Abstract
eIF4A is a key component in eukaryotic translation initiation; however, it has not been clear how auxiliary factors like eIF4B and eIF4G stimulate eIF4A and how this contributes to the initiation process. Based on results from isothermal titration calorimetry, we propose a two-site model for eIF4A binding to an 83.5 kDa eIF4G fragment (eIF4G-MC), with a high- and a low-affinity site, having binding constants KD of ∼50 and ∼1000 nM, respectively. Small angle X-ray scattering analysis shows that the eIF4G-MC fragment adopts an elongated, well-defined structure with a maximum dimension of 220 Å, able to span the width of the 40S ribosomal subunit. We establish a stable eIF4A–eIF4B complex requiring RNA, nucleotide and the eIF4G-MC fragment, using an in vitro RNA pull-down assay. The eIF4G-MC fragment does not stably associate with the eIF4A–eIF4B–RNA-nucleotide complex but acts catalytically in its formation. Furthermore, we demonstrate that eIF4B and eIF4G-MC act synergistically in stimulating the ATPase activity of eIF4A.
INTRODUCTION
In eukaryotes, translation initiation relies on spatial and temporal interactions between many initiation factors; however, their interaction map is still poorly understood. Initiation begins with eIF1, eIF1A, and eIF3 stimulated recruitment of the ternary complex (eIF2–GTP–Met-tRNAiMet) to the 40S ribosomal subunit forming the 43S pre-initiation complex (PIC). The following steps involve the initiation factor eIF4F that consists of three initiation factors: the cap-binding protein eIF4E, the archetypical DEAD-box helicase eIF4A and the large scaffold protein eIF4G that contains binding sites for the two other factors. Upon assembly of the 43S PIC, mRNA bound to eIF4E in the eIF4F complex is recruited to the 43S PIC through interaction with eIF3, forming the 48S PIC. Next, the search for the AUG start codon begins (the scanning process) involving eIF1 and eIF1A along with the eIF4F complex. Recognition of the start codon triggers the eIF5 stimulated GTPase activity of eIF2, and the subsequent release of inorganic phosphate (Pi) irreversibly commits the PIC to initiation at this start codon. This is followed by the eIF5B-dependent joining of the 60S ribosomal subunit resulting in a competent 80S ribosome ready for elongation [for recent reviews, see refs (1,2)].
Both mammalian and yeast eIF4G contain binding sites for the poly(A)-binding protein (PABP), eIF4A and eIF4E, while only mammalian eIF4G contains binding sites for eIF3 and the Mnk1 kinase. The mammalian eIF4G has two eIF4A binding domains: one in the central part and the other in the carboxy-terminal part of eIF4G (3) (Figure 1A). Mutations in the carboxy-terminal eIF4A binding domain were found to decrease 48S PIC formation 3- to 4-fold using toe-print analysis; while mutations in the central eIF4A binding domain abolished 48S PIC formation (4). Yeast eIF4G has only a single binding domain for eIF4A (5) that is homologous to the central part in the mammalian eIF4G (3).
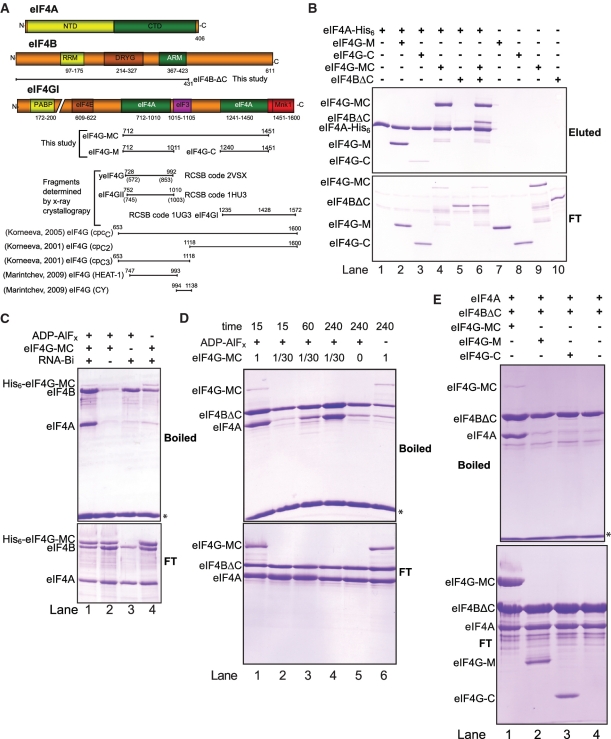
Figure 1.
Interactions between eIF4A, eIF4B and eIF4G fragments. (A) Schematic representation of the constructs used in this study or mentioned in the text. Colored bars show motifs expected to be involved in protein–protein interactions (interacting partner indicated) and motifs involved in dimerization (DRYG) or RNA binding (RRM and ARM). Poly(A) binding protein (PAPB). yeIF4G is the yeast homolog of mammalian eIF4G. The numbering in parenthesis for yeIF4G and eIF4GII correspond to the amino acid position, while the numbering above refer to the corresponding position in human eIF4GI based on sequence alignment using Clustal W. (B) Cobalt pull-down experiments using eIF4A-His6 (lanes 1–6) or negative controls without (lanes 7–10) and the indicated proteins. Proteins eluted using 400 mM Imidazole from the TALON beads are shown in the upper gel (eluted) and 15% of the flow through (FT) is shown in the lower gel. All eIF4A-His6 were bound to the TALON beads. (C) Streptavidin pull-down experiments using a pU30 biotinylated RNA. All reactions contain eIF4B and eIF4A and where indicated His6-eIF4G-MC, nucleotide analogs or biotinylated RNA. All reactions were incubated for 1 h at 4°C. The upper gel shows the proteins that bound to the RNA (boiled) and the lower gel shows 15% of the flow through (FT). (D) As in (C) but reactions contain eIF4A, eIF4BΔC and pU30-biotin along with the indicated amount of eIF4G-MC and ADP-AlFx as nucleotide analog. Each reaction was incubated at 4°C for the indicated time (min). (E) As in (D) but using fixed amount of either eIF4G-MC, eIF4G-M or eIF4G-C where indicated. Each reaction was incubated for 1 hr at room temperature. The asterisk indicates the position of the streptavidin moiety eluted by boiling. Due to the similar position of eIF4G-MC and eIF4B on a SDS–PAGE gel, eIF4BΔC was used in (B), (D) and (E). Note that eIF4B has a strong nucleotide-independent RNA binding leading to a basal level of eIF4B while eIF4G-MC has a low nucleotide-independent RNA binding. eIF4A has a low nucleotide-dependent RNA binding.
It has been demonstrated that eIF4B stimulates the ATPase and helicase activities of eIF4A (6–8), in agreement with genetic data from Saccharomyces cerevisiae (9), and that human eIF4G stimulates the ATPase activity of eIF4A (10), although the interplay between the factors is not known. The molecular mechanism behind activation of eIF4A by eIF4B is not known; however, a soft-clamp model has been proposed for human eIF4G's stimulation of eIF4A (11) in which eIF4G stabilizes an eIF4A conformation that stimulates its activity. Accordingly, the crystal structure of the yeast eIF4G–eIF4A complex demonstrated that eIF4A was kept in a slightly open conformation (5) compared to the closed conformation known from various DEAD-box ATPases bound to RNA and nucleotide analogs (12–15). In this manner eIF4G orients the different conserved motifs in eIF4A so that they are ready to bind RNA and ATP (5). In this structure eIF4G interacts with both the N-terminal domain (NTD) and the C-terminal domain (CTD) of eIF4A. However, the NTD interaction was predicted to be broken upon binding of RNA and ATP to eIF4A (5), suggesting that eIF4G does not stably bind the closed active conformation of eIF4A. A fragment of human eIF4G (cpcC) that includes the two eIF4A binding sites, but not the N-terminal part and the eIF4E binding site (residues 653–1600, see Figure 1A), activated the ATPase activity of eIF4A to nearly the same extent as eIF4F itself (10). In fact, eIF4G can replace eIF4F in an in vitro assay if both eIF4A and eIF4B are present (16). It has also been demonstrated that there is a dynamic exchange between free eIF4A and eIF4A incorporated into eIF4F (17,18). It is therefore advantageous to study the interaction between eIF4A and eIF4G outside of the context of the eIF4F complex. Even though eIF4B has been shown to stimulate the helicase activity of eIF4A, a stable reconstituted complex between these proteins has never been firmly demonstrated. Electrophoretic mobility shift assay has been the only way to detect the complex to date (19).
