M4
M1M3 conferred post weaning expression to oligodendrocytes at levels similar to that realized by the 9.5-kb 5′ flanking sequence; a result indicating that M1 with M3 is sufficient for maximal expression at P21 and P90 (Figure 2). Interestingly, at P21, a comparison of M1M3 to M1M2M3 or M1M2M3M4 demonstrates that M1 and M3 not only suffice for maximal expression, but they also express to higher levels than constructs containing the additional modules (Figure 2, P21). Similarly, deletion of M4 from 9.5 kb (9.5ΔM4) increased levels of expression in oligodendrocytes by 39% at P21 and 82% at P90 (Cervical spinal cord Figure 2). These data show that additional module sequences interfere with maximal oligodendrocyte expression thus revealing negative functional interactions active in post weaning samples. In contrast, at P14, the M1M2M3M4 and the intact 9.5-kb constructs expressed in oligodendrocytes to levels higher than all other constructs tested, demonstrating that M2 and/or M4 contribute positively to oligodendrocyte expression at this early age (Figure 2: 9.5 kb, M1M3, M1M2M3, M1M3M4, P14). Consistent with the positive activity realized in P14 samples, constructs lacking M4 expressed in oligodendrocytes at levels significantly lower than that realized by the 9.5-kb construct. Thus, M4, first recognized as an enhancer with targeting activity limited to Schwann cells, also modulates expression in oligodendrocytes in a developmentally contextual manner.
The proximal promoter plays a silencing role in Schwann cells
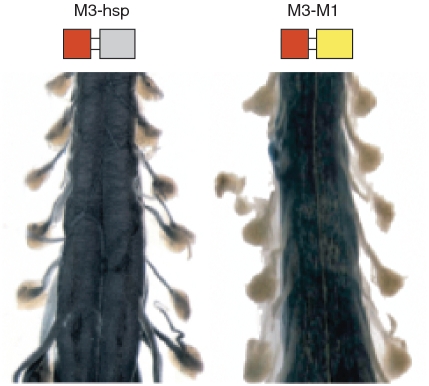
The first 6 kb of mouse mbp 5′-flanking sequence includes M1, M2 and M3, and demonstrates no activity in Schwann cells (31). Consequently, it was unanticipated that a reporter in which M3 alone was ligated to a 300 bp hsp minimal promoter would acquire the ability to drive expression in Schwann cells during myelin elaboration (29). As the minimal promoter itself does not target Schwann cells, this observation requires that cryptic M3 Schwann cell targeting activity normally is suppressed through interactions with another 5′-flanking sequence. To probe the potential role of promoter-enhancer interactions in conferring this suppression, we compared the expression programs realized from the M1M3 and hspM3 constructs (Figure 3) and show that the cryptic Schwann cell targeting activity is silenced entirely when M3 is associated with M1, the 377-bp mbp proximal promoter.
Figure 3.
M1 restricts expression of M3 to oligodendrocytes. Whole mount histochemical preparations of mouse spinal cords at post-natal Day 9 show that M3 ligated to the hsp minimal promoter drives lacZ expression in both spinal cord (oligodendrocytes) and spinal roots (Schwann cells). In contrast, expression is limited to oligodendrocytes when M3 is ligated to the mbp M1 promoter.
M4 alone drives high levels of expression in adult Schwann cells but its activity is enhanced by M3 during myelin biogenesis
Deleting M4 from 9.5 kb of 5′-flanking sequence (9.5ΔM4, Figure 4) silences reporter gene expression in Schwann cells at all ages. Similarly, constructs regulated by M1M2 and M1M3 fail to express above background levels in Schwann cells, at all ages. These observations are consistent with the autonomous and robust enhancer activity that M4 exerts in Schwann cells, demonstrated previously in the context of both randomly inserted transgenes and hsp promoted lacZ reporter genes inserted at HPRT (27,29,31).
Figure 4.
Expression phenotypes of mbp reporter constructs in Schwann cells. Graphs show levels of β-galactosidase activity realized from mbp regulated reporter constructs in the sciatic nerves of transgenic mice at P14, P21 and P90. Significant differences (when P < 0.01) are represented by asterisk or hash compared to 9.5 kb or M1M2M3M4, respectively. Comparisons between all constructs are shown in Supplementary Table S2.
To explore the potential effects of other modules on M4 mediated Schwann cell expression, the quantitative expression programs realized by all M4 bearing constructs were compared. At P21 and P90, the M1M4 construct expressed in Schwann cells in the same range as the 9.5-kb regulated construct (Figure 4) indicating that, at these ages, M4 confers Schwann cell expression without modulation arising through interactions with other regulatory sequences. However, at P14, when mbp mRNA accumulation nears peak levels, constructs containing M3 (M1M2M3M4 and M1M3M4) expressed at levels significantly higher than M1M4 alone (Figure 4). Similarly, the 9.5-kb construct expressed at higher levels than 9.5ΔM3 (460 versus 381, respectively P < 0.02).
