Abstract
Several ribosomal protein families contain paralogues whose roles may be equivalent or specialized to include extra-ribosomal functions. RpL22e family members rpL22 and rpL22-like are differentially expressed in Drosophila melanogaster: rpL22-like mRNA is gonad specific whereas rpL22 is expressed ubiquitously, suggesting distinctive paralogue functions. To determine if RpL22-like has a divergent role in gonads, rpL22-like expression was analysed by qRT-PCR and western blots, respectively, showing enrichment of rpL22-like mRNA and a 34 kDa (predicted) protein in testis, but not in ovary. Immunohistochemistry of the reproductive tract corroborated testis-specific expression. RpL22-like detection in 80S/polysome fractions from males establishes a role for this tissue-specific paralogue as a ribosomal component. Unpredictably, expression profiles revealed a low abundant, alternative mRNA variant (designated ‘rpL22-like short’) that would encode a novel protein lacking the C-terminal ribosomal protein signature but retaining part of the N-terminal domain. This variant results from splicing of a retained intron (defined by non-canonical splice sites) within rpL22-like mRNA. Polysome association and detection of a low abundant 13.5 kDa (predicted) protein in testis extracts suggests variant mRNA translation. Collectively, our data show that alternative splicing of rpL22-like generates structurally distinct protein products: ribosomal component RpL22-like and a novel protein with a role distinct from RpL22-like.
INTRODUCTION
In several ribosomal protein (RP) gene families [most notable in certain yeast species and plant systems—reviewed by ref. (1)], paralogous proteins exist, presumably derived from duplication events in the evolutionary history of the gene. Paralogous RPs may have functionally redundant roles within the ribosome, or in some instances, their roles may be specialized in ribosome biogenesis or translation, contributing to heterogeneity within the ribosome cycle [e.g. (2)]. Alternatively, specialized roles for paralogous RPs may include extra-ribosomal or extra-translational functions [see review by Warner and McIntosh (3) for some discussion on this issue]. Specialized roles may be indicated particularly if a paralogue is expressed in a cell-, tissue- or developmental stage-specific manner.
Recent studies in Saccharomyces cerevisiae have revised the previously held view that many RP paralogues dually expressed in that species, are functionally equivalent (4). Instead, some paralogues are specialized for differential functions or cellular locations (4,5), leading Komili et al. (4) to propose a ‘ribosome code’ that regulates translation of specific mRNAs in different physiological states. Although less than that reported in yeast and plant systems, tissue-specific ribosome heterogeneity due to assembly of RP structural variants into ribosomes has also recently been reported in rodent mammary gland and liver for RpL22-like1 and in testis for RpL10- and RpL39-like (2).
In Drosophila melanogaster, RpL22 and RpL22-like are members of the conserved RpL22e family specific to eukaryotes. Unlike most fly RP paralogues that display between 65% and 100% amino acid identity (6), RpL22 and RpL22-like are instead only 37% identical (6), suggesting considerable ‘opportunity’ for disparate functions between family members. RpL22 family members in Drosophila also exhibit unique structural features at the N-terminus compared to orthologues in other species. Fly RpL22e family members contain an N-terminal extension of unknown function that is homologous to the C-terminal end of histone H1 [previously described only for RpL23a and RpL22 by ref. (7)]. Structural divergence between RpL22 and RpL22-like is most prominent within the N-terminal extension. Over time the novel domain may have specified new functions for these proteins in addition to their functions in the ribosome cycle.
In addition to considerable amino acids divergence between these paralogues in D. melanogaster, their expression patterns are also dissimilar. Transcripts for rpL22 are ubiquitously expressed. Previous studies have revealed rpL22-like mRNA expression in embryonic gonads, adult ovary and germline stem cells by in situ hybridization or RT-PCR (8–10). Recent microarray analyses showed enrichment of rpL22-like in adult testis, but not in adult ovary [FlyAtlas; (11)]. Shotgun mass spectrometric data support the existence of RpL22-like protein in fly embryos (www.ebi.ac.uk/pride/Q8T3X3), but no protein expression data for other developmental stages and/or specific tissues have been established. Tissue-specificity of rpL22-like expression suggests that RpL22-like may have a distinct role compared to its paralogue RpL22, at least in the embryonic gonad.
Although its position on the 60S subunit has recently been mapped by cryoEM to the base of the subunit on the most recently published 80S ribosome model (12), the cellular role for RpL22 has not been completely characterized (13). Interestingly, partially reconstituted ribosomes that lack RpL22 are still translation competent, suggesting that the protein may have a regulatory or non-ribosomal role (13), or alternatively, function under different physiological conditions. In Drosophila, additional roles and interactions for RpL22 have been proposed [based on high-throughput yeast two hybrid screens assembled in the Drosophila Interactions Database version 2010_10 (DroID: http://www.droidb.org)], awaiting further characterization. Among these interactions are several putative extra-ribosomal roles for RpL22, including interactions with a transcriptional repressor complex in Kc cells (14) and with nuclear enzyme poly-ADP ribose polymerase [mediated through the N-terminal histone H1-like domain (7)], for example.
Based on C-terminal homology to RpL22 and its tissue-specific expression pattern, it is reasonable to hypothesize that RpL22-like has a gonad-specific ribosomal function, although other functions cannot be excluded. In fact, several protein–protein interactions are also catalogued for RpL22-like in the Drosophila Interactions Database and none overlap with those proposed for RpL22, including those that are likely to be non-ribosomal in nature. Together, this information suggests that RpL22 and RpL22-like have distinct functions, either within the ribosomal cycle and/or in non-ribosomal pathways. That one of the functions of RpL22-like is as a ribosomal component had not been previously investigated prior to this study. Such developmental or tissue-specific regulation of a putative RP is not widely known in animal systems and is more commonplace in plants (1).
To explore the possibility that RpL22-like functions as a tissue-specific ribosomal component, we first refined its developmental and tissue-specific expression pattern to facilitate its biochemical characterization. By quantitative (q) RT-PCR, we determined that rpL22-like mRNA is highly enriched in adult testes compared to ovaries. Using paralogue-specific antibodies (Abs) in western blots, we detected a highly abundant protein of the predicted molecular weight (MW) for RpL22-like in testes. A higher MW immunoreactive species was also detected in fly heads. Immunohistochemical (IHC) analysis of the male reproductive tract shows that RpL22-like is exclusively found within testes and not within seminal vesicles or accessory glands. We further demonstrate that RpL22-like is a ribosomal component (80S and polysomes), suggesting its incorporation into actively translating ribosomes.
These studies also led to a novel finding that rpL22-like is alternatively spliced using non-canonical splice sites to remove an intron that generates a short form designated rpL22-like short, found in lower abundance than the full-length mRNA isoform. Surprisingly, the most abundant rpL22-like mRNA isoform retains the previously uncharacterized intron. rpL22-like short mRNA would encode a protein consisting nearly exclusively of amino acid residues in the N-terminal domain of RpL22-like fused in frame to residues at the very end of the C-terminus, thereby eliminating the majority of the conserved L22e RP signature. Detection of rpL22-like short mRNA on polysomes and the presence of a low abundant protein of the predicted MW in testis extracts suggest that the spliced variant may be translated. This study provides the first experimental confirmation that RpL22-like is a ribosomal component that is enriched in testis, and that its gene through alternative splicing may also encode a novel protein (RpL22-like short) with a novel non-ribosomal function based on its predicted amino acid structure.
METHODS
Fly stocks
Wild-type Canton S D. melanogaster were used for polysome profiling (a kind gift from Todd Laverty, HHMI-JFRC). All other experiments used wild-type Oregon R D. melanogaster obtained from Carolina Biological.
Primers
For a list of primers and oligonucleotides (Integrated DNA Technologies) used in this study, see Table 1.
Table 1.
