Abstract
Various kinds of stress on human cells induce the formation of endogenous stress granules (SGs). Human Argonaute 2 (hAgo2), the catalytic core component of the RNA-induced silencing complex (RISC), can be recruited to SGs as well as P-bodies (PBs) indicating that the dynamic intracellular distribution of hAgo2 in SGs, in PBs or at other sub-cellular sites could be related to the efficiency of the RNA interference (RNAi) machinery. Here, we studied the influence of heat shock, sodium arsenite (NaAsO2), cycloheximide (CHX) and LipofectamineTM 2000-mediated transfection of phosphorothioate (PS)-modified oligonucleotides (ON) on the intracellular localization of hAgo2 and the efficiency of RNAi.
Fluorescence microscopy and sedimentation analysis of cell fractions indicate stress-induced accumulation of hAgo2 in SGs and the loss of distinctly composed complexes containing hAgo2 or their sub-cellular context. Transfection of cells with PS-ON induces cell stress that is phenotypically similar to the established inducers heat shock and NaAsO2. The intracellular re-distribution of hAgo2 is related to its increased metabolic stability and to decreased RNAi directed by microRNA or by short interfering RNA. Here, we propose a functional model of the relationship between cell stress, translocation of hAgo2 to SGs providing a depot function, and loss of RNAi activity.
INTRODUCTION
The Argonaute protein family constitute a highly conserved family of nucleic acid-binding proteins whose members have been implicated in RNA interference (RNAi) and related phenomena in several organisms (1–5). In humans eight Argonaute proteins have been identified, which can be subdivided into the Ago subfamily and the Piwi (P-element-induced wimpy testis) subfamily (6,7). The expression of Piwi proteins (HIWI1, HIWI2, HIWI3 and HILI) is mostly restricted to the germ line where they associate with piRNAs to facilitate silencing of mobile genetic elements (6,8–10). The Ago subfamily consists of four ubiquitously expressed members, hAgo1-4. Despite their high sequence similarity endonuclease activity is restricted to hAgo2 (5,11,12). Human Ago2 can bind short interfering RNA (siRNA) as well as microRNA (miRNA). As the effector molecule of the RNA induced silencing complex (RISC) it represses target RNA either by site-specific cleavage or by inhibition of translation. In addition, hAgo2 seems to be involved in distinct steps of small RNA maturation (13).
To accomplish gene regulatory processes hAgo2 needs to interact with diverse proteins and protein complexes. A recent study showed that most of these proteins are RNA-binding proteins that are involved in distinct steps of RNA processing, maturation, transport and the regulation of RNA stability and translation (14). Some of these interactions are likely to be mediated by RNA whereas some proteins may bind directly to hAgo2 or associate with it through other protein components (14,15). It seems to be reasonable to speculate on a dynamic network of hAgo2-complexes, which vary in composition and localization at distinct cellular sites of action. In addition, miRNA components of the RNAi machinery are thought to be involved in the control of gene expression of up to 30% of all human genes (16,17), which regulate essential developmental processes such as embryogenesis and cell differentiation as well as cell proliferation and programmed cell death (18–20). Further, miRNAs are thought to play an essential role in human diseases, in particular in malignant cell proliferation (20).
Because of its central role in gene regulation processes the RNAi machinery itself needs to be regulated under certain cellular conditions by post-translational modifications (21,22). For instance, a very recent study described that phosphorylation of TRBP at four serine residues (serine-142, -152, -283 and -286) is mediated by the mitogen-activated protein kinase (MAPK) Erk leading to enhanced miRNA production by increasing the stability of the miRNA-generating complex. In addition this post-translational modification was shown to be important in effecting the mitogenic signalling (23). In mouse and Drosophila it was shown that their respective Piwi proteins underlie post-translational modifications, more precisely arginine methylations, which have an impact on the sub-cellular localization and stability of these proteins (24–26).
A post-translational modification of hAgo2 was reported by Qi et al. (27) who described that hydroxylation of hAgo2 at proline-700 mediated by type I collagen prolyl-4-hydroxylase [C-P4H(I)] is important for hAgo2 stability and effective siRNA-mediated RNAi. In addition this hydroxylation had an impact on hAgo2 PB-localization. Putative hydroxylation sites were also found in other human Ago proteins as well as in mouse and Drosophila Ago2 proteins (21).
In line with those findings it was reported by two independent laboratories that hAgo2-mediated gene silencing might be linked to MAPK signalling pathways that are activated in response to cellular stress (28,29). Zeng et al. (29) showed that hAgo2 is post-translationally modified by phosphorylation of serine-387 through the p38/MAPK signalling pathway. Mutating serine-387 to alanine decreased the localization of hAgo2 to PBs. Adams et al. showed that the hAgo2 protein stability, i.e. the hAgo2 expression level, is regulated by the epidermal growth factor receptor (EGFR)/MAPK signalling pathway. Over-expression of EGFR increased cellular hAgo2 protein levels, which were correlated with enhanced miRNA activity in malignant cell growth of breast cancer cells. MAPKs are members of signalling pathways that transfer extracellular signals from the cell membrane to the nucleus allowing the cell to respond to extracellular stimuli. Thus induction of post-translational modifications of hAgo2 and/or other proteins of the RNAi machinery may represent mechanisms how cellular systems regulate RNAi, thereby co-ordinating gene expression in context of the cellular environment.
The sub-cellular localization of hAgo2 is thought to occur mainly cytoplasmatically visualized by distinct foci but also nuclear localization of hAgo2 has been reported (30–32). In the cytoplasm, hAgo2 associates with two dynamic, transient structures known as stress granules (SGs) and processing bodies (PBs) (15,33–36). SGs are composed of stalled pre-initiation complexes which contain mRNA, small ribosomal subunits and a subset of translation initiation factors including eIF3, eIF4E, eIF4G and PABP (37,38). PBs also contain mRNA but lack other pre-initiation factors and rather contain a number of proteins associated with mRNA decay such as Dcp1A and Dcp2 (37,38). SGs and PBs share some protein components such as the cap binding protein eIF4E and the translational repressor RAP-55, however, not all PB components are found in SGs, or vice versa (37). It might well be that SGs and PBs are spatially, compositionally, or functionally linked (39).
It is consistently reported that hAgo2 associates with components of both, SGs and PBs such as HuR for SGs and Dcp1A for PBs (33,40). However, it is controversially discussed whether these mRNP structures have an implication for miRNA and/or siRNA-mediated gene silencing processes. For instance it is an ongoing matter of debate whether PBs are required for small RNA-mediated gene silencing or whether they simply form as a consequence of silencing (40–43). Furthermore, hAgo2 re-localization to PBs seems to be highly regulated by cellular signalling pathways suggesting that recruitment of hAgo2 to PBs may play a yet unknown function in the RNA-mediated gene regulation (27,29). It is even less well understood why and how hAgo2 is enriched in SGs, which can be stimulated by oxidative and translational stress (35,36,44). This re-localization might be dependent on the presence of mature miRNA since no enrichment of hAgo2 in SGs was observed in Dicer−/− cells upon induction of translational stress by hippuristanol treatment (35). In addition Pare et al. (36) described that the recruitment of hAgo2 to both SGs and PBs is dependent on heat shock protein (Hsp) 90 activity.
In this study we provide experimental evidence that induction of cellular stress and accumulation of hAgo2 in SGs results in decreased siRNA- and miRNA-mediated RNAi and we propose a model describing the spatial and functional sub-cellular localization of hAgo2. This model assumes that SGs represent non-functional RNAi compartments and suggests that a dynamic and reversible exchange of hAgo2 between SGs, PBs and other cellular compartments may occur in human cells.
Materials and Methods
Oligonucleotides, plasmids and chemicals
All oligonucleotides were purchased from Biomers (Ulm, Germany) or IBA (Göttingen, Germany) HPLC purified. PS-modified internucleotide linkages are depicted by ‘s’, bold letters represent 2′-O-methyl substitutions. Deoxyribonucleotides are indicated by capital letters and lower case letters indicate ribonucleotides.
| siLAM | Passenger | 5′-gccucagcacguaccucuaTT-3′ |
| Guide | 5′-uagagguacgugcugaggcTT-3′ | |
| mlet-7A | Passenger | 5′-acuauacaaucuacuggcguucc-3′ |
| Guide | 5′-ugcguuaguagguuguauaguuu-3′ | |
| siAgo2 | Passenger | 5′-gcacggaaguccaucugaauu-3′ |
| Guide | 5′-uucagauggacuuccgugcuu-3′ | |
| Ctrl-RNA | Passenger | 5′-cgaacucacuggucugaccTT-3′ |
| Guide | 5′-ggucagaccagugaguucgTT-3′ | |
| asAgo2PS | 5′-GsTsGsTsTsTsTsGsTsGsTsTsGsCsTsTsTsCsAsCsTsCsTsC-3′ | |
| asAgo2OMe | 5′-GTGTTTTGTGTTGCTTTCACTCTC-3′ | |
| asAgo2PS/OMe | 5′-GsTsGsTsTsTsTsGsTsGsTsTsGsCsTsTsTsCsAsCsTsCsTsC-3′ | |
| asAgo2mutPS | 5′-GsTsGsTsTsCsCsGsTsGsTsTsAsAsTsTsTsCsAsAsGsCsTsC-3′ | |
| asAgo2mutOMe | 5′-GTGTTCCGTGTTAATTTCAAGCTC-3′ | |
| asAgo2mutPS/OMe | 5′-GsTsGsTsTsCsCsGsTsGsTsTsAsAsTsTsTsCsAsAsGsCsTsC-3′ | |
The hAgo2-directed asON, termed asAgo2, is directed against nucleotide positions 3024–3047 within the open reading frame of the hAgo2 mRNA (45). The mutated form of this asON, termed asAgo2mut, carries point mutations at position 6&7, 13&14 and 20&21 numbered 5′ to 3′.
| Ctrl-ON1 | 5′-TsAsCsCsGsCsTsCsTsTsTsTsGsAsCsTsTsTsTsA-3′ |
| Ctrl-ON2 | 5′-GsGsTsGsTsCsAsAsCsAsGsAsAsCsTsGsGsG-3′ |
| Ctrl-ON3 | 5′-CsTsGsAsCsTsTsGsAsTsGsGsTsCsCsAsTsG-3′ |
| Ctrl-ON4 | 5′-GsTsGsAsCsCsTsTsCsTsGsGsAsCsCsTsTsG-3′ |
The Renilla luciferase-encoding plasmid RL-Hmga2m7 has been described by Mayr et al. (46) and carries seven binding sites for mlet-7A. The firefly luciferase-encoding plasmid pGL3-Control was purchased from Promega (Mannheim, Germany).