M2: a role in oligodendrocytes during myelin biogenesis
We showed previously that M1, the 377 bp 5′ of the translation start site, is sufficient to drive expression in oligodendrocytes and that by extending the sequence to −794 bp, to include M2, expression levels increased in oligodendrocytes (29). However, when M2 was ligated to an hsp minimal promoter in a lacZ-reporter construct, it lacked autonomous targeting activity (28). To expose further modulating activities, M2 was deleted from M1M2M3M4. Consistent with our earlier observations, M1M3M4 drove expression in the P14 cervical spinal cord at only 64% of that realized by the parent construct (Figure 2: 222 ± 22 versus 348 ± 55, respectively). Thus, during myelin biogenesis, M2 contributes significantly to oligodendrocyte expression even in the presence of M3 and M4. However, at later ages, the M1M3M4 and the M1M2M3M4 constructs expressed at similar levels (P21: 136 ± 32 and 148 ± 20; P90: 90 ± 8.3 and 75 ± 14, respectively) demonstrating that M2 activity is maximal during, and perhaps limited to, the period of CNS myelin biogenesis (Figure 2). In the PNS, the M1M3M4 and M1M2M3M4 constructs expressed to similar levels at all ages examined suggesting that M2 plays no significant role in Schwann cells (Figure 4).
Inter-module sequence
Deletion of all inter-modular sequences present in the 9.5-kb construct yielding the M1M2M3M4 construct had no effect on the expression program realized in maturing oligodendrocytes (Figure 2). As predicted by this observation, removal of only the inter-modular sequence lying between M2 and M3 (9.5ΔI) also had no affect on the oligodendrocyte expression program (Figure 2). In contrast, deletion of inter-enhancer sequences had significant consequences on the expression program realized in Schwann cells. Deletion of the sequence lying between M2 and M3 reduced expression at P21 to 68% of the 9.5-kb value (9.5ΔI, 320 ± 69 versus 9.5 475 ± 73) while deletion of all other inter-modular sequences had little additional effect reducing expression to 61% of the 9.5-kb value (M1M2M3M4 289.8 ± 91 versus 9.5-kb 475 ± 73) (Figure 4). In contrast, at P90, the expression levels realized by both deletion constructs revealed no significant differences (Figure 4). Similarly, at P14, the M1M2M3M4 construct expressed in the same range as the 9.5-kb construct (479 ± 81 versus 460 ± 69, respectively). Notably, at this young age, the construct deleted of the non-conserved sequence between M2 and M3 demonstrated expression levels markedly higher than the intact 9.5-kb construct (9.5: 460 ± 69 versus 9.5ΔI: 764 ± 102).
M3 function in the endogenous golli-mbp gene
To evaluate potential regulatory interactions with sequences extending beyond the 9.5 kb of 5′-flanking sequence evaluated in reporter genes, and to determine the extent to which regulatory activity revealed by reporter constructs docked at the HPRT locus reflects function within the context of the endogenous mbp locus, we derived mice bearing a mbp allele deleted of M3 (Figure 5). M3 is located within the 5′-flanking sequence of mbp and within intron 3 of the overlapping golli transcriptional unit. Mice homozygous for the M3 knockout allele (M3KO) demonstrated no obvious behavioral abnormalities, and had normal growth and reproductive performance (up to 1 year of age examined).
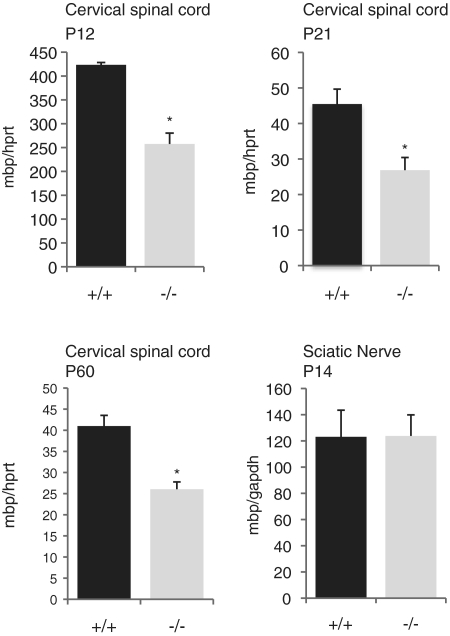
To evaluate the potential effects of M3 deletion on transcription initiated by the mbp promoter in oligodendrocytes, cervical spinal cord samples from homozygous M3KO and wild-type litter-mates were analyzed for relative levels of accumulated mbp mRNA by real-time PCR. At all ages examined mbp mRNA accumulated to 60% of wild-type values (Figure 6). Although in the predicted direction, the effect of the M3 deletion on the endogenous locus was not as strong as that revealed by the 9.5-kb reporter construct deleted of M3 (at P14, 9.5ΔM3 = 18.3% of 9.5 kb; at P21, 43% of 9.5 kb and at P90, 45% of 9.5 kb). Similarly, comparison between other reporter constructs with and without M3 showed that the presence of M3 had a greater effect on expression of reporter genes than on the endogenous locus (see Figure 2: M1M2 versus M1M2M3; M1M4 versus M1M3M4).
