Abstract
Cyclin E supports pre-replication complex (pre-RC) assembly, while cyclin A-associated kinase activates DNA synthesis. We show that cyclin E, but not A, is mounted upon the nuclear matrix in sub-nuclear foci in differentiated vertebrate cells, but not in undifferentiated cells or cancer cells. In murine embryonic stem cells, Xenopus embryos and human urothelial cells, cyclin E is recruited to the nuclear matrix as cells differentiate and this can be manipulated in vitro. This suggests that pre-RC assembly becomes spatially restricted as template usage is defined. Furthermore, failure to become restricted may contribute to the plasticity of cancer cells.
INTRODUCTION
Cyclin E is a G1 cyclin that associates preferentially with cyclin-dependent protein kinase 2 (CDK2). There are two mammalian E type cyclins, with similar expression, function and structure in complex with CDK2 (1). Cyclin E rises in mid G1, peaks after passage through the restriction point (R), and falls around G1/S (2). Initiation of mammalian DNA replication is controlled by CDK2, which, in association with cyclins E and A, acts on proteins that assemble at replication origins. Cyclin E plays a role in pre-replication complex (pre-RC) formation in higher eukaryotes, cooperating with CDC6 and CDT1 to support assembly of the mini chromosome maintenance (MCM) complex (3–7). Cyclin E is also required for centrosome duplication (8), where its role also appears to be distinct from that of cyclin A (3,9).
Despite increasingly well-defined roles for cyclin E, fibroblasts isolated from mice deficient in E1 and 2 are able to proliferate, indicating that cyclin E is redundant in the mitotic cell cycle. In fact, cyclin E appears to be essential for MCM assembly only in cells preparing for S phase that is not preceded by mitosis, such as those re-entering cycle from G0 or undergoing endoreduplication (5,10). By contrast, CDK2-deficient fibroblasts do not encounter similar problems (11), suggesting that cyclin E may act independently of CDK2 under some circumstances. Consistent with this, a kinase-deficient mutant partially corrects the phenotype of cyclin E null cells, restoring MCM complex assembly (12). Similarly, the centrosomal function of cyclin E may also be CDK2 independent (8). Thus, while CDK2 is the preferred binding partner of cyclin E and cyclin E can clearly activate CDK2, evidence points to additional CDK-independent roles for cyclin E.
DNA replication is constrained in nuclear space to discrete sub-nuclear foci that are associated with a non-chromatin protein/RNA structure, that is defined as ‘nuclear matrix’ or ‘nucleoskeleton’, and can be visualized by electron microscopy (13,14). Nuclear matrix-associated DNA is enriched with replication origins during G1 phase, suggesting that recruitment plays a role in initiation (15), while the pre-RC proteins CDC6 and ORC1 both undergo transient association in mammalian cells (16). However, there is little evidence to indicate at what point in pre-RC assembly DNA replication origins are recruited, or what the nuclear matrix-associated receptors might be.
In Xenopus egg extracts cyclin E is reported to bind to chromatin, most likely via CDC6, which acts as a chromatin-associated receptor (4). Similarly, work with mouse embryonic fibroblasts and HeLa cells indicates that cyclin E is loaded onto chromatin and that this involves direct interaction with both CDT1 and MCM proteins (12). Thus, immobilization of cyclin E appears to occur in association with pre-RC formation and the prevailing view is that this reflects association with chromatin. However, the protocols used in most of these studies do not discriminate between chromatin and nuclear matrix, and the picture is further complicated by analyses of cell types of distinct species, differentiation and pathological state.
Here we show that (i) cyclin E is immobilized in the nucleus by association with the nuclear matrix, (ii) that recruitment occurs as cells differentiate and (iii) that cyclin E is not attached to the nuclear matrix in cancer cells. This is the first time that either of the replication-associated cyclins has been linked with the nuclear matrix. These novel findings point to an important role for cyclin E in genome organization. They also argue that despite the relative ease with which cancer cells can be manipulated, they are a questionable choice for analysis of nuclear matrix function or the spatial organization of DNA replication.