Unwinding of double-stranded (ds) nucleic acid or rearrangement of RNA–protein complexes is required in virtually all steps of the gene expression pathway and involves the coordinated action of RNA helicases (20–23). eIF4A belongs to helicase superfamily 2, and the helicase activity of eIF4A is believed to be required to unwind mRNA secondary structure both during the formation of the 48S PIC and the scanning process. The involvement of eIF4A has been shown to be related to the degree of secondary structure in the 5′-untranslated region (5′-UTR) of the mRNA and it is required for translation of mRNAs containing even weak secondary structures in the 5′-UTR (16,24). It was recently demonstrated that the DEAH/RHA helicase DHX29 is also required for translation if the 5′-UTR of the mRNA is highly structured (25). The action of DHX29 is not necessarily mediated by helicase activity acting on the mRNA but instead could be remodeling of the 48S PIC (25). Another helicase involved in translation initiation is Ddx3/Ded1 (26,27). It is not clear how or if these three different helicases function in concert.
The 40S ribosomal subunit does not contain a motor for the movement along the mRNA during scanning. Unless diffusion can account for this movement, this force must be provided by some external factor. Historically, eIF4A is believed to act as a helicase downstream of the ribosome allowing unhindered scanning. Based on a cryo electron microscopy reconstruction of the 40S ribosomal subunit in complex with eIF3 and eIF4G, eIF4G was placed near the exit site (28). This observation led to the following suggestion (29): eIF4A is positioned downstream of the 48S PIC, eIF4G makes contact with mRNA upstream of the scanning 48S PIC, while these two factors remain in a complex together. Another recent model for the scanning process (30) proposed a Brownian ratchet model, where eIF4B serves as the pawl and eIF4A as the clamp allowing only forward movement at the trailing edge of the ribosomal subunit. Even low-resolution structural information of larger fragments of eIF4G could provide valuable insights into the function of these initiation factors during the formation of the 48S PIC, as well as their position relative to the 40S subunit.
We are interested in the interactions between eIF4A, eIF4B and eIF4G and how these might be integrated in the scanning process. Using purified recombinant proteins for our study, we report on how the activity of eIF4A is regulated through its interactions with eIF4B and eIF4G. Furthermore, we report the shape of a large eIF4G fragment and its complex with eIF4A determined by small-angle X-ray scattering (SAXS).
MATERIALS AND METHODS
Plasmids and purifications
eIF4A, eIF4B full length, eIF4BΔC and eIF4G fragments were all expressed from pET30Ek/LIC plasmids (Novagen). DNA constructs were made by Ligation Independent Cloning using primers listed in Supplementary Table S1. The 5′-primers were modified by having the sequence encoding a TEV (tobacco etch virus) protease recognition site inserted in front of the open reading frame, allowing proteolytic removal of the histidine-tag. PCR was performed using DNA encoding wild-type human eIF4A, eIF4B or eIF4G1 as template, respectively. To construct eIF4A with a 6xHis tag in the C-terminus a stop codon (TGA) was mutated to a glycine (GGA) by site-directed mutagenesis (QuikChange® Lightning, Stratagene) allowing read-through to the C-terminal histidine tag in the pET30 vector, using the oligos listed in Supplementary Table S1. All constructs that involved PCR were sequenced to verify that no mutations were introduced during the PCR. Plasmids were transformed into the BL21 Rosetta Escherichia coli strain and cultures were grown at 37°C to an OD600 of approximately 0.8 and then induced with 0.5 mM IPTG and incubated over night at 20°C. Cells were harvested by centrifugation and the cell pellet was resuspended in lysis buffer containing 20 mM Tris–HCl pH 7.6, 500 mM KCl, 20 mM Imidazole pH 8, 5 mM MgCl2, 0.5 mM β-mercaptoethanol (β-ME), 1 mM PMSF, Complete Protease Inhibitor (Roche Diagnostics GmbH) and 10% glycerol. The cell suspension was sonicated and then centrifuged at 14 000 rpm for 30 min. The supernatant was filtered through a 0.45 μm filter prior to loading on a Ni2+ charged immobilized metal affinity chromatography (IMAC) column (GE Healthcare). Bound protein was eluted from the Ni2+ affinity column with a 20–500 mM imidazole gradient in lysis buffer without protease inhibitors. The histidine affinity tag was removed by addition of 1/300 (w/w) His-tagged TEV protease and incubation on ice for 2 h followed by dialysis over night against 20 mM Tris–HCl pH 7.6, 500 mM KCl, 5 mM MgCl2, 0.5 mM β-ME and 10% glycerol, after which the dialyzed mixture was re-loaded on a Ni2+ affinity column. The proteolytic cleavage with TEV leaves a glycine upstream of the start methionine. The flow through contained essentially pure protein while the TEV protease and the cleaved tag remained bound to the column. eIF4G-MC was in some cases further purified on a Source 15S column equilibrated in 20 mM HEPES pH 6.8, 10% glycerol, 50 mM NaCl, 0.5 mM dithiotretiol (DTT) and eluted by increasing the concentration of NaCl to 200 mM in a linear gradient. This resulted in pure protein with an OD260/OD280 ratio of 0.6 indicating the absence of RNA.
Cobalt affinity pull-down
Cobalt charged TALON beads (TALON Superflow, BD Bioscience) were prepared (20 μl of a 50% slurry per reaction) by washing three times with 50 μl buffer A (150 mM NaCl, 20 mM Tris–HCl pH 7.6, 5 mM MgCl2, 10% glycerol, 0.1% NP40, 0.5 mM β-ME) per reaction. All centrifugation steps were done at 3000 rpm for 2 min, after which the Eppendorf tubes containing the beads were turned 180° and briefly centrifuged again for 3–4 s to allow for more complete removal of the supernatant. During the last wash, the beads were resuspended in the number of Eppendorf tubes equal to the number of experiments. After the last centrifugation, the buffer was removed leaving behind the beads. About 5 μg of the His-tagged protein was used together with a surplus of the non tagged proteins as indicated and the mixture was incubated at 4°C for 1 h in a total volume of 100 μl buffer A. Where indicated, ADP, AlCl3 and BeSO4 were added at a final concentration of 1 mM while NaF was added to a concentration of 10 mM. The protein mixture and the beads were incubated for 30 min at 4°C on a roller. Following incubation, the beads were gently pelleted and 15 μl supernatant was removed as ‘flow through’. This was followed by three washes of 100 µl ice cold buffer A. After the last wash the supernatant was carefully removed and the beads were resuspended in 17 μl buffer A containing 400 mM Imidazole to elute bound protein. After incubation for 10–15 min at 4°C, the beads were pelleted by centrifugation at 13.4 k rpm for 1 min and 15 μl supernatant containing the bound protein were removed as ‘Eluted’. SDS–PAGE loading buffer was added to samples, which were boiled for 3 min and analyzed by SDS–PAGE. Coomassie blue staining was used to visualize the protein after electrophoresis.