Figure 6.
Expression of mbp is reduced in oligodendrocytes of M3KO mice. At P12, 21 and 60, real-time RT–PCR evaluation of mbp mRNA from the cervical spinal cords of M3KO homozygotes revealed accumulation to 60% of the wild-type level (*P < 0.01). Number of mice analyzed: P12 (2+/+ and 5−/−); P21 (6+/+ and 5−/−); P60 (5+/+ and 5−/−). In P14 sciatic nerves, no difference between WT and M3KO mbp expression was observed.
To examine the role of M3 in the Schwann cell lineage, mbp mRNA accumulation in sciatic nerve samples from homozygous M3KO mice at P14 was evaluated. No significant alteration in mbp mRNA accumulation was detected (Figure 6).
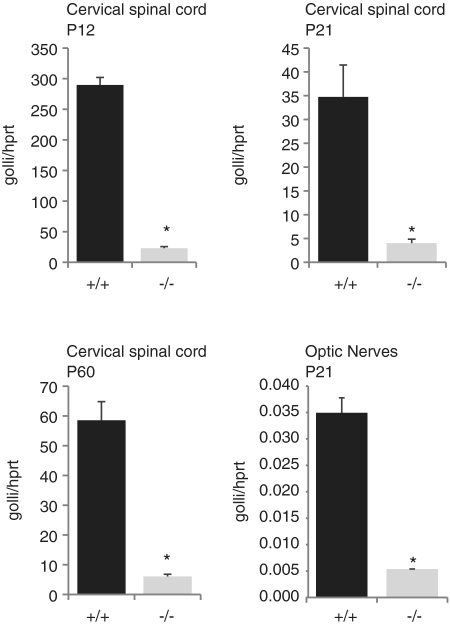
As noted above, the mbp gene lies within the more widely expressed golli transcriptional unit and we therefore investigated the potential effect of its deletion on golli transcription. We first compared golli mRNA accumulation in the cervical spinal cord of homozygous M3KO mice. At P12, P21 and P60, M3KO mice accumulated only 10% of wt golli mRNA levels (Figure 7). Golli transcripts accumulate in a developmental program distinct from mbp extending to multiple neuronal populations in addition to oligodendrocytes (42,43). Thus, to evaluate the possibility that golli expression was completely silenced in spinal cord oligodendrocytes, while still expressed in neurons, we next examined optic nerves that are richly invested with oligodendrocytes but contain no neuronal cell bodies. Optic nerves of M3KO mice accumulated golli mRNA at levels similar to that observed in the spinal cord (15% of wt, Figure 7) leading to the conclusion that golli expression in oligodendrocytes is massively down regulated, but not entirely silenced, when M3 is absent; an observation suggesting that M3 engages both of the widely separated (>79 kb in mouse) mbp and golli promoters.
Figure 7.
Expression of golli is reduced in M3KO mice. Real-time PCR analysis of golli mRNA from the cervical spinal cord showed reduced accumulation in M3KO homozygous mice (−/−) compared to wild-type (+/+). Similar relative reductions were observed at all ages investigated. The same dramatic reduction in golli mRNA accumulation also was observed in the optic nerves of M3KO (−/−) mice where no neuronal cell bodies are located.
DISCUSSION
The regulatory roles played by specific enhancers and the promoter of the mbp gene were shown here to be modulated through partnerships elaborated among them. Such modulation was either positive or negative and extended to both qualitative and quantitative expression phenotypes. Furthermore, modulation was contextual on both the glial lineage and stage of maturation. Myelin elaboration takes place over a short time span during which glial cells support intense membrane and myelin protein synthesis. Upregulated expression of numerous genes accompanies this process and at P14, active myelination is ongoing and near peak levels of myelin gene expression are realized in both spinal cord oligodendrocytes and sciatic nerve Schwann cells. During the period when myelin elaboration is most active, the interactions we encountered typically led to upregulated reporter expression. Also at that stage, enhancers demonstrating autonomous targeting activity in only one glial lineage were nonetheless capable of contributing to reporter expression in the other. In contrast, by P21 when most myelin has been elaborated and many myelin genes, including mbp, are downregulated, regulatory interactions tended to be less dramatic or resulted in negative effects. Consistent with this overall program, the reporter regulated by M3hsp expressed in Schwann cells only during the period of most intense myelin elaboration. Notably, interaction between M3 and the proximal promoter silences even this transient activity in Schwann cells. In the related M3 knockout experiment, we show that M3 up-regulates expression of the endogenous mbp locus in oligodendrocytes. Although that role was predicted by its oligodendrocyte targeting capacity, unexpectedly it also functions as the major enhancer of the golli promoter in oligodendrocytes. Finally, non-conserved inter-enhancer sequence was shown to affect regulatory activity, although only in myelin elaborating Schwann cells. These combined observations demonstrate that an additive model of enhancer activity cannot account for the integrated activity of mbp/golli regulatory sequences and highlights a seemingly ubiquitous role for regulatory sequence interactions in the mechanism controlling transcription.