NaAsO2 and CHX were purchased from Sigma-Aldrich (Taufkirchen, Germany). All other chemicals were purchased from Roth (Karlsruhe, Germany) or Sigma-Aldrich (Taufkirchen, Germany).
Cell culture and induction of cell stress
The cell line ECV-304 was described as an endothelial cell line derived from spontaneously immortalized human umbilical vein cells. However, the German Collection of Microorganisms and Cell Cultures (DSMZ) showed by DNA fingerprinting that this cell line is a derivative of the human urinary bladder carcinoma cell line T-24 (47). ECV-304 cells were cultured in Medium 199 (Lonza, Verviers, Belgium) supplemented with 10% fetal calf serum (PAA, Pasching, Austria) and routinely splitted 2–3 times a week after trypsinization. Cells were incubated at 37°C in an atmosphere of 5% CO2.
For induction of cellular stress ECV-304 cells were treated with 250 µM NaAsO2 for 90 min, incubated with 20 µg/ml cycloheximide for 24 h, transfected with LipofectamineTM 2000/100 nM PS-ON (asAgo2mutPS unless mentioned otherwise) for 4 h, or maintained overnight at 42°C (heat shock) and 5% CO2.
For investigating the hAgo2 stability we used constitutively firefly luciferase-expressing ECV-GL3 cells, which were treated with CHX (20 µg/ml) for up to 48 h. As a control for the translational suppression by CHX we monitored firefly luciferase protein expression and activity.
Transfection of cells
ECV-304 cells were seeded into 12-well culture plates (Greiner, Frickenhausen, Germany) at a density of 1.5 × 105 cells/well, 16 h prior to treatment with ON. The next day, cells were washed twice with PBS. For transfection we used LipofectamineTM 2000 (Invitrogen, Karlsruhe, Germany) according to manufacturer’s instructions. Transfection mixes were prepared in a final volume of 400 μl/well OptiMEM medium (Invitrogen) containing the indicated amounts of ON and 10 µg/ml of LipofectamineTM 2000. Cells were incubated for 4 h at 37°C and 5% CO2. Subsequently, the transfection medium was replaced by complete medium and cells were incubated for 20 h at 37°C and 5% CO2.
RNAi activity assay
For testing the influence of cell stress on the RNAi activity, induction of cell stress was performed as described above followed by the transfection of ECV-304 cells either with 50 nM mlet-7A, 100 ng RL-Hmga2m7 and 25 ng pGL3-Control when testing the miRNA-mediated gene regulation or with 50 pM siLAM when the siRNA-mediated target suppression was investigated. All transfections were performed in a final transfection volume of 400 µl per well of a 12 well plate. After 24 h mlet-7A-mediated knockdown of Renilla luciferase was measured by a dual luciferase assay (Fermentas, Burlington, Canada). The siLAM-mediated degradation of lamin A/C mRNA was monitored via RT–qPCR. In order to be able to detect even small changes of the extent of RNAi, siRNA and miRNA concentrations were chosen such that they were in the range of half maximal target inhibition [(48) and data not shown].
Dual luciferase assay
The dual luciferase assay (Promega) was performed following manufacturer’s instructions. The results were expressed as the ratio of Renilla luciferase to firefly luciferase activity (Rluc/Fluc) with firefly luciferase as the transfection control. The values obtained from cells treated with mlet-7A were normalized to those from cells transfected with Ctrl-RNA and the same amount of Renilla and firefly luciferase encoding plasmids, respectively.
Western analysis
For quantification of hAgo2 or firefly luciferase protein levels ECV-304 and ECV-GL3 cells, respectively were washed twice with PBS, harvested and lysed/denatured in 30 µl lysis buffer [250 mM Tris–HCl pH 6.8, 2% (w/v) SDS, 10% (v/v) glycerine, 10 mM DTT, 0.025% (w/v) bromphenol blue] for 5 min at 95°C. Samples were separated by 8% SDS–PAGE and transferred onto PVDF membrane (Millipore, Schwalbach, Germany). Membranes were blocked for 30 min in 1 × TBST buffer [30 mM Tris–HCl (pH 7.5), 150 mM NaCl, 0.25% (v/v) Tween-20] containing 10% (w/v) skim milk and subsequently incubated with the corresponding primary antibodies overnight at 4°C. The rat anti-Ago2 (11A9) antibody was purchased from Ascenion (München, Germany) and diluted 1:50 in blocking buffer. The mouse anti-firefly luciferase antibody (ab7358) was purchased from Abcam (Cambridge, UK) and diluted 1:2000 in blocking buffer. The loading control β-actin was detected by rabbit anti-β-actin antibody (ab8227, 1:5000 dilution; Abcam). Blots were washed three times with 1 × TBST buffer and incubated with the respective HRP-conjugated secondary antibodies [1:2000 goat anti-rabbit IgG, 1:1000 goat anti-mouse IgG (both from Dako, Hamburg, Germany), 1:5000 goat anti-rat IgG from Jackson ImmunoResearch (Suffolk, UK)] for 1 h at room temperature. Then blots were washed five times with 1 × TBST buffer and incubated with ECL Western Blotting Substrate (Pierce, Rockford, IL, USA). Chemiluminescence was detected on Amersham HyperfilmTM (GE Healthcare, Chalfont St Gilles, UK) and quantified using Image Quant 5.2 software (Amersham Pharmacia Biotech, Freiburg, Germany). In case of hAgo2 the results were expressed as the ratio of hAgo2 to β-actin signal intensities and values obtained from ECV-304 cells treated with siAgo2 were normalized to those obtained from cells transfected with Ctrl-RNA. In case of firefly luciferase the results obtained from ECV-GL3 cells treated with CHX were normalized to those obtained from cells incubated in the absence of CHX.
RT–qPCR
For the quantification of lamin A/C mRNA, ECV-304 cells were washed twice with PBS, harvested and lysed with 200 µl PBS containing 1% (v/v) NP40. The total RNA was extracted using phenol-chloroform followed by ethanol precipitation. A random pool of hexanucleotides primed the synthesis of cDNA by a cDNA first strand synthesis kit (Fermentas, Burlington, Canada) according to the manufacturer’s specifications. RT–qPCR was accomplished with Platinum SYBR Green qPCR Supermix (Invitrogen) on the GeneAmp 5700 thermal cycler (Applied Biosystems, Freiburg, Germany). For the detection of lamin A/C cDNA, the following primers were used: forward primer, 5′-AATGATCGCTTGGCGGTCTA-30; reverse primer, 5′-GCCCTGCGTTCTCCGTTT-3′. The results were expressed as the ratio of lamin A/C to β-glucoronidase cDNA with β-glucoronidase (forward primer, 5′-TTTGGAATTTTGCCGATTTCAT-3′; reverse primer, 5′-GCCGAGTGAAGATCCCCTTT-3′) used as the internal control. The values obtained from cells treated with siLAM were normalized to those obtained from cells transfected with Ctrl-RNA.
Fluorescence microscopy
Initially 7.5 × 104 ECV-304 cells per well were grown on a 24 well Imaging Plate FC (zell-kontakt, Nörten-Hardenberg, Germany) over night. Induction of cell stress was performed as described above followed by immunostaining of hAgo2, Dcp1A and TIA1 which was performed as described recently by Rüdel et al. (31). DNA was stained with Hoechst 33342 dye (Invitrogen) at a final concentration of 2 µg/ml for 10 min at room temperature. Microscopic analyses were carried out by semi-confocal Apotome microscopy (Axiovert 200M) using AxioVision software (Zeiss, Jena, Germany).
The rabbit anti-Dcp1A antibody (1:200 dilution, kindly provided by Jens Lykke-Andersen) was used to detect PBs. The goat anti-TIA1 antibody (1:200 dilution; Santa Cruz Biotechnology, Heidelberg, Germany) was applied for staining SGs. Human Ago2 was detected by the rat anti-Ago2 (11A9) antibody purchased from Ascenion (München) and diluted 1:10. The respective Alexa fluorophore conjugated secondary antibodies were from Invitrogen and used in a 1:400 dilution.
Density gradient centrifugation
A number of 3 × 106 ECV-304 cells per petri dish (92 × 16 mm; Sarstedt, Nümbrecht, Germany) were seeded 16 h prior to induction of cell stress. Subsequently cells were washed twice with PBS, harvested and resuspended in 500 µl cell fractionation buffer (250 mM sucrose, 140 mM NaCl, 1 mM EDTA, 20 mM Tris–HCl, pH 8.0, 2 mM DTT). Cell lysis was performed by 15 passes with a Dounce homogenizer plus 10 passes through a 21G syringe. The nuclei of total cell extracts were removed by centrifugation at 1000g for 10 min at 4°C. The post-nuclear supernatants were layered onto a linear gradient of 10–25% (w/v) iodixanol in cell fractionation buffer and centrifuged at 48 000g for 18 h at 4°C. Gradients were eluted and the linearity of the gradients was confirmed by refractometry. Proteins were precipitated from the gradient fractions with 20% (w/v) trichloroacetic acid. The content of hAgo2 protein in each fraction was determined by western analysis. In order to determine the distribution of siRNA within gradient fractions ECV-304 cells were transfected with 50 nM siLAM in a final volume of 5 ml/petri dish OptiMEM medium (Invitrogen) containing 5 µg/ml of LipofectamineTM 2000 prior to cell fractionation. Total RNA was isolated from each fraction with phenol:chloroform:isoamyl alcohol (25:24:1) followed by ethanol precipitation. The content of siLAM in each fraction was determined via hybridization to a 32P-labeled siLAM-specific probe according to established methodology (49), separation of the hybridization product on a 20% (w/v) acrylamide non-denaturing PAGE, and quantification in the use of a PhosphoImager and Image Quant 5.2 software (Amersham Pharmacia Biotech).
Statistic analysis
The ANOVA test and the program SPSS 12.0 were used to test the significance of differences of siRNA- or miRNA-inducted RNAi activity (Figure 4) as well as hAgo2 protein stability (Figure 3B) in ECV-304 cells, which were left untreated or stressed by different means.