MATERIALS AND METHODS
Cell culture
Cell lines were sourced from JCRB or ATCC and maintained as recommended. Mouse cardiac fibroblasts expressing Col1A1 (not shown) were derived from atria dissected from a C57/BL6 mouse heart. Mouse ES cells (KES1) were differentiated to embryoid bodies or embryoid body outgrowths as described (17). Xenopus laevis embryos were produced as described (18). NHU cells were established from surgical samples collected with ethical review and informed patient consent and induced to differentiate as described (19). NIH3T3 cells were synchronised and labelled with Bromodeoxyuridine (BrdU) as described (3).
Nuclear fractionation
Nuclear fractionation was carried out as described (20). Typically, 25% of cell lysate in CSK buffer was removed (total) and the rest a diluted 2-fold with CSK, 0.2% Triton X-100 and separated by centrifugation into detergent supernatant (SN) and pellet (P). Pellet was resuspended in CSK with 0.5 M NaCl to produce salt wash or ‘w’. Pellet was further re-suspended in digestion buffer and incubated with or without DNase 1 (Roche), supplemented with 0.5 M NaCl and separated by centrifugation. Cell equivalents of each fraction were analysed by western blot. Mouse cyclin E was detected with rabbit polyclonal antibody 07-687 (Upstate) in Figure 1B and F, and verified with ab7959 (not shown); and with ab7959 (Abcam) in Figures 1C and 3A. Human cyclin E was detected with mouse monoclonal antibody sc247 HE12 (Santa Cruz). Xenopus cyclin E was detected with a rabbit polyclonal antibody that was a kind gift from Tim Hunt. Cyclin A was detected with C4710 (Sigma), CDK2 with 7954-1 (Abcam), CDC6 with sc9964 (Santa Cruz), lamin B2 with E3 (invitrogen) and histone H3 with 1791 (Abcam).
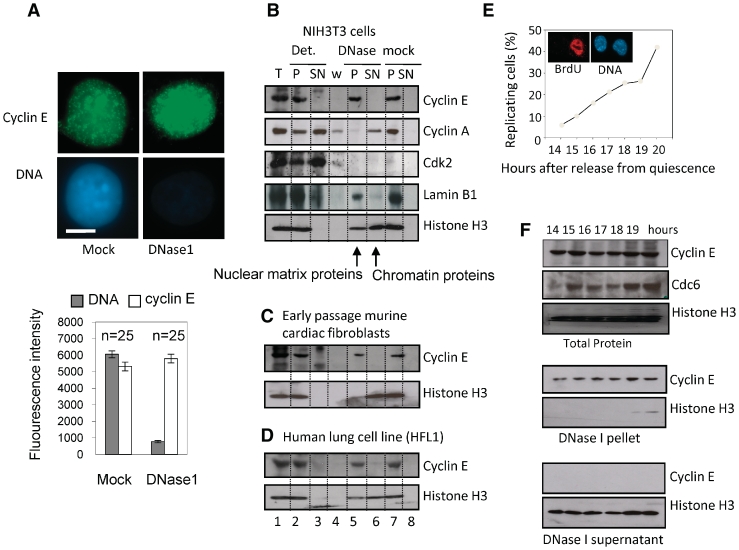
Figure 1.
Cyclin E is present in nuclear matrix-associated foci. (A) Immunofluorescent detection of cyclin E (green) and DNA (blue) in cycling NIH3T3 cells after extraction with or without DNase. Bar is 5 μm. Histogram shows average fluorescence intensity derived from multiple images with SEM. (B) Western blot showing cyclin E and other proteins as indicated, in fractions generated by serial extraction of NIH3T3 cells. T = total protein. Cells were lysed in detergent containing buffer to generate pellet (P) and supernatant (SN). Pellet was washed with 0.5 M NaCl to generate ‘w’ and extracted with or without DNase. All fractions are derived from cell equivalents. (C and D) As in B, showing cyclin E in early passage murine cardiac fibroblasts and human lung cell line HFL1, respectively. (E) The proportion of NIH3T3 cells engaged in DNA synthesis between 14 and 19 h after release from quiescence, measured by incorporation of BrdU. Inset shows BrdU (red) in nuclei (blue) at 19 h. (F) Western blots showing total cyclin E, CDC6 and histone H3 14–19 h after release from quiescence (upper panel). After extraction with DNase cyclin E remained in the pellet (middle panel), despite complete release of histone H3 (lower panel), at all time points.