List of primers and oligonucleotides used in experiments described in this article
| Name | Sequence | Experimental use |
|---|---|---|
| FDmL22likeBamHI | 5′-GTCACGGATCCATGAGTTCCCAGACGCAGAAAAAGAATGCTTCCAA-3′ | RT-PCR and cloning; bridge RT-PCR; FLAG-tagged constructs |
| RDmL22likeBamHI | 5′-GTCACGGATCCTTAGGCAAAGGTTTTTCCGCCATTGTCGTCGGCAA-3′ | RT-PCR and cloning; in vitro transcription |
| RDmL22likeBamHI_shortened | 5′-GTCACGGATCCTTAGGCAAAGGTTTTTCCGCCATTG-3′ | RT-PCR and cloning; bridge RT-PCR |
| FL22like_exon1/2 | 5′-GATACTAATTTCTTTGGAGAATG-3′ | Bridge RT-PCR |
| RL22like_novel exon2/3 | 5′-TTAGGCAAAGGTTTTTCCGCCATTGTCGTCGGCAAGAGG-3′ | Bridge RT-PCR; northern analysis |
| RDmL22-likeFLAG-BamH1 | 5′-GTCACGGATCCTTACTTGTCATCGTCATCCTTGTAGTCGCCGCGGCCGATGGCAAAGGTTTTTCCGCCATTGTCGTCGGCAA-3′ | FLAG-tagged constructs |
| FpEXP5-T7 | 5′-CCGCGAAATTAATACGACTCACTATAGGGAGACACACGACGG-3′ | in vitro transcription |
| FL22likeFull | 5′-GCTCAACCAGCTGAAGGATCA-3′ | qRT-PCR |
| RL22likeFull | 5′-GAAGTAGCGCTTGGAGAAGTGC-3′ | qRT-PCR |
| FL22likeShort | 5′-CAGAATGCTTCACGCAAGAACTT-3′ | qRT-PCR |
| RL22likeShort | 5′-AATGTCGTCGGCAAGAAGC-3′ | qRT-PCR |
| FBeta2Tubulin | 5′-GTGCTGAACTGGTGGATTCCGT-3′ | qRT-PCR |
| RBeta2Tubulin | 5′-GGTCAGCTGGAAGCCCTGAA-3′ | qRT-PCR |
| FVasa | 5′-CTGTACGAAAACGAGGATGGTGA-3′ | qRT-PCR |
| RVasa | 5′-ACCACCGTCCCCTCTTTCA-3′ | qRT-PCR |
| FrpL32 | 5′-CTAAGCTGTCGCACAAATGG-3′ | qRT-PCR |
| RrpL32 | 5′-ACGCACTCTGTTGTCGATACC-3′ | qRT-PCR |
| FL22-like_startcodon | 5′-ATGAGTTCCCAGACGCAGAAAAAGAATGCTTCCAAGGCC-3′ | Recomb. expression |
| FL22 | 5′-AAGATGGCTCCTACCGCCAAGACCAACAAGGG-3′ | Recomb. expression |
| RL22 | 5′-TTACTCGGCATCGTCGTCCTCATCG-3′ | Recomb. expression |
| RL22like_exon1 | 5′-GGAAGCATTCTTTTTCTGCGTCTGGGAACTC-3′ | Northern analysis |
| RL22like_exon3 | 5′-TTAGGCAAAGGTTTTTCCGCCATTGTCGTCGGC-3′ | Northern analysis |
| RL22like full specific | 5′-GGCCGTGGACACTACCCGCAACCAATCACGCAGGC-3′ | Northern analysis |
Antibodies
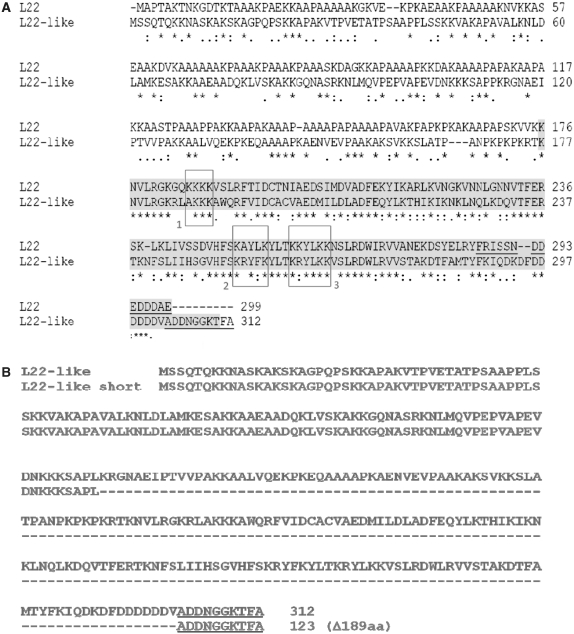
The following peptide sequences were obtained from FlyBase (15) and used for polyclonal Ab generation from GenScript: RpL22: FRISSNDDEDDDAE; RpL22-like: ADDNGGKTFA. Polyclonal Abs were protein A purified from rabbit and mouse, respectively. HRP-conjugated secondary Abs (Promega) were used in western analysis. Antibody specificity was tested using bacterially expressed recombinant RpL22 and RpL22-like (Supplementary Figure S2). Preimmune sera from rabbit and mouse were used to confirm the absence of immunoreactive proteins in fly extracts (Supplementary Figure S2). Anti-mouse Alexa 488 Fluor and anti-rabbit Alexa 568 Fluor secondary Abs (Invitrogen) were used for IHC.
RNA isolation from fly tissue and cultured cells
Tissues were dissected from wild-type adult flies of mixed age (2- to 7-day old) in sterile 1X PBS and immediately frozen on dry ice. S2 cells were cultured under standard conditions in S2 media (Invitrogen) and collected during log phase. RNA isolation was completed using TRIzol reagent (Invitrogen) following manufacturer’s guidelines. RNase-free glycogen (10 µg; Invitrogen) was used as a carrier to increase yield. Samples with A260/280 and A260/230 ratios <1.8 were discarded. Qualitative analysis of ∼300 ng total RNA via gel electrophoresis ensured RNA integrity. Only samples that passed both spectrophotometric and qualitative analyses were used for further experiments.
RT-PCR Analysis
Total RNA (500 ng) from adult tissues and cultured S2 cells or 100 ng of embryo, larval and adult polyA+ RNA (Clontech) was used in RT-PCR analyses. SuperScript One-Step RT-PCR System with Platinum Taq DNA Polymerase (Invitrogen) was used following manufacturer’s guidelines.
Cloning and Sequencing of rpL22-like cDNAs
Gel-purified RT-PCR products were cloned into pMT/V5-His-TOPO® (Invitrogen), transformed into One Shot TOP10 Chemically Competent E. coli (Invitrogen), plated onto selective media (LB, ampicillin 100 µg/ml) and plasmid DNAs purified using the miniPrep system (Qiagen). Multiple clones for rpL22-like cDNAs were sequenced using an ABI 310 Genetic Analyzer (Applied Biosystems) or by GeneWay Research Laboratories.
Bacterial recombinant protein expression for western analysis
Gel-purified RT-PCR amplicons were cloned into E. coli expression vector pEXP5-CT/TOPO (Invitrogen) following manufacturer’s guidelines. Recombinant expression in transformed OneShot BL21 Star (DE3) Chemically Competent E. coli (Invitrogen) was induced with 0.5 mM IPTG.
cDNA synthesis and qRT-PCR analysis
Total RNA (1 µg) was used in cDNA synthesis using the SuperScript III First-Strand Synthesis System (Invitrogen) following manufacturer’s guidelines and cDNAs stored at −80°C. One microliter of cDNA was used for qRT-PCR using the ABI 7300 real-time PCR system (Applied Biosystems) with SYBR green power master mix reagent (Applied Biosystems). Primers were designed using Primer Express software (Applied Biosystems). Primers (200 nM final concentration) were annealed at 55°C in a three-step amplification stage. Average ΔCT values obtained for all genes were normalized to rpL32 (rpL32/rp49) for comparison to data by Shigenobu et al. (8). All samples were run in triplicate with an n = 3. Standard deviations for each sample were calculated using the average ΔCT of three runs. ΔΔCT values were calculated using the average ΔCT from each sample. Fold differences were calculated using the comparative CT method (ΔΔCT), using fold difference = 2−ΔΔCT, as directed by the ABI 7300 qRT-PCR manual.
Protein extract preparation, SDS–PAGE and western blotting
Drosophila tissues were dissected in 0.7× PBS supplemented with Mini Complete protease inhibitor cocktail without EDTA (Roche) and immediately frozen on dry ice. S2 cells were cultured as described above. Pelleted S2 cells were homogenized and lysed in RIPA buffer supplemented with Mini Complete protease inhibitor cocktail without EDTA (Roche) and microcentrifuged at maximum speed for 5 min. Soluble fractions were quantitated using the DC protein assay system (BioRad).
Soluble extract (10–15 µg) or insoluble fractions were mixed with reducing SDS-sample buffer and proteins separated by SDS–PAGE (5% stacking gel; 12.5% separating gel) at 200 V and electro-transferred onto 0.2 µm Westran S PVDF membrane (Whatman) at 100 V for 1 h. After blocking with 5% non-fat dry milk (NFDM) in 1× PBS with 0.1% Tween-20 (5% NFDM) for 1 h, membranes were incubated overnight with primary Ab (1:1000 in 3% NFDM) at 4°C. HRP-conjugated secondary Abs (1:50 000) were incubated at 4°C for 2 h. Chemilluminescent detection was achieved using ECL-Plus (GE Healthcare) and BioMax Chemilluminescence film (Kodak).
Northern analysis
Embryonic, larval, and adult poly(A+) mRNAs (15 µg; Clontech) were resolved on a 1.5% formaldehyde agarose gel in MOPS buffer and blotted onto a 0.2 μm Optitran nitrocellulose membrane. Filters were hybridized with 32P-labelled oligonucleotide probes complementary to coding regions within rpL22-like (Table 1) according to hybridization conditions in Sambrook et al. (16). RNAs were visualized by phosphorimaging. Filters were stripped in boiling 0.1% SDS and re-hybridized as above. Size estimates for detected RNAs were determined using RNA markers (Promega or Invitrogen).
Immunohistochemistry
Testis squashes and IHC preparation was performed as previously reported (17,18) with minor modifications. Testes from mature adult wild-type flies of mixed age (2–7-day old) were dissected in 0.7% saline, squashed and quickly frozen on dry ice. Tissues were fixed and permeablized in ice-cold ethanol for 10 min and in 4% formaldehyde (in 1× PBS) for 7 min at room temperature (RT), followed by 2× 15 min washes in PBS with 0.3% Triton X-100 and 0.3% sodium deoxycholate and a single wash of PBT (PBS with 0.1% Triton X-100). Blocking occurred for 1 h with PBTB (PBT with 3% BSA) at RT. Squashes were incubated overnight at 4°C with primary Ab (1:100 in PBTB) in a humid chamber and washed four times for 15 min in PBTB at RT. Alexa Fluor-conjugated secondary Abs (1:200 in PBTB) were incubated at 4°C in a humid chamber for 1 h and then washed four times for 10 min in PBTB at RT. DAPI staining (0.2 µg/ml in PBS) was performed after final Ab washes for 5 min, washed twice for 1 min in PBS and mounted in Cytoseal 60 (Richard-Allan Scientific). Fluorescent micrographs were taken using Nikon Eclipse TE200U inverted fluorescence microscope coupled to a digital CCD camera.
Polysome analysis
Ribosome extracts were prepared using modified procedures from Qin et al. (19), Houmani and Ruf (20) and Pelczar and Filipowicz (21). Adult male Canton S flies (2.37g) were homogenized in lysis buffer [1:5 w/v; (19)] and homogenates clarified by centrifugation at 10K rpm for 10 min. Approximately 100 OD260 U/µl were loaded onto a 10–50% linear sucrose gradient [prepared by the horizontal method described in ref. (20)] and spun at 35K rpm for 160 min in a SW-41 rotor. Gradient fractions (0.5 ml) were collected and read at OD260 using a DU-800 spectrophotometer. Fractions were pooled and pelleted with equal volumes of 20 mM Tris–HCl, pH 7.2 and spun at 40K rpm for 12 h in an SW41 rotor. Pellets were resuspended in 20 mM Tris–HCl, pH 7.2 and subjected to SDS–PAGE and western analysis.