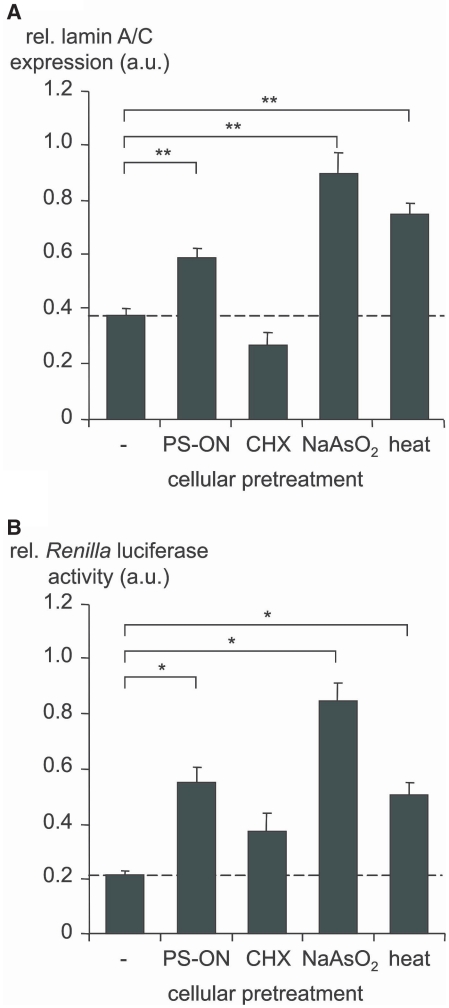
Figure 4.
Translocation of hAgo2 to SGs is accompanied by decreased siRNA- and miRNA-induced RNAi. ECV-304 cells were stressed by different means as indicated below the bars (for detail see the ‘Material and Methods’ section). Subsequently, (A) lamin A/C-directed siRNA (50 pM siLAM) or (B) Renilla luciferase-directed miRNA (50 nM mlet-7A), RL-Hmga2m7 target vector and pGL3 control vector, respectively were transfected. The siLAM-mediated suppression of lamin A/C mRNA was quantified by RT–qPCR. The mlet-7A-mediated inhibition of Renilla luciferase activity was determined by dual-luciferase assay. The siRNA- or miRNA-mediated gene silencing activity in the absence of cellular stress is indicated by a dashed line in each panel. Levels greater than the dashed line indicate loss of RNAi. As control, Ctrl-RNA transfected ECV-304 cells were used and set 1. For each data point three to five independent experiments, each carried out in duplicates, were analyzed using the ANOVA test and the Program SPSS 12.0. Indicated are mean values ± standard error of the mean (SEM); *P < 0.05, **P < 0.01.
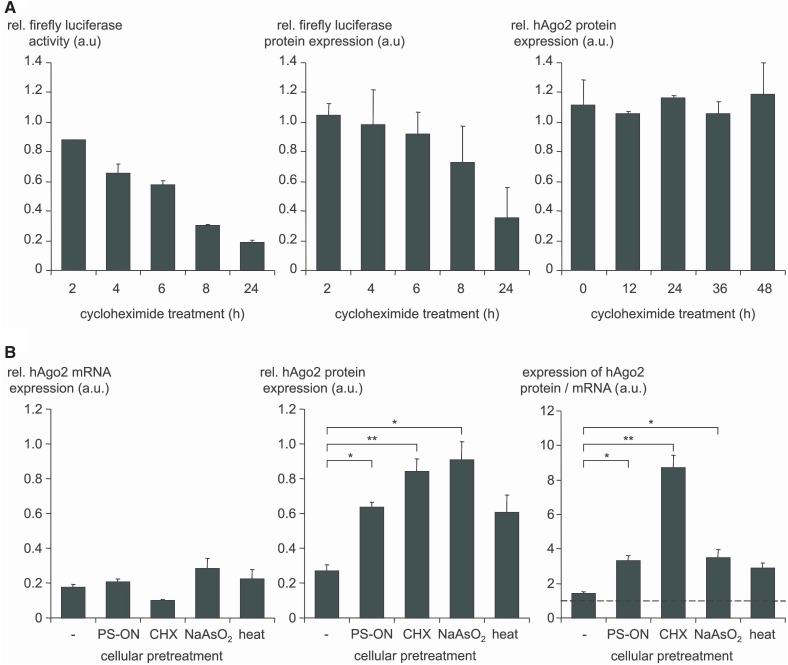
Figure 3.
Translocation of hAgo2 to SGs is accompanied by its increased metabolic stability and by resistance to its siRNA-mediated knockdown. (A) CHX treatment (20 µg/ml) of ECV-GL3 cells, which blocks translation, shows suppression of stable firefly luciferase activity (left panel) as well as firefly luciferase protein expression (middle panel) whereas hAgo2 protein was not affected for up to 48 h of treatment (right panel). As control, ECV-GL3 cells, which were cultured in the absence of CHX were used and set 1. Indicated are mean values ± standard deviations. (B) ECV-304 cells were first stressed by different means as indicated below the bars (for details see the ‘Material and Methods’ section) and then transfected with 100 nM hAgo2-directed siAgo2 in order to test whether the hAgo2 gene expression can be suppressed under cellular stress conditions. After 24 h the gene silencing activity of siAgo2 was determined on the level of hAgo2 mRNA (RT–qPCR) and on the level of Ago2 protein (western analysis). The results are depicted in the left (hAgo2 mRNA) and the middle (hAgo2 protein) panel. As control, Ctrl-RNA transfected ECV-304 cells were used and set 1. The ratio of the hAgo2 protein and mRNA upon siAgo2 treatment is shown on the right panel. A value of 1 (dashed line) indicates a comparable suppression at the mRNA and protein level whereas a value greater than 1 means that less suppression at the protein level was observed. For each data point three to five independent experiments, each carried out in duplicates, were analyzed using the ANOVA test and the Program SPSS 12.0. Indicated are mean values ± standard error of the mean (SEM); *P < 0.05, **P < 0.01.
Results
In this study the human cell line ECV-304 was used as a model system to investigate the relationship between sub-cellular localization of endogenous hAgo2 and the efficiency of RNAi because ECV-304 cells have been well characterized with regard to siRNA-induced RNAi, to microscopic analysis, and they have been analyzed by organelle fractionation via density gradient centrifugation (48,50).
Different kinds of cell stress direct hAgo2 to SGs
Cell stress on ECV-304 cells was induced by three different means which included heat shock, i.e. a transient shift to 42°C. Secondly, cells were exposed transiently to NaAsO2, an inducer of oxidative stress that has been described with regard to re-localization of hAgo2 to SGs (27,29,35). Thirdly, we included a widely used transfection method, i.e. LipofectamineTM 2000-based transfection of PS-ON, which is known for being related to partial cellular toxicity (data not shown).
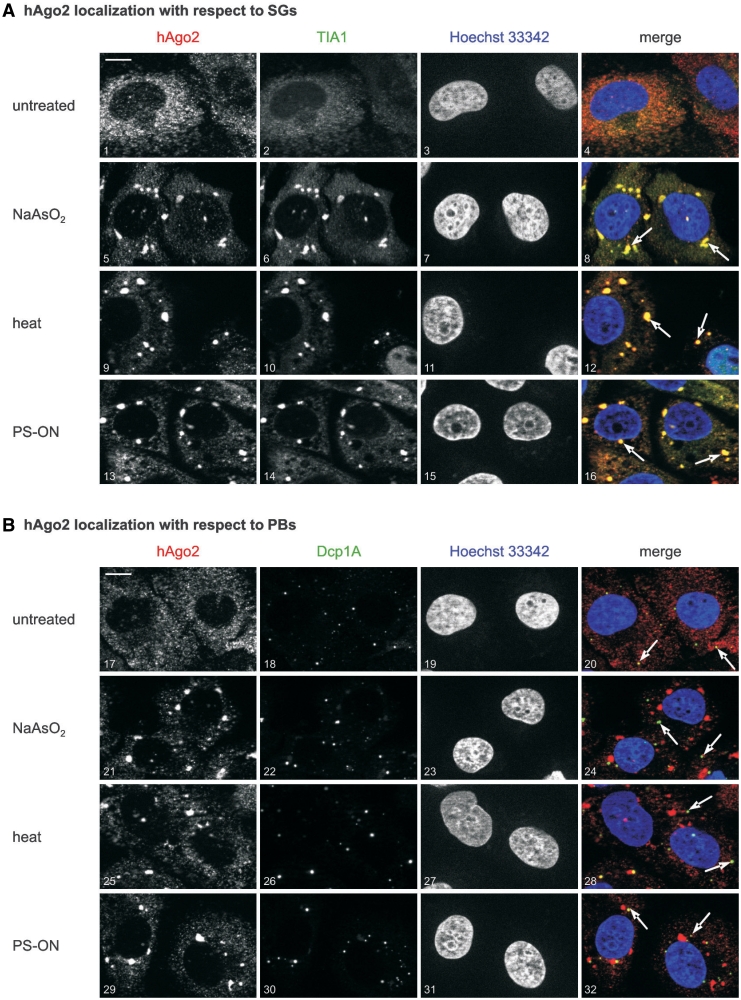
Immunofluorescence analyses of the localization of endogenous hAgo2 in untreated ECV-304 cells showed diffuse signals indicating that the majority of hAgo2 protein is distributed throughout the cytoplasm (Figure 1A, upper panel). A clear re-localization of hAgo2 to granular structures was observed after induction of oxidative stress by treatment of cells with NaAsO2. Co-staining of ECV-304 cells with antibodies against the SG-specific marker TIA-1 confirmed aggregation of hAgo2 in SGs (Figure 1A, second upper panel). A comparable although less sharp localization of hAgo2 was observed when ECV-304 cells were incubated at 42°C prior to immunostaining (Figure 1A, second lower panel). These observations are consistent with previous reports as outlined in the introduction section (35,36,44). Interestingly, transfection of ECV-304 cells with PS-ON gave rise to a similar pattern of the intracellular localization of hAgo2 and indicates that this kind of cell stress is phenotypically similar to the established inducers of stress, NaAsO2 and heat (Figure 1A, lower panel).
Figure 1.