Biotin-RNA pull-down
For RNA pull-down 20 μl of a 50% slurry of High-Capacity Streptavidin agarose Resin (Thermo Scientific) and 100 pmol of pU30-biotin (Dharmacon) were used per reaction. All centrifugation steps were done as described above. Prior to experiments the beads were washed three times in buffer B (20 mM Tris–HCl pH 7.6, 150 mM NaCl, 5 mM MgCl2, 1 mM DTT). All components in the RNA pull-down (protein, nucleotide, analogs and biotinylated RNA) were mixed in a total volume of 100 μl of buffer B and incubated for the indicated times on a roller. Where indicated ADP, AlCl3, NaF and BeSO4 were added as described above. Following incubation the beads were gently pelleted and 15 μl solution was removed as ‘flow through’. The beads were washed three times using 100 µl buffer B. After the last centrifugation, supernatant was removed as much as possible and due to the extreme affinity of streptavidin for biotin, we eluted the bound protein by boiling the beads with 15 μl 2.5 × SDS loading buffer. The samples were boiled for 3 min and analyzed by SDS–PAGE. Coomassie blue staining was used to visualize the protein after electrophoresis. The boiling of the beads resulted in a more pronounced background binding to the beads in RNA pull-down experiments.
ATPase assay
ATPase activity was monitored by quantifying the released Pi according to Baginsky (31) in an assay optimized for a plate reader (Victor3, Perkin Elmer) and 96-wells plates. Each reaction contained 45 µl assay buffer B with the indicated proteins and poly(U) (Sigma) [1µg p(U)/1 µg eIF4A] and was started by the addition of 5 µl 30 mM ATP. The plate was incubated at RT for 60 min, after which the color was developed by adding 50 µl freshly made solution prepared by mixing 25 ml (170 mM ascorbic acid, 0.5 M HCl, 0.1% SDS) and 5 ml (28 mM ammonium heptamolybdate). Both of these solutions were prepared in advance and placed on ice for at least 1 h. After incubation for 10 min, 75 µl stop solution (154 mM Sodium Meta arsenic, 86 mM trisodium citrate, 2% acetic acid) was added. After 30 min the color was stable and the absorption at λ = 860 nm was measured using the plate reader.
SAXS
The eIF4A-4G complex was isolated by gel filtration chromatography with a Superdex 200 10/300 GL column (GE healthcare) equilibrated in buffer C (20 mM Tris–HCl pH 7.6, 150 mM NaCl, 5 mM MgCl2, 1 mM EDTA, 1 mM DTT) and the complex was concentrated to 5 mg/ml. The SAXS measurements were carried out on a laboratory-based instrument at Aarhus University (32). Measurements were done at concentrations c and c/2 for 3 h each at 4°C. Background subtraction and all necessary normalizations were carried out with the SUPERSAXS package (Oliveira,C.L.P. and Pedersen,J.S., unpublished data). The data were normalized to absolute scale by use of water as a primary standard. The final intensity is displayed as a function of the modulus of the scattering vector, q = 4π sinθ/λ, where λ = 1.54 Å and is the X-ray wavelength and 2θ is the angle between the incoming and scattered X-rays. Indirect Fourier Transformation (IFT) was performed (33) implemented in a home-written program (34) (Oliveira, C.L.P. and Pedersen,J.S., unpublished data). The ab initio modeling was carried out using the program DAMMIN (35) that uses a search space filled with close-packed spheres, regarded as dummy atoms. Using a simulated annealing technique and constraints that gives globular protein-like structures, the subset of dummy atoms that gives the best fit of the experimental data is obtained. Owing to the Monte Carlo nature of the optimization procedure, the generated models are not unique. Thus in order to obtain the most probable shape, 10 individual runs were performed, and compared and averaged using the DAMAVER program package (36).
Isothermal titration calorimetry
Prior to isothermal titration calorimetry (ITC) analysis proteins were dialyzed overnight against buffer C and their concentrations were determined by total amino acid analysis. The protein solutions were degassed using a MicroCal ThermoVac with stirring prior to the experiment. The ITC measurements were carried out on a Microcal VP-ITC instrument at 30°C. The reference power was set at ∼15 μcal/s and the stirrer at 312 rpm. All handling of the Microcal VP-ITC and ThermoVac was done according to manufacturer’s user manual.
Surface plasmon resonance
Surface plasmon resonance (SPR) experiments were carried out on a Biacore T100 instrument at 25°C using a Series S Sensor Chip NTA Certified (Biocore AB) allowing binding of His-tagged proteins. The preprogrammed NI-Kin method was used with the modification that both channels were injected with running buffer [10 mM HEPES pH 7.4, 150 mM NaCl, 50 µM EDTA, 0.05% Tween-20, 0.1% BSA (Sigma)] containing 0.5 mM NiCl2. Regeneration of the chip was performed after each measurement according to the NI-Kin method program [10 mM HEPES pH 8.3, 150 mM NaCl, 350 mM EDTA, 0.05% Tween-20, 0.1% BSA (Sigma)]. Buffers were filtrated using a 0.22 μm filter (Advantec) before use. The SPR experiments were done according to manufacturer’s user manual.
RESULTS
Complex formation between eIF4A and eIF4G-MC
To investigate the interaction between eIF4A and three eIF4G fragments, eIF4G-MC, eIF4G-M or eIF4G-C and the C-terminal truncated eIF4B (eIF4BΔC) (Figure 1A), a cobalt affinity pull-down was employed using C-terminal His-tagged eIF4A (eIF4A-His6). The pull-down experiments were carried out in the absence of nucleotide and RNA. eIF4BΔC was used in the cobalt affinity experiments since full-length eIF4B migrates at nearly the same position as eIF4G-MC on an SDS–PAGE gel, and is more prone to degradation, but otherwise appears to behave similarly to eIF4BΔC with respect to complex formation with eIF4A (see below). eIF4A-His6 formed a stable complex with either eIF4G-MC or eIF4G-M (Figure 1B, lanes 2 and 4), while only a very weak interaction was detected with eIF4G-C and no interaction was detected with eIF4BΔC, (Figure 1B, lanes 3 and 5). Interestingly, if both eIF4G-MC and eIF4BΔC were added together, the interaction between eIF4A and eIF4G-MC remained intact; however, eIF4BΔC could now also be detected (Figure 1B, lane 6). eIF4BΔC could also be detected in a pull-down experiment with eIF4A-His6 in the presence of eIF4G-M, however eIF4G-C did not support eIF4BΔC interaction (data not shown). Control reactions without eIF4A-His6 in the pull-down experiments were negative (Figure 1B, lanes 7–10).