RESULTS
rpL22-like mRNA transcripts are enriched in the testis
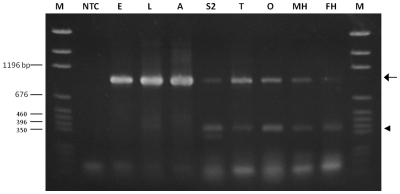
Gene expression profiling of embryonic germline stem cells (8) and adult ovary germline stem cells (10) previously identified a novel and possible germline stem cell-specific gene known as rpL22-like (due to its similarity to rpL22; Figure 1A). An important goal was to refine previously reported rpL22-like mRNA expression profiles (8–10) in order to initiate studies on RpL22-like protein expression and function. By RT-PCR analysis, we determined that rpL22-like is expressed not only in embryos, larvae and adults, but also in an embryonic-derived S2 cell line, as well as within gonads and heads of both sexes (Figure 2). The expected amplicon of ∼939 bp was present in all samples using primers determined by BLAST analysis to target only rpL22-like in the D. melanogaster genome. Interestingly, in addition to the expected RT-PCR product for rpL22-like there was a prominent lower MW amplicon of ∼392 bp that was reproducibly amplified in multiple experiments for all RNA samples analysed (Figure 2). Molecular characterization (cloning and sequencing) of both amplicons indicated that both are derived from the rpL22-like gene. We refer to this lower MW amplicon as ‘rpL22-like short’ (described in detail below).
Figure 1.
(A) Clustal W alignment of D. melanogaster RpL22 and RpL22-like sequences. Aligned sequences (RpL22: FBgn0015288; RpL22-like: FBgn0034837) show conservation at the C-terminus (46% sequence identity; 73% sequence similarity), but divergence at the N-terminus (sequence identity 36%; sequence similarity 47%). Overall amino acid identity is 37% (6). Shaded amino acid residues mark conservation within the RpL22e superfamily. Other residues define the N-terminal extension with homology to histone H1 [(7) for RpL22 and RpL23a]. Boxes highlight functional residues involved in nuclear localization (box 1), rRNA binding (boxes 2 and 3) and nucleolar localization (boxes 2 and 3) (20). Underlined residues in the C-termini were used to make polyclonal peptide Abs. (B) Translation of rpL22-like short mRNA (GenBank accession no. HM756190) using Translation Tool (expasy.org) revealed a putative protein of 123 amino acid, consisting primarily of the histone H1-like domain (N-terminus) and not the RpL22-like domain (C-terminus). Underlined residues in the C-termini were used to make polyclonal peptide Abs.
Figure 2.
RT-PCR analysis of rpL22-like in different developmental stages, tissues, and in S2 cells. RT-PCR using rpL22-like primers to amplify the coding sequence resulted in the expected 939 bp amplicon (rpL22-like, arrow) in all samples. An additional smaller amplicon (rpL22-like short, arrowhead) of ∼390 bp was present in all samples. Variability in the intensity of rpL22-like short was noted in numerous replicates of the RT-PCR for various samples as noted here for embryonic and adult samples. NTC: no template control; E: embryo; L: larva; A: adult; S2: S2 cells; T: testis; O: ovary; MH: male heads; FH: female heads; M: pGEM marker.
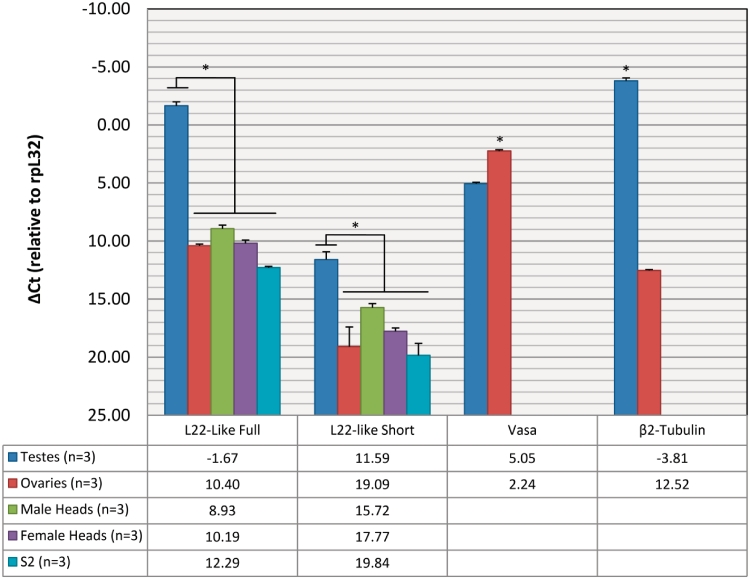
Based on RT-PCR data, rpL22-like isoforms were detected in a variety of developmental stages and tissues. To determine isoform abundance in fly gonads, heads and S2 cells, we used qRT-PCR and isoform-specific primers on different RNA samples (Figure 3; Supplementary Table S2), showing that both rpL22-like and rpL22-like short mRNAs are enriched in testis compared with other tissues examined. These data corroborate previously published microarray analyses (11) and high-throughput expression analyses (FlyAtlas: flyatlas.org), showing that rpL22-like mRNA is highly enriched (∼4300-fold in this study) in testis compared with ovary (Supplementary Table S2). Using the testis-specific β2-tubulin gene (22) and the germline-specific Vasa gene [males: (18); females: (23)] for comparative purposes in the fly gonad, mRNA levels seen for rpL22-like short (and for rpL22-like) in tissues other than in testis may be due to basal level transcription (Figure 3) as β2-tubulin mRNA [widely regarded as a testis-specific protein, e.g. (22,24)] has some level of detection even in ovaries.
Figure 3.
qRT-PCR reveals rpL22-like mRNA enrichment in testis. Using isoform-specific primers, qRT-PCR shows that both isoforms are more highly expressed in testis compared with other tissues. Vasa and β2-tubulin serve as germ cell- and testis-specific controls, respectively. Numbers in table represent CT values. *P < 0.01.
While found at ∼9800-fold lower levels compared with rpL22-like in testes, all samples showed that the shorter isoform is not the prominent isoform, with highest and lowest amounts in testes and in S2 cells, respectively. Steady-state levels of both rpL22-like mRNA isoforms are therefore highest in the testis.
RpL22-like protein is differentially expressed and found in active ribosomes
Based on the relative abundance of rpL22-like mRNA, we determined that RpL22-like protein expression would best be analysed in testes compared with other tissues. Since no RpL22-like Abs were previously available, paralogue-specific polyclonal Abs targeting C-terminal amino acid residues were designed for recognition of RpL22 or RpL22-like (Figure 1A). The C-terminal peptide Ab for RpL22 is identical to that used successfully by Ni et al. (14) in IHC experiments and ChIP analysis; therefore, we anticipated that this Ab would be useful in our protein blots and IHC studies to detect RpL22. The C-terminal Ab for RpL22-like recognition in protein blots and IHC studies was similarly based on the location of the Ab epitope for paralogue RpL22.
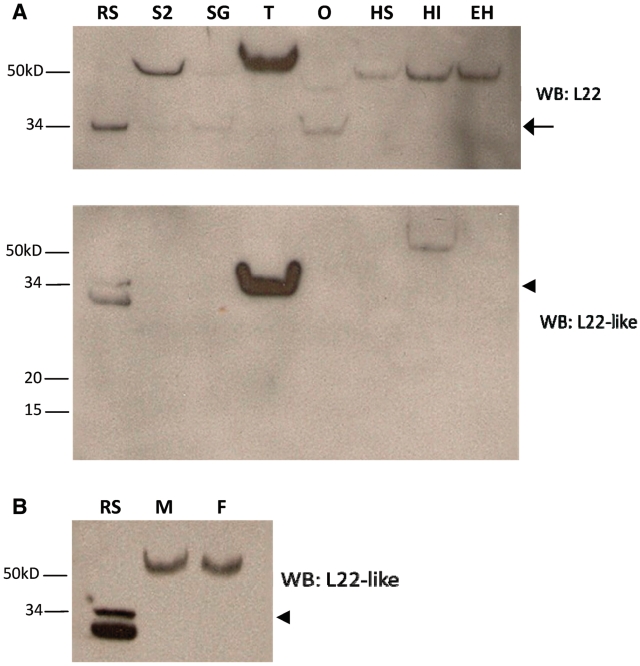
We first confirmed that our Abs were specific for the proteins of interest by preimmune sera analysis and detection of recombinant tagged proteins (Supplementray Figures S2 and S3). Western blot analysis was used to screen adult gonads, larval salivary glands, S2 cells and fly heads for the presence of RpL22. As RpL22 is ubiquitously expressed, we expected that RpL22 would be detected in all tissues. Two prominent immunoreactive species, one at the predicted MW (∼33 kDa) for RpL22 and the other at ∼50 kDa (the latter seen in all tissues), were identified (Figure 4A). The amount of 50 kDa species varied in different tissues in multiple experiments and may represent incorporation of RpL22 into an SDS-resistant complex. Interestingly, relatively little, if any RpL22 of the expected size is found in the testis and in heads.
Figure 4.
Western blot analysis confirms differential expression of RpL22-like. (A) Compared to the recombinant standard (RS), western analysis for RpL22 shows immunodetection at either the expected MW of 32.9 kDa (arrow) and/or at a higher MW at ∼50 kDa in all tissues. Immunodetection of RpL22-like at the expected MW of 34.3 kDa (arrowhead) is solely visible in testes. Insoluble extracts from male and female heads contain a higher MW species of RpL22-like [whole intact heads of mixed sex with eyes (HI)]. No immunoreactive species is seen in eyeless heads (eyes are dissected out; EH) or in a soluble head extract. The additional lower band in the RS sample is endogenous bacterial protein recognized by mouse antisera (see Supplementary Figure S2 for additional explanation). (B) The electrophoretic shift in RpL22-like is not sex specific as seen by western analysis of whole head tissue from males (M) and females (F). S2: S2 cells; SG: larval salivary glands; T: testis; O: ovary; HS: soluble head extract.