Different kinds of cell stress direct hAgo2 to SGs. As indicated on the left margin, ECV-304 cells were left untreated (picture 1–4 and 17–20) or stressed by 250 μM NaAsO2 for 90 min at 37°C (picture 5–8 and 21–24), incubated at 42°C for 16 h (picture 9–12 and 25–28), or transfected with LipofectamineTM 2000 and 100 nM PS-modified ON (asAgo2mutPS) 24 h prior to fixation (picture 13–16 and 29–32). Subsequently, cells were co-stained with a rat anti-hAgo2 antibody (red color, left column in A and B) and (A) a goat anti-TIA-1 antibody (stress granule marker, green color) or (B) a rabbit anti-Dcp1A (P-body marker, green color). All preparations were also co-stained with Hoechst 33342 dye (blue color). Merged images are shown on the right column. Co-staining of hAgo2 and the SG marker TIA1 can be seen in (A) whereas no co-localization could be observed when hAgo2 and the PB-specific marker Dcp1A were co-stained. The white arrows in panel (A) indicate the staining of SGs and in panel (B) the staining of PBs in an exemplary manner. Optical sections are displayed either as merged images or as greyscale images of the respective red, green or blue channel. The white bar in the upper left of panel (A) and (B) represents a 10 µm scale bar.
The hAgo2 protein is known for its localization in PBs (27,33,34) which is partly compatible with own observations that indicate a PB-specific localization of endogenous hAgo2 in untreated cells in the use of a hAgo2-specific monoclonal antibody. In its use we and others (31) observed that signals specific for endogenous hAgo2 are mainly homogenously dispersed but indicate some foci, which are positive for PB marker proteins. With regard to this kind of localization of hAgo2 we investigated whether cell stress has any influence on the PB association of hAgo2. In the use of Dcp1A as an established marker for PBs (51), we observed no detectable accumulation of hAgo2 in PBs upon induction of cellular stress with NaAsO2, heat, nor transfection of PS-ON (Figure 1B). In order to shed some light on the general meaning of these observations we investigated primary human endothelial cells (HUVEC) and studied their hAgo2 localization under cellular stress conditions. We found a similar stress-related intracellular distribution of endogenous hAgo2 in HUVEC cells (Supplementary Figure S1). This is consistent with the view that the phenomena described for ECV-304 cells are not restricted to this tumor cell line but might rather point to a more general stress response mechanism. It should be noted that the ratio of apparently stressed phenotypes observed for HUVEC is somewhat smaller than found for ECV-304 cells under same cellular conditions, which may reflect a cell-type specific extent of resistance to stress induction. In summary, we conclude that the induction of cellular stress is related to the accumulation of hAgo2 in SGs but not in PBs.
Human Argonaute 2 is contained in at least three distinguishable complexes
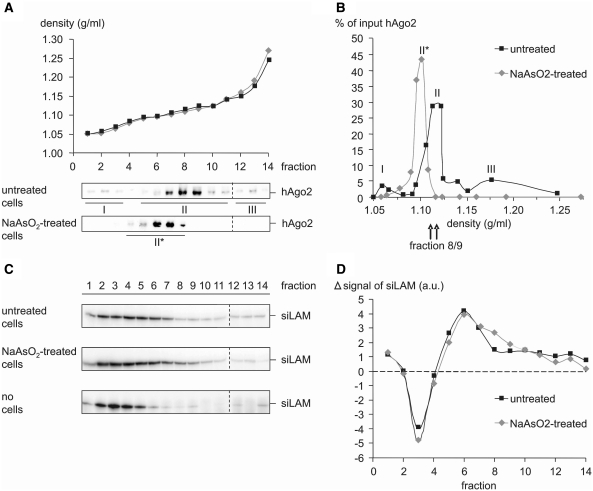
In order to provide independent evidence for the re-localization of hAgo2 upon cell stress we performed cell fractionation by density gradient centrifugation. The post-nuclear supernatant of lysates of untreated or NaAsO2-stressed ECV-304 cells was separated on continuous 10–25% (w/v) iodixanol gradients. The presence of hAgo2 in density fractions was monitored by immunoblotting and visualization with rat anti-Ago2 antibody (Figure 2A). These data indicate that in untreated ECV-304 cells hAgo2 is present in three distinct complexes or compartments that differ by density (Figure 2B), which is reminiscent of a distribution of hAgo2 that was recently described (14). Conversely, a similar analysis of the intracellular distribution of hAgo2 subsequent to NaAsO2 treatment of cells shows only one major hAgo2-positive fraction whereas complexes I and III (Figure 2B) were absent. We cannot decide whether complex II of untreated cells was shifted to lower density fractions (Figure 2A and B; complex II*) or whether this signal monitors an independent site or structure containing hAgo2.
Figure 2.
Density gradient fractionation indicates a NaAsO2-induced re-distribution of hAgo2 to different complexes or organelles. (A and B) ECV-304 cells were either left untreated or incubated with 250 μM NaAsO2 for 90 min at 37°C. The post-nuclear supernatant of total cell extracts was separated on a continuous 10–25% (w/v) iodixanol gradient and fractions were analyzed by western analysis with rat anti-Ago2 [(A), lower panels] (B) A chart of the hAgo2 content of fractions of untreated cells shows three distinct populations (I–III) with the main peak in fraction 8 and 9. In NaAsO2-treated cells hAgo2 is primarily located in one fraction (II*) with the main peak in fraction 6 and 7. The distribution of hAgo2 relative to the sedimentation density is expressed as percentage of the respective signal in each fraction to the sum of all fractions. (C and D) ECV-304 cells were transfected with 50 nM siLAM and then left untreated or stressed by incubation with 250 μM NaAsO2 for 90 min at 37°C. (C) Cell fractionation was performed as described above and total RNA was isolated from each fraction. The siLAM content in each fraction was determined via hybridization to a 32P-labeled siLAM-specific probe. As a further control naked siLAM was separated in the absence of a total cell extract on a continuous 10–25% (w/v) iodixanol gradient (lower panel). A comparison of the distribution of transfected siRNA in untretaed versus NaAsO2-stressed cells with signals normalized to the distribution of naked siRNA is shown (D) and indicates a ‘shoulder’ in fraction 8/9 of NaAsO2-stressed cells compared to untreated cells.
In order to link the gradient sedimentation data to RNAi-related functions we performed siRNA-mediated cleavage assays with pools of fractions representing all of the four peaks shown in Figure 2B (complex 1: fraction 1–3, complex 2*: fraction 6/7, complex 2: fraction 8/9, complex 3: fraction 12–14). Target RNA cleavage assays were performed as described by Höck et al. (14) using ICAM1-directed siRNA [si2B, (52)], which was transfected prior to cell fractionation, and 32P-labeled in vitro transcribed cleavage substrate [ICAM1-IVT, 140 nt in length, position 53–124 represent position 1783 – 1854 of the endogenous ICAM1-mRNA (NM_000201.2)]. A control cleavage reaction with cleavage competent S100 extract prepared from HeLa cells and supplemented with iodixanol (that was used for density gradient centrifugation) showed no influence of iodixanol on cleavage efficiency (data not shown). Subsequently we performed cleavage reactions (90 min at 37°C) in which pooled fractions were tested either directly or after immunoprecipitation with the rat anti-Ago2 (11A9) antibody (31). After anti-hAgo2 immunoprecipitation a weak target RNA cleavage activity was observed in fractions representing peak II* (fraction 6/7) and peak II (fraction 8/9), respectively in case of untreated cells but not in case of NaAsO2-stressed cells (data not shown). It should be noted, however, that it was described by Höck et al. (14) that only complex I is able to induce microRNA-induced target RNA cleavage. The supposed discrepancy between these (miRNA-mediated RNAi) and our observations (siRNA-mediated RNAi) might be explained by the different test systems since Höck et al. used lysates of Flag/HA–hAgo2-transfected HEK 293 cells, which were separated using gradients from 15–55% (w/v) sucrose whereas we monitored endogenous hAgo2 and used a 10–25% (w/v) iodixanol gradient for separation. Further it should be noted that for functional tests we used target RNA cleavage by transfected siRNA. Conversely Höck et al. used target RNA cleavage mediated by endogenous miR-19b.
Furthermore we investigated a potential influence of cell stress on the intracellular distribution of siRNA. ECV-304 cells were transfected with 50 nM of lamin A/C directed siRNA [siLAM, (53)] and then left untreated or stressed by NaAsO2 treatment. The post-nuclear supernatant of total cell extracts was separated on a continuous 10–25% (w/v) iodixanol gradient and total RNA was isolated from each fraction. The siLAM content in each fraction was determined via hybridization to a 32 P-labeled siLAM-specific probe (Figure 2C, upper versus second upper panel). As a further control naked siLAM was separated in the absence of a total cell extract on a continuous 10–25% (w/v) iodixanol gradient and its distribution within the density gradient was determined as described above (Figure 2C, lower panel). The comparison of the distribution of transfected siLAM versus naked siLAM (Figure 2C, the two upper panels versus the lower panel) shows a shift of a substantial amount of siRNA from low to high density fractions. This is consistent with the assumption that the amount of siRNA contained in higher density fractions is complexed with cellular material, including cellular components of the RNAi machinery. A comparison of the distribution of transfected siRNA under normal conditions versus stress conditions with signals normalized to the distribution of naked siRNA indicates that the RNAi-competent fractions 6–9, which include peak II* and II contain siRNA in either case (Figure 2D). A closer look at the siLAM content of fractions of untreated versus NaAsO2-treated cells indicates a shoulder in fractions 8/9 of NaAsO2-treated cells. This observation is somewhat surprising since in NaAsO2-treated cells fraction 8 contained only 10% of the input hAgo2 and in fraction 9 no hAgo2 signal above the detection limit could be observed (Figure 2A). Therefore we tested other RNAi associated proteins with respect to their sedimentation density in a 10–25% (w/v) iodixanol gradient. Preliminary results indicate that the distribution of Dicer is not affected by the induction of cellular stress using NaAsO2. In both cases Dicer shows a main peak in fraction 8 (data not shown). Therefore one might speculate that in stressed cells siRNA still associates with the RNAi machinery, i.e. the RISC loading complex but cannot be handed over to hAgo2 and therefore accumulate in fraction 8.
In summary, we conclude that induction of cell stress lead to altered hAgo2-containing complexes or a change of the sub-cellular localization of hAgo2 or both, which might also effect the cleavage activity of hAgo2 and/or the loading of siRNA to hAgo2.
Human Argonaute 2 shows increased metabolic stability under cell stress
Next, we investigated whether the metabolic stability of hAgo2 is affected by cellular stress, which is indicated by recent studies that describe a relationship between post-translational modifications of hAgo2 induced by cellular stress and its stability. Based on pulse-chase experiments we and Weinmann et al. (54) estimated that a half-life of hAgo2 in normally growing human cells of ∼4 h (Supplementary Figure S6) . In this study, we did not observe a decrease of the hAgo2 protein within 48 h after block of translation by CHX, an inhibitor of protein biosynthesis in eukaryotic cells. As a control for CHX activity, i.e. the block of protein synthesis we monitored firefly luciferase enzyme activity, which was clearly decreased (Figure 3A, left versus right panel). In order to exclude the possibility that under cellular stress conditions most proteins are stabilized but protein activity may be lost we measured the decrease of firefly luciferase protein expression upon CHX treatment (Figure 3A, middle panel). The firefly luciferase protein level decreased somewhat slower than enzyme activity which we cannot explain. However, after 24 h of CHX treatment firefly luciferase was decreased on both levels. Since CHX is known to induce translational stress this might imply a link between cellular stress, hAgo2 metabolism and/or its localization.