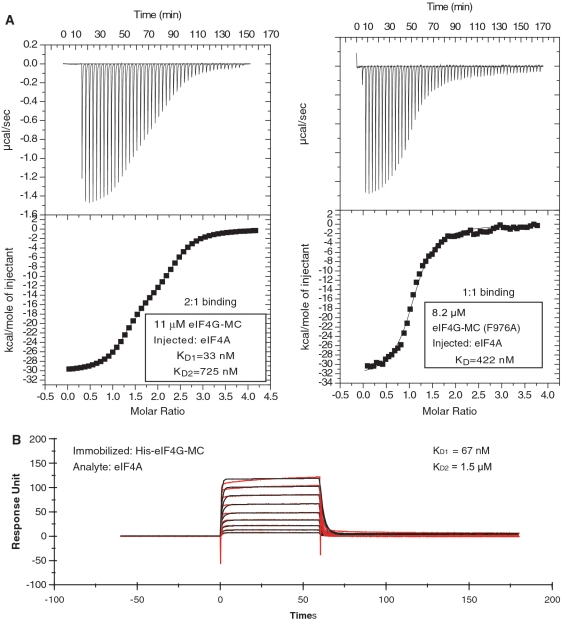
To further investigate the interaction between eIF4A and eIF4G-MC, ITC and SPR were used. In an ITC experiment with eIF4A being added to eIF4G-MC in the chamber, the best fit obtained was with a two-site binding model, suggesting a high-affinity site (KD = 50 ± 15 nm) [standard deviation (n = 4)] and a low-affinity site (KD = 775 ± 181 nM) [standard deviation (n = 4)] (Figure 2A, left panel displays a representative experiment). Performing the ITC experiment in reverse, with eIF4G-MC being added to eIF4A in the chamber, gave similar results, KD ∼ 42 nM for the high-affinity site and 1.6 μM for the low-affinity site (data not shown). SPR results, with immobilized eIF4G-MC and eIF4A as analyte, could also be fitted to a heterologous two-state binding model characterized by a high-affinity site (KD ∼ 67 nM) and a low-affinity site (KD ∼ 1.5 μM) (Figure 2B). To investigate the location of the high-affinity site we mutated the conserved phenylalanine 978 to alanine in eIF4G-MC. This mutant has previously been shown to affect binding between eIF4A and the middle region of eIF4G containing the high-affinity site (4,29). In the yeast structure, Phe 838 (equivalent to 978 in human eIF4GI) fits into a hydrophobic groove formed by four residues from eIF4A (5). ITC data obtained by adding eIF4A to eIF4G-MC (F978A) in the chamber was best fitted to a 1:1 binding model (KD = 417 ± 41 nM) [standard deviation (n = 2)], suggesting that the F978A mutant disrupted the high-affinity binding site (Figure 2A, right panel displays a representative experiment).
Figure 2.
Two eIF4A molecules bind one eIF4G-MC molecule. (A) Left panel, ITC experiment demonstrating a 2:1 binding of eIF4A to eIF4G-MC. The continuous line is a fit of the experimental data to a two-site model. Right panel, ITC experiment demonstrating a 1:1 binding of eIF4A to eIF4G-MC (F976A). The continuous line is a fit of the experimental data to a one-site model. (B) SPR experiment shows similar results as obtained by ITC of the interaction between eIF4A and eIF4G-MC with a best-fit of a two-state heterologous binding with one site displaying a high affinity and one site low affinity.
SAXS investigation of the complex between eIF4A and eIF4G-MC
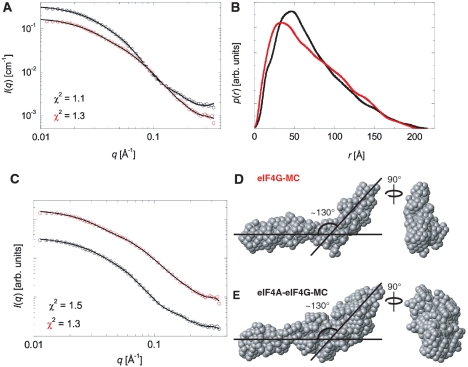
The complex between eIF4A and eIF4G-MC demonstrated by cobalt affinity pull-down could also be isolated by gel filtration chromatography performed in the absence of RNA and nucleotide (Supplementary Figure S1). SAXS analysis was used to determine the overall shape of the complex purified in this manner. The scattering curves were measured for the complex between eIF4A and eIF4G-MC and for eIF4G-MC alone (Figure 3A). Indirect Fourier Transformation (IFT) was used for determining the pair distance distribution function p(r) that is a histogram of distances between pair of points within the complexes and thus gives information on particle shape and maximum particle diameter (Figure 3B). By comparison with p(r) functions of known structures (37), it was concluded that the particles have an elongated shape. The p(r) functions approach zero at the maximum diameter, which is around 200–220 Å for both particles. Ab initio shape determination was carried out and averaged final models were derived, each based on 10 individual runs. The resulting model of eIF4G-MC by itself displays an elongated shape with a distinct 130° bend near the middle (Figure 3D). The model of the complex between eIF4A and eIF4G-MC was qualitatively similar but indicated extra mass located preferentially to one side of the bend, which we believe may represent eIF4A in the complex (Figure 3E). The mass of the eIF4A and eIF4G-MC complex calculated from the SAXS data suggested a 1:1 complex. Densitometric analysis (Supplementary Figure S1) also indicated a 1:1 complex. Overall, the molecular shape revealed by SAXS most likely represents one molecule of eIF4A bound to the high-affinity site of eIF4G-MC, while eIF4A bound to the low-affinity site dissociates due to the dilution during the gel filtration.
Figure 3.
SAXS analysis of the complex between eIF4A and eIF4G-MC and eIF4G-MC alone. (A) Indirect Fourier transformation (IFT) fits of the scattering data for eIF4G-MC (red) and the complex between eIF4A and eIF4G-MC (black). (B) Pair distances distribution functions from the IFT fitting in (A) with functions scaled for comparison. (C) Fits to the scattering data for the ab initio shape determination with DAMMIN. (D) Ab initio model of eIF4G-MC. (E) Ab initio model of the complex between eIF4A and eIF4G-MC.
The crystal structures of two fragments of mammalian eIF4G overlapping with our eIF4G-MC fragment have previously been determined (Figure 1A). The so-called middle fragment of eIF4GII (corresponding to residues 752–1010 within our eIF4G-MC fragment of eIF4GI) contains a high-affinity eIF4A binding site (38). A C-terminal fragment of eIF4GI consisting of residues 1235–1572 contains a low-affinity eIF4A binding site and a Mnk1 kinase site (39). These two eIF4G fragments consist, nearly exclusively, of α-helical HEAT repeats (HR). It was suggested that large portions of the remaining eIF4G were unstructured (29,39). Accordingly, residues 1011–1234 in our eIF4G-MC fragment, which are not covered by any of the two α-helical fragments, might be unstructured. However, since the ab initio models fit the SAXS data very well out to the highest scattering vector q (Figure 3C), this indicates that this portion of eIF4G is not unstructured and flexible. To investigate the secondary structure of the eIF4G-MC fragment, we performed synchrotron circular dichroism (CD) measurements. Taking into account that the known portions of eIF4G-MC do not contain any β-sheets, our CD measurements suggest that approximately 50% of the residues 1011–1234 are folded into β-sheets (Supplementary Figure S2), supporting a well-defined conformation of these residues, in accordance with our SAXS analysis.