Further, western analysis confirmed that RpL22-like expression within the gonad is confined to males, as a protein of the expected MW (∼34 kDa) is highly enriched in testis tissue and not ovary (Figure 4A). We also noted an immunoreactive product (found in lower abundance than in testes) in an insoluble head extract (only a very limited amount of this product is seen in a soluble head fraction in some preparations). Surprisingly in head tissue, immunodetection is seen at a higher MW (∼60 kDa). What factors contribute to the electrophoretic shift for both RpL22 and RpL22-like are unknown, but may include post-translational modifications or assembly into SDS-resistant complexes. In fact, in silico analysis for both RpL22 and RpL22-like predicts several sites for post-translational modifications, particularly for phosphorylation [Eukaryotic Linear Motif resource for functional sites in proteins (ELM; http://elm.eu.org/)]. Unlike the protein expression pattern in the gonad, RpL22-like is detected in both male and female heads (Figure 4B). Detection of RpL22-like in soluble and insoluble fractions in different tissues, coupled with a difference in some aspect of structural configuration (accounting for the higher MW) may indicate that RpL22-like has a different function in different tissues. Alternatively, the protein may have the same function in different tissues, but its subcellular distribution may be subject to specific regulation.
The absence of RpL22-like detection in an extract from heads in which eyes were excised (Figure 4A) suggests that RpL22-like may be expressed in the eye; however, removal of eyes from fly heads sometimes removes underlying brain medulla tissue as well. We were unable to resolve this issue by analysing extracts from isolated eyes due to eye pigment interference in protein fractionation and western blot analyses. Instead, we analysed the amount of RpL22-like in eyeless (ey) mutant heads where the amount of eye tissue is significantly reduced compared to wild-type, and determined that there is a significant decrease in the amount of RpL22-like in quantitatively similar amounts of head protein (Supplementary Figure S4). Resolution of RpL22-like expression in the head/eye awaits IHC analyses currently in progress.
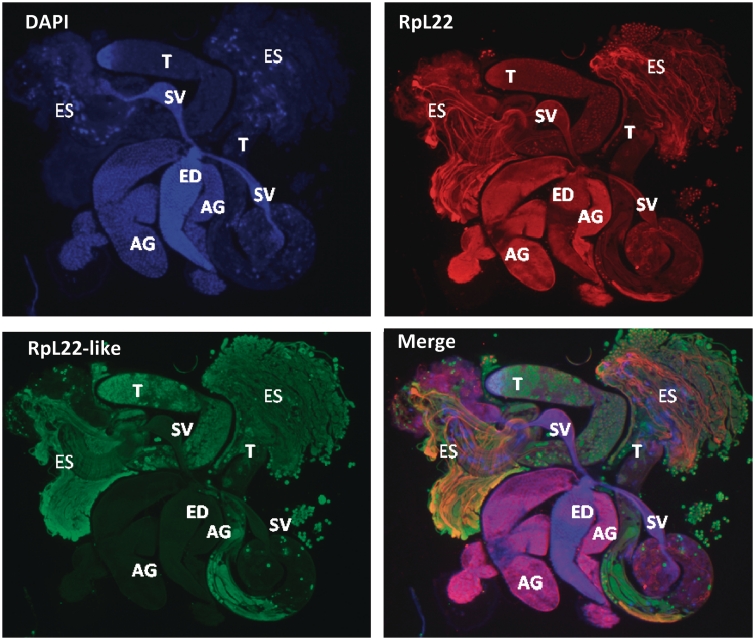
Given that RpL22-like is enriched in testis, we used IHC to confirm its presence in the testis and to determine its localization relative to RpL22 by using the same paralogue-specific Abs as were used for protein blots. Immunohistochemistry on the adult male reproductive tract confirmed that RpL22-like expression is confined to testes and is not detected within seminal vesicles, accessory glands or the ejaculatory duct (Figure 5). RpL22-like is present within all stages of sperm cells, contained within testes [see extruded sperm (ES) and testis internal contents]. In contrast, RpL22 is expressed throughout the tract, including sperm cells. A more detailed description of the staining patterns for RpL22 and RpL22-like within different cell types in testes and the reproductive tract is forthcoming in a separate report (M.G. Kearse and V.C. Ware, in preparation).
Figure 5.
Immunofluorescent staining of RpL22 and RpL22-like in adult male reproductive tract using paralogue-specific Abs. Extruded sperm (ES) were released from the testes during dissection. Spermatogenesis is initiated at the apical tip of the testis (T) and progresses through the testis coils. The most mature sperm are, therefore, located distal to the apical end of the testis. Mature sperm pass from the testis into the seminal vesicle (SV). Seminal fluid is added to sperm from the accessory gland (AG), and sperm are released through the ejaculatory duct (ED). RpL22 (red) is ubiquitously expressed in the reproductive system. RpL22-like (green) is expressed exclusively within the testis and within sperm cells. DNA is visualized by DAPI staining (blue).
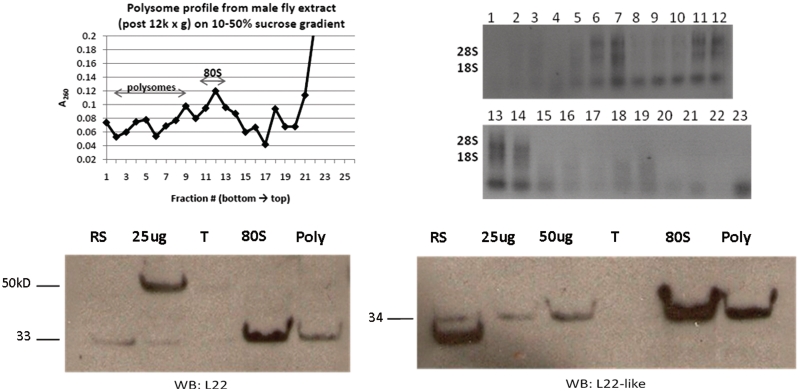
Comparison of the amino acid sequence of RpL22-like with that of RpL22 shows conservation of documented functional residues [(20); Figure 1A] though their functionality within RpL22-like has not been confirmed. Yet, we might predict that RpL22-like is an RNA binding RP competent for nucleolar import. In order to resolve this fundamental question about RpL22-like function, we performed western analysis on pooled fractions (from whole male flies) containing polysomes and 80S ribosomes isolated on 10–50% sucrose density gradients (Figure 6). Both RpL22 and RpL22-like are detected in 80S ribosome and polysome fractions, indicating that both proteins are stably associated with ribosomes. As RpL22-like is highly abundant in testis, we infer that the protein is a component of testis ribosomes. The presence of RpL22 in ribosome fractions from male extracts, however, does not verify that RpL22 functions as an RP in the testis; this determination will require additional analyses. Interestingly, only RpL22 of the expected MW is detected in ribosome and polysome fractions. The higher MW protein detected in Western blots is not an apparent component of ribosomes, suggesting that this RpL22-containing component may have a non-ribosomal role.
Figure 6.
Density gradient ultra-centrifugation showing L22-like association with active translational machinery. Soluble extracts from adult male flies were fractionated on 10–50% sucrose gradients. The top left panel shows the absorbance profile of the sucrose gradient. Extracted RNAs from fractions are shown on the top right. Fractions containing polysomes (poly) and 80S ribosomes were pooled, pelleted separately and subjected to western analysis (bottom panels). Both paralogues were detected at the expected MW in fractions containing 80S ribosomes and polysomes, indicating that both are stable components of translating ribosomes. No sizeable amount of either protein was detected at the top (T) of the gradient. Input: 25 µg and/or 50 µg whole male extract.
rpL22-like short is an alternatively spliced mRNA variant
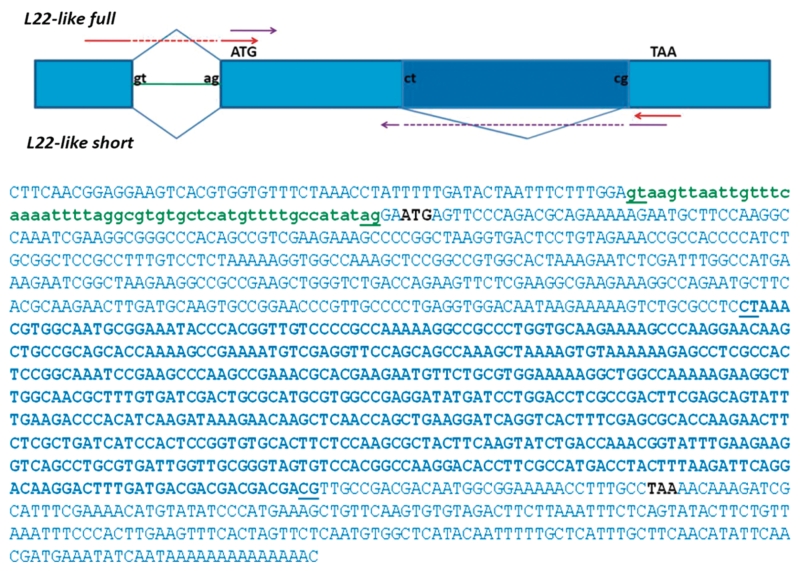
Only one annotated transcript has previously been reported in Flybase for rpL22-like; therefore, the identification of additional amplicons was not anticipated. To rule out non-specific or off-target amplification, the lower MW amplicon was cloned and sequenced. Sequence analysis of 29 different clones (2: embryo; 2: larvae; 6: testis; 19: ovary) derived from PCR products using different RNA samples and primer sets (Table 1) confirmed that the lower MW amplicon was derived from the rpL22-like gene and therefore presumed to be a previously unidentified spliced variant of rpL22-like mRNA (Figure 2). We have named this novel mRNA product ‘rpL22-like short’ to reflect its truncated structure. Its deduced sequence of 123 amino acid would consist of the fly-specific histone H1-like N-terminal extension fused in frame to the last 10 amino acid in the C-terminus (Figure 1B). The proposed protein sequence lacks 189 amino acid that comprises the majority of the conserved RpL22e family signature at the C-terminal end.
