Abstract
We investigated the allele- and strand-specific transcriptional landscape of a megabase-wide genomic region of mouse Ube3a (ubiquitin protein ligase E3A) by means of a highly parallel SNP genotyping platform. We have successfully identified maternal-specific expression of Ube3a and its antisense counterpart (Ube3a-ATS) in brain, but not in liver. Because of the use of inter-subspecies hybrid mice, this megabase-wide analysis provided high-resolution picture of the transcriptional patterns of this region. First, we showed that brain-specific maternal expression of Ube3a is restricted to the second half part of the locus, but is absent from the first half part. Balance of allelic expression is altered in the middle of the locus. Second, we showed that expression of the brain-specific Ube3a-ATS appeared to be terminated in the region upstream to the Ube3a transcription start site. The present study highlights the importance of locus-wide competition between sense and antisense transcripts.
INTRODUCTION
Recent genome-wide studies have identified many mono-allelically expressed genes among mammalian genomes (1–5). Mono-allelically expressed genes are those primarily expressed from one allele, and the choice of the allele is determined in a stochastic or parent-of-origin-specific manner. In the mouse genome, more than one hundred autosomal genes exhibit parent-of-origin-specific gene expression, the so-called imprinted genes. The expression of imprinted genes is primarily governed by the imprint control element (ICE), which shows differential DNA methylation status between two parental alleles.
Non-coding antisense transcription is thought to regulate mono-allelic gene expression because it is frequently observed at imprinted gene loci; 15% of imprinted murine genes are associated with the antisense transcript (6). One of the best-known examples of an antisense transcript arising from an imprinted gene locus is Air, which is paternally transcribed from the antisense strand of Igf2r and represses expression of neighboring genes on the same chromosome (7). Kcnq1ot1 is another paternally expressed antisense transcript, and it suppresses the surrounding imprinted domain controlled by KvDMR (8). Antisense transcripts arising from other imprinted genes have been reported (9–13), but their function is not known. Some antisense transcripts arise from imprinted loci spanning 100–1000 kb, suggesting that antisense-mediated regulation of genomic imprinting is achieved in a locus-wide manner.
In order to fully understand the relationship between allele specificity and locus-wide competition between sense and antisense transcripts, we examined the gene locus of Ube3a (ubiquitin protein ligase E3A) whose orthologous partner, UBE3A, in human is known to cause Angelman syndrome, a neurogenetic disorder, when mutated (14,15). Ube3a is expressed from the maternal allele specifically in brain, but is bi-allelically expressed in most other tissues. Ube3a-ATS (also known as LNCAT), an antisense counterpart of Ube3a, is also specifically expressed in the brain from a paternal allele (16,17). Transcription of Ube3a-ATS spans a 1 Mb genomic region (≥460 kb in the case of the human genome), starting from alternative U-exons upstream of Snurf/Snrpn (16), and is detected only in neuronal cells concomitantly with maternal expression of Ube3a.
To examine the megabase-wide landscape of strand- and allele-specific transcriptional activity in the Ube3a–Snurf/Snrpn region, we exploited a highly parallel SNP genotyping platform—the Illumina GoldenGate Genotyping Assay—to target the transcriptome and to detect transcriptional output. This highly parallel SNP genotyping platform enables the simultaneous genotyping of hundreds of SNPs (18,19). Here, we made custom-panels of GoldenGate Assay covering murine SNPs within the ≥1Mb Ube3a–Snurf/Snrpn region. We identified SNPs for the standard laboratory mouse strain, C57BL/6J (B6), and MSM/Ms (MSM), an inbred strain derived from the Japanese wild mouse (Mus musculus molossinus) (20). Commonly used inbred mouse strains are primarily derived from the Mus musculus domesticus subspecies, with some genomic contributions from molossinus, musculus and castaneus subspecies (21). Inbred strains derived from molossinus have a higher frequency of polymorphims than the standard laboratory mouse strains derived from M. musculus domesticus, when they are compared with the B6 reference genome. A high frequency of polymorphisms is necessary to examine allele-specific transcriptional activity at a high resolution.
MATERIALS AND METHODS
Computer-based detection of SNPs and genotyping experiments
We sequenced five MSM BAC clones (MSMg01-118E18, MSMg01-106D06, MSMg01-352C08, MSMg01-480H06 and MSMg01-407I16) covering the Ube3a–Snurf/Snrpn genomic region. BAC clones were selected according to the alignment between the BAC-end sequences (22) and the mouse genome assembly UCSC mm6 (NCBI build 34). Nucleotide sequence alignment between BAC sequences and the C57BL/6J reference genome, UCSC mm8 (NCBI build 36), identified 5698 candidate SNPs. Because the mm8 assembly has a large insertion within the Ube3a–Snurf/Snrpn intergenic region by comparison with the mm6 build, many intergenic SNPs might not be included in our list. A total of 2,304 SNPs were selected, and these candidate SNPs were validated by the Illumina GoldenGate Assay targeting genomic DNA from the mouse strains C57BL/6J, MSM, and their reciprocal crosses. A total of 1420 SNPs were selected for transcriptome-targeting analyses: 712 for detecting plus-strand expression and 708 for detecting minus-strand expression from the Ube3a–Snurf locus (Supplementary Figure S1). Because Illumina GoldenGate Assay’s allele-specific primer is designed for either strand of the genome sequence, one SNP site can only detect the transcriptional output from either strand, if the assay targets cDNA. Plus-strand denotes those that detect the expression from plus strand of the UCSC mouse genome assembly, whereas minus-strand indicates the expression from minus-strand.