Complex formation between eIF4B and eIF4A is promoted by eIF4G-MC
The cobalt affinity pull-down experiments described above were performed in the absence of RNA and nucleotide; to investigate the effects of RNA and nucleotide we performed streptavidin–agarose pull-down assays utilizing a biotinylated 30mer uracil RNA. Using this assay a stable complex was detected between eIF4A and eIF4B in a reaction containing ADP-AlFx and eIF4G-MC (Figure 1C, lane 1). ADP-AlFx is an analog that resembles the transition state during ATP hydrolysis in DEAD-box ATPases (14). No eIF4A–eIF4B complex was detected in the absence of either eIF4G-MC (Figure 1C, lane 3) or ADP-AlFx (Figure 1C, lane 4), only eIF4B was bound to RNA in these reactions most likely through its two RNA binding motifs. In the absence of biotinylated RNA (Figure 1C, lane 2) only a weak non-specific binding of eIF4B was detected. Interestingly, even though eIF4G-MC was required for the formation of the eIF4A–eIF4B–RNA–ADP–AlFx complex, it was not present in the complex (Figure 1C–E). We reasoned that eIF4G might be acting catalytically in the formation of the eIF4A–eIF4B–RNA–ADP–AlFx complex. This idea was tested by comparing the amount of eIF4A–eIF4BΔC complex formed after 15 min incubation, with approximately stoichiometric amount of eIF4G-MC present (Figure 1D, lane 1) to the amount of eIF4A–eIF4BΔC complex formed after 15, 60 and 240 min incubation with a 30-fold lower concentration of eIF4G-MC (Figure 1D, lanes 2–4). The build-up of eIF4A–eIF4BΔC complex over time strongly indicated the catalytic effect of eIF4G-MC (Figure 1D, lanes 2–4). As controls, either eIF4G-MC or the nucleotide was omitted from the reaction (Figure 1D, lanes 5 and 6). Full-length eIF4B also formed a complex with RNA, nucleotide and eIF4A that was dependent on eIF4G-MC (Supplementary Figure S3A); however, the complex was not formed as efficiently as with eIF4BΔC. The eIF4A–eIF4BΔC complex can be formed with both an ATP analog (ADP–BeF3) and a transition state analog (ADP–AlFx) (Figure 1D and Supplementary Figure S3B) and to a lesser degree with ADPNP (data not shown) but not with ADP, AlFx, BeF3 or NaF alone (Supplementary Figure S3B and data not shown). Furthermore, the eIF4A–eIF4BΔC–RNA–ADP–BeF3 complex was stable enough to be purified on an ion exchange column while no complex was formed if RNA was omitted (Supplementary Figure S4). Importantly, in the experiment in Supplementary Figure S4, which contained poly(U) as RNA, the complex was treated with RNase A prior to ion exchange chromatography, strongly suggesting that eIF4A and eIF4B interact directly. To investigate if both eIF4A-binding domains in eIF4G were required for eIF4A–eIF4B complex formation, RNA pull-downs using either eIF4G-MC, eIF4G-M or eIF4G-C with eIF4A, eIF4BΔC and ADP-AlFx were carried out. An eIF4A–eIF4BΔC complex could only be identified using eIF4G-MC (Figure 1E).
The complex between eIF4A and eIF4BΔC was also isolated using cobalt affinity pull-down in the presence of a His-tagged eIF4A, eIF4BΔC, eIF4G-MC, a U25 oligo and ADP–BeF3 (Supplementary Figure S3C, lane 2), but the complex was absent when eIF4G-MC was not added (Supplementary Figure S3C, lane 3). We used a 25 mer uracil oligo since an RNA of this length most likely will not accommodate binding of both eIF4A and eIF4B independently. Concomitant with eIF4A–eIF4BΔC complex formation, a reduction in the amount of eIF4G-MC isolated in complex with eIF4A was apparent (Supplementary Figure S3C, compare lanes 1 and 2 in the upper panel). The flow-through contained more eIF4G-MC and less eIF4BΔC as expected when eIF4A formed a complex with eIF4BΔC (Supplementary Figure S3C, compare lanes 1 and 2 in the lower panel).
Synergistic activation of the ATPase activity of eIF4A requires eIF4B and eIF4G-MC
Based on the data presented above we hypothesized that eIF4B and eIF4G-MC synergistically stimulate the ATPase activity of eIF4A. A Baginsky ATPase assay (31), modified for a plate reader that measures the amount of inorganic phosphate produced when ATP is hydrolyzed, was used to test this hypothesis. The ATPase activity of eIF4A could be stimulated by poly(U), as expected for an RNA dependent ATPase (Figure 4A, reactions 1 and 2). Under the experimental conditions used, near stoichiometric amounts of eIF4B had no effect (Figure 4A, reaction 3) whereas eIF4G-MC in sub-stoichiometric amounts (1/10) only slightly, but reproducibly, enhanced the ATPase activity (Figure 4A, reaction 4). However, when both eIF4B and eIF4G-MC were present, more than 3-fold stimulation was observed (Figure 4A, reaction 5). Increasing the amount of eIF4A by 2-fold nearly doubled the activity (Figure 4A, reaction 6), suggesting that eIF4A is the only rate-limiting factor. The activity was dependent on RNA and eIF4A (Figure 4A, reactions 7 and 8) and the stimulation by eIF4B and eIF4G-MC was specific since an unrelated DEAD-box ATPase, Dbp5, did not respond to these factors but was stimulated by poly(U) as expected (Figure 4B, reactions 1–3). Only the eIF4G-MC fragment, but not eIF4G-M or eIF4G-C, led to a synergistic activation of eIF4A with either eIF4B or eIF4BΔC (Supplementary Figure S5). All control reactions were done with the addition of buffer instead of ATP (Figure 4 and Supplementary Figure S5). An RNA binding motif of 40 residues, suspected to be involved in scanning (40), is located just upstream of the eIF4G-MC region. We therefore tested whether a longer eIF4G-MC protein containing residues 682–1451 including this RNA binding motif would further stimulate eIF4A. With respect to stimulation of the ATPase activity this longer version of eIF4G-MC gave similar results to those obtained with eIF4G-MC (data not shown) but this protein did indeed bind the U30-Biotin two to three times stronger than eIF4G-MC (data not shown).
Figure 4.
Synergistic activation of eIF4A by eIF4B and eIF4G-MC. (A) The ATPase activity was measured using Baginsky’s method for detection of inorganic phosphate. The results were recalculated to μM Pi/h using a standard curve of known Pi concentrations. Reactions containing the indicated proteins were initiated by the addition of ATP or buffer as controls. Amount of proteins are: eIF4A = 2 μM, eIF4BΔC = 1 μM, eIF4G-MC = 0.2 μM. In reaction six, 4 μM of eIF4A was added. The amount of added proteins can be seen below the graph on a SDS–PAGE gel stained with Coomassie blue (representative from one experiment). Error bars display standard error (n = 4). (B) Same as in (A) except that Dbp5 replaced eIF4A to investigate the specificity of eIF4B/eIF4G-MC stimulation. Dbp5 is inherently more active than eIF4A. Error bars display the standard error (n = 4). The absolute OD860 values were in all cases at least two to three times below saturation of the ATPase assay.
DISCUSSION
In order to clarify the role of various regions in mammalian eIF4G, we produced the eIF4G-MC protein delimited in either end by two well-defined structural domains established by structures of smaller human eIF4G fragments and the structure of yeast eIF4A in complex with a fragment of yeast eIF4G (5,38,39). A stable complex between eIF4A and eIF4G-MC, independent of RNA and nucleotide, was identified and ITC and SPR analysis demonstrated a two-site binding model with a low and a high-affinity site. Furthermore, a stable complex between eIF4A and eIF4B, dependent on RNA, nucleotide and catalytic amounts of eIF4G-MC, was also reconstituted. In agreement with these data, a synergistic stimulation of eIF4A’s ATPase activity by eIF4B and eIF4G-MC was established.