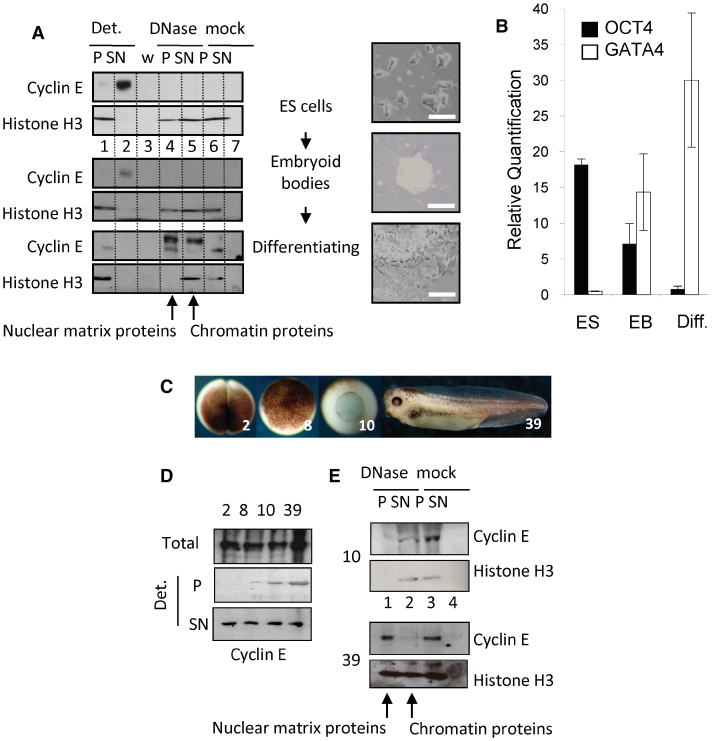
Figure 3.
Cyclin E is recruited to the nuclear matrix during embryonic stem cell differentiation and Xenopus laevis development. (A) Western blot showing protein fractions derived from murine ES cells before and after embryoid body formation, and after 25 days in culture under differentiation conditions. Images show cell populations before extraction. Bar = 50 μm. Cyclin E changes from detergent soluble in ES cells and embryoid bodies to mostly DNase resistant in differentiated cells despite efficient extraction of histone H3. (B) Quantitative RT–PCR of differentiation markers Oct4 and GATA4 in cells in A, with SEM. (C) Xenopus embryos at stages 2, 8, 10 and 39. (D) Western blots showing total cyclin E and the proportion that is detergent-resistant (P) and detergent soluble (SN) at each stage. (E) Western blot showing detergent-resistant cyclin E after further incubation without (mock) or with DNase. Histone H3 was used to control for chromatin digestion. By stage 39 the detergent resistant fraction of cyclin E is DNase resistant.
Immunofluorescence
Cells on coverslips were washed in PBS and CSK, 0.1% Triton X-100 (detergent washed). For DNase 1 treatment, coverslips were washed as above with digestion buffer plus DNase 1 and incubated at 25°C for 45 min. Cells were fixed in 4% paraformaldehyde for 20 min and blocked with antibody buffer (10% BSA, 0.02% SDS, 0.1% Triton X-100 in PBS). Human cyclin E was detected with mouse monoclonal antibody sc247 HE12 (Santa Cruz) and mouse cyclin E with E4 (Santa Cruz). DNA was counterstained with Hoechst 33258 (Sigma). Images were collected using a Zeiss Axiovert 200 M and Openlab image acquisition software. Fluorescence intensity was quantified from raw images acquired under identical imaging parameters.
Gene expression
RNA was extracted using Trizol (Ambion) and reverse-transcribed with SuperScript III (Invitrogen). Primers and probes for quantitative RT–PCR are given in Supplementary Table S2. GAPDH was used to normalize results, which are expressed as relative quantification (RQ).