Physical association of regulatory modules
The mechanism through which regulatory sequence interaction modulates gene expression is unknown. However, 3C and 4C studies with other loci strongly support looping models in which enhancer function is conferred through close physical association with the proximal promoter (24). Beyond the demonstrated association of promoters and enhancers, evidence has emerged for the existence of transcription factories where co-localization occurs (25,44,45). In this model, PolII and its related factors dynamically associate with active genes at nuclear foci. An extension of this model proposes that specialized factories enriched in a particular combination of transcription factors may self-organize by the preferential co-localization of co-regulated genes sharing those transcription factors (46). Consistent with this, erythroid Klf1-dependent co-regulated genes appear to preferentially associate (47). Other active erythroid genes that were not dependent on Klf1 did not show similar co-localization supporting the notion that the specificity of these factories is based on shared regulatory factors. They also observed that randomly inserted YAC transgenes carrying the Hbb locus co-localized with the endogenous Hbb locus indicating that an active transgene locus can bring its ectopic chromosomal site to the relevant specialized factory. In like manner, the rapid and co-ordinate up-regulation of the myelin gene family during myelin biogenesis could be greatly facilitated in an environment rich in specific factors. Co-localization of oligodendrocyte and Schwann cell enhancers (e.g. M3 and M4) could be one factor contributing to their ability to modulate each other’s activity. Thus, it will be important to determine the extent to which the functionally interacting golli/mbp sequences, along with the known regulatory sequences of other co-regulated myelin genes, co-localize during the maturation of myelin forming cells (23).
If the interactions at specialized transcription factories followed a stoichiometric model, deletion of M3 could affect the expression of other myelin genes. However, it is thought unlikely that the deletion of one enhancer would affect transcription of co-regulated genes at a specialized factory formed in its absence (47). Consistent with this notion, we observed no change in the accumulated mRNA levels of the myelin genes plp and mag in the cervical spinal cord of M3KO mice (data not shown).
Intervening DNA
Looping models of enhancer function assign little or no functional significance to inter-modular DNA. However, in a recent review, Bulger and Groudine (48) suggested that enhancer-promoter co-localization might be established through a process requiring the intervening DNA sequences (49–51). In the present investigation, close juxtaposition of all four mbp regulatory modules had no significant effect on either the qualitative or quantitative expression programs realized by any reporter gene expressed in oligodendrocytes at any age. By experimentally bringing the modules into close proximity, it is possible that loops and the related function associated with the intervening DNA become unnecessary. Alternatively, any requirement for inter-enhancer sequence might be accommodated by the short, non-conserved, enhancer flanking sequences remaining within such constructs.
In contrast to oligodendrocytes, deletion of inter-enhancer sequences significantly affected the expression programs realized in Schwann cells. Specifically, when compared to the 9.5-kb construct, the expression levels realized in Schwann cells by the M1M2M3M4 module-only construct were reduced at P21 and P90 while at P14, the 9.5 kb construct deleted only of inter-enhancer I (9.5ΔI) revealed an up-regulated expression program. Whether this signifies a lineage specific role for inter-enhancer sequences or the presence of currently unrecognized TFBS in the deleted sequence remains unresolved.
Enhancer sharing: mbp and golli
Previously, 1.1 kb of golli 5′-flanking sequence was shown to drive golli expression in fetal neurons but not in the oligodendrocyte lineage (52,53). We show here, in addition to its well-characterized role in regulating transcription of mbp, M3 enhances expression from the upstream golli promoter where it accounts for 90% of its output in oligodendrocytes. Several loci configured with multiple promoters regulated by common enhancers have been described previously (14,26,54–58). The mechanism through which an enhancer simultaneously affects the activity of multiple promoters in the same cell is not known but a dynamic physical interaction among the three sequences appears likely. This could take the form of a ‘flip-flop’ model, as proposed for the globin locus where regulatory sequence switches back and forth between two genes (59) or a hit-and-run (60) mechanism. Alternatively, the enhancer may be able to form a complex with both promoters concurrently, a circumstance potentially facilitated in the context of a common transcription factory (26).