In order to test whether hAgo2 could be metabolically stabilized or protected from catabolic decrease in SGs or at other sites under cellular stress conditions we applied the different kinds of cell stress and determined the level of hAgo2 mRNA and protein upon transfection of ECV-304 cells with saturating amounts (f.c. 100 nM, f.v. 400 μl) of a hAgo2-directed siRNA (Figure 3B, left and middle panel). The levels of mRNA showed no significant differences in all cases. By contrast the hAgo2 protein level were significantly increased upon induction of cell stress by CHX, NaAsO2 and PS-ON transfection. Thus, the ratio of both hAgo2 protein and mRNA levels shows that all different kinds of stress induction are related to an apparent increase of hAgo2 protein (Figure 3B, right panel). It should be noted that this phenomenon is statistically significant for CHX, NaAsO2 and PS-ON transfection while the effect on the hAgo2 protein level induced by heat shock is only moderate.
Taken together these observations indicate that cellular stress could be related to stabilization of hAgo2 protein. So we asked whether increased levels of hAgo2 protein in stressed cells are related to the extent of siRNA- or miRNA-induced RNAi.
Translocation of hAgo2 to SGs is related to decreased RNAi
We observed an accumulation of hAgo2 in SGs upon induction of cellular stress (Figure 1) and a re-arrangement of hAgo2-containing complexes or their re-localization or both (Figure 2A and B). In normally growing cells we observed hAgo2 in at least three distinguishable complexes (complex 1, 2, 3), whereas in NaAsO2-stressed ECV-304 cells only one major hAgo2-positive complex (complex 2*) could be observed. Moreover we did not detect target RNA cleavage activity in anti-hAgo2 immunoprecipitated density gradient fractions of NaAsO2-stressed cells (data not shown) but did observe an apparent accumulation of intracellular siRNA in fraction 8/9 (Figure 2C and D).
The absence of complex I and III in fractionation experiments of NaAsO2-stressed cells (Figure 2) prompted us to ask whether the efficiency of small RNA-mediated gene silencing is influenced by cellular stress conditions. To study the effect of cellular stress on the siRNA-mediated pathway, we used the robustly working and potent lamin A/C-directed siRNA termed siLAM (53). Prior to transfection of ECV-304 cells with siRNA we induced cellular stress by treatment with NaAsO2, heat, CHX or PS-ON. The concentration of siLAM was chosen such that the most sensitive range of the dose-response relationship was covered, i.e. in the range of the IC50 concentration (48). The extent of siRNA-induced RNAi in the lamin A/C system was studied after 24 h at the mRNA level via RT–qPCR. It is striking that those kinds of cell stress, which were shown to induce accumulation of hAgo2 in SGs (compare Figure 1A) were also related to a statistically significant decrease in siLAM-mediated target gene suppression (Figure 4A). In case of stress induction of ECV-304 cells by CHX treatment we did observe an even slightly increased siLAM-mediated suppression of lamin A/C mRNA. This finding is difficult to interpret since CHX has been shown to resolve PBs as well as SGs (38).
In order to investigate whether the miRNA-mediated gene silencing pathway is similarly affected by induction of cellular stress we used an established miRNA-regulated system termed ‘mutated let-7A’ (mlet-7A). This miRNA is not naturally expressed in ECV-304 cells. This allowed us to observe the miRNA-induced phenotype despite the background of endogenously expressed miRNAs. Moreover it is not clear whether cell stress does or does not influence the expression and loading of endogenous small RNA. Therefore we decided to use transient assay systems for both, siRNA as well as miRNA. First, cellular stress was induced, subsequently; cells were transfected with all components assembling the siRNA- and miRNA-induced RNAi system, respectively. Thereby we by-pass effects by the involvement of hAgo2 pre-loading with endogenous miRNA because miRNA as well as siRNA have to be newly loaded on hAgo2 after their transfection. Thus a comparison of both systems seems to be permitted. Moreover the transient transfection of both systems (siRNA-mediated RNAi or miRNA-mediated RNAi) assured constant amounts of delivered and presumably intracellular amounts of siRNA and miRNA, respectively. This assumption was confirmed by comparing the amounts of intracellular siLAM detectable in stressed versus unstressed cells after transfection, which was performed by liquid hybridization in the use of 32P-labeled siLAM-specific probe (Figure 2C).
The mlet-7A targets the Renilla luciferase mRNA, which carries the mutated 3′UTR of the high mobility group AT-hook 2 gene (RL-Hmga2m7) with seven binding sites for mlet-7A (46). The efficiency of miRNA-induced RNAi was measured at the protein level. A decrease of mlet-7A-mediated suppression of the Renilla luciferase activity was observed after induction of cellular stress in all cases (Figure 4B). While NaAsO2-induced cell stress led to a complete loss of miRNA function, heat shock and LipofectamineTM 2000/PS-ON were related to an intermediate but statistically significant loss of miRNA-induced RNAi. It is important to note that induction of translational stress by CHX treatment also had an impact on the miRNA-mediated gene silencing pathway (Figure 4B). Again, the basis of this phenomenon might not be easy to reveal because of the disassembling effect of CHX on PBs and SGs (38), which suggests a complex functional interference of CHX with the miRNA-pathway at levels, which cannot be further dissected here.
In summary, these experiments indicate a correlation between hAgo2 aggregation in SGs and decreased siRNA- and miRNA-induced gene silencing activity.
Avoiding the phosphorothioate chemistry of ON increases the extent of RNAi
The LipofectamineTM 2000-mediated transfection of cells with PS-modified oligonucleotides is known for being related to a certain extent of cellular toxicity. In this study we further showed this is related to characteristics of cell stress with respect to sub-cellular localization and RNAi activity of hAgo2.
The phosphorothioate chemistry was the first kind of chemical modification of antisense and immunostimulatory oligonucleotides. Still, this so-called first generation chemistry is widely used and it can be found in clinical development of antisense oligonucleotides. Thus, we decided to have a closer look to this phenomenon since this is common transfection methodology as well as ON chemistry and the results described in our study might generally have great impact on the use of PS-ON.
The analysis depicted in Figure 1A shows the formation of hAgo2-positive SGs upon LipofectamineTM 2000-mediated transfection of cells with PS-ON which is accompanied by reduced siRNA- and miRNA-mediated RNAi (Figure 4).
To study the sequence-dependency of this phenomenon we tested the hAgo2-directed PS-modified asON, asAgo2, and its mutated form, asAgo2mut, in parallel to other four fully PS-modified asONs, which do not have an endogenous target. From these experiments we concluded that the asAgo2 used in this study induces a loss of RNAi activity by the sum of two pathways, an asON-specific suppression of hAgo2 and a stress-related inhibition of hAgo2 (Supplementary Figure S2). The phenomenon seems to be partly sequence-dependent since fully PS-modified ON with different nucleotide sequences show varying levels of reduced RNAi induced by siRNA or by miRNA. We think that this variability could be due to sequence motifs or to different base composition.
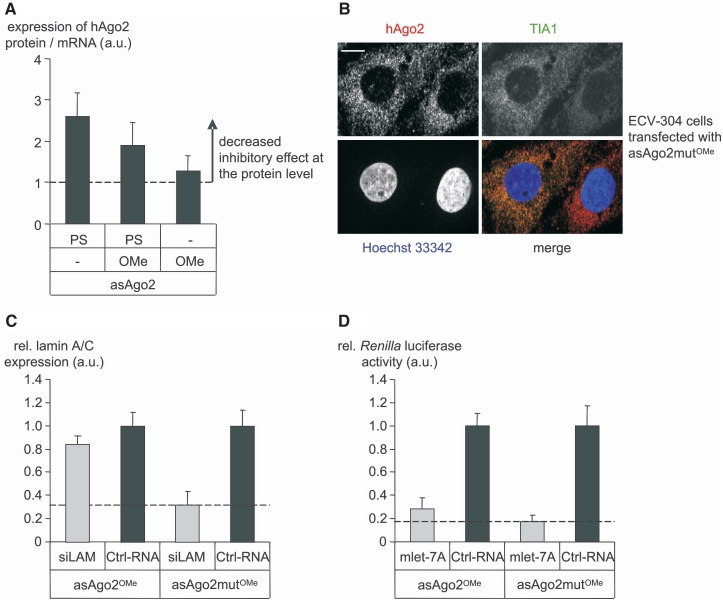
To study the contribution of the chemistry of ON to stress we tested stress-related parameters in the use of the asAgo2 and its control asAgo2mut as fully modified PS-ON, OMe-modified PS-ON and OMe-modified ON. As readout we first used the ratio of hAgo2 mRNA and protein level after transfection of asAgo2 and asAgo2mut, respectively. This ratio indicates an increased hAgo2 protein level upon transfection with the fully PS-modified version of asAgo2 (see also Figure 3B). Progressively replacing the PS modification by the OMe chemistry avoids cell stress and allows suppression of hAgo2 protein (Figure 5A and B). It should be noted that OMe-modified asON did not lead to sequence-independent decrease of RNAi (data not shown) as it was shown for their PS-modified counterpart (Supplementary Figure S2). Further, the mis-matched asAgo2 version, termed asAgo2mutOMe, did not affect hAgo2 mRNA or protein at conditions of preserved activity of asAgo2OMe (data not shown). ECV-304 cells transfected with asAgo2mutOMe, which contains OMe substitutions but avoids the PS-modification did not induce intracellular accumulation of hAgo2 in TIA1-positive SGs (Figure 5B), i.e. large irregularly shaped aggregates as can be seen in Figure 1A (lower panel). Both experiments strongly suggest that specific chemical modifications of oligonuclotides, here the PS chemistry, can interfere with the metabolism and function of hAgo2 as a key component of the RNAi machinery. These experiments also suggest to by-pass cell stress in the use of alternatively modified oligonucleotide-based tools, e.g. the OMe-gapmer chemistry used here.
Figure 5.