RESULTS AND DISCUSSION
In murine NIH3T3 cells, cyclin E resists extraction with DNase despite efficient extraction of the majority of nuclear DNA and is located within punctate nuclear foci (Figure 1A). Thus, cyclin E is associated with nuclease-resistant, non-chromatin nuclear structures that are consistent with the nuclear matrix. Analysis of protein fractions generated by sequential extraction with detergent, salt and nuclease confirm that none of the immobilized cyclin E is released by DNase, despite release of more than half of the histone H3. Lamin B1, a nuclear protein, predicted to fractionate with the nuclear matrix, is detected in the DNase pellet fraction, confirming the validity of the fractionation protocol (Figure 1B, lanes 5 and 6). Similar results were obtained with other differentiated cells, including primary murine cardiac fibroblasts (Figure 1C), human lung cell line HFL1 (Figure 1D), in vitro differentiated mouse embryonic stem cells and normal human urothelial (NHU) cells (see later). Four independent cyclin E antibodies gave similar results (not shown). Therefore, immobilization of cyclin E on the nuclear matrix occurs in primary and established cell lines of both human and murine origin. Under the same conditions we did not detect CDK2 or cyclin A in the nuclear matrix fraction (Figure 1B) and there have been no other reports of nuclear matrix-association for either cyclin E or cyclin A. Therefore, although both cyclins have been localized to sub-nuclear foci (21), our analysis suggests that cyclins E and A undergo fundamentally different types of interaction.
NIH3T3 cells synchronised by release from quiescence pass through R after ∼15 h and enter S phase at 18–19 h (3) (Figure 1E). Cyclin E protein levels remained similar throughout this period, while CDC6 expression increased between 14 and 15 h, indicating passage through R (Figure 1F). Throughout, cyclin E remained resistant to high salt (not shown) and DNase (Figure 1F), suggesting that its association with the nuclear matrix is not regulated during passage through R, or entry to S phase.
Much of the evidence identifying cyclin E as chromatin-associated protein stems from analysis of activated Xenopus egg extracts that contain cyclin E but little DNA or nuclear structure on which to bind, or from cancer cells, usually HeLa cells. Consistent with the prevailing view, our analysis indicates that cyclin E is not DNase resistant in embryonic and undifferentiated lineages, or in cancer cells (Figure 2 and Supplementary Table S1). In some cases, such as the prostate cancer line LNCAP or the urothelial cancer line RT4, cyclin E resists extraction with detergent (Figure 2A), but in all but one of the eight cancer cell lines we studied cyclin E was not resistant to DNase and is therefore not mounted on the nuclear matrix. Notably, cyclin E remained organized into sub-nuclear foci (Figure 2B), showing that attachment to an underlying architecture is not required to specify foci.