In this investigation, functional interactions realized by multiple mbp/golli regulatory sequences have been revealed through the output of HPRT docked constructs and an endogenous allele deleted of M3. Although M3 deletion in the endogenous locus led to downregulation of mbp, the extent of the downregulation was not as extensive as that predicted by HPRT docked constructs. Thus, the large golli/mbp locus may contain additional mbp regulatory sequence/s partially compensating for the loss of the M3 enhancer. Alternatively, the different chromatin environments, inter-genic interactions involving the regulatory sequences controlling HPRT expression, or competition for different transcription factories might all contribute to such quantitative differences.
Finally, the present understanding of transcriptional regulation and regulatory sequence interactions has been achieved through insights obtained with multiple highly diverse experimental approaches (61). Here, using in vivo functional analysis we demonstrate that functional interactions between regulatory modules are commonplace extending to both qualitative and quantitative expression phenotypes. Although the positive or negative consequences of such interactions are currently unpredictable, this investigation has assigned their origin to specific sequence domains within the mbp 5′-flanking sequence thus providing insight that should facilitate future investigations.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Genome Canada/Genome Quebec as part of the Regulatory Genetics and Gene Regulators in Disease (GRID) programs and by the Multiple Sclerosis Society of Canada; Pre-doctoral fellowship support from Genome Canada/Genome Quebec and the Research Institute of the MUHC (to S.D.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors are grateful to F. Bourdeau, C. Dy, T. Fernandez, I. Tretjakoff and P. Valera for excellent technical support and to Dr R. Sladek, Dr D. Fulton and Dr E. Harmsen for helpful comments on the article.
REFERENCES
- 1.Ferretti V, Poitras C, Bergeron D, Coulombe B, Robert F, Blanchette M. PReMod: a database of genome-wide mammalian cis-regulatory module predictions. Nucleic Acids Res. 2007;35:D122–D126. doi: 10.1093/nar/gkl879. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ho Sui SJ, Fulton DL, Arenillas DJ, Kwon AT, Wasserman WW. oPOSSUM: integrated tools for analysis of regulatory motif over-representation. Nucleic Acids Res. 2007;35:W245–W252. doi: 10.1093/nar/gkm427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Meng H, Banerjee A, Zhou L. BLISS 2.0: a web-based tool for predicting conserved regulatory modules in distantly-related orthologous sequences. Bioinformatics. 2007;23:3249–3250. doi: 10.1093/bioinformatics/btm368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Loots GG, Locksley RM, Blankespoor CM, Wang ZE, Miller W, Rubin EM, Frazer KA. Identification of a coordinate regulator of interleukins 4, 13, and 5 by cross-species sequence comparisons. Science. 2000;288:136–140. doi: 10.1126/science.288.5463.136. [DOI] [PubMed] [Google Scholar]
- 5.Pennacchio LA, Rubin EM. Genomic strategies to identify mammalian regulatory sequences. Nat. Rev. Genet. 2001;2:100–109. doi: 10.1038/35052548. [DOI] [PubMed] [Google Scholar]
- 6.Pennacchio LA, Ahituv N, Moses AM, Prabhakar S, Nobrega MA, Shoukry M, Minovitsky S, Dubchak I, Holt A, Lewis KD, et al. In vivo enhancer analysis of human conserved non-coding sequences. Nature. 2006;444:499–502. doi: 10.1038/nature05295. [DOI] [PubMed] [Google Scholar]
- 7.Ho L, Crabtree GR. Chromatin remodelling during development. Nature. 2010;463:474–484. doi: 10.1038/nature08911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J, Casaccia P. Epigenetic regulation of oligodendrocyte identity. Trends Neurosci. 2010;33:193–201. doi: 10.1016/j.tins.2010.01.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Li Q, Peterson KR, Fang X, Stamatoyannopoulos G. Locus control regions. Blood. 2002;100:3077–3086. doi: 10.1182/blood-2002-04-1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Stamatoyannopoulos G. Control of globin gene expression during development and erythroid differentiation. Exp. Hematol. 2005;33:259–271. doi: 10.1016/j.exphem.2004.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rubio ED, Reiss DJ, Welcsh PL, Disteche CM, Filippova GN, Baliga NS, Aebersold R, Ranish JA, Krumm A. CTCF physically links cohesin to chromatin. Proc. Natl Acad. Sci. USA. 2008;105:8309–8314. doi: 10.1073/pnas.0801273105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Capelson M, Corces VG. Boundary elements and nuclear organization. Biol. Cell. 2004;96:617–629. doi: 10.1016/j.biolcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 13.Fromm G, Bulger M. A spectrum of gene regulatory phenomena at mammalian beta-globin gene loci. Biochem. Cell Biol (Biochimie et biologie cellulaire) 2009;87:781–790. doi: 10.1139/O09-048. [DOI] [PubMed] [Google Scholar]