Alignment of the coding sequences for rpL22-like short (GenBank accession no. HM756190) and full-length rpL22-like (GenBank accession no. HQ190956) revealed a surprising finding that the proposed splice sites surrounding the uncharacterized intron (0.567 kb) were non-canonical in sequence [5′ splice site (SS): CT; 3′ SS: CG] compared to typical sequences found in most introns (5′ SS: GT; 3′ SS: AG), including the annotated intron within the rpL22-like gene (Figure 7). Non-canonical splice sites are indeed rare, but have been described in other eukaryotic genes as well [e.g. (25)]. In all 29 rpL22-like short sequenced clones, proposed 5′ splice donor and 3′ splice acceptor sites were identical to those shown in Figure 7, strongly supporting new intron–exon definitions within rpL22-like.
Figure 7.
RpL22-like coding region showing novel splice site junctions for rpL22 like-short. Exon sequences (capitalized) and intronic sequences (lowercase) were derived from Flybase (FBgn0034837). The coding region of rpL22-like is shown in blue caps. The previously annotated intron (FlyBase FB2010_06) is shown in green lowercase. The novel intron is represented in bold with non-canonical splice sites underlined (5′SS: CT, 3′SS: CG). The rpL22-like short sequence (GenBank accession no. HM756190) was derived from sequencing multiple (29 total) cloned cDNAs from RT-PCR analyses (Figure 2). Red and purple arrows in the splicing diagram represent primer pairs used in RT-PCR analyses in Figure 8A and Supplementary Figure S1.
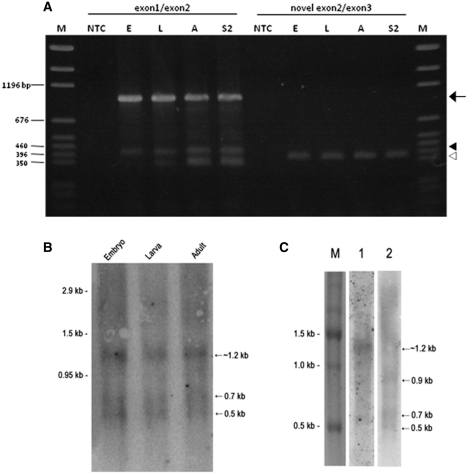
Template switching artifacts can occur in some cases where RT-PCR is performed on templates that have direct repeats flanking a proposed intron, thereby giving false signals of truncated, alternatively spliced products (26). Although direct repeats do not flank the proposed retained intron in this case, we used additional RT-PCR analyses with primers that specifically hybridize to proposed exon–exon junctions and overlap the junction by 4 nt, expecting that an rpL22-like short amplicon would be produced (using stringent conditions) only if the proposed exon–exon junction was present. In all cases, RT-PCR amplicons were consistent with expected product sizes: ∼950 bp for rpL22-like and ∼392 bp for rpL22-like short using an exon1/2 bridge primer, and ∼370 bp derived from rpL22-like short using the novel exon2/3 bridge primer (Figure 8A).
Figure 8.
(A) RT-PCR analysis of rpL22-like transcripts using primers that bridge exons in rpL22-like and rpL22-like short. RT-PCR products using the exon1/2 bridge primer should hybridize to rpL22-like and rpL22-like short, producing bands of ∼950 bp (arrow) and 392 bp (closed arrowhead), respectively (red primer set in Figure 7). All samples show such products in multiple experiments. Other amplicons (<350 bp) were cloned and sequenced and determined to be the result of non-specific amplification. The novel rpL22-like short exon2/3 bridge primer should specifically hybridize only to rpL22-like short (purple primer set in Figure 7). The expected amplicon of ∼370 bp (open arrowhead) is seen in all samples, confirming the presence of rpL22-like short. E: embryo; L: larval; A: adult; S2: S2 cells; M: pGEM marker. (B) Northern blot analysis of rpL22-like mRNAs from different developmental stages. PolyA+ RNA from embryonic, larval and adult fly stages was probed with 32P-labelled rpL22-like- specific cDNA oligomers. Estimated transcript sizes based on RNA markers are shown in kilobases (kb). rpL22-like mRNA is predicted to be 1.194 kb (Flybase). rpL22-like short mRNA is predicted to be a minimum size of ∼0.625 kb. Arrows highlight prominent transcripts detected in all stages. (C) Embryonic PolyA+RNA was probed with 32P-labelled rpL22-like-specific cDNA oligomers [(full specific—lane1) or (flanking—lane 2)]. RNA sizes were determined relative to an RNA marker.
Additional control RT-PCR experiments to rule out artifactual amplification of a shortened transcript were performed using in vitro synthesized full-length and short transcripts derived from cloned cDNA templates and flanking or bridge primer sets (Supplementary Figure S1). With flanking primers and full-length in vitro synthesized RNA, a single amplicon of ∼939 bp was generated, as expected. Notably low MW amplicons representing rpL22-like short cDNAs were not produced. Using in vitro short RNA as a template, an expected amplicon of ∼392 bp was generated. Only when bridge primer sets (novel exon2/exon3) were used with short in vitro RNA templates were amplified products of the expected size generated; no amplicons were produced with bridge primers and full-length in vitro RNA. We conclude that the low MW amplicon generated from in vivo polyA+ mRNA templates from different developmental stages as well as from various tissues is an rpL22-like mRNA variant and not an artifact of aberrant RT-PCR amplification.
Further evidence for the presence of lower MW rpL22-like mRNA isoforms was shown in northern blot analyses using embryonic, larval and/or adult polyA+ RNA and 5′ and 3′ flanking rpL22-like oligonucleotide probes that would detect both full-length and short mRNAs or an intron-specific probe that would detect full-length mRNA and the ‘retained’ intron (if sufficiently stable). It should be noted that the bridge primer used in RT-PCR experiments described above would not be expected to detect rpL22-like short mRNA exclusively since the majority of sequences in that probe would also hybridize to full-length mRNA under the standard hybridization conditions used.
Using probes that should detect both full- and short-mRNAs, we identified in each developmental stage, two prominent hybridization signals at ∼0.7 and 0.5 kb (in addition to a signal at ∼1.2 kb) that are smaller in size than the length expected for full-length rpL22-like mRNA. The 0.7 kb species falls within the range of the minimum size for an rpL22-like short transcript at ∼0.625 kb, not accounting for possible variation in polyadenylation (Figure 8B). The lower MW RNA species are less abundant than the RNA species at ∼1.2 kb (which likely represents full-length rpL22-like mRNA)—an expected quantitative result for rpL22-like short mRNA based on qRT-PCR results (Figure 3). The identity of the ∼0.5 kb species is unknown; however, it is unlikely to represent the excised intron itself since flanking rpL22-like probes would not hybridize exclusively to retained intron sequences.
Interestingly, in northern blot experiments where embryo polyA+ RNA (different sample than used in Figure 8B) was initially probed with an intron-specific probe, we detected an RNA of the size expected for full-length rpL22-like mRNA and a lower MW species that may represent the excised intron of 0.57 kb (Figure 8C). The putative intron species is less abundant than the full-length species (expected based on the relative amount of spliced variant compared to full-length mRNA from qRT-PCR data; Figure 3) and would only be detected as such if rpL22-like mRNA is alternatively spliced and the intron is stable. Alternatively, this species may represent a specific degradation product derived from rpL22-like mRNA. In general, detection of introns would be rare, unless the proposed intron encodes a stable, functional small RNA, as has been shown for small nucleolar RNAs (Psi18S-531; Psi28S-2179; Flybase) encoded by rpL22 introns.
When the same embryo RNA blot was stripped and re-probed with flanking probes, the pattern of hybridization differed from the intron-specific hybridization pattern in that the ∼0.7 and 0.5 kb species were clearly present as previously noted in the developmental RNA blot. The presumptive intron species was not apparent in this blot using flanking probes. An unidentified RNA species of ∼0.9 kb is also prominent, and may be faintly represented in all stages in the developmental blot (Figure 8B). Importantly, the northern blot data demonstrate the presence of smaller RNA species not previously predicted for rpL22-like based on genome annotation in Flybase and may represent alternatively spliced mRNA variants although we cannot conclusively discount the possibility that smaller RNA species might represent specific rpL22-like mRNA degradation products detected with our probes. When northern blot data, together with RT-PCR amplification data using in vivo and in vitro RNA templates are considered, we favour the conclusion that rpL22-like short mRNA is a bona fide transcript produced by alternative splicing of the retained intron found within rpL22-like mRNA.
rpL22-like short mRNA is associated with polysomes and may be translated
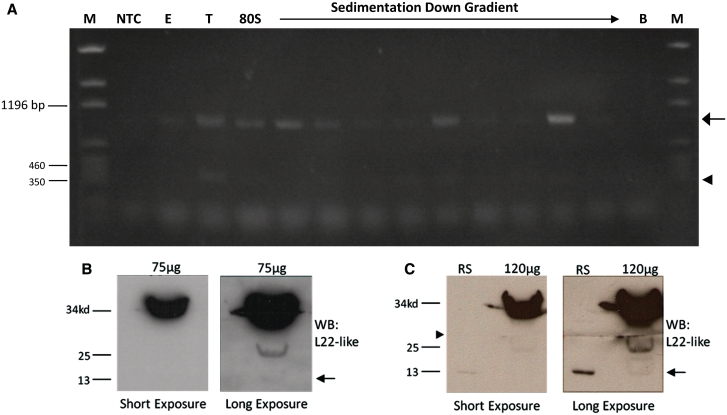
Initial western analysis of testis extracts (at protein concentrations sufficient to detect RpL22-like) did not detect a protein product of the expected MW for RpL22-like short (Figure 4A). Based on the low abundance of the rpL22-like short mRNA within testes, we speculated that a putative short protein product may be equally rare. To determine if the short mRNA might be translated, we analysed RNA isolated from polysomes (from male extracts) by RT-PCR (Figure 9A). rpL22-like short (and rpL22-like) mRNA was detected in polysomes (Figure 9A), suggesting that the short mRNA is translated.