Figure 2.
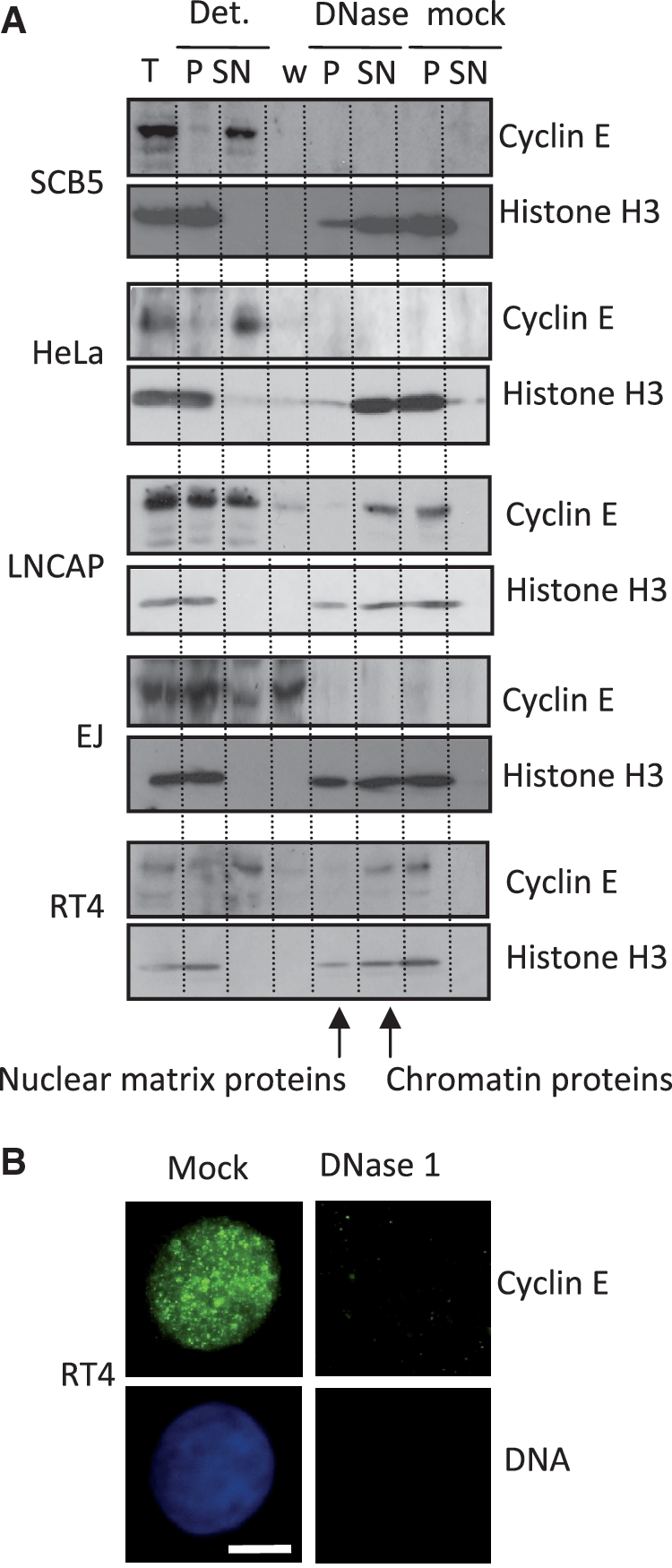
Cyclin E is not immobilized on the nuclear matrix in cancer cells. (A) Western blot showing absence of cyclin E from the DNase resistant fraction in representative cancer cell lines fractionated by the procedure used in Figure 1. (B) Immunofluorescence images of RT4 cells showing cyclin E (green) and DNA (blue). Cyclin E occupies a focal pattern in mock treated cells, but is fully extracted by DNase. Bar is 5 μm.
To reconcile these two sets of observations and test whether both states are in fact encountered in vivo, we used three different experimental systems. These yielded consistent results and show clearly that recruitment to the nuclear matrix is established as cells differentiate.
In undifferentiated murine embryonic stem (ES) cells the majority of cyclin E was not resistant to DNase (Figure 3A, lane 2 upper panels), but shifted into the DNase-resistant fraction (lane 4, lower panels) upon induction of differentiation (17). Reduced OCT4 and increased GATA4 expression confirmed endodermal differentiation (Figure 3B), although markers for other lineages were also present (not shown). Similar results were obtained with X. laevis embryos during development from fertilized egg to tadpole. The embryo is transcriptionally silent until the mid blastula transition (MBT) when zygotic transcription begins in a process that involves chromatin remodelling and reorganisation of replication origins (22). Prior to the MBT (embryo stages 2 and 8, Figure 3C), cyclin E was fully solubilised by detergent (Figure 3D), while after the MBT (stages 10 and 39) a resistant fraction of cyclin E was evident. Further analysis showed that in gastrulating embryos (stage 10), this was released by DNase (Figure 3E), while in the tadpoles (stage 39) it was completely resistant (lane 1). Therefore, cyclin E is recruited to the nuclear matrix during Xenopus development. If cyclin E recruitment is interpreted as a surrogate for replication origin recruitment, the data imply that this occurs sometime after the MBT. The matrix-association of some key housekeeping genes, such as the rRNA genes, is reorganised at the MBT as they become activated. Others such as the keratin domain, which encodes genes expressed only in adult keratinocytes, are not specified until later in development (23). Thus the formation of functional links with the nuclear matrix may be a staged process that occurs as template usage is defined.