The interaction between eIF4A and its two binding domains in human eIF4GI was demonstrated earlier (3,4). Using SPR analysis, the so-called middle fragment [residues 653–1118 of human eIF4GI (cpC3)] and a C-terminal fragment [residues 1118–1600 of human eIF4GI (cpC2)] were shown to have KD values of 17 and 330 nM, respectively, for their eIF4A complexes (41). Our data, obtained using either ITC or SPR to characterize the interaction of eIF4G-MC with eIF4A, were best-fitted with a two binding site model containing a low- and a high-affinity site, with the two binding constants in the same range as identified earlier in the cpC2 and cpC3 eIF4G fragments (41); (see Figure 1A and Table 1 for an overview of eIF4G fragments and their corresponding KD for eIF4A interaction). A shorter fragment of the middle domain of human eIF4GI [residues 747–993 eIF4G1 (HEAT-1)] has much lower affinity for eIF4A, KD = 12 μM (29), suggesting that the binding site may span a longer region (see Figure 1A and Table 1). Indeed, weak interaction between eIF4A-CTD and the eIF4GI CY fragment (spanning residues 994–1138, see Figure 1A) have been demonstrated by NMR (29). The CY fragment of eIF4GI is included within the middle fragment (cpC3) that has a KD of 17 nM (41) with the exception of 20 amino acids (residues 653–1118, see Figure 1A). This observation suggests that the binding site for eIF4A-CTD found in the 994–1138 region may contribute to the nM affinity of the high-affinity site for eIF4A we measured using the eIF4G-MC fragment (Figure 2) and that was likewise observed for the middle binding site of the eIF4GI fragment, cpC3 (41). The stoichiometry of two molecules of eIF4A binding to one molecule of eIF4G-MC suggested by both our ITC and SPR experiments is in accordance with earlier results (41). However, our results conflict with data from Fujita and coworkers which was also obtained using ITC indicating a 1:1 complex (42). The reason for this discrepancy is not clear, but the concentrations of eIF4A and eIF4G used in ref. (42) may have been too low to detect a 2:1 complex. Our results also conflicts with data from Li et al. (43) that used an alternative approach for determining the ratio of eIF4A:eIF4G. Specific antibodies against either eIF4AI or eIF4AII were used to immunoprecipitate protein eluted from m7GTP-sepharose incubated with mammalian cell extracts. This demonstrated that only eIF4AI could be detected after eIF4AI precipitation and like-wise for eIF4AII, suggesting that only one eIF4A molecule binds eIF4G. If two eIF4A molecules bind the same eIF4G molecule, western blotting of material from immunoprecipitation (IP) with eIF4AI antibody should detect eIF4AI and some eIF4AII and vice versa. The difference in binding affinity between the two sites could result in only one eIF4A bound to the high-affinity site after washing during the IP experiment. However, the same study suggested that the binding between eIF4A and either the middle fragment or the C-terminal fragment of eIF4G were similar (43). The reason for the discrepancy between our data demonstrating that eIF4G-C has low-affinity for eIF4A and the data showing that both eIF4A binding sites are of comparable affinity in IP experiments (43) is not clear. However, the use of mammalian cell extracts (43) instead of purified proteins might mask the low-affinity site by having another component stabilizing the complex between eIF4A and the C-terminal eIF4A binding domain. Alternatively, we cannot completely rule out that under in vivo like conditions, binding to the high-affinity site may exclude binding to the low-affinity site, although we in vitro clearly observe simultaneous binding to both sites. Whatever the reason for the discrepancy, it is noteworthy that we get the same qualitative results using very different assays: ITC, SPR and cobalt affinity pull-down. The consequence of having two eIF4A molecules bound to eIF4G is not known but it may induce conformational changes in eIF4G promoting the recycling of eIF4A and stimulation of eIF4A binding to eIF4B. If eIF4G in vivo contains two eIF4A binding sites with KD values of ∼50 and 1000 nM, respectively, and we further assume eIF4A and eIF4G have cellular concentrations of 50 and 2 μM, respectively, as determined for the yeast factors (44), then both sites are likely to be fully occupied.
Table 1.
Dissociation constants for the complexes of eIF4A with different fragments of human eIF4GI
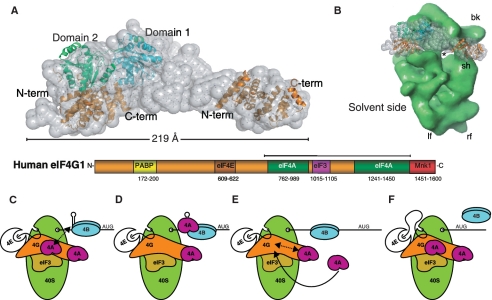
Our SAXS analysis demonstrated that both eIF4G-MC and its complex with eIF4A have an elongated structure with a bend (Figure 3). By comparing our SAXS envelope for the complex between eIF4A and eIF4G-MC with the crystal structure of yeast eIF4G in complex with eIF4A (5) and the structure of the C-terminal fragment of eIF4G (39), we can propose a qualitative model for the complex between eIF4A and eIF4G-MC (Figure 5A). This elongated complex has a length of ∼220 Å, which is even more remarkable considering that the eIF4G-MC protein constitutes only about half of the full-length eIF4G. Such a dimension justifies the description of eIF4G as a scaffold protein spanning the width of the 40S ribosomal subunit (Figure 5B). eIF4G, bound to the 40S ribosomal subunit through eIF3, most likely has dimensions that would allow eIF4A to be placed at both the exit and entry of the mRNA channel of the 40S ribosomal subunit. Further research is therefore needed to address the precise position of eIF4A relative to the entry and exit sites.
Figure 5.
Docking of known structural elements of eIF4G in our SAXS envelope and a model for the synergistic activation of eIF4A by eIF4B and eIF4G. (A) Docking of the structure of yeast eIF4A–eIF4G (PDB entry 2SVO) and the first HEAT domain from the structure of the C-terminal fragment of human eIF4G1 (PDB entry 1UG3) into the SAXS envelope of the complex between eIF4A and eIF4G-MC. (B) Comparison of the size of the 40S ribosomal subunit [3D EM databank #1376 (46)] and the SAXS envelope of the eIF4A–eIF4G complex. (C–F) Model of eIF4A activation by eIF4G and eIF4B. (C) The 48 PIC is stalled by a hairpin and eIF4G holds eIF4A in a conformation ready for binding to eIF4B. (D) eIF4B in the presence of RNA and ATP stimulates the dissociation of the eIF4A–eIF4G complex so that eIF4A can form a complex with eIF4B, RNA and ATP and induce unwinding after which eIF4A leaves the complex. (E) The PIC moves along the RNA and eIF4G rebinds a molecule of eIF4A. (F) A second eIF4A binding site may be involved in recruitment of another eIF4A. See ‘Discussion’ section for further details.
eIF4A and eIF4B cannot replace eIF4F during scanning in a reconstituted translation initiation system analyzed using toe-printing, whereas eIF4G could replace eIF4F if both eIF4A and eIF4B were present (16). This agrees with our data showing that eIF4G is necessary and sufficient for recruitment of eIF4A into an eIF4A–RNA–eIF4B-nucleotide complex, and that eIF4B and eIF4G activate the ATPase activity of eIF4A synergistically. Our results also suggested a mutually exclusive binding between eIF4A and either eIF4G or eIF4B, RNA and nucleotide (Supplementary Figure S3C). This indicates that the eIF4A–eIF4G complex and the eIF4A–eIF4B–RNA-nucleotide complex are two separate and consecutive steps in translation initiation that both are required for eIF4A function. We therefore propose the following model for how eIF4B and eIF4G regulate the function of eIF4A during translation initiation (Figure 5C–F). Having brought eIF4A to its place of action through eIF4G (Figure 5C), we suggest that eIF4G induces a conformational change of eIF4A preparing it for the interaction with eIF4B, ATP and RNA (Figure 5D) as already suggested by the soft clamp model (11) and the crystal structure of the yeast eIF4A–eIF4G complex (5). This may also explain the weak stimulation by eIF4G of the ATPase activity (Figure 4). In vitro RNA is required for eIF4A–eIF4B complex formation as well as an ATP analog suggesting that eIF4B also needs to be in an activated state bound to RNA. In fact, we observe binding of both eIF4B and eIF4BΔC to RNA without concomitant binding of eIF4A (Figure 1C–E). When the eIF4A–eIF4B–RNA–ATP complex is formed, eIF4B stimulates the ATPase and helicase activity of eIF4A (Figure 5D–E). eIF4B may stimulate unwinding solely by promoting the closed conformation of eIF4A known from the structure of its paralogue eIF4AIII in complex with RNA and ATP-analogs (12–14). If true, the following eIF4B stimulated ATP hydrolysis may only be required for the release of eIF4A from the RNA as earlier described for the DEAD-box ATPase Ded1p (45). After eIF4A dissociates from the mRNA and eIF4B, it may then rebind eIF4G and be ready for a new round of unwinding/remodeling (17,18). Whether the low-affinity eIF4A site functions as a recruitment site for binding to the high-affinity site or only serves to regulate the binding of eIF4A to the high-affinity site remains to be settled (Figure 5E). After unwinding the PIC may now move further along the mRNA towards the AUG codon (Figure 5F).