- 14.Sharpe J, Nonchev S, Gould A, Whiting J, Krumlauf R. Selectivity, sharing and competitive interactions in the regulation of Hoxb genes. EMBO J. 1998;17:1788–1798. doi: 10.1093/emboj/17.6.1788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tsytsykova AV, Rajsbaum R, Falvo JV, Ligeiro F, Neely SR, Goldfeld AE. Activation-dependent intrachromosomal interactions formed by the TNF gene promoter and two distal enhancers. Proc. Natl Acad. Sci. USA. 2007;104:16850–16855. doi: 10.1073/pnas.0708210104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schwartz RJ, Olson EN. Building the heart piece by piece: modularity of cis-elements regulating Nkx2-5 transcription. Development. 1999;126:4187–4192. doi: 10.1242/dev.126.19.4187. [DOI] [PubMed] [Google Scholar]
- 17.Yuh CH, Bolouri H, Davidson EH. Genomic cis-regulatory logic: experimental and computational analysis of a sea urchin gene. Science. 1998;279:1896–1902. doi: 10.1126/science.279.5358.1896. [DOI] [PubMed] [Google Scholar]
- 18.Yuh CH, Bolouri H, Davidson EH. Cis-regulatory logic in the endo16 gene: switching from a specification to a differentiation mode of control. Development. 2001;128:617–629. doi: 10.1242/dev.128.5.617. [DOI] [PubMed] [Google Scholar]
- 19.Yuh CH, Davidson EH. Modular cis-regulatory organization of Endo16, a gut-specific gene of the sea urchin embryo. Development. 1996;122:1069–1082. doi: 10.1242/dev.122.4.1069. [DOI] [PubMed] [Google Scholar]
- 20.Dekker J, Rippe K, Dekker M, Kleckner N. Capturing chromosome conformation. Science. 2002;295:1306–1311. doi: 10.1126/science.1067799. [DOI] [PubMed] [Google Scholar]
- 21.Amano T, Sagai T, Tanabe H, Mizushina Y, Nakazawa H, Shiroishi T. Chromosomal dynamics at the Shh locus: limb bud-specific differential regulation of competence and active transcription. Dev. Cell. 2009;16:47–57. doi: 10.1016/j.devcel.2008.11.011. [DOI] [PubMed] [Google Scholar]
- 22.Jhunjhunwala S, van Zelm MC, Peak MM, Cutchin S, Riblet R, van Dongen JJ, Grosveld FG, Knoch TA, Murre C. The 3D structure of the immunoglobulin heavy-chain locus: implications for long-range genomic interactions. Cell. 2008;133:265–279. doi: 10.1016/j.cell.2008.03.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Fraser J, Rousseau M, Shenker S, Ferraiuolo MA, Hayashizaki Y, Blanchette M, Dostie J. Chromatin conformation signatures of cellular differentiation. Genome Biol. 2009;10:R37. doi: 10.1186/gb-2009-10-4-r37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kagey MH, Newman JJ, Bilodeau S, Zhan Y, Orlando DA, van Berkum NL, Ebmeier CC, Goossens J, Rahl PB, Levine SS, et al. Mediator and cohesin connect gene expression and chromatin architecture. Nature. 2010;467:430–435. doi: 10.1038/nature09380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Osborne CS, Chakalova L, Brown KE, Carter D, Horton A, Debrand E, Goyenechea B, Mitchell JA, Lopes S, Reik W, et al. Active genes dynamically colocalize to shared sites of ongoing transcription. Nat. Genet. 2004;36:1065–1071. doi: 10.1038/ng1423. [DOI] [PubMed] [Google Scholar]
- 26.Xu M, Cook PR. Similar active genes cluster in specialized transcription factories. J. Cell Biol. 2008;181:615–623. doi: 10.1083/jcb.200710053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Denarier E, Forghani R, Farhadi HF, Dib S, Dionne N, Friedman HC, Lepage P, Hudson TJ, Drouin R, Peterson A. Functional organization of a Schwann cell enhancer. J Neurosci. 2005;25:11210–11217. doi: 10.1523/JNEUROSCI.2596-05.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Dionne N. PhD Thesis. McGill University; 2006. Structure and function of Module 3, a conserved enhancer of the myelin basic protein gene. [Google Scholar]