Figure 9.
rpL22-like short mRNA may be translated. (A) Whole male extract was subjected to 10–50% linear sucrose gradient ultra-centrifugation. RT-PCR analysis of gradient fractions representing 80S subunits and polysomes shows association of rpL22-like (arrow) and rpL22-like short (arrowhead) mRNAs with active translating ribosomes, suggesting rpL22-like short is translated. (B) An initial western analysis of 75 µg of testis extract shows immunodetection at the expected MW of ∼34 kDa and lower at ∼25 and ∼13 kDa (faint band; arrow). ‘Short exposure’ was for 3 s and ‘long exposure’ was for 30 min. (C) Increased protein loading to 120 µg of testis extract enhanced immunodetection of the ∼13 kDa band (arrow) with a longer exposure (30 min compared to 4 s for short exposure). Given the abundance of RpL22-like as a ‘sink’ for antiRpL22-like Ab, the membrane was cut to maximize immunodetection of smaller MW proteins (arrowhead). The ∼13 kDa band (arrow) aligns with the recombinant RpL22-like short protein standard. M: pGEM marker; NTC: no template control; E: male extract; B: bottom of gradient; RS: recombinant standard.
Based on our qRT-PCR results that indicated a large quantitative imbalance in the two mRNA isoform levels in the testis with rpL22-like short being ∼9800-fold less abundant (Figure 3), we increased significantly (by nearly an order of magnitude) the amount of protein loaded for western analysis (100–120 µg compared with 15 µg) in an attempt to detect a protein that putatively might be RpL22-like short. With this protein loading strategy, additional immunodetection is seen at ∼25 and ∼13 kDa (Figure 9B and C). Notably, recombinant RpL22-like short migrates at the identical position with the ∼13 kDa reference band. The ∼25 kDa (and ∼13 kDa) protein may be a degradation product, only visualized with protein overloading or alternatively, the ∼25 kDa protein may be a post-translationally modified outcome of rpL22-like short expression. Whether or not these bands represent degradation products derived from RpL22-like is unclear; however, a computational investigation of proteolytic sites that would be found in the Drosophila RpL22-like amino acid sequence (FlyBase ID: FBgn0034837) does not predict degradation products of either MW (PeptideCutter, Expasy.org). A definitive resolution of this issue was not addressed in this study, but will be addressed by MALDI-TOF analysis (in progress). The relatively low abundance of the ∼13 kDa protein (estimated based on exposure times required to visualize RpL22-like short compared to RpL22-like; Figure 9B and C) is similar to what might be expected if RpL22-like short protein expression (when compared to RpL22-like expression) is approximately proportional to the amounts of rpL22-like short and rpL22-like mRNAs in testis.
DISCUSSION
RpL22-like is a tissue-specific RP
As a strategy for biochemical characterization of RpL22 from tissues in which the protein is abundant, we refined the expression pattern for rpL22-like in several tissues and developmental stages, showing that both rpL22-like mRNA and its protein product are highly enriched in adult testes compared to other tissues as analysed by qRT-PCR and western analysis, respectively. Within the male reproductive tract, RpL22-like expression is limited to testes, as no other reproductive organs showed expression, as visualized by IHC.
In a microarray study evaluating testis-specific paralogue gene expression in general, Mikhaylova et al. (27) identified RpL22 as one of 12 down-regulated RP genes. In this study, RpL22-like escaped identification as a paralogue and as an up-regulated gene in testes because its homology to RpL22 fell below the minimum 50% homology threshold. Microarray data from FlyAtlas suggest that the levels of rpL22 and rpL22-like mRNAs in testis are comparable. RpL22-like may augment a function(s) of RpL22 by providing a testis-specific ribosomal role under specific developmental or physiological conditions.
Outside of the reproductive system, RpL22-like is found within head tissue in possibly the eye (but in lower abundance compared with testes) from both sexes in a structural conformation that is distinct from the testis conformation, suggesting a possible alternative functional role for RpL22-like in the head. Although rpL22-like mRNA levels in male and female heads are low based on qRT-PCR results (Figure 3), higher levels of rpL22-like expression in the relevant tissue in the head may be masked by the abundance of other tissue types. Interestingly, the quantity of rpL22-like mRNA measured in heads and ovaries is similar (Figure 3); yet, protein expression differs considerably with no protein detected in ovaries.
We have confirmed the prediction that RpL22-like is a ribosomal component based on its co-sedimentation with gradient-purified ribosomes and polysomes from male extracts. Taken together with quantitative expression data from testis, we infer that RpL22-like is a component of testis ribosomes. The majority of RpL22-like in male extracts was found in association with ribosomes or polysomes (and little, if any, was detected at the top of gradient profiles), suggesting that RpL22-like is a more permanent ribosomal component than has been determined for RACK1 [see review by ref. (3)]. Yet, not necessarily a protein with an exclusively ribosomal function, RpL22-like may have an alternate role(s) as well. Though it is known that RpL22 is ubiquitously expressed (e.g. shown in the entire male reproductive tract by IHC), it is unclear if RpL22 functions exclusively as an RP within the testis.
Differences in RpL22-like structure compared with its paralogue RpL22 are most apparent in the N-terminal domain; however, there are also C-terminal amino acid differences that may contribute to functional differences between RpL22-like and RpL22. A small C-terminal extension of nine amino acid is apparent at the very C-terminal end of RpL22-like. The recently published 5.5 Å model of the eukaryotic 80S ribosome shows the position of RpL22 on the 60S subunit surface near its base (12). In this model, the N- and C-terminal segments of RpL22 are positioned near each other, extending from the subunit surface, which may allow for interactions with other components. If paralogue binding is mutually exclusive (though our data do not discount a model in which both RpL22 and RpL22-like are present within the same ribosome), it is reasonable to position RpL22-like similarly, yet propose that its interactions may differ.
Alternative splicing of rpL22-like through intron retention
This study demonstrated that alternative splicing of rpL22-like generates two structurally distinct mRNA isoforms that are enriched within testes. A rare novel mRNA transcript called rpL22-like short results from splicing of an intron that is retained within the more abundant, full-length rpL22-like. Basal levels of rpL22-like short mRNA are detectable by RT-PCR within heads and ovaries suggesting that the alternative splicing machinery is not limited to testis; however, the process may be subject to specific regulation within the testis where rpL22-like short mRNA levels are comparatively more abundant.
It is unknown if alternative splicing of rpL22-like occurs in all stages of spermatogenesis or if splicing is cell-type specific. The low abundance of rpL22-like short mRNA relative to the full-length variant may be consistent with a specialized role in a subset of spermatocytes. Stage-specific gene expression during fly spermatogenesis has been the subject of intense study, showing that some testis-specific genes are activated in primary spermatocytes while others (comet and cup genes) are transcribed post-meiotically, even within mid-to-late stage elongating spermatids in Drosophila [reviewed by ref. (28)]. Although unique splicing phenomena of this type have not been described in this system, stage-specific splicing in spermatogenesis remains an intriguing possibility,
Intron retention is among the rarest forms of splicing in vertebrates and invertebrates; yet, it is the most prevalent type of alternative splicing found in protozoa, fungi and plants [reviewed by ref. (29)]. This phenomenon has been described in several other Drosophila genes [e.g. Suppressor-of-white-apricot (30), c-cam3 (31), Sxl (32), erect wing (33), transformer-2 (34), nuclear export factor 1 (35)] and the consequences of retaining an intron can have profound effects on gene expression leading to an unequal accumulation of mRNA variants. For rpL22-like, the major form retains the intron.
Sakabe and de Souza (36) identified several features that support a higher incidence of intron retention: weak splice sites, genes with overall short intron lengths and higher expression levels, and specific densities of splicing regulatory elements. Non-canonical splice sites were eliminated from the analysis, although it was apparent that bona fide examples of splicing using non-canonical splice site recognition were present in their human gene data sets.
Other studies have further evaluated non-canonical splice site usage in a number of organisms (e.g. 25,37,38). Sheth et al. (38) categorized sub-types of splice sites based on U2- and U12-dependent spliceosomes and described additional rare splice site types. In at least one case in Drosophila, an intron within the rudimentary gene is defined by a CT donor SS (Flybase.org). Although originally noted in Mount et al. (37) that the fly perB intron E has a 3′SS of CG, the most recent version of Flybase (2010_08) has revised this proposal to reflect a canonical splice site.
The proposed retained intron is defined by a set of non-canonical splice site signals (5′SS: CT; 3′SS: CG) that may impact not only the mechanism of splicing, but also the kinetics of splicing regulation. It is noted that both 5′ and 3′ splice site sequence motifs are weak (MAXENT scores: −15.93 and −15.67, respectively) compared to the 5′ and 3′ scores for the upstream intron in rpL22-like (MAXENT scores: 8.57 and 7.23, respectively) when analysed using a human splice site model to predict splice site strength [MaxEntScan: (39)]. These data are consistent with an intron retention model (36) and support the conclusion that the splicing rates for the upstream intron and the retained intron are different.
Intron retention often results in premature stop codon insertion, thereby directing the alternative transcript to the nonsense mediated decay (NMD) pathway (40). The absence of premature termination codons in the short variant sequence likely eliminates this variant from NMD, and favours the interpretation that the short variant has functional significance by generating a novel protein, as proposed in this study.