It was demonstrated that eIF4H, as a close homolog of eIF4B, can partially displace the CTD of eIF4A from the eIF4GI CY fragment (residues 994–1138) (29), an area which most likely is contained within the high-affinity eIF4A binding site in eIF4G-MC (See above). This may partially explain how eIF4H/eIF4B can induce the dissociation of eIF4A from eIF4G to allow the assembly of an eIF4A–eIF4B/4H–RNA-nucleotide complex according to our model. A compelling question would be whether eIF4H could also form a complex with eIF4A that required eIF4G. This may indeed be the case, as we suggest that the data presented in the Supplementary Figure S4 by Marintchev et al. (29) could be interpreted as an eIF4A–eIF4H complex dependent on eIF4G, RNA and nucleotide but not containing eIF4G, agreeing with our results, and not as suggested a trimeric eIF4A–eIF4H–eIF4G complex.
eIF4A has generally been considered the canonical RNA helicase; however, other models are emerging where eIF4A does not function as a helicase. For example, instead of having eIF4A depicted in Figure 5C–F as a helicase, the model could also have been adapted to the framework of the ratchet and pawl model (30) having eIF4A and eIF4B playing a role upstream instead of downstream of the 40S ribosomal subunit. Downstream unwinding may also be carried out by the Dhx29 or Ddx3 helicase. eIF4A on its own is not a very active enzyme; however, our studies clearly demonstrate how eIF4G and eIF4B in collaboration can significantly enhance the activity of eIF4A. Our finding that eIF4G acts catalytically in the formation of the complex between eIF4A, RNA, ATP and eIF4B is most likely crucial for the enzymatic activity of eIF4A during translation initiation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Alfred Benson Foundation (to K.H.N.); The Danish Council for Independent Research, Natural Sciences (G.R.A.); a Hallas-Møller stipend from the Novo Nordic foundation (G.R.A.); Danish National Research Foundation (G.R.A.); The Danish Council for Independent Research, Natural Sciences (J.S.P. and M.A.B.) and Danish National Research Foundation (J.S.P. and C.L.P.O.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Gitte Hartvigsen for technical assistance, Peter Andreasen for help with Biacore experiments and Lise Kvisgaard for help with the Baginsky assay. We would like to thank William Merrick and Alan Hinnebusch for suggestions and critical reading of the article and Simon Morley and Nahum Sonenberg for constructs and Ulrich Baumann for providing the coordinates of the yeast eIF4A–eIF4G structure prior to publication.
REFERENCES
- 1.Sonenberg N, Hinnebusch AG. Regulation of translation initiation in eukaryotes: mechanisms and biological targets. Cell. 2009;136:731–745. doi: 10.1016/j.cell.2009.01.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pestova TV, Lorsch JR, Hellen CUT. The Mechanism of Translation Initiation in Eukaryotes. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2007. [Google Scholar]
- 3.Imataka H, Sonenberg N. Human eukaryotic translation initiation factor 4G (eIF4G) possesses two separate and independent binding sites for eIF4A. Mol. Cell. Biol. 1997;17:6940–6947. doi: 10.1128/mcb.17.12.6940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Morino S, Imataka H, Svitkin YV, Pestova TV, Sonenberg N. Eukaryotic translation initiation factor 4E (eIF4E) binding site and the middle one-third of eIF4GI constitute the core domain for cap-dependent translation, and the C-terminal one-third functions as a modulatory region. Mol. Cell. Biol. 2000;20:468–477. doi: 10.1128/mcb.20.2.468-477.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Schütz P, Bumann M, Oberholzer AE, Bieniossek C, Trachsel H, Altmann M, Baumann U. Crystal structure of the yeast eIF4A-eIF4G complex: an RNA-helicase controlled by protein-protein interactions. Proc. Natl Acad. Sci. USA. 2008;105:9564–9569. doi: 10.1073/pnas.0800418105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rozen F, Edery I, Meerovitch K, Dever TE, Merrick WC, Sonenberg N. Bidirectional RNA helicase activity of eucaryotic translation initiation factors 4A and 4F. Mol. Cell. Biol. 1990;10:1134–1144. doi: 10.1128/mcb.10.3.1134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rogers GWJ, Richter NJ, Merrick WC. Biochemical and kinetic characterization of the RNA helicase activity of eukaryotic initiation factor 4A. J. Biol. Chem. 1999;274:12236–12244. doi: 10.1074/jbc.274.18.12236. [DOI] [PubMed] [Google Scholar]
- 8.Grifo JA, Abramson RD, Satler CA, Merrick WC. RNA-stimulated ATPase activity of eukaryotic initiation factors. J. Biol. Chem. 1984;259:8648–8654. [PubMed] [Google Scholar]
- 9.Altmann M, Muller PP, Wittmer B, Ruchti F, Lanker S, Trachsel H. A Saccharomyces cerevisiae homologue of mammalian translation initiation factor 4B contributes to RNA helicase activity. EMBO J. 1993;12:3997–4003. doi: 10.1002/j.1460-2075.1993.tb06077.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Korneeva NL, First EA, Benoit CA, Rhoads RE. Interaction between the NH2-terminal domain of eIF4A and the central domain of eIF4G modulates RNA-stimulated ATPase activity. J. Biol. Chem. 2005;280:1872–1881. doi: 10.1074/jbc.M406168200. [DOI] [PubMed] [Google Scholar]
- 11.Oberer M, Marintchev A, Wagner G. Structural basis for the enhancement of eIF4A helicase activity by eIF4G. Genes Dev. 2005;19:2212–2223. doi: 10.1101/gad.1335305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Andersen CBF, Ballut L, Johansen JS, Chamieh H, Nielsen KH, Oliveira CLP, Pedersen JS, Séraphin B, Le Hir H, Andersen GR. Structure of the exon junction core complex with a trapped DEAD-box ATPase bound to RNA. Science. 2006;313:1968–1972. doi: 10.1126/science.1131981. [DOI] [PubMed] [Google Scholar]
- 13.Bono F, Ebert J, Lorentzen E, Conti E. The crystal structure of the exon junction complex reveals how it maintains a stable grip on mRNA. Cell. 2006;126:713–725. doi: 10.1016/j.cell.2006.08.006. [DOI] [PubMed] [Google Scholar]
- 14.Nielsen KH, Chamieh H, Andersen CBF, Fredslund F, Hamborg K, Le Hir H, Andersen GR. Mechanism of ATP turnover inhibition in the EJC. RNA. 2009;15:67–75. doi: 10.1261/rna.1283109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sengoku T, Noreki O, Nakamura A, Kobayashi S, Shigeyuki Y. Structural basis for RNA unwinding by the DEAD-Box protein Drosophila Vasa. Cell. 2006;125:287–300. doi: 10.1016/j.cell.2006.01.054. [DOI] [PubMed] [Google Scholar]
- 16.Pestova TV, Kolupaeva VG. The roles of individual eukaryotic translation initiation factors in ribosomal scanning and initiation codon selection. Genes Dev. 2002;16:2906–2922. doi: 10.1101/gad.1020902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoder-Hill J, Pause A, Sonenberg N, Merrick WC. The p46 subunit of eukaryotic initiation factor (eIF)-4F exchanges with eIF-4A. J. Biol. Chem. 1993;268:5566–5573. [PubMed] [Google Scholar]