Target preparation for the Illumina GoldenGate genotyping assay
Genomic DNA was isolated by the standard protocol. Briefly, the dissected tissues (kidney) were treated with proteinase K overnight at 55°C, and the genomic DNA was precipitated with ethanol after phenol/chloroform extraction. Isolated genomic DNA was subjected to the Illumina GoldenGate genotyping assay by following the manufacturer’s procedures. For transcriptome-targeted assays, total RNA from brain and liver was isolated from 8- to 10-week-old C57BL/6J and MSM mice by using Trizol reagent (Invitrogen), and was reverse-transcribed by using random hexamers. The resultant cDNA was then subjected to the Illumina GoldenGate Assay.
Pre-processing of raw data
The signal intensity values from the Cy3 and Cy5 channels were subjected to loess normalization by using the limma package from R (23). The efficacy of two-color normalization in a transcriptome-targeted GoldenGate Assay has been described previously (24). Unsupervised thresholding based on the discriminant analysis method (25) was conducted for each Cy3 and Cy5 signal distribution to discriminate expressed/non-expressed SNP sites. When both the Cy3 and Cy5 signals were under the assigned threshold values, the SNP site was marked as non-expressed. The theta value [(2/π)tan−1(SM/SB)], where SM and SB denote signals representing MSM and B6 alleles, respectively (19), was then calculated for each SNP site to discriminate between the expressed allele type, i.e. B6 (<0.15), MSM (0.82<) or bi-allelic (intermediates). Threshold values were determined by the same method as for expressed/non-expressed SNP sites. The raw data were deposited in the NCBI GEO under accession number GSE21667.
Sequence analyses of the expressed alleles of Ube3a and Ube3a-ATS
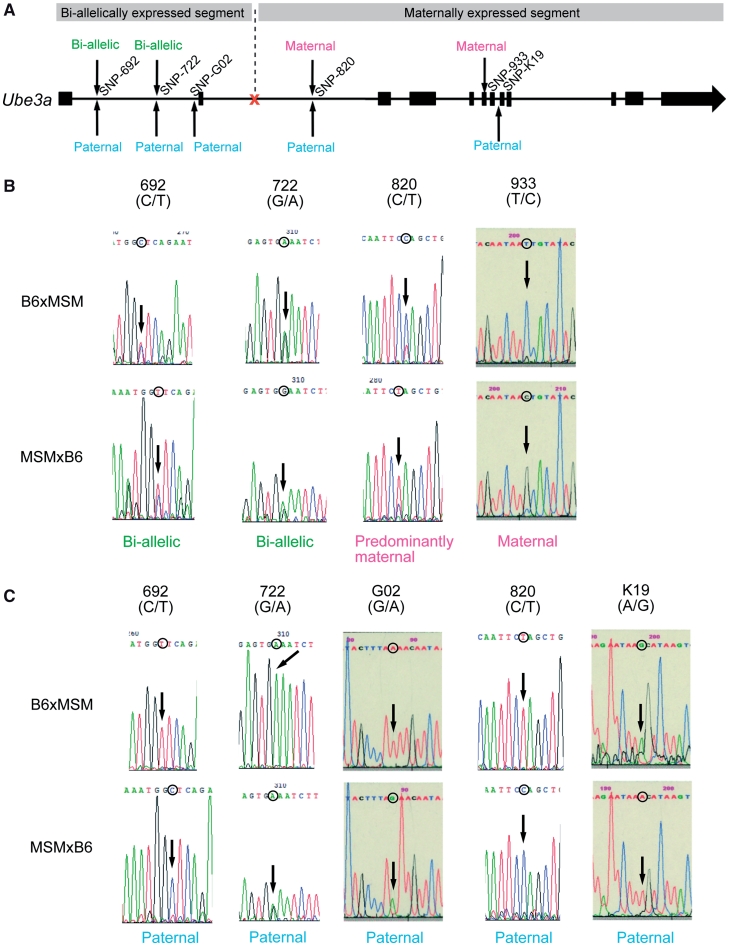
Total RNA from the brains of hybrid mice (BM and MB, 8- to 10-weeks old) was prepared by using Trizol reagent (Invitrogen) according to the manufacturer’s protocol. RNA concentration was determined with a NanoDrop ND-1000 spectrophotometer (NanoDrop Technologies). Total RNA (5 µg) was reverse transcribed with 50 U of Super ScriptTM reverse transcriptase (Invitrogen) using specific forward and reverse primers. Strand-specific cDNA synthesis was performed by using reverse primers for the sense cDNA strand of Ube3a and forward primers for the antisense cDNA strand of Ube3a. The primer sequences were SNP-692, 5′-CCAACCATGAAAGGCTTGAAAT-3′ (forward) and 5′-TCCTACAAATTTCCTGGGCAAG-3′ (reverse); SNP-722, 5′-ATAAATTTGGGCGCCTGTCAAT-3′ (forward) and 5′-GAGGCATCACTGAACTAGCAAGG-3′ (reverse); SNP-G02, 5′-TAAGTATTAAGAAGACTGGAG-3′ (forward) and 5′-ACAGTGGAAGAAACAGGTCAC-3′; SNP-820, 5′-GGGCATTGGATCCTATTACAGA-3′ (forward) and 5′-GGAAACAGCAAAATCATCCTCA-3′ (reverse); SNP-933, 5′-ATCCTGCAGACTTGAAGAAGC-3′ (forward) and 5(′-ATCATACATCATTGGGTTACC-3′ (reverse); SNP-K19, 5′-TTCTGGTTTTCTCAAGTTCAG-3′ (forward) and 5′-AGATTTATTGAGAATGTAGTC-3′ (reverse). The forward and reverse primer pairs were used to amplify the cDNA products for 35 cycles. PCR products were extracted from agarose gels using the MinElute Gel Extraction Kit (Qiagen) and were then subjected to sequence analysis.