Transfection of cells with LipofectamineTM 2000 and PS-modified ON shows stress characteristics. First, ECV-304 cells were transfected with 100 nM of hAgo2-directed asON (asAgo2) or control ON (asAgo2mut) carrying the indicated modifications. The effect of the above mentioned pre-treatment of cells with asON on the hAgo2 expression, localization and RNAi activity was determined after 24 h. (A) The ratio of hAgo2 mRNA and hAgo2 protein indicates cell stress in case of PS-ON but not for 2′-OMe-modified ON. (B) Pre-treatment of cells with 2′-OMe-modified ON (asAgo2mutOMe) is not related to co-localization of hAgo2 (upper left panel) and TIA1 (upper right panel) in large irregularly shaped SGs. The lack of co-localization (lower panel right) indicates the absence of stress and was interpreted as a prerequisite for the capability of a hAgo2-directed asON to suppress hAgo2 effectively at the protein level. The white bar in the upper left panel represents a 10 µm scale bar. (C and D) The effect of asAgo2OMe-mediated knockdown of hAgo2 on siRNA- or miRNA-induced gene silencing activity was investigated. (C) To test the siRNA pathway ECV-304 cells were transfected with 50 pM lamin A/C directed siLAM. (D) The miRNA pathway was studied with 50 nM Renilla luciferase directed mlet-7A, RL-Hmga2m7 target vector, and pGL3 control vector, respectively. The siLAM-mediated suppression of lamin A/C mRNA was quantified by RT–qPCR. The mlet-7A-mediated inhibition of Renilla luciferase activity was determined by dual-luciferase assay. In both cases (C and D) as controls, Ctrl-RNA transfected ECV-304 cells were used and set 1. Indicated are mean values ± standard deviations. A dashed line in each panel indicates the full siRNA- or miRNA-mediated gene silencing activity in the absence of hAgo2 knockdown.
In order to test whether the OMe-gapmer design allows investigating functional RNAi activity in the absence of induced cell stress we used two monitor systems (Figure 5C and D). Firstly, siRNA-mediated suppression of lamin A/C (53) and, secondly, the activity of the miRNA mlet-7A in a recombinant mlet-7A-dependent luciferase system (46). As a control for the hAgo2-directed asON asAgo2OMe we used the derivative asAgo2mutOMe that contains mis-matches at position 6/7, 13/14 and 20/21 and was shown to have no effect on hAgo2 gene expression (data not shown). In this experiment, down-regulation of hAgo2 by asAgo2OMe leads to the loss of siRNA-induced suppression of lamin A/C gene expression (Figure 5C). In the mlet-7A-regulated transient system, the asAgo2OMe–mediated inhibition of hAgo2 shows only slight effects on the miRNA pathway (Figure 5D). This might reflect a less crucial role of hAgo2 in the miRNA pathway (45) or a bypass mechanism for hAgo2 in case of its limited availability. Further we cannot exclude that other RNAi associated proteins or even protein networks are affected under cellular stress conditions. This could explain different effects of the hAgo2 knockdown on siRNA- and miRNA-mediated gene silencing activity in the use of PS-modified versus OMe-modified hAgo2-directed asON (Figure 5C and D; Supplementary Figure S2).
In summary, we conclude that the OMe chemistry rather than the PS chemistry used in transfection experiments does not direct hAgo2 protein to SGs and leaves hAgo2 accessible for RNAi machineries.
Cell stress-induced decrease of RNAi activity is reversible
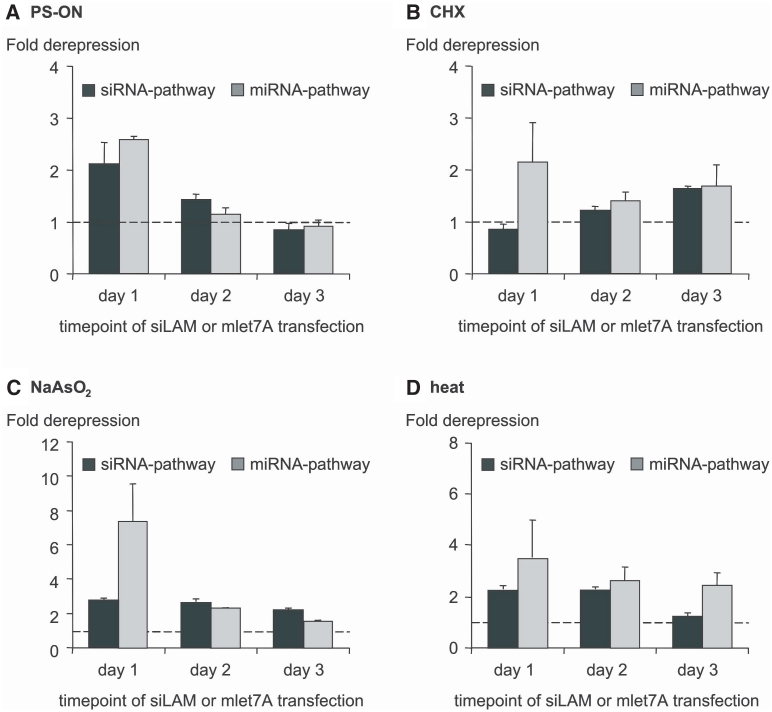
Next, we investigated whether the loss of RNAi activity could be reversible, as one would predict in a typical stress-like scenario. The experimental design was such that first, cell stress was induced transiently by different means as described in the ‘Material and Methods’ section. Subsequently, one, two or three days after release of stress, cells were tested for their performance in siRNA- or miRNA-mediated RNAi by the siLAM or the mlet-7A system as described before. Reversibility of the apparent loss of function of hAgo2 in siRNA- and miRNA-induced RNAi was observed in both systems. In case of siRNA-mediated gene regulation the recovery of RNAi activity was most pronounced for PS-ON-mediated cell stress followed by heat shock and NaAsO2 (Figure 6). In case of miRNA-mediated gene regulation, most obvious reversibility of stress-induced loss of RNAi was observed for PS-ON-mediated stress, followed by CHX, NaAsO2 and heat shock in this order (Figure 6). These findings are compatible with the hypothesis that the hAgo2 protein is involved in continuous spatial re-localization between sites at which it is involved in RNAi and sites at which it is captured and not available for active RNAi though being physically intact. So far, we have no data concerning the question whether this presumed steady state involves reversible post-translational modifications of hAgo2 but this should be explored in future studies.
Figure 6.
Cell stress-induced decrease of RNAi activity is reversible. Cell stress was transiently induced by the means indicated in the top of each panel [(A)–(D), for detail see the ‘Material and Methods’ section]. One, 2 or 3 days after release of stress, ECV-304 cells were tested for their capability to perform siRNA- or miRNA-mediated RNAi. Briefly, ECV-304 cells were transfected either with 50 pM lamin A/C directed siLAM or with 50 nM Renilla luciferase directed mlet-7A, RL-Hmga2m7 target vector and pGL3 control vector at day 1–3 after induction of cell stress. The extent of siRNA- and miRNA-mediated target inhibition in the absence of cell stress was normalized to Ctrl-RNA transfected ECV-304 cells and set to 1 (indicated by a dashed line in each panel). Higher numbers indicate derepression of target expression, i.e. loss of efficacy of siRNA or miRNA, respectively. Indicated are mean values ± standard deviations.
Discussion
This study strongly suggests that various kinds of cell stress are related to translocation of hAgo2 protein to SGs (Figure 1A). It is important to note that we decided to monitor the localization of endogenous hAgo2 rather than using transient or stable over-expression of recombinant and possibly tagged hAgo2 since over-expression of proteins associated with SGs or PBs are known to induce ‘spontaneous’ SG or PB formation even in the absence of cellular stress (38). This kind of unintended effect in the use of recombinant systems could partially explain inconsistencies in the literature on the sub-cellular localization of hAgo2, which has been described as mainly cytoplasmatic (31,35,55) while some reports indicate an almost exclusive association of hAgo2 with PBs (34,56).
Intracellular stress-related re-distribution of hAgo2 is also indicated by density gradient analysis of cell fractions. This analysis indicates at least three sub-populations of hAgo2 in normal growing cells, which is consistent with a recent study of Höck et al. (14), and only one major hAgo2-positive population in stressed ECV-304 cells. Target RNA cleavage assays with pools of anti-hAgo2 immuno-enriched gradient fractions were performed and the distribution of transfected siLAM under normal conditions versus stress conditions within density gradient fractions was measured. Both analyses indicated that the induction of cell stress also affects the cleavage activity of hAgo2 and/or the loading of siRNA to hAgo2. This observation is compatible with suppressed RNAi in stressed cells. It is important to note that in either case (unstressed versus stressed cells) the RNAi-competent fractions 6–9 contain siRNA. Therefore one might hypothesize that the observed lack of siRNA-induced RNAi in stressed cells is not due to the absence of siRNA at critical sub-cellular sites.
Furthermore fluorescence microscopic analysis showed no evidences that cellular stress affects neither transfection efficacy nor the sub-cellular localization of siRNA or miRNA (Supplementary Figures S3 and S4, pictures 5–20). In this context it should be noted that the kind of microscopic studies as performed here do not distinguish between active and non-active siRNA or miRNA molecules. It has recently been suggested that a number of a few hundred molecules of siRNA or less than that is necessary for half maximal target suppression (57,58). If one considers that 10 000 molecules of siRNA or more than that are delivered to cells after transfection it is obvious that the intracellular localization of active species cannot be identified in front of the background of a hundred-fold excess of inactive species.
The inverse relationship between increased abundance of hAgo2 after induction of cell stress and the loss of RNAi is striking. We speculate on increased metabolic stability of hAgo2 by its translocation to SGs or to other sites where protein degradation is presumably limited. This is reminiscent of a depot-like accumulation of potentially active hAgo2 under stress conditions. We do not speculate on increased translation of hAgo2 because hAgo2 mRNA levels remain unaltered under cellular stress conditions (data not shown) although we have no experimental evidence excluding increased mRNA translation. It is important to note that microscopic studies as performed here do not provide quantitative data on the sub-cellular distribution of hAgo2.