- 18.Pause A, Methot N, Svitkin Y, Merrick WC, Sonenberg N. Dominant negative mutants of mammalian translation initiation factor eIF-4A define a critical role for eIF-4F in cap-dependent and cap-independent initiation of translation. EMBO J. 1994;13:1205–1215. doi: 10.1002/j.1460-2075.1994.tb06370.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Rozovsky N, Butterworth AC, Moore MJ. Interactions between eIF4AI and its accessory factors eIF4B and eIF4H. RNA. 2008;14:2136–2148. doi: 10.1261/rna.1049608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jankowsky E, Gross CH, Shuman S, Pyle AM. Active disruption of an RNA-protein interaction by a DExH/D RNA helicase. Science. 2001;291:121–125. doi: 10.1126/science.291.5501.121. [DOI] [PubMed] [Google Scholar]
- 21.Fairman ME, Maroney PA, Wang W, Bowers HA, Gollnick P, Nilsen TW, Jankowsky E. Protein displacement by DExH/D ‘RNA helicases’ without duplex unwinding. Science. 2004;304:730–734. doi: 10.1126/science.1095596. [DOI] [PubMed] [Google Scholar]
- 22.Cordin O, Banroques J, Tanner NK, Linder P. The DEAD-box protein family of RNA helicases. Gene. 2006;367:17–37. doi: 10.1016/j.gene.2005.10.019. [DOI] [PubMed] [Google Scholar]
- 23.Fairman-Williams ME, Guenther U, Jankowsky E. SF1 and SF2 helicases: family matters. Curr. Opin. Struct. Biol. 2010;20:313–324. doi: 10.1016/j.sbi.2010.03.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Svitkin YV, Pause A, Haghighat A, Pyronnet S, Witherell G, Belsham GJ, Sonenberg N. The requirement for eukaryotic initiation factor 4A (elF4A) in translation is in direct proportion to the degree of mRNA 5′ secondary structure. Rna. 2001;7:382–394. doi: 10.1017/s135583820100108x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pisareva VP, Pisarev AV, Komar AA, Hellen CU, Pestova TV. Translation initiation on mammalian mRNAs with structured 5′ UTRs requires DExH-box protein DHX29. Cell. 2008;135:1237–1250. doi: 10.1016/j.cell.2008.10.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tarn WY, Chang TH. The current understanding of Ded1p/DDX3 homologs from yeast to human. RNA Biol. 2009;6:17–20. doi: 10.4161/rna.6.1.7440. [DOI] [PubMed] [Google Scholar]
- 27.Lee M, Lee SJ, Choi HJ, Jung YW, Frokiaer J, Nielsen S, Kwon TH. Regulation of AQP4 protein expression in rat brain astrocytes: role of P2X7 receptor activation. Brain Res. 2008;1195:1–11. doi: 10.1016/j.brainres.2007.12.023. [DOI] [PubMed] [Google Scholar]
- 28.Siridechadilok B, Fraser CS, Hall RJ, Doudna JA, Nogales E. Structural roles for human translation factor eIF3 in initiation of protein synthesis. Science. 2005;310:1513–1515. doi: 10.1126/science.1118977. [DOI] [PubMed] [Google Scholar]
- 29.Marintchev A, Edmonds KA, Marintcheva B, Hendrickson E, Oberer M, Suzuki C, Herdy B, Sonenberg N, Wagner G. Topology and regulation of the human eIF4A/4G/4H helicase complex in translation initiation. Cell. 2009;136:447–460. doi: 10.1016/j.cell.2009.01.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Spirin AS. The ribosome as a conveying thermal ratchet machine. J. Biol. Chem. 2009;284:21103–21119. doi: 10.1074/jbc.X109.001552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Baginsky ML, Rodwell VW. Metabolism of pipecolic acid in a Pseudomonas species. V. Pipecolate oxidase and dehydrogenase. J. Bacteriol. 1967;94:1034–1039. doi: 10.1128/jb.94.4.1034-1039.1967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pedersen J. A flux- and background-optimized version of the NanoSTAR small-angle X-ray scattering camera for solution scattering. J. Appl. Cryst. 2004;37:369–380. [Google Scholar]
- 33.Glatter O. A new method for the evaluation of small-angle scattering data. J. Appl. Crystallogr. 1977;10:415–421. [Google Scholar]
- 34.Pedersen JS, Hansen S, Bauer R. The aggregation behaviour of zinc-free insulin studied by small-angle neutron scattering. Eur. Biophys. J. 1994;22:379–389. doi: 10.1007/BF00180159. [DOI] [PubMed] [Google Scholar]
- 35.Svergun DI. Restoring low resolution structure of biological macromolecules from solution scattering using simulated annealing. Biophys. J. 1999;76:2879–2886. doi: 10.1016/S0006-3495(99)77443-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kozin MB, Svergun DI. Automated matching of high- and low-resolution structural models. J. Appl. Crystallogr. 2001;34:33–41. [Google Scholar]
- 37.Glatter O. Interpretation of real-space information from small-angle scattering experiments. J. Appl. Crystallogr. 1979;12:166–175. [Google Scholar]
- 38.Marcotrigiano J, Lomakin IB, Sonenberg N, Pestova TV, Hellen CU, Burley SK. A conserved HEAT domain within eIF4G directs assembly of the translation initiation machinery. Mol. Cell. 2001;7:193–203. doi: 10.1016/s1097-2765(01)00167-8. [DOI] [PubMed] [Google Scholar]
- 39.Bellsolell L, Cho-Park PF, Poulin F, Sonenberg N, Burley SK. Two structurally atypical HEAT domains in the C-terminal portion of human eIF4G support binding to eIF4A and Mnk1. Structure. 2006;14:913–923. doi: 10.1016/j.str.2006.03.012. [DOI] [PubMed] [Google Scholar]
- 40.Prevot D, Darlix JL, Ohlmann T. Conducting the initiation of protein synthesis: the role of eIF4G. Biol. Cell. 2003;95:141–156. doi: 10.1016/s0248-4900(03)00031-5. [DOI] [PubMed] [Google Scholar]
- 41.Korneeva NL, Lamphear BJ, Hennigan FL, Merrick WC, Rhoads RE. Characterization of the two eIF4A-binding sites on human eIF4G-1. J. Biol. Chem. 2001;276:2872–2879. doi: 10.1074/jbc.M006345200. [DOI] [PubMed] [Google Scholar]
- 42.Fujita Y, Oe M, Tutsumino T, Morino S, Imataka H, Tomoo K, Ishida T. Domain-dependent interaction of eukaryotic initiation factor eIF4A for binding to middle and C-terminal domains of eIF4G. J. Biochem. 2009;146:359–368. doi: 10.1093/jb/mvp078. [DOI] [PubMed] [Google Scholar]
- 43.Li W, Belsham GJ, Proud CG. Eukaryotic initiation factors 4A (eIF4A) and 4G (eIF4G) mutually interact in a 1:1 ratio in vivo. J. Biol. Chem. 2001;276:29111–29115. doi: 10.1074/jbc.C100284200. [DOI] [PubMed] [Google Scholar]
- 44.von der Haar T, McCarthy JEG. Intracellular translation initiation factor levels in Saccharomyces cerevisiae and their role in cap-complex function. Mol. Microbiol. 2002;46:531–544. doi: 10.1046/j.1365-2958.2002.03172.x. [DOI] [PubMed] [Google Scholar]
- 45.Liu F, Putnam A, Jankowsky E. ATP hydrolysis is required for DEAD-box protein recycling but not for duplex unwinding. Proc. Natl Acad. Sci. USA. 2008;105:20209–20214. doi: 10.1073/pnas.0811115106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Passmore LA, Schmeing TM, Maag D, Applefield DJ, Acker MG, Algire MA, Lorsch JR, Ramakrishnan V. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007;26:41–50. doi: 10.1016/j.molcel.2007.03.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.