- 29.Farhadi HF, Lepage P, Forghani R, Friedman HC, Orfali W, Jasmin L, Miller W, Hudson TJ, Peterson AC. A combinatorial network of evolutionarily conserved myelin basic protein regulatory sequences confers distinct glial-specific phenotypes. J. Neurosci. 2003;23:10214–10223. doi: 10.1523/JNEUROSCI.23-32-10214.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forghani F, Peterson AC. Glial Cells in Health and Disease. Cambridge University Press; 2007. The proximal promoter of the myelin basic protein (MBP) gene restricts expression to myelinating oligodendrocytes. 24 Aug 2007 page S93, London. [Google Scholar]
- 31.Forghani R, Garofalo L, Foran DR, Farhadi HF, Lepage P, Hudson TJ, Tretjakoff I, Valera P, Peterson A. A distal upstream enhancer from the myelin basic protein gene regulates expression in myelin-forming schwann cells. J. Neurosci. 2001;21:3780–3787. doi: 10.1523/JNEUROSCI.21-11-03780.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Barbarese E, Carson JH, Braun PE. Accumulation of the four myelin basic proteins in mouse brain during development. J. Neurochem. 1978;31:779–782. doi: 10.1111/j.1471-4159.1978.tb00110.x. [DOI] [PubMed] [Google Scholar]
- 33.de Ferra F, Engh H, Hudson L, Kamholz J, Puckett C, Molineaux S, Lazzarini RA. Alternative splicing accounts for the four forms of myelin basic protein. Cell. 1985;43:721–727. doi: 10.1016/0092-8674(85)90245-4. [DOI] [PubMed] [Google Scholar]
- 34.Bronson SK, Plaehn EG, Kluckman KD, Hagaman JR, Maeda N, Smithies O. Single-copy transgenic mice with chosen-site integration. Proc. Natl Acad. Sci. USA. 1996;93:9067–9072. doi: 10.1073/pnas.93.17.9067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Palais G, Nguyen Dinh Cat A, Friedman H, Panek-Huet N, Millet A, Tronche F, Gellen B, Mercadier JJ, Peterson A, Jaisser F. Targeted transgenesis at the HPRT locus: an efficient strategy to achieve tightly controlled in vivo conditional expression with the tet system. Physiol. Genomics. 2009;37:140–146. doi: 10.1152/physiolgenomics.90328.2008. [DOI] [PubMed] [Google Scholar]
- 36.Hooper M, Hardy K, Handyside A, Hunter S, Monk M. HPRT-deficient (Lesch-Nyhan) mouse embryos derived from germline colonization by cultured cells. Nature. 1987;326:292–295. doi: 10.1038/326292a0. [DOI] [PubMed] [Google Scholar]
- 37.O'Gorman S, Dagenais NA, Qian M, Marchuk Y. Protamine-Cre recombinase transgenes efficiently recombine target sequences in the male germ line of mice, but not in embryonic stem cells. Proc. Natl Acad. Sci. USA. 1997;94:14602–14607. doi: 10.1073/pnas.94.26.14602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Benjamins JA, Morell P. Proteins of myelin and their metabolism. Neurochem. Res. 1978;3:137–174. doi: 10.1007/BF00964057. [DOI] [PubMed] [Google Scholar]
- 39.Foran DR, Peterson AC. Myelin acquisition in the central nervous system of the mouse revealed by an MBP-Lac Z transgene. J. Neurosci. 1992;12:4890–4897. doi: 10.1523/JNEUROSCI.12-12-04890.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Raine CS. Morphology of myelin and myelination. In: Morell P, editor. Myelin. 2nd edn. New York: Plenum Publishing Corp.; 1984. pp. 1–50. [Google Scholar]
- 41.Zeller NK, Hunkeler MJ, Campagnoni AT, Sprague J, Lazzarini RA. Characterization of mouse myelin basic protein messenger RNAs with a myelin basic protein cDNA clone. Proc. Natl Acad. Sci. USA. 1984;81:18–22. doi: 10.1073/pnas.81.1.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marty MC, Alliot F, Rutin J, Fritz R, Trisler D, Pessac B. The myelin basic protein gene is expressed in differentiated blood cell lineages and in hemopoietic progenitors. Proc. Natl Acad. Sci. USA. 2002;99:8856–8861. doi: 10.1073/pnas.122079599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pribyl TM, Campagnoni CW, Kampf K, Kashima T, Handley VW, McMahon J, Campagnoni AT. The human myelin basic protein gene is included within a 179-kilobase transcription unit: expression in the immune and central nervous systems. Proc. Natl Acad. Sci. USA. 1993;90:10695–10699. doi: 10.1073/pnas.90.22.10695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Iborra FJ, Pombo A, Jackson DA, Cook PR. Active RNA polymerases are localized within discrete transcription ‘factories’ in human nuclei. J. Cell Sci. 1996;109(Pt 6):1427–1436. doi: 10.1242/jcs.109.6.1427. [DOI] [PubMed] [Google Scholar]