RESULTS
Highly parallel genotyping of C57BL/6J and MSM/Ms
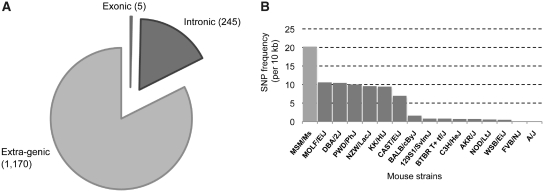
Prior to transcriptome-targeted SNP genotyping, we selected a set of measurable SNPs between C57BL/6J (B6) and MSM/Ms (MSM), by sequencing five MSM BAC clones covering Ube3a–Snurf/Snrpn genomic region and assayed 2304 candidate SNP sites with the Illumina GoldenGate Assay, targeting genomic DNA from B6, MSM, and the two reciprocal crosses. A total of 1420 validated SNPs were selected for transcriptome-targeted analyses (Supplementary Figure S1). Most SNPs (82.3%) were located within the extra-genic region of Ube3a and Snurf/Snrpn, whereas five were exonic and 245 were intronic (Figure 1A). Because of the unprocessed nascent-like nature of endogenous antisense transcripts (26), we generated cDNA targets by random hexamers priming and used for the assay. This cDNA targets should represent not only the poly(A)-plus–processed mRNAs but also the poly(A)-minus RNA population. The SNP frequency detected between B6 and MSM in the corresponding region is the highest among the combination of B6 and other mouse strains (Figure 1B). Thus, analyses using crosses between B6 and MSM provide a higher resolution picture of locus-wide transcriptional activity together with insights into strand- and allele-specificity.
Figure 1.
Statistics of measurable SNP sites. (A) SNP sites were classified as exonic, intronic, or extra-genic. (B) SNP frequencies between B6 and other strains. The number of SNPs between B6 and MSM within the Ube3a locus (including the 5 kb up- and downstream regions) was counted. The SNP frequencies between B6 and the other strains were obtained from the Perlegen Mouse SNP catalogue (21).
Mono-allelic Ube3a expression associated with antisense transcription
Because the Illumina GoldenGate Assay is primarily designed for DNA-targeted genotyping experiments, pre-processing of the raw data derived from the transcriptome-targeted assay is required for accurate data analyses. Therefore, we used loess normalization to adjust signal intensity distribution of two channels and set threshold values based on the discriminant analysis method (25) to distinguish expressed sites from non-expressed sites and also to distinguish between the three different types of allelic expression (B6 mono-allelic, MSM mono-allelic and bi-allelic). Of the 1420 validated SNPs selected in the genotyping assays, 712 were used for detecting plus-strand expression and 708 were selected for detecting minus-strand expression. To examine the reproducibility of the results, we performed transcriptome-targeted assays for the two reciprocal hybrids B6xMSM (BM) and MSMxB6 (MB). Using this approach, we successfully generated a high-resolution strand- and allele-specific transcriptional landscape of the Ube3a–Snurf/Snrpn genomic region in brain and liver tissues (Figure 2).
Figure 2.
Transcriptional landscape of the Ube3a–Snurf/Snrpn region. Status of strand- and allele-specific expression at Ube3a–Snurf locus in brain (A), and in liver (B). Each color indicates the expression status for every SNP site: B6 allele (magenta), MSM allele (green), both alleles (black) and non-expressed (light gray). Intensity value thresholds were set by the discriminant analysis method (25) to distinguish between expressed and non-expressed SNP sites and to distinguish between the three types of expressed allele. Assays were performed for both of the reciprocal hybrids, BM (B6xMSM) and MB (MSMxB6). The former indicates maternally contributed strain. Expression strands are distinguished by plus and minus. ‘Plus’ denotes those that detect the expression from plus-strand of the UCSC mouse genome assembly, whereas ‘Minus’ indicates the expression from minus-strand. Gene location is denoted in the upper panel: thick and narrow bars colored navy indicate exonic and intronic regions, respectively.