Using D. melanogaster nucleotide and amino acid sequences for rpL22- like in a BLAST search for homologous sequences in other sequenced Drosophila species, we show that five other species (D. sechellia, D. simulans, D. erecta, D. ananassae and D. yakuba) contain an orthologous gene (Supplementary Figures S5 and S6). The other six species (D. pseudoobscura, D. persimilis, D. wilistoni, D. mojavensis, D. virilis and D. grimshawi) lack an rpL22-like orthologue. Within species that contain an rpL22-like orthologue, production of an rpL22-like short orthologue is also theoretically possible, generated by alternative splicing using conserved non-canonical sequences (5′SS: CT; 3′SS: CG) at the retained intron/exon boundaries (Supplementary Figures S5 and S6). Evolutionary conservation of the alternative splicing pattern in the melanogaster lineage would lend further support that the alternative transcript encodes a functional product with a structure that is generally conserved in all members of the melanogaster group except D. ananassae (see sequence alignment Supplementray Figure S6).
Protein structural diversity generated through alternative splicing of rpL22-like
Alternative splicing contributes to the enormous amount of protein diversity observed within eukaryotic cells. The rarer mRNA isoform (rpL22-like short) encodes a protein in which the majority of the C-terminal RP signature has been eliminated. Most of the structure of the short protein variant is comprised of the majority of the divergent N-terminal domain. Functional residues previously identified in RpL22 that are required for nuclear and nucleolar localization as well as RNA binding (20) are absent from this short protein variant, possibly restricting its subcellular compartmentalization and RNA binding ability. Alternatively, other regions of the protein may have redundant functions that would replace missing functional segments. In fact, a computational prediction using ELM (http://elm.eu.org/) highlights a bipartite variant of the classical NLS containing basic residues at RpL22-like short amino acid positions 90–111 (and also predicted for RpL22-like) that may prove functional for nuclear import and subnuclear compartmentalization.
Collectively, our data support the conclusion that rpL22-like encodes not only a RP, but an extra-RP as well. In silico analyses may provide clues about a putative extra-ribosomal function for the short protein. Although no specific DNA-binding motifs are apparent (41), 51 of 123 amino acid residues have DNA binding capacity, with 18 of those residues clustered within the first 24 amino acid at the very N-terminal end (42,43). Given this prediction (and its limited structural similarity to histone H1), we speculate that the short protein variant may interact with testis chromatin and have a role in chromatin repackaging and condensation during the transition from a nucleosome-based to a protamine-based configuration, as occurs in maturing sperm cells during spermiogenesis [e.g. reviewed by ref. (44)].
Perspectives on RpL22e paralogue function and evolution
Paralogous members of the RpL22e family have been described in other animal genomes [e.g. rpL22-like1 in Mus musculus (NP_080793.1), Danio rerio (NP_001038800), Xenopus tropicalis (Q5I0R6), Homo sapiens (AAH62731), Rattus norvegicus (NP_001102018.1)] and their tissue-specific expression often varies considerably compared with the fly pattern described here (45). Relatively little is known about functional redundancy or specificity of other rpL22e paralogues in other species; however, in at least one case, an rpL22 knockout mouse only exhibited a mild phenotype in T cell development, but was otherwise viable and fertile (46), suggesting that mouse paralogue rpL22-like1 could rescue critical functions lost by rpL22 disruption.
Proteins of the RpL22e family in Caenorhabditis elegans [RpL22; (47)] and Drosophila are essential [RpL22: (48,49)]. However, this is not the case for RpL22 in yeast (50) or in mice as discussed above (46). Flybase reports that a P-element chromosomal insertion located 150 nts upstream of rpL22-like is lethal in fly development, suggesting that the rpL22-like gene is also essential. The nature of the essential RpL22-like function within the fly is intriguing, since one would predict that disruption of RP function in the testis would affect male fertility, and not viability. This suggests that rpL22-like has an essential role (ribosomal or non-ribosomal) in another tissue(s) (head or eye) at some stage in fly development. In this case, it appears that RpL22 cannot replace the function lost by RpL22-like disruption, either because of a lack of co-expression in the appropriate tissue in time and space or because the role of RpL22-like is unique. Similarly, RpL22-like is unable to replace functions lost by RpL22 disruption, correspondingly suggesting that essential functions prescribed by each protein are not redundant.
Do the essential roles of these paralogues include non-ribosomal roles? The presence of anti-L22-like and anti-L22 immunoreactive products in high-MW complexes within eye tissue and the testis, respectively, raises the possibility that each paralogue may be post-translationally modified or bound within detergent-resistant complexes and function in a role(s) that is distinct from its ribosomal role, since neither high-MW species was found in association with ribosomes.
From an evolutionary perspective, gene duplication is the likely mechanism by which paralogous genes arise and generate tissue- or lineage-specific genes [e.g. (51)]. Several inferences about RpL22e family evolutionary history become apparent from genomic analyses of the Drosophilidae. We and others (8) propose that rpL22 is the ancestral gene that was duplicated in a series of complex events whose extant outcome is the ubiquitous expression of rpL22 and cell lineage-specific expression of rpL22-like. The rpL22-like gene is only found in the melanogaster group. One hypothesis addressing why obscura lineage genomes lack the rpL22-like gene requires that gene duplication occurred in the melanogaster group, allowing for essential functions to be shared between paralogues. Over time gene divergence may have directed changes in paralogue function. The absence of the rpL22-like gene from the obscura lineage may indicate that ‘essential’ functions were retained in the ancestral rpL22 gene or are provided by another gene(s). No doubt comparative analyses of paralogue expression and function in different Drosophila species will be instrumental in providing further insights into the evolutionary history and functional diversity displayed within the RpL22e family.
ACCESSION NUMBERS
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Lehigh University Faculty Research Grants (607233, 607277 to V.W., partial); Pennsylvania Department of Health (Block Grant). The work described here is in partial fulfilment of the requirements for the PhD degree for M.K., who is a Nemes Fellow. Alex Chen is an undergraduate research assistant, supported in part as a Lehigh University-Howard Hughes Medical Institute student from a grant to Lehigh University from the Howard Hughes Medical Institute through the Precollege and Undergraduate Science Education Programme. Funding for open access charge: Lehigh University Faculty Research Grant.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Dr Anna Gumpert for microscopy assistance, Dr Lynne Cassimeris for Alexa 2° Ab, Kandiss Schrader for S2 cells, Lee Graham (Lehigh) and Todd Laverty (HHMI-JFRC) for flies, Dr Brian Chen for advice about proteomics tools, and Maria Brace for figure preparation assistance. Dr Michael Kuchka and Dr Barry Bean are acknowledged for stimulating discussions about the work reported here. Dr Michael Kuchka and Dr Lee Hughes are acknowledged for comments about the manuscript.
REFERENCES
- 1.McIntosh KB, Bonham-Smith PC. Ribosomal protein gene regulation: what about plants? Can. J. Bot. 2006;84:342–362. [Google Scholar]
- 2.Sugihara Y, Honda H, Iida T, Morinaga T, Hino S, Okajima T, Matsuda T, Nadano D. Proteomic analysis of rodent ribosomes revealed heterogeneity including ribosomal proteins L10-like, L22-like 1, and L39-like. J. Proteome Res. 2010;9:1351–1366. doi: 10.1021/pr9008964. [DOI] [PubMed] [Google Scholar]
- 3.Warner JR, McIntosh KB. How common are extraribosomal functions of ribosomal proteins? Mol. Cell. 2009;34:3–11. doi: 10.1016/j.molcel.2009.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Komili S, Farny NG, Roth FP, Silver PA. Functional specificity among ribosomal proteins regulates gene expression. Cell. 2007;131:557–571. doi: 10.1016/j.cell.2007.08.037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kim TY, Ha CW, Huh WK. Differential subcellular localization of ribosomal protein L7 paralogs in Saccharomyces cerevisiae. Mol. Cells. 2009;27:539–546. doi: 10.1007/s10059-009-0077-0. [DOI] [PubMed] [Google Scholar]
- 6.Marygold SJ, Roote J, Reuter G, Lambertsson A, Ashburner M, Millburn GH, Harrison PM, Yu Z, Kenmochi N, Kaufman TC, et al. The ribosomal protein genes and Minute loci of Drosophila melanogaster. Genome Biol. 2007;8:R216. doi: 10.1186/gb-2007-8-10-r216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koyama Y, Katagiri S, Hanai S, Uchida K, Miwa M. Poly(ADP-ribose) polymerase interacts with novel Drosophila ribosomal proteins, L22 and L23a, with unique histone-like amino-terminal extensions. Gene. 1999;226:339–345. doi: 10.1016/s0378-1119(98)00529-0. [DOI] [PubMed] [Google Scholar]
- 8.Shigenobu S, Arita K, Kitadate Y, Noda C, Kobayashi S. Isolation of germline cells from Drosophila embryo by flow cytometry. Dev. Growth Differ. 2006;48:49–57. doi: 10.1111/j.1440-169X.2006.00845.x. [DOI] [PubMed] [Google Scholar]