Regarding the loss of miRNA- and siRNA-induced RNAi we speculate on functional trapping of hAgo2 in SGs. Since it was described by Kedersha et al. (38) that SGs are transient structures which may only be present for a limited time following the application or release of stress we investigated the time-dependent localization of hAgo2 upon NaAsO2 treatment and confirmed the above mentioned observation of Kedersha et al. (Supplementary Figure S5A). From our data we estimated a half life time of NaAsO2-induced SGs of ∼45 min and observed a complete re-localization of hAgo2 to a diffuse cytoplasmatic distribution after 3 h. Surprisingly the recovery of the RNAi activity upon removal of the stress-inducing agent takes clearly longer (1–3 days) than the relocalization of hAgo2 to a diffuse cytoplasmatic localization (Figure 6C). In order to shed light on this apparent discrepancy we investigated the siRNA-mediated RNAi activity of hAgo2 at shorter time points upon removal of the NaAsO2 (2–6 h) and observed even greater phenotypes (Supplementary Figure S5B). Up to 4 h upon removal of the NaAsO2 no RNAi activity was measurable, 6 h upon the NaAsO2 treatment a 8-fold derepression of target gene expression in NaAsO2-treated cells compared to untreated cells was detected, which decreased to a 2-fold derepression after 24 h (Supplementary Figure S5B) but remained measurable up to 3 days (Figure 6C). There are many possibilities to speculate about reasons for the delayed RNAi recovery. For example, one might hypothesize that hAgo2 is converted to a modified and RNAi-incompetent version when accumulating in SGs and that this presumed modified hAgo2 does not re-enter the RNAi pathway upon its release, i.e. its re-conversion to the normal state is slow.
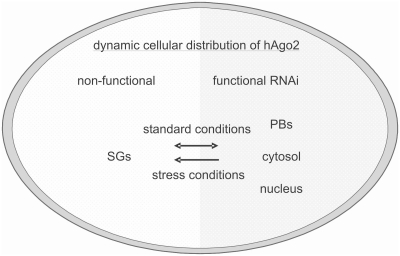
Based on the observations described in this study we postulate a minimal model that relates cell stress to the sub-cellular distribution of hAgo2 and to the efficiency of RNAi (Figure 7). According to this model the cell is sub-divided into two functionally different spaces, one allows active RNAi because hAgo2 is present in a biologically active form and the second does not allow functional RNAi because hAgo2 is present in a non-active state. The functionally active space includes PBs as well as nuclear and cytosolic portions. Conversely, we assign SGs to the functionally inactive space. Further we postulate a dynamic exchange of hAgo2 between both compartments. Upon the induction of cell stress by NaAsO2, PS-ON or heat shock major amounts of hAgo2 are translocated from functionally active sites to inactive sites, which include SGs. Even though fluorescence microscopy does not provide quantitative data their fluorescence intensity at cellular stress conditions is remarkably high. Thus, one might speculate that this is the major compartment of inactive hAgo2. After release of stress, however, the system seems to act in a reversible manner because hAgo2 is redistributed to the non-stressed state (Supplementary Figure S5A) and RNAi is restored (Supplementary Figure S5B and Figure 6).
Figure 7.
Schematic depiction of a proposed model of the relationship between cell stress, translocation of hAgo2 to SGs and loss of RNAi. Active siRNA- and miRNA-induced RNAi occur in the functional compartment (right side), which includes the cytosol, the nucleus and PBs under non-stress conditions. The non-functional compartment includes SGs (left side) to which hAgo2 migrates under stress conditions. Since the hAgo2-related extent of RNAi is reversibly linked with cell stress we assume exchange of hAgo2 between both compartments, i.e. a kind of steady state. The non-functional compartment contains transient structures composed of stalled preinitiation complexes and untranslated mRNA, which are known as SGs. Under cellular stress conditions the equilibrium is perturbed and hAgo2 accumulates in SGs. This may circumvent the functional interactions of hAgo2 with other RNAi involved proteins which induces the exclusion of hAgo2 from the RNAi pathway.
In this context it should be noted that recently Kedersha et al. (39) proposed that mRNA released from disassembled polysomes is sorted and remodeled at SGs, from which selected transcripts are than delivered to PBs for degradation. Similarly, Leung et al. (35,59) proposed that the localization of miRNPs to PBs and SGs reflects different functional states of miRNPs. Further studies should focus on the complex interplay between stress, RNAi mechanism and cellular signaling pathways.
One might further speculate that hAgo2 provides important cellular functions that require its localization in SGs, which is adverse to the cellular capability of RNAi. This opposing situation might occur without any causative relationship between cell stress and typical known RNAi-based processes. It might occur coincidently, which would be consistent with the findings of Kedersha et al. (39). Alternatively, one might speculate that the cellular capability of active RNAi could be in conflict with crucial stress response mechanisms, i.e. an inverse coupling of both cellular response options might exist. This would imply that the cellular response to a severe stress situation has higher priority than executing RNAi including miRNA-facilitated regulation of gene expression.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Institute’s budget.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We cordially thank Jens Lykke-Andersen [University of California, San Diego (UCSD), La Jolla, USA] for providing the Dcp1A-directed antibody, Dirk Grimm (University of Heidelberg, Germany) for providing the stably FLAG/HA-hAgo2 expressing HeLa cell line, Johanna Klement (UK S-H, Lübeck, Germany) for her help with the statistical analysis of the data, Sandra D. Laufer for providing HeLa S100 extract and Ulrike Weirauch (University of Marburg, Germany) for initial experiments.
REFERENCES
- 1.Carmell MA, Xuan Z, Zhang MQ, Hannon GJ. The Argonaute family: tentacles that reach into RNAi, developmental control, stem cell maintenance, and tumorigenesis. Genes Dev. 2002;16:2733–2742. doi: 10.1101/gad.1026102. [DOI] [PubMed] [Google Scholar]
- 2.Faehnle CR, Joshua-Tor L. Argonautes confront new small RNAs. Curr. Opin. Chem. Biol. 2007;11:569–577. doi: 10.1016/j.cbpa.2007.08.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Okamura K, Ishizuka A, Siomi H, Siomi MC. Distinct roles for Argonaute proteins in small RNA-directed RNA cleavage pathways. Genes Dev. 2004;18:1655–1666. doi: 10.1101/gad.1210204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Peters L, Meister G. Argonaute proteins: mediators of RNA silencing. Mol. Cell. 2007;26:611–623. doi: 10.1016/j.molcel.2007.05.001. [DOI] [PubMed] [Google Scholar]
- 5.Tolia NH, Joshua-Tor L. Slicer and the argonautes. Nat. Chem. Biol. 2007;3:36–43. doi: 10.1038/nchembio848. [DOI] [PubMed] [Google Scholar]
- 6.Höck J, Meister G. The Argonaute protein family. Genome Biol. 2008;9:210. doi: 10.1186/gb-2008-9-2-210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hutvagner G, Simard MJ. Argonaute proteins: key players in RNA silencing. Nat. Rev. Mol. Cell Biol. 2008;9:22–32. doi: 10.1038/nrm2321. [DOI] [PubMed] [Google Scholar]
- 8.Girard A, Sachidanandam R, Hannon GJ, Carmell MA. A germline-specific class of small RNAs binds mammalian Piwi proteins. Nature. 2006;442:199–202. doi: 10.1038/nature04917. [DOI] [PubMed] [Google Scholar]
- 9.Thomson T, Lin H. The biogenesis and function of PIWI proteins and piRNAs: progress and prospect. Annu. Rev. Cell Dev. Biol. 2009;25:355–376. doi: 10.1146/annurev.cellbio.24.110707.175327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 11.Liu J, Carmell MA, Rivas FV, Marsden CG, Thomson JM, Song JJ, Hammond SM, Joshua-Tor L, Hannon GJ. Argonaute 2 is the catalytic engine of mammalian RNAi. Science. 2004;305:1437–1441. doi: 10.1126/science.1102513. [DOI] [PubMed] [Google Scholar]
- 12.Meister G, Landthaler M, Patkaniowska A, Dorsett Y, Teng G, Tuschl T. Human Argonaute2 mediates RNA cleavage targeted by miRNAs and siRNAs. Mol. Cell. 2004;15:185–197. doi: 10.1016/j.molcel.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 13.Diederichs S, Haber DA. Dual role for argonautes in microRNA processing and posttranscriptional regulation of microRNA expression. Cell. 2007;131:1097–1108. doi: 10.1016/j.cell.2007.10.032. [DOI] [PubMed] [Google Scholar]
- 14.Höck J, Weinmann L, Ender C, Rudel S, Kremmer E, Raabe M, Urlaub H, Meister G. Proteomic and functional analysis of Argonaute-containing mRNA-protein complexes in human cells. EMBO Rep. 2007;8:1052–1060. doi: 10.1038/sj.embor.7401088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Meister G, Landthaler M, Peters L, Chen PY, Urlaub H, Luhrmann R, Tuschl T. Identification of novel argonaute-associated proteins. Curr. Biol. 2005;15:2149–2155. doi: 10.1016/j.cub.2005.10.048. [DOI] [PubMed] [Google Scholar]
- 16.Lewis BP, Burge CB, Bartel DP. Conserved seed pairing, often flanked by adenosines, indicates that thousands of human genes are microRNA targets. Cell. 2005;120:15–20. doi: 10.1016/j.cell.2004.12.035. [DOI] [PubMed] [Google Scholar]
- 17.Xie X, Lu J, Kulbokas EJ, Golub TR, Mootha V, Lindblad-Toh K, Lander ES, Kellis M. Systematic discovery of regulatory motifs in human promoters and 3′ UTRs by comparison of several mammals. Nature. 2005;434:338–345. doi: 10.1038/nature03441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Martinez J, Busslinger M. Life beyond cleavage: the case of Ago2 and hematopoiesis. Genes Dev. 2007;21:1983–1988. doi: 10.1101/gad.1591407. [DOI] [PubMed] [Google Scholar]
- 19.O’Carroll D, Mecklenbrauker I, Das PP, Santana A, Koenig U, Enright AJ, Miska EA, Tarakhovsky A. A Slicer-independent role for Argonaute 2 in hematopoiesis and the microRNA pathway. Genes Dev. 2007;21:1999–2004. doi: 10.1101/gad.1565607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Alvarez-Garcia I, Miska EA. MicroRNA functions in animal development and human disease. Development. 2005;132:4653–4662. doi: 10.1242/dev.02073. [DOI] [PubMed] [Google Scholar]