Ube3a is expressed from the maternal allele in a brain-specific manner (27–29), whereas Snurf/Snrpn shows paternal-specific gene expression in most tissues (30). We observed concordance trends, showing paternal-specific expression of Snurf/Snrpn in both brain and liver (Figure 2). Snurf/Snrpn expression in the liver was restricted to the region as in the UCSC Known Genes Track (31), whereas it in the brain was widespread and included extra-genic regions of the Snurf/Snprn locus (from 100 kb upstream to 200 kb downstream). We did not obtain data for the ∼400 kb region between Ube3a and Snurf/Snrpn because of the lack of measurable SNPs to assay (see ‘Materials and Methods’ section); however, brain-specific paternal expression of the minus strand is likely to extend to the Ube3a peripheral region, which spans nearly 1 Mb of the genome and thus is antisense to Ube3a. This widespread transcription initiated from the upstream region of Snurf/Snrpn (producing Ube3a-ATS) has been described in the previous studies (16,17). We also found that the transcription of the minus strand in liver was not widespread like in brain. Since the antisense transcription is only seen in brain where Ube3a shows mono-allelic expression, we suggest that the widespread transcription originated from Snurf/Snrpn locus might be involved in the control of mono-allelic Ube3a expression. This transcriptional status was detected for both of the reciprocal hybrids (BM and MB), indicating that the expression is parent-of-origin-specific.
Allelic imbalance of Ube3a expression in brain
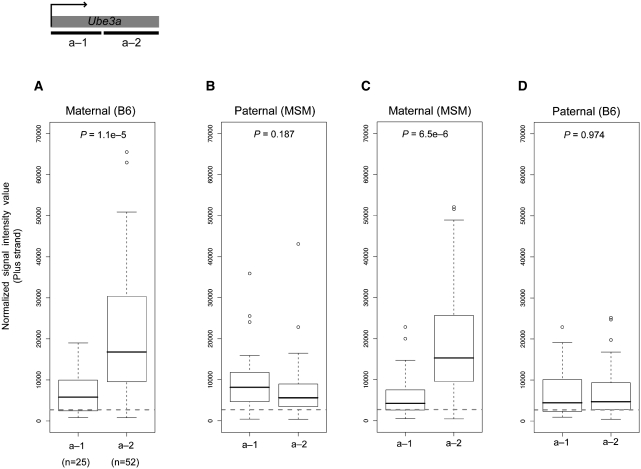
Our analysis can achieve locus-wide parallel genotyping of the expressed alleles and can also measure transcript abundance. Thus, the method can provide information not possible with assays targeting a small number of SNPs. As described, plus-strand expression of the Ube3a locus was observed specifically in the brain, consistent with known Ube3a expression pattern. However, we found that mono-allelic Ube3a expression was not detected along the entire locus; there is a shift from bi-allelic expression to mono-allelic expression within the locus. Circular binary segmentation (CBS) method (32) of the paternal and maternal signal intensity values identified a shift of allelic expression dividing the genomic locus into subregions showing bi-allelic expression and mono-allelic expression, respectively (Supplementary Figure S2). The ‘bi-allelic’ region corresponds to about ∼25 kb region including the 1st and 2nd exon. The ‘mono-allelic’ region corresponds to the rest of the locus (Figure 3A), and the transition appeared to occur in the middle of the 2nd intron. Such disparity was observed for both of the reciprocal hybrids. Individual SNP genotyping by strand-specific RT–PCR and the nucleotide sequencing also confirmed bi-allelic expression of the 5′ part of Ube3a in the brain tissue (Figure 3B and C). The result led to the question whether this transition is caused by a reduction of paternal Ube3a expression in the latter half, or by an increase in maternal expression. To clarify these possibilities, we compared the intensity of allele-specific expression for the two segmental regions. We found that maternal expression in both reciprocal hybrids is notably increased in the 3′ part, whereas paternal expression does not differ between the two regions (Figure 4). This result indicates that allelic imbalance of Ube3a expression is caused by an increased maternal expression in the latter half of the locus.
Figure 3.
Expressed alleles within the Ube3a locus. (A) Sequence analysis summary. SNP sites within the Ube3a locus for sequence analysis are indicated by arrows (thick lines and narrow lines indicate exonic and intronic regions, respectively). Expressed allele decisions are denoted for the plus and minus strand. Expression from plus strand means Ube3a sense expression. An orange mark denotes computationally estimated site which shows allelic balance alteration. (B) Sequence analysis results and decisions of expressed allele from plus strand for B6xMSM (upper) and MSMxB6 (lower). (C) Sequence analysis results and decisions of expressed allele from minus strand for B6xMSM (upper) and MSMxB6 (lower).
Figure 4.
Comparison of maternal and paternal expression of Ube3a in brain. Comparison of maternal and paternal expression of Ube3a in brain for the two regions, denoted a-1 and a-2, estimated by genomic segmentation analysis. (A) Maternal expression in the BM hybrid, (B) paternal expression in the BM hybrid, (C) maternal expression in the MB hybrid and (D) paternal expression in the MB hybrid. Statistical significance (P-values) was tested with the Wilcoxon test. The number of SNPs in the corresponding regions is indicated in the parenthesis. Dashed line indicates the threshold value to distinguish between expressed and non-expressed SNP sites.