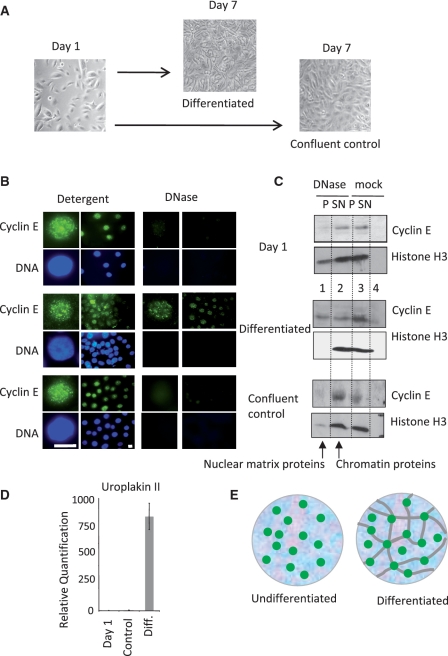
Complementary analysis in a well-characterised model of urothelial cell differentiation (19), revealed a similar picture. In undifferentiated cycling and confluent populations of NHU cells (Figure 4A) cyclin E was fully extracted with DNase (Figure 4B and C, upper and lower panels). However in cells induced to differentiate, approximately half of the cyclin E was recruited to the nuclear matrix (Figure 4B and C, middle panels). The transition to differentiated urothelium was verified visually (Figure 4A) and by analysis of uroplakin 2 expression (Figure 4D).
Figure 4.
Recruitment of cyclin E to the nuclear matrix during differentiation of NHU cells (A) Cycling NHU cells were grown to confluence without differentiation or induced to differentiate over 7 days. (B) Cyclin E (green) and DNA (blue) in nuclei from cycling cells (Day 1, upper panels), differentiated (Day 7, middle panels) and confluent control cells (Day 7, lower panels), after extraction with detergent or DNase as indicated. Bar is 10 μm. (C) Detergent and DNase extracted protein fractions from the populations shown in A. Cyclin E changes from DNase sensitive in undifferentiated NHU and confluent control cells to partly DNase resistant in differentiated NHU cells, despite efficient extraction of histone H3. (D) Quantitative RT-PCR showing uroplakin 2 expression with SEM. (E) Model of cyclin E foci (green) associated with chromatin (blue background), unconstrained by attachment to the nuclear matrix (gray lines) in undifferentiated cells, but mounted upon a nuclear matrix in differentiated cells.
These analyses in three diverse differentiating systems show that cyclin E is a conditional nuclear matrix protein that is recruited during differentiation and imply a conserved matrix-associated function. We propose that recruitment to the nuclear matrix reflects changes in the location at which cyclin E executes its function in pre-RC assembly, and that this switches from being unconstrained in nuclear space in undifferentiated cells, to being specified by immobilization on an underlying nuclear architecture (Figure 4E), potentially restricting the genome to cell type-specific patterns of DNA replication.
Attachment of DNA to the nuclear matrix occurs via sequences that anchor the genome in loops, while additional transient associations are thought to give rise to loops that are reeled through factories as DNA replicates (24). Changes in loop size have been observed directly and linked to transcriptional and developmental decisions, suggesting that their selection may contribute to restriction of gene expression and limit the plasticity of differentiating cells compared to stem and progenitor cells. Remodelling of attachments and establishment of replicon size occurs during passage through mitosis in Xenopus (25), but little is known about how pre-RCs are recruited to the matrix. Moreover, in the absence of a mitotic transition the recruitment process may be subtly different. It seems likely that the long G1 phase that cells experience upon exit from quiescence involves some rebuilding of the nuclear architecture, but it remains to be seen whether this underlies the specific requirement for cyclin E in non-mitotic cycles.
The difference between differentiated adult cells and cancer cells with respect to cyclin E has implications for the functional organization of the nucleus and its relationship with disease. Deregulation of cyclin E confers genome instability (26), possibly through defects in pre-RC assembly (27). Thus, the link between cyclin E and transformation may partly reflect failure to correctly assemble on the nuclear matrix, helping to support an undifferentiated state.