- 45.Xu M, Cook PR. The role of specialized transcription factories in chromosome pairing. Biochim. Biophys. Acta. 2008;1783:2155–2160. doi: 10.1016/j.bbamcr.2008.07.013. [DOI] [PubMed] [Google Scholar]
- 46.Schoenfelder S, Clay I, Fraser P. The transcriptional interactome: gene expression in 3D. Current Opin. Genet. Dev. 2010;20:127–133. doi: 10.1016/j.gde.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 47.Schoenfelder S, Sexton T, Chakalova L, Cope NF, Horton A, Andrews S, Kurukuti S, Mitchell JA, Umlauf D, Dimitrova DS, et al. Preferential associations between co-regulated genes reveal a transcriptional interactome in erythroid cells. Nat. Genet. 2010;42:53–61. doi: 10.1038/ng.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Bulger M, Groudine M. Enhancers: the abundance and function of regulatory sequences beyond promoters. Dev. Biol. 2010;339:250–257. doi: 10.1016/j.ydbio.2009.11.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Demers C, Chaturvedi CP, Ranish JA, Juban G, Lai P, Morle F, Aebersold R, Dilworth FJ, Groudine M, Brand M. Activator-mediated recruitment of the MLL2 methyltransferase complex to the beta-globin locus. Mol. Cell. 2007;27:573–584. doi: 10.1016/j.molcel.2007.06.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dorsett D. Cohesin, gene expression and development: lessons from Drosophila. Chromosome Res. 2009;17:185–200. doi: 10.1007/s10577-009-9022-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cockerill PN. NFAT is well placed to direct both enhancer looping and domain-wide models of enhancer function. Sci. Signaling. 2008;1:pe15. doi: 10.1126/stke.113pe15. [DOI] [PubMed] [Google Scholar]
- 52.Givogri MI, Kampf K, Schonmann V, Campagnoni AT. Identification of a novel silencer that regulates the myelin basic protein gene in neural cells. Gene. 2000;252:183–193. doi: 10.1016/s0378-1119(00)00223-7. [DOI] [PubMed] [Google Scholar]
- 53.Landry CF, Pribyl TM, Ellison JA, Givogri MI, Kampf K, Campagnoni CW, Campagnoni AT. Embryonic expression of the myelin basic protein gene: identification of a promoter region that targets transgene expression to pioneer neurons. J. Neurosci. 1998;18:7315–7327. doi: 10.1523/JNEUROSCI.18-18-07315.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Gould A, Morrison A, Sproat G, White RA, Krumlauf R. Positive cross-regulation and enhancer sharing: two mechanisms for specifying overlapping Hox expression patterns. Gene Dev. 1997;11:900–913. doi: 10.1101/gad.11.7.900. [DOI] [PubMed] [Google Scholar]
- 55.Zammit PS, Cohen A, Buckingham ME, Kelly RG. Integration of embryonic and fetal skeletal myogenic programs at the myosin light chain 1f/3f locus. Dev. Biol. 2008;313:420–433. doi: 10.1016/j.ydbio.2007.10.044. [DOI] [PubMed] [Google Scholar]
- 56.Swamynathan SK, Piatigorsky J. Regulation of the mouse alphaB-crystallin and MKBP/HspB2 promoter activities by shared and gene specific intergenic elements: the importance of context dependency. Int. J. Dev. Biol. 2007;51:689–700. doi: 10.1387/ijdb.072302ss. [DOI] [PubMed] [Google Scholar]
- 57.Chang TH, Primig M, Hadchouel J, Tajbakhsh S, Rocancourt D, Fernandez A, Kappler R, Scherthan H, Buckingham M. An enhancer directs differential expression of the linked Mrf4 and Myf5 myogenic regulatory genes in the mouse. Dev. Biol. 2004;269:595–608. doi: 10.1016/j.ydbio.2004.02.013. [DOI] [PubMed] [Google Scholar]
- 58.Xu X, Scott MM, Deneris ES. Shared long-range regulatory elements coordinate expression of a gene cluster encoding nicotinic receptor heteromeric subtypes. Mol. Cell. Biol. 2006;26:5636–5649. doi: 10.1128/MCB.00456-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Wijgerde M, Grosveld F, Fraser P. Transcription complex stability and chromatin dynamics in vivo. Nature. 1995;377:209–213. doi: 10.1038/377209a0. [DOI] [PubMed] [Google Scholar]
- 60.Rigaud G, Roux J, Pictet R, Grange T. In vivo footprinting of rat TAT gene: dynamic interplay between the glucocorticoid receptor and a liver-specific factor. Cell. 1991;67:977–986. doi: 10.1016/0092-8674(91)90370-e. [DOI] [PubMed] [Google Scholar]
- 61.Birney E, Stamatoyannopoulos JA, Dutta A, Guigo R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis ET, Thurman RE, et al. Identification and analysis of functional elements in 1% of the human genome by the ENCODE pilot project. Nature. 2007;447:799–816. doi: 10.1038/nature05874. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.