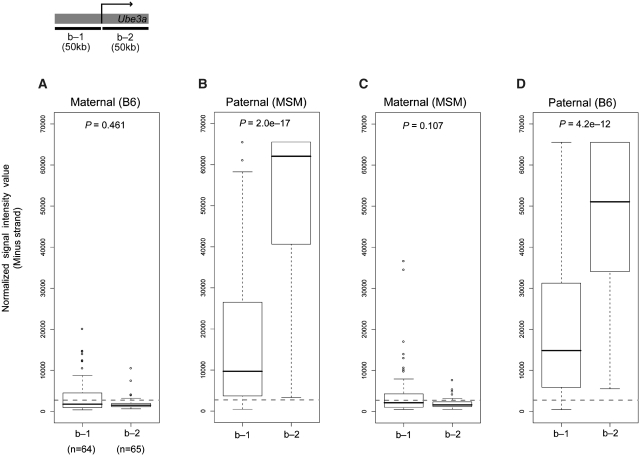
We also observed a notable difference in paternal Ube3a-ATS expression in the vicinity of the Ube3a transcription start site. CBS showed clear disparity of paternal antisense expression between the two regions separated by Ube3a transcription start site (Supplementary Figure S3). In the region upstream to the Ube3a transcription start site, antisense transcription still occurs from a paternal allele, but the ratio between the two alleles was significantly reduced compared to that from the Ube3a gene body. Indeed, paternal antisense expression was significantly higher in the Ube3a gene body than in the upstream region of Ube3a transcription start site (P = 2.0e–17, Wilcoxon test) (Figure 5B and D). By comparison, maternal antisense transcript was hardly detected in the two regions (Figure 5A and C). This finding suggests that Ube3a-ATS expression is preferentially suppressed in the vicinity of the Ube3a transcription start site.
Figure 5.
Comparison of maternal and paternal expression of Ube3a-ATS. Comparison of maternal and paternal expression of Ube3a in brain for the two regions separated by the transcription start site of Ube3a, denoted b-1 and b-2, estimated by genomic segmentation analysis. (A) maternal expression in the BM hybrid, (B) paternal expression in the BM hybrid, (C) maternal expression in the MB hybrid and (D) paternal expression in the MB hybrid. Statistical significance (P-values) was tested with the Wilcoxon test. The number of SNPs in the corresponding regions is indicated in the parenthesis. Dashed line indicates the threshold value to distinguish between expressed and non-expressed SNP sites.
DISCUSSION
We used a highly parallel SNP genotyping system to successfully characterize the transcriptional landscape of the Ube3a–Snurf/Snrpn region. We used a combinatorial approach involving expressed allele genotyping together with genomic tiling-array-like high-resolution transcriptome analysis. SNP frequency of inter-subspecies hybrid is twice as high compared to other strain combination, enabling high-resolution analyses of allele-specific transcriptional activity within the megabase-wide region. Although we ourselves sequenced the MSM BAC contigs of the Ube3a–Snurf/Snrpn locus, the entire MSM genome sequences are now publicly available (http://molossinus.lab.nig.ac.jp/msmdb/index.jsp), thus facilitating the same analysis of the other gene loci.
We used cDNA targets representing whole transcriptome of both the poly(A)+ processed mRNA and non-poly(A) RNA population. We did this because of the unprocessed nascent-like nature of endogenous antisense transcription. In the case of Air, an antisense counterpart of Igf2r, transcription spans a 100-kb genomic region, and the transcript appears to be unspliced (7,33). Also, recent micorarray-based transcriptome analyses and expression analyses of individual genes showed that endogenous antisense transcripts in mammals tend to be poly(A)-negative (26,34). Thus, SNP genotyping targeting the whole transcriptome is essential to full understanding of locus-wide competition between sense and antisense transcripts.
One of the interesting observations from our analysis is that brain-specific maternal expression of Ube3a occurs only from the second half part of the locus, but not from the first half. This ‘allelic-shift’ in Ube3a expression is the consequence of the marked difference in maternal Ube3a expression between the two regions (5′ and 3′ half part). Although this observation might reflect the existence of an alternative maternal-specific variant that lacks first and second exons, we could not find any evidences supporting the existence of such variant(s) in the public datasets; there are no enrichment of histone H3K4 tri-methylation active mark (35,36) or CAGE tags that indicates transcription start site in the second intron (data not shown). No ESTs that aligned in the vicinity are found as well. Meanwhile, there is a possibility that ‘allelic-shift’ might reflect the situation in the nuclear transcriptome, because our analysis uses cDNA target generated from the total RNA, not mere poly(A)-selected RNA. As we also detected considerable level of poly(A)-minus Ube3a sense and antisense transcript with heterogeneous nature in its molecular weight by northern hybridization (data not shown), allelic imbalance of Ube3a expression might be regulated in a competitive manner with antisense transcription in the nucleus.
In humans, strand-specific RT–PCR analysis has previously suggested that another maternally transcribed unit exists within the downstream region to UBE3A (37). In addition, alternatively terminated variants as well as several poly-adenylation signals have been identified for the UBE3A locus (38). Since we observed predominant maternal expression not only from the Ube3a genic region, but also from the extra-genic downstream region, murine Ube3a might express maternal transcripts from its downstream region.