Changes in the form and texture of the nucleus have long been used to diagnose and classify tumours and changes in nuclear matrix composition have been observed in cancer cells (28). If, as we suggest, recruitment of cyclin E to the nuclear matrix influences genome organization, possibly conferring reduced plasticity, failure to become immobilized could have prognostic value. However, it will be necessary to distinguish between defects in cyclin E that impair recruitment, and wider problems with the internal organization of the nucleus in cancer cells.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Biotechnology and Biosciences Research Council (BBSRC); Yorkshire Cancer Research; York Against Cancer (to J.S.); British Heart Foundation (to J.F.-X.A.). Funding for the open access charge: BBSRC; Yorkshire Cancer Research, partial.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank T. Hunt and J. Gannon for Xenopus cyclin E antibody, A. Surani and K. Hilton for murine ES cells and R. Wilson for critical comment.
REFERENCES
- 1.Honda R, Lowe ED, Dubinina E, Skamnaki V, Cook A, Brown NR, Johnson LN. The structure of cyclin E1/CDK2: implications for CDK2 activation and CDK2-independent roles. EMBO J. 2005;24:452–463. doi: 10.1038/sj.emboj.7600554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ekholm SV, Zickert P, Reed SI, Zetterberg A. Accumulation of cyclin E is not a prerequisite for passage through the restriction point. Mol. Cell. Biol. 2001;21:3256–3265. doi: 10.1128/MCB.21.9.3256-3265.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Coverley D, Laman H, Laskey RA. Distinct roles for cyclins E and A during DNA replication complex assembly and activation. Nat. Cell Biol. 2002;4:523–528. doi: 10.1038/ncb813. [DOI] [PubMed] [Google Scholar]
- 4.Furstenthal L, Kaiser BK, Swanson C, Jackson PK. Cyclin E uses Cdc6 as a chromatin-associated receptor required for DNA replication. J. Cell. Biol. 2001;152:1267–1278. doi: 10.1083/jcb.152.6.1267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Geng Y, Yu Q, Sicinska E, Das M, Schneider JE, Bhattacharya S, Rideout WM, Bronson RT, Gardner H, Sicinski P. Cyclin E ablation in the mouse. Cell. 2003;114:431–443. doi: 10.1016/s0092-8674(03)00645-7. [DOI] [PubMed] [Google Scholar]
- 6.Mailand N, Diffley JF. CDKs promote DNA replication origin licensing in human cells by protecting Cdc6 from APC/C-dependent proteolysis. Cell. 2005;122:915–926. doi: 10.1016/j.cell.2005.08.013. [DOI] [PubMed] [Google Scholar]
- 7.Su TT, O’Farrell PH. Chromosome association of minichromosome maintenance proteins in Drosophila endoreduplication cycles. J. Cell. Biol. 1998;140:451–460. doi: 10.1083/jcb.140.3.451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matsumoto Y, Maller JL. A centrosomal localization signal in cyclin E required for Cdk2-independent S phase entry. Science. 2004;306:885–888. doi: 10.1126/science.1103544. [DOI] [PubMed] [Google Scholar]
- 9.Hemerly AS, Prasanth SG, Siddiqui K, Stillman B. Orc1 controls centriole and centrosome copy number in human cells. Science. 2009;323:789–793. doi: 10.1126/science.1166745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Parisi T, Beck AR, Rougier N, McNeil T, Lucian L, Werb Z, Amati B. Cyclins E1 and E2 are required for endoreplication in placental trophoblast giant cells. EMBO J. 2003;22:4794–4803. doi: 10.1093/emboj/cdg482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Berthet C, Aleem E, Coppola V, Tessarollo L, Kaldis P. Cdk2 knockout mice are viable. Curr. Biol. 2003;13:1775–1785. doi: 10.1016/j.cub.2003.09.024. [DOI] [PubMed] [Google Scholar]