- 21.Heo I, Kim VN. Regulating the regulators: posttranslational modifications of RNA silencing factors. Cell. 2009;139:28–31. doi: 10.1016/j.cell.2009.09.013. [DOI] [PubMed] [Google Scholar]
- 22.Rüdel S, Meister G. Phosphorylation of Argonaute proteins: regulating gene regulators. Biochem.J. 2008;413:e7–e9. doi: 10.1042/BJ20081244. [DOI] [PubMed] [Google Scholar]
- 23.Paroo Z, Ye X, Chen S, Liu Q. Phosphorylation of the human microRNA-generating complex mediates MAPK/Erk signaling. Cell. 2009;139:112–122. doi: 10.1016/j.cell.2009.06.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kirino Y, Kim N, Planell-Saguer M, Khandros E, Chiorean S, Klein PS, Rigoutsos I, Jongens TA, Mourelatos Z. Arginine methylation of Piwi proteins catalysed by dPRMT5 is required for Ago3 and Aub stability. Nat. Cell Biol. 2009;11:652–658. doi: 10.1038/ncb1872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Reuter M, Chuma S, Tanaka T, Franz T, Stark A, Pillai RS. Loss of the Mili-interacting Tudor domain-containing protein-1 activates transposons and alters the Mili-associated small RNA profile. Nat. Struct. Mol. Biol. 2009;16:639–646. doi: 10.1038/nsmb.1615. [DOI] [PubMed] [Google Scholar]
- 26.Vagin VV, Wohlschlegel J, Qu J, Jonsson Z, Huang X, Chuma S, Girard A, Sachidanandam R, Hannon GJ, Aravin AA. Proteomic analysis of murine Piwi proteins reveals a role for arginine methylation in specifying interaction with Tudor family members. Genes Dev. 2009;23:1749–1762. doi: 10.1101/gad.1814809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Qi HH, Ongusaha PP, Myllyharju J, Cheng D, Pakkanen O, Shi Y, Lee SW, Peng J, Shi Y. Prolyl 4-hydroxylation regulates Argonaute 2 stability. Nature. 2008;455:421–424. doi: 10.1038/nature07186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Adams BD, Claffey KP, White BA. Argonaute-2 expression is regulated by epidermal growth factor receptor and mitogen-activated protein kinase signaling and correlates with a transformed phenotype in breast cancer cells. Endocrinology. 2009;150:14–23. doi: 10.1210/en.2008-0984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Zeng Y, Sankala H, Zhang X, Graves PR. Phosphorylation of Argonaute 2 at serine-387 facilitates its localization to processing bodies. Biochem. J. 2008;413:429–436. doi: 10.1042/BJ20080599. [DOI] [PubMed] [Google Scholar]
- 30.Robb GB, Brown KM, Khurana J, Rana TM. Specific and potent RNAi in the nucleus of human cells. Nat. Struct. Mol. Biol. 2005;12:133–137. doi: 10.1038/nsmb886. [DOI] [PubMed] [Google Scholar]
- 31.Rüdel S, Flatley A, Weinmann L, Kremmer E, Meister G. A multifunctional human Argonaute2-specific monoclonal antibody. RNA. 2008;14:1244–1253. doi: 10.1261/rna.973808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ohrt T, Mutze J, Staroske W, Weinmann L, Hock J, Crell K, Meister G, Schwille P. Fluorescence correlation spectroscopy and fluorescence cross-correlation spectroscopy reveal the cytoplasmic origination of loaded nuclear RISC in vivo in human cells. Nucleic Acids Res. 2008;36:6439–6449. doi: 10.1093/nar/gkn693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Liu J, Valencia-Sanchez MA, Hannon GJ, Parker R. MicroRNA-dependent localization of targeted mRNAs to mammalian P-bodies. Nat. Cell Biol. 2005;7:719–723. doi: 10.1038/ncb1274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sen GL, Blau HM. Argonaute 2/RISC resides in sites of mammalian mRNA decay known as cytoplasmic bodies. Nat. Cell Biol. 2005;7:633–636. doi: 10.1038/ncb1265. [DOI] [PubMed] [Google Scholar]
- 35.Leung AK, Calabrese JM, Sharp PA. Quantitative analysis of Argonaute protein reveals microRNA-dependent localization to stress granules. Proc. Natl Acad. Sci. USA. 2006;103:18125–18130. doi: 10.1073/pnas.0608845103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pare JM, Tahbaz N, Lopez-Orozco J, LaPointe P, Lasko P, Hobman TC. Hsp90 regulates the function of argonaute 2 and its recruitment to stress granules and P-bodies. Mol. Biol. Cell. 2009;20:3273–3284. doi: 10.1091/mbc.E09-01-0082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Anderson P, Kedersha N. RNA granules. J. Cell Biol. 2006;172:803–808. doi: 10.1083/jcb.200512082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kedersha N, Anderson P. Mammalian stress granules and processing bodies. Methods Enzymol. 2007;431:61–81. doi: 10.1016/S0076-6879(07)31005-7. [DOI] [PubMed] [Google Scholar]
- 39.Kedersha N, Stoecklin G, Ayodele M, Yacono P, Lykke-Andersen J, Fritzler MJ, Scheuner D, Kaufman RJ, Golan DE, Anderson P. Stress granules and processing bodies are dynamically linked sites of mRNP remodeling. J. Cell Biol. 2005;169:871–884. doi: 10.1083/jcb.200502088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Bhattacharyya SN, Habermacher R, Martine U, Closs EI, Filipowicz W. Relief of microRNA-mediated translational repression in human cells subjected to stress. Cell. 2006;125:1111–1124. doi: 10.1016/j.cell.2006.04.031. [DOI] [PubMed] [Google Scholar]
- 41.Eulalio A, Behm-Ansmant I, Schweizer D, Izaurralde E. P-body formation is a consequence, not the cause, of RNA-mediated gene silencing. Mol. Cell Biol. 2007;27:3970–3981. doi: 10.1128/MCB.00128-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pillai RS, Bhattacharyya SN, Artus CG, Zoller T, Cougot N, Basyuk E, Bertrand E, Filipowicz W. Inhibition of translational initiation by Let-7 MicroRNA in human cells. Science. 2005;309:1573–1576. doi: 10.1126/science.1115079. [DOI] [PubMed] [Google Scholar]
- 43.Chu CY, Rana TM. Translation repression in human cells by microRNA-induced gene silencing requires RCK/p54. PLoS Biol. 2006;4:e210. doi: 10.1371/journal.pbio.0040210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Gallois-Montbrun S, Kramer B, Swanson CM, Byers H, Lynham S, Ward M, Malim MH. Antiviral protein APOBEC3G localizes to ribonucleoprotein complexes found in P bodies and stress granules. J. Virol. 2007;81:2165–2178. doi: 10.1128/JVI.02287-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Mescalchin A, Detzer A, Weirauch U, Hahnel MJ, Engel C, Sczakiel G. Antisense tools for functional studies of human Argonaute proteins. RNA. 2010 doi: 10.1261/rna.2204610. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Mayr C, Hemann MT, Bartel DP. Disrupting the pairing between let-7 and Hmga2 enhances oncogenic transformation. Science. 2007;315:1576–1579. doi: 10.1126/science.1137999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dirks WG, MacLeod RA, Drexler HG. ECV304 (endothelial) is really T24 (bladder carcinoma): cell line cross-contmination at source. In Vitro Cell Dev. Biol. Anim. 1999;35:558–559. doi: 10.1007/s11626-999-0091-8. [DOI] [PubMed] [Google Scholar]
- 48.Detzer A, Overhoff M, Wunsche W, Rompf M, Turner JJ, Ivanova GD, Gait MJ, Sczakiel G. Increased RNAi is related to intracellular release of siRNA via a covalently attached signal peptide. RNA. 2009;15:627–636. doi: 10.1261/rna.1305209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Overhoff M, Wunsche W, Sczakiel G. Quantitative detection of siRNA and single-stranded oligonucleotides: relationship between uptake and biological activity of siRNA. Nucleic Acids Res. 2004;32:e170. doi: 10.1093/nar/gnh168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Detzer A, Overhoff M, Mescalchin A, Rompf M, Sczakiel G. Phosphorothioate-stimulated cellular uptake of siRNA: a cell culture model for mechanistic studies. Curr. Pharm. Des. 2008;14:3666–3673. doi: 10.2174/138161208786898770. [DOI] [PubMed] [Google Scholar]
- 51.Kedersha N, Tisdale S, Hickman T, Anderson P. Real-time and quantitative imaging of mammalian stress granules and processing bodies. Methods Enzymol. 2008;448:521–552. doi: 10.1016/S0076-6879(08)02626-8. [DOI] [PubMed] [Google Scholar]
- 52.Kretschmer-Kazemi FR, Sczakiel G. The activity of siRNA in mammalian cells is related to structural target accessibility: a comparison with antisense oligonucleotides. Nucleic Acids Res. 2003;31:4417–4424. doi: 10.1093/nar/gkg649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Elbashir SM, Harborth J, Weber K, Tuschl T. Analysis of gene function in somatic mammalian cells using small interfering RNAs. Methods. 2002;26:199–213. doi: 10.1016/S1046-2023(02)00023-3. [DOI] [PubMed] [Google Scholar]
- 54.Weinmann L, Hock J, Ivacevic T, Ohrt T, Mutze J, Schwille P, Kremmer E, Benes V, Urlaub H, Meister G. Importin 8 is a gene silencing factor that targets argonaute proteins to distinct mRNAs. Cell. 2009;136:496–507. doi: 10.1016/j.cell.2008.12.023. [DOI] [PubMed] [Google Scholar]
- 55.Tan GS, Garchow BG, Liu X, Yeung J, Morris JP, Cuellar TL, McManus MT, Kiriakidou M. Expanded RNA-binding activities of mammalian Argonaute 2. Nucleic Acids Res. 2009;37:7533–7545. doi: 10.1093/nar/gkp812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Savas JN, Ma B, Deinhardt K, Culver BP, Restituito S, Wu L, Belasco JG, Chao MV, Tanese N. A role for Huntington's disease protein in dendritic RNA granules. J. Biol. Chem. 2010;285:13142–13153. doi: 10.1074/jbc.M110.114561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Detzer A, Sczakiel G. Phosphorothioate-stimulated uptake of siRNA by mammalian cells: a novel route for delivery. Curr. Top. Med. Chem. 2009;9:1109–1116. doi: 10.2174/156802609789630884. [DOI] [PubMed] [Google Scholar]
- 58.Laufer SD, Restle T. Peptide-mediated cellular delivery of oligonucleotide-based therapeutics in vitro: quantitative evaluation of overall efficacy employing easy to handle reporter systems. Curr. Pharm. Des. 2008;14:3637–3655. doi: 10.2174/138161208786898806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Leung AK, Sharp PA. Function and localization of microRNAs in mammalian cells. Cold Spring Harb. Symp. Quant. Biol. 2006;71:29–38. doi: 10.1101/sqb.2006.71.049. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.