Because we used whole brain as a source of RNA, our observations reflect the situation in the mixture of brain tissue cell types, consists of neurons and glial cells. Previous studies using primary brain cell cultures revealed that expression of Ube3a is parental-of-origin-specific in neurons, but bi-allelic in glial cells (39,40). Thus, our findings using the entire brain transcriptome can only provide an ‘average’ of Ube3a expression for the two major brain cell types. Indeed, we found that a certain level of Ube3a expression was detected not only from the maternal allele but also from the paternal allele (Figure 4B and D). This is most likely the result of the bi-allelic nature of Ube3a expression in glial cells.
Our results for the expression of the antisense transcript of Ube3a (Ube3a-ATS) most likely reflect expression in neurons because Ube3a-ATS is not expressed in glial cells (39,40). We did not investigate the molecular basis for the reduction in Ube3a-ATS expression, but our observations suggest that overlap of the antisense transcript with the transcription start site or a CpG island that covers the Ube3a transcription start site might be essential for repression of paternal Ube3a expression. It has shown previously that abnormal antisense transcription generated by genetic mutation induces DNA methylation in the promoter region of the HBA2 locus, resulting in repression of gene expression (41). Silencing of paternal Ube3a expression might also be induced by similar mechanism. Intriguingly, a slight, but not statistically significant (P > 0.05, Wilcoxon test), upregulation of maternal Ube3a-ATS is observed from the upstream region of the transcription start site of Ube3a (Figure 5A and C). This suggests that leakage of maternal antisense transcription near the promoter might arise as a result of Ube3a expression from the maternal allele. Another experimental approach is therefore required to examine the molecular mechanism underlying the control of gene expression mediated by Ube3a-ATS. Overall, our data clearly showed that allele-specific expression of Ube3a is variable depending on the genomic position.
Recently, several studies using high-throughput technology have identified many examples of mono-allelically expressed genes (1–5), including random mono-allelic genes and imprinted genes, with some of these associated with sense and antisense transcription. In addition, many imprinted genes (or imprinted regions) show expression of the antisense transcript (6). Although high-throughput sequencing also can generate the data that distinguish strand- and allele-specificity of the transcripts, it requires huge number of sequence reads to retrieve sufficient depth for each SNP sites, and also requires target enrichment step to analyze particular gene locus. Here, we demonstrated that array-based approach of megabase-wide region can provide high-resolution insight into allele-specific sense and antisense transcriptional dynamics, which is the foundation for understanding the mechanisms of antisense-mediated gene regulation. The highly parallel SNP genotyping approach we undertook in this study is also applicable to the analysis of immunoprecipitated DNA, including methylated DNA, and histone modifications. We have now succeeded to establish B6–MSM hybrid ES cells and its neuronal differentiation in vitro (Kohama, C. et al., manuscript in preparation). Analyses targeting immunopreciptated DNA coupled with the transcriptome analysis of the hybrid ES cell differentiation have the potential to provide a deeper understanding of the association between epigenetic regulation and strand-specific transcription.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
President's Discretionary Fund by RIKEN (to H.K.); Grant-in-Aid for Scientific Research on Innovative Areas by the Japan Society for the Promotion of Science (JSPS) (to H.K.); Research Fellowship for Young Scientists by JSPS (to K.N.). Funding for open access charge: Grant-in-Aid for Scientific Research on Innovative Areas by JSPS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors acknowledge Rieko Ikeda (RIKEN BioResource Center) for technical suggestions, and Dr Atsushi Toyoda (National Institute of Genetics, Japan) for MSM BAC sequencing.
REFERENCES
- 1.Babak T, Deveale B, Armour C, Raymond C, Cleary MA, van der Kooy D, Johnson JM, Lim LP. Global survey of genomic imprinting by transcriptome sequencing. Curr. Biol. 2008;18:1735–1741. doi: 10.1016/j.cub.2008.09.044. [DOI] [PubMed] [Google Scholar]