- 12.Geng Y, Lee YM, Welcker M, Swanger J, Zagozdzon A, Winer JD, Roberts JM, Kaldis P, Clurman BE, Sicinski P. Kinase-independent function of cyclin E. Mol. Cell. 2007;25:127–139. doi: 10.1016/j.molcel.2006.11.029. [DOI] [PubMed] [Google Scholar]
- 13.Berezney R, Coffey DS. Nuclear protein matrix: association with newly synthesized DNA. Science. 1975;189:291–293. doi: 10.1126/science.1145202. [DOI] [PubMed] [Google Scholar]
- 14.Hozak P, Hassan AB, Jackson DA, Cook PR. Visualization of replication factories attached to the nucleoskeleton. Cell. 1993;73:361–373. doi: 10.1016/0092-8674(93)90235-i. [DOI] [PubMed] [Google Scholar]
- 15.Djeliova V, Russev G, Anachkova B. Dynamics of association of origins of DNA replication with the nuclear matrix during the cell cycle. Nucleic Acids Res. 2001;29:3181–3187. doi: 10.1093/nar/29.15.3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fujita M, Ishimi Y, Nakamura H, Kiyono T, Tsurumi T. Nuclear organization of DNA replication initiation proteins in mammalian cells. J. Biol. Chem. 2002;277:10354–10361. doi: 10.1074/jbc.M111398200. [DOI] [PubMed] [Google Scholar]
- 17.Turksen K. Stem Cell Protocols. 2006 doi: 10.1007/978-1-4939-2668-8. Methods in Molecular Biology. Humana Press Inc., Totowa, New Jersey. [DOI] [PubMed] [Google Scholar]
- 18.Fisher ME, Isaacs HV, Pownall ME. eFGF is required for activation of XmyoD expression in the myogenic cell lineage of Xenopus laevis. Development. 2002;129:1307–1315. doi: 10.1242/dev.129.6.1307. [DOI] [PubMed] [Google Scholar]
- 19.Cross WR, Eardley I, Leese HJ, Southgate J. A biomimetic tissue from cultured normal human urothelial cells: analysis of physiological function. Am. J. Physiol. Renal. Physiol. 2005;289:F459–468. doi: 10.1152/ajprenal.00040.2005. [DOI] [PubMed] [Google Scholar]
- 20.Ainscough JF, Rahman FA, Sercombe H, Sedo A, Gerlach B, Coverley D. C-terminal domains deliver the DNA replication factor Ciz1 to the nuclear matrix. J. Cell. Sci. 2007;120:115–124. doi: 10.1242/jcs.03327. [DOI] [PubMed] [Google Scholar]
- 21.Erlandsson F, Martinsson-Ahlzen HS, Wallin KL, Hellstrom AC, Andersson S, Zetterberg A. Parallel cyclin E and cyclin A expression in neoplastic lesions of the uterine cervix. Br. J. Cancer. 2006;94:1045–1050. doi: 10.1038/sj.bjc.6603038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hyrien O, Maric C, Mechali M. Transition in specification of embryonic metazoan DNA replication origins. Science. 1995;270:994–997. doi: 10.1126/science.270.5238.994. [DOI] [PubMed] [Google Scholar]
- 23.Vassetzky Y, Hair A, Mechali M. Rearrangement of chromatin domains during development in Xenopus. Genes. Dev. 2000;14:1541–1552. [PMC free article] [PubMed] [Google Scholar]
- 24.Ottaviani D, Lever E, Takousis P, Sheer D. Anchoring the genome. Genome Biol. 2008;9:201. doi: 10.1186/gb-2008-9-1-201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Lemaitre JM, Danis E, Pasero P, Vassetzky Y, Mechali M. Mitotic remodeling of the replicon and chromosome structure. Cell. 2005;123:787–801. doi: 10.1016/j.cell.2005.08.045. [DOI] [PubMed] [Google Scholar]
- 26.Spruck CH, Won K-A, Reed SI. Deregulated cyclin E induces chromosome instability. Nature. 1999;401:297–300. doi: 10.1038/45836. [DOI] [PubMed] [Google Scholar]
- 27.Ekholm-Reed S, Mendez J, Tedesco D, Zetterberg A, Stillman B, Reed SI. Deregulation of cyclin E in human cells interferes with prereplication complex assembly. J. Cell. Biol. 2004;165:789–800. doi: 10.1083/jcb.200404092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Zink D, Fischer AH, Nickerson JA. Nuclear structure in cancer cells. Nat. Rev. 2004;4:677–687. doi: 10.1038/nrc1430. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.