- 2.Bjornsson HT, Albert TJ, Ladd-Acosta CM, Green RD, Rongione MA, Middle CM, Irizarry RA, Broman KW, Feinberg AP. SNP-specific array-based allele-specific expression analysis. Genome Res. 2008;18:771–779. doi: 10.1101/gr.073254.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Palacios R, Gazave E, Goni J, Piedrafita G, Fernando O, Navarro A, Villoslada P. Allele-specific gene expression is widespread across the genome and biological processes. PLoS One. 2009;4:e4150. doi: 10.1371/journal.pone.0004150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Serre D, Gurd S, Ge B, Sladek R, Sinnett D, Harmsen E, Bibikova M, Chudin E, Barker DL, Dickinson T, et al. Differential allelic expression in the human genome: a robust approach to identify genetic and epigenetic cis-acting mechanisms regulating gene expression. PLoS Genet. 2008;4:e1000006. doi: 10.1371/journal.pgen.1000006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Zhang K, Li JB, Gao Y, Egli D, Xie B, Deng J, Li Z, Lee JH, Aach J, Leproust EM, et al. Digital RNA allelotyping reveals tissue-specific and allele-specific gene expression in human. Nat. Methods. 2009;6:613–618. doi: 10.1038/nmeth.1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Reik W, Walter J. Genomic imprinting: parental influence on the genome. Nat. Rev. Genet. 2001;2:21–32. doi: 10.1038/35047554. [DOI] [PubMed] [Google Scholar]
- 7.Sleutels F, Zwart R, Barlow DP. The non-coding Air RNA is required for silencing autosomal imprinted genes. Nature. 2002;415:810–813. doi: 10.1038/415810a. [DOI] [PubMed] [Google Scholar]
- 8.Mancini-Dinardo D, Steele SJ, Levorse JM, Ingram RS, Tilghman SM. Elongation of the Kcnq1ot1 transcript is required for genomic imprinting of neighboring genes. Genes Dev. 2006;20:1268–1282. doi: 10.1101/gad.1416906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Choo JH, Kim JD, Kim J. Imprinting of an evolutionarily conserved antisense transcript gene APeg3. Gene. 2008;409:28–33. doi: 10.1016/j.gene.2007.10.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hernandez A, Martinez ME, Croteau W, St Germain DL. Complex organization and structure of sense and antisense transcripts expressed from the DIO3 gene imprinted locus. Genomics. 2004;83:413–424. doi: 10.1016/j.ygeno.2003.08.024. [DOI] [PubMed] [Google Scholar]
- 11.Joh K, Yatsuki H, Higashimoto K, Mukai T, Soejima H. Antisense transcription occurs at the promoter of a mouse imprinted gene, commd1, on the repressed paternal allele. J. Biochem. 2009;146:771–774. doi: 10.1093/jb/mvp147. [DOI] [PubMed] [Google Scholar]
- 12.Jong MT, Carey AH, Caldwell KA, Lau MH, Handel MA, Driscoll DJ, Stewart CL, Rinchik EM, Nicholls RD. Imprinting of a RING zinc-finger encoding gene in the mouse chromosome region homologous to the Prader-Willi syndrome genetic region. Hum. Mol. Genet. 1999;8:795–803. doi: 10.1093/hmg/8.5.795. [DOI] [PubMed] [Google Scholar]
- 13.Lee YJ, Park CW, Hahn Y, Park J, Lee J, Yun JH, Hyun B, Chung JH. Mit1/Lb9 and Copg2, new members of mouse imprinted genes closely linked to Peg1/Mest(1) FEBS Lett. 2000;472:230–234. doi: 10.1016/s0014-5793(00)01461-7. [DOI] [PubMed] [Google Scholar]
- 14.Kishino T, Lalande M, Wagstaff J. UBE3A/E6-AP mutations cause Angelman syndrome. Nat. Genet. 1997;15:70–73. doi: 10.1038/ng0197-70. [DOI] [PubMed] [Google Scholar]
- 15.Matsuura T, Sutcliffe JS, Fang P, Galjaard RJ, Jiang YH, Benton CS, Rommens JM, Beaudet AL. De novo truncating mutations in E6-AP ubiquitin-protein ligase gene (UBE3A) in Angelman syndrome. Nat. Genet. 1997;15:74–77. doi: 10.1038/ng0197-74. [DOI] [PubMed] [Google Scholar]
- 16.Landers M, Bancescu DL, Le Meur E, Rougeulle C, Glatt-Deeley H, Brannan C, Muscatelli F, Lalande M. Regulation of the large (approximately 1000 kb) imprinted murine Ube3a antisense transcript by alternative exons upstream of Snurf/Snrpn. Nucleic Acids Res. 2004;32:3480–3492. doi: 10.1093/nar/gkh670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Le Meur E, Watrin F, Landers M, Sturny R, Lalande M, Muscatelli F. Dynamic developmental regulation of the large non-coding RNA associated with the mouse 7C imprinted chromosomal region. Dev. Biol. 2005;286:587–600. doi: 10.1016/j.ydbio.2005.07.030. [DOI] [PubMed] [Google Scholar]
- 18.Fan JB, Chee MS, Gunderson KL. Highly parallel genomic assays. Nat. Rev. Genet. 2006;7:632–644. doi: 10.1038/nrg1901. [DOI] [PubMed] [Google Scholar]
- 19.Fan JB, Gunderson KL, Bibikova M, Yeakley JM, Chen J, Wickham Garcia E, Lebruska LL, Laurent M, Shen R, Barker D. Illumina universal bead arrays. Methods Enzymol. 2006;410:57–73. doi: 10.1016/S0076-6879(06)10003-8. [DOI] [PubMed] [Google Scholar]
- 20.Moriwaki K, Miyashita N, Mita A, Gotoh H, Tsuchiya K, Kato H, Mekada K, Noro C, Oota S, Yoshiki A, et al. Unique inbred strain MSM/Ms established from the Japanese wild mouse. Exp. Anim. 2009;58:123–134. doi: 10.1538/expanim.58.123. [DOI] [PubMed] [Google Scholar]
- 21.Frazer KA, Eskin E, Kang HM, Bogue MA, Hinds DA, Beilharz EJ, Gupta RV, Montgomery J, Morenzoni MM, Nilsen GB, et al. A sequence-based variation map of 8.27 million SNPs in inbred mouse strains. Nature. 2007;448:1050–1053. doi: 10.1038/nature06067. [DOI] [PubMed] [Google Scholar]
- 22.Abe K, Noguchi H, Tagawa K, Yuzuriha M, Toyoda A, Kojima T, Ezawa K, Saitou N, Hattori M, Sakaki Y, et al. Contribution of Asian mouse subspecies Mus musculus molossinus to genomic constitution of strain C57BL/6J, as defined by BAC-end sequence-SNP analysis. Genome Res. 2004;14:2439–2447. doi: 10.1101/gr.2899304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Smyth GK. Limma: linear models for microarray data. In: Robert Gentleman VJC, Huber W, Irizarry RA, Dudoit S, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 24.Ritchie ME, Forrest MS, Dimas AS, Daelemans C, Dermitzakis ET, Deloukas P, Tavare S. Data analysis issues for allele-specific expression using Illumina’s GoldenGate assay. BMC Bioinformatics. 2010;11:280. doi: 10.1186/1471-2105-11-280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Otsu N. A threshold selection method from gray-level histograms. IEEE Trans. Sys. Man. Cybern. 1979;9:62–66. [Google Scholar]
- 26.Kiyosawa H, Mise N, Iwase S, Hayashizaki Y, Abe K. Disclosing hidden transcripts: mouse natural sense-antisense transcripts tend to be poly(A) negative and nuclear localized. Genome Res. 2005;15:463–474. doi: 10.1101/gr.3155905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Albrecht U, Sutcliffe JS, Cattanach BM, Beechey CV, Armstrong D, Eichele G, Beaudet AL. Imprinted expression of the murine Angelman syndrome gene, Ube3a, in hippocampal and Purkinje neurons. Nat. Genet. 1997;17:75–78. doi: 10.1038/ng0997-75. [DOI] [PubMed] [Google Scholar]
- 28.Rougeulle C, Glatt H, Lalande M. The Angelman syndrome candidate gene, UBE3A/E6-AP, is imprinted in brain. Nat. Genet. 1997;17:14–15. doi: 10.1038/ng0997-14. [DOI] [PubMed] [Google Scholar]
- 29.Vu TH, Hoffman AR. Imprinting of the Angelman syndrome gene, UBE3A, is restricted to brain. Nat. Genet. 1997;17:12–13. doi: 10.1038/ng0997-12. [DOI] [PubMed] [Google Scholar]
- 30.Shemer R, Birger Y, Riggs AD, Razin A. Structure of the imprinted mouse Snrpn gene and establishment of its parental-specific methylation pattern. Proc. Natl Acad. Sci. USA. 1997;94:10267–10272. doi: 10.1073/pnas.94.19.10267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Venkatraman ES, Olshen AB. A faster circular binary segmentation algorithm for the analysis of array CGH data. Bioinformatics. 2007;23:657–663. doi: 10.1093/bioinformatics/btl646. [DOI] [PubMed] [Google Scholar]
- 33.Lyle R, Watanabe D, te Vruchte D, Lerchner W, Smrzka OW, Wutz A, Schageman J, Hahner L, Davies C, Barlow DP. The imprinted antisense RNA at the Igf2r locus overlaps but does not imprint Mas1. Nat. Genet. 2000;25:19–21. doi: 10.1038/75546. [DOI] [PubMed] [Google Scholar]
- 34.Numata K, Osada Y, Okada Y, Saito R, Hiraiwa N, Nakaoka H, Yamamoto N, Watanabe K, Okubo K, Kohama C, et al. Identification of novel endogenous antisense transcripts by DNA microarray analysis targeting complementary strand of annotated genes. BMC Genomics. 2009;10:392. doi: 10.1186/1471-2164-10-392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Meissner A, Mikkelsen TS, Gu H, Wernig M, Hanna J, Sivachenko A, Zhang X, Bernstein BE, Nusbaum C, Jaffe DB, et al. Genome-scale DNA methylation maps of pluripotent and differentiated cells. Nature. 2008;454:766–770. doi: 10.1038/nature07107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Rougeulle C, Cardoso C, Fontes M, Colleaux L, Lalande M. An imprinted antisense RNA overlaps UBE3A and a second maternally expressed transcript. Nat. Genet. 1998;19:15–16. doi: 10.1038/ng0598-15. [DOI] [PubMed] [Google Scholar]
- 38.Kishino T, Wagstaff J. Genomic organization of the UBE3A/E6-AP gene and related pseudogenes. Genomics. 1998;47:101–107. doi: 10.1006/geno.1997.5093. [DOI] [PubMed] [Google Scholar]
- 39.Kishino T. Imprinting in neurons. Cytogenet. Genome Res. 2006;113:209–214. doi: 10.1159/000090834. [DOI] [PubMed] [Google Scholar]
- 40.Yamasaki K, Joh K, Ohta T, Masuzaki H, Ishimaru T, Mukai T, Niikawa N, Ogawa M, Wagstaff J, Kishino T. Neurons but not glial cells show reciprocal imprinting of sense and antisense transcripts of Ube3a. Hum. Mol. Genet. 2003;12:837–847. doi: 10.1093/hmg/ddg106. [DOI] [PubMed] [Google Scholar]
- 41.Tufarelli C, Stanley JA, Garrick D, Sharpe JA, Ayyub H, Wood WG, Higgs DR. Transcription of antisense RNA leading to gene silencing and methylation as a novel cause of human genetic disease. Nat. Genet. 2003;34:157–165. doi: 10.1038/ng1157. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.