- 9.Shigenobu S, Kitadate Y, Noda C, Kobayashi S. Molecular characterization of embryonic gonads by gene expression profiling in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 2006;103:13728–13733. doi: 10.1073/pnas.0603767103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kai T, William D, Spradling AC. The expression profile of purified Drosophila germline stem cells. Dev. Biol. 2005;283:486–502. doi: 10.1016/j.ydbio.2005.04.018. [DOI] [PubMed] [Google Scholar]
- 11.Chintapalli VR, Wang J, Dow JA. Using FlyAtlas to identify better Drosophila melanogaster models of human disease. Nat. Genet. 2002;39:715–720. doi: 10.1038/ng2049. [DOI] [PubMed] [Google Scholar]
- 12.Armache J-P, Jarasch A, Anger AM, Villa E, Becker T, Bhushan S, Jossinet F, Habeck M, Dindar G, Franckenberg S, et al. Localization of eukaryote-specific ribosomal proteins in a 5.5-Å cryo-EM map the 80S eukaryotic ribosome. Proc. Natl Acad. Sci. USA. 2010;107:19754–19759. doi: 10.1073/pnas.1010005107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lavergne JP, Conquet F, Reboud JP, Reboud AM. Role of acidic phosphoproteins in the partial reconstitution of the active 60 S ribosomal subunit. FEBS Lett. 1987;216:83–88. doi: 10.1016/0014-5793(87)80761-5. [DOI] [PubMed] [Google Scholar]
- 14.Ni J-Q, Liu L-P, Hess D, Rietdorf J, Sun F-L. Drosophila ribosomal proteins are associated with linker histone H1 and suppress gene transcription. Genes Dev. 2006;20:1959–1973. doi: 10.1101/gad.390106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Crosby MA, Goodman JL, Strelets VB, Zhang P, Gelbart WM FlyBase Consortium. FlyBase: genomes by the dozen. Nucleic Acids Res. 2007;35(Database issue):D486–D491. doi: 10.1093/nar/gkl827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning: A Laboratory Manual. NY: Cold Spring Harbor Laboratory Press; 1989. [Google Scholar]
- 17.Hime GR, Brill JA, Fuller MT. Assembly of ring canals in the male germ line from structural components of the contractile ring. J. Cell Sci. 1996;109:2779–2788. doi: 10.1242/jcs.109.12.2779. [DOI] [PubMed] [Google Scholar]
- 18.Tazuke SI, Schulz C, Gilboa L, Fogarty M, Mahowald AP, Guichet A, Ephrussi A, Wood CG, Lehmann R, Fuller MT. A germline-specific gap junction protein required for survival of differentiating early germ cells. Development. 2002;129:2529–2539. doi: 10.1242/dev.129.10.2529. [DOI] [PubMed] [Google Scholar]
- 19.Qin X, Ahn S, Speed TP, Rubin GM. Global analyses of mRNA translational control during early Drosophila embryogenesis. Genome Biol. 2007;8:R63. doi: 10.1186/gb-2007-8-4-r63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Houmani JL, Ruf IK. Clusters of basic amino acids contribute to RNA binding and nucleolar localization of ribosomal protein L22. PLoS ONE. 2009;4:e5306. doi: 10.1371/journal.pone.0005306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Pelczar P, Filipowicz W. The host gene from intronic U17 small nucleolar RNAs in mammals has no protein-coding potential and is a member of the 5′-terminal oligopyrimidine gene family. Mol. Cell. Biol. 1998;18:4509–4518. doi: 10.1128/mcb.18.8.4509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kemphues KJ, Raff RA, Kaufman TC, Raff EC. Mutation in a structural gene for a beta-tubulin specific to testis in Drosophila melanogaster. Proc. Natl Acad. Sci. USA. 1979;76:3991–3995. doi: 10.1073/pnas.76.8.3991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sano H, Nakamura A, Kobayashi S. Identification of a transcriptional regulatory region for germline-specific expression of vasa gene in Drosophila melanogaster. Mech. Dev. 2002;112:129–139. doi: 10.1016/s0925-4773(01)00654-2. [DOI] [PubMed] [Google Scholar]
- 24.Rudolph JE, Kimble M, Hoyle HD, Subler MA, Raff EC. Three Drosophila beta-tubulin sequences: a developmentally regulated isoform (beta 3), the testis-specific isoform (beta 2), and an assembly-defective mutation of the testis-specific isoform (B2t8) reveal both an ancient divergence in metazoan isotypes and structural constraints for beta-tubulin function. Mol. Cell Biol. 1987;7:2231–2242. doi: 10.1128/mcb.7.6.2231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Burset M, Seledtsov IA, Solovyev VV. Analysis of canonical and non-canonical splice sites in mammalian genomes. Nucleic Acids Res. 2000;28:4364–4375. doi: 10.1093/nar/28.21.4364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cocquet J, Chong A, Zhang G, Veitia RA. Reverse transcriptase template switching and false alternative transcripts. Genomics. 2006;88:127–131. doi: 10.1016/j.ygeno.2005.12.013. [DOI] [PubMed] [Google Scholar]
- 27.Mikhaylova LM, Nguyen K, Nurminsky DI. Analysis of the Drosophila melanogaster testes transcriptome reveals coordinate regulation of paralogous genes. Genetics. 2008;179:305–315. doi: 10.1534/genetics.107.080267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.White-Cooper H. Molecular mechanisms of gene regulation during Drosophila spermatogenesis. Reproduction. 2010;139:11–21. doi: 10.1530/REP-09-0083. [DOI] [PubMed] [Google Scholar]
- 29.Keren H, Lev-Maor G, Ast G. Alternative splicing and evolution: diversification, exon definition and function. Nat. Rev. Genet. 2010;11:345–355. doi: 10.1038/nrg2776. [DOI] [PubMed] [Google Scholar]
- 30.Zachar Z, Chou TB, Bingham PM. Evidence that a regulatory gene autoregulates splicing of its transcript. EMBO J. 1987;6:4105–4111. doi: 10.1002/j.1460-2075.1987.tb02756.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Cheung PH, Culic O, Qiu Y, Earley K, Thompson N, Hixson DC, Lin SH. The cytoplasmic domain of C-CAM is required for C-CAM-mediated adhesion function: studies of a C-CAM transcript containing an unspliced intron. Biochem. J. 1993;295:427–435. doi: 10.1042/bj2950427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Samuels ME, Schedl P, Cline TW. The complex set of late transcripts from the Drosophila sex determination gene sex-lethal encodes multiple related polypeptides. Mol. Cell Biol. 1991;11:3584–3602. doi: 10.1128/mcb.11.7.3584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Koushika SP, Soller M, DeSimone SM, Daub DM, White K. Differential and inefficient splicing of a broadly expressed Drosophila erect wing transcript results in tissue-specific enrichment of the vital EWG protein isoform. Mol. Cell Biol. 1999;19:3998–34007. doi: 10.1128/mcb.19.6.3998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mattox W, Baker BS. Autoregulation of the splicing of transcripts from the transformer-2 gene of Drosophila. Genes Dev. 1991;5:786–796. doi: 10.1101/gad.5.5.786. [DOI] [PubMed] [Google Scholar]
- 35.Ivankova N, Tretyakova I, Lyozin GT, Avanesyan E, Zolotukhin A, Zatsepina OG, Evgen'ev MB, Mamon LA. Alternative transcripts expressed by small bristles, the Drosophila melanogaster nxf1 gene. Gene. 2010;458:11–19. doi: 10.1016/j.gene.2010.02.013. [DOI] [PubMed] [Google Scholar]
- 36.Sakabe NJ, de Souza SJ. Sequence features responsible for intron retention in human. BMC Genomics. 2007;8:59. doi: 10.1186/1471-2164-8-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Mount SM, Burks C, Hertz G, Stormo GD, White O, Fields C. Splicing signals in Drosophila: intron size, information content, and consensus sequences. Nucleic Acids Res. 1992;20:4255–4262. doi: 10.1093/nar/20.16.4255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Sheth N, Roca X, Hastings ML, Roeder T, Krainer AR, Sachidanandam R. Comprehensive splice-site analysis using comparative genomics. Nucleic Acids Res. 2006;34:3955–3967. doi: 10.1093/nar/gkl556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yeo G, Burge CB. Maximum entropy modeling of short sequence motifs with applications to RNA splicing signals. J. Comput. Biol. 2004;11:377–394. doi: 10.1089/1066527041410418. [DOI] [PubMed] [Google Scholar]
- 40.Lareau LF, Green RE, Bhatnagar RS, Brenner SE. The evolving roles of alternative splicing. Curr. Opin. Struct. Biol. 2004;14:273–282. doi: 10.1016/j.sbi.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 41.Wilson D, Charoensawan V, Kummerfeld SK, Teichmann SA. DBD – taxonomically broad transcription factor predictions: new content and functionality. Nucleic Acids Res. 2008;36(Database issue):D88–D92. doi: 10.1093/nar/gkm964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hwang S, Gou Z, Kuznetsov IB. DP-Bind: a web server for sequence-based prediction of DNA-binding residues in DNA-binding proteins. Bioinformatics. 2007;23:634–636. doi: 10.1093/bioinformatics/btl672. [DOI] [PubMed] [Google Scholar]
- 43.Kuznetsov IB, Gou Z, Li R, Hwang S. Using evolutionary and structural information to predict DNA-binding sites on DNA-binding proteins. Proteins. 2006;64:19–27. doi: 10.1002/prot.20977. [DOI] [PubMed] [Google Scholar]
- 44.Hennig W. Chromosomal proteins in the spermatogenesis of Drosophila. Chromosoma. 2003;111:489–494. doi: 10.1007/s00412-003-0236-6. [DOI] [PubMed] [Google Scholar]
- 45.Bastian F, Parmentier G, Roux J, Moretti S, Laudet V, Robinson-Rechavi M. Bgee: integrating and comparing heterogeneous transcriptome data among species. DILS: LNCS. 2008;5109:124–131. [Google Scholar]
- 46.Anderson SJ, Lauritsen JP, Hartman MG, Foushee AM, Lefebvre JM, Shinton SA, Gerhardt B, Hardy RR, Oravecz T, Wiest DL. Ablation of ribosomal protein L22 selectively impairs alphabeta T cell development by activation of a p53-dependent checkpoint. Immunity. 2007;26:759–772. doi: 10.1016/j.immuni.2007.04.012. [DOI] [PubMed] [Google Scholar]
- 47.Kamath RS, Fraser AG, Dong Y, Poulin G, Durbin R, Gotta M, Kanapin A, Le Bot N, Moreno S, Sohrmann M, et al. Systematic functional analysis of the Caenorhabditis elegans genome using RNAi. Nature. 2003;421:231–237. doi: 10.1038/nature01278. [DOI] [PubMed] [Google Scholar]
- 48.Bourbon HM, Gonzy-Treboul G, Peronnet F, Alin MF, Ardourel C, Benassayag C, Cribbs D, Deutsch J, Ferrer P, Haenlin M, et al. A P-insertion screen identifying novel X-linked essential genes in Drosophila. Mech. Dev. 2002;110:71–83. doi: 10.1016/s0925-4773(01)00566-4. [DOI] [PubMed] [Google Scholar]
- 49.Boutros M, Kiger AA, Armknecht S, Kerr K, Hild M, Koch B, Haas SA, Paro R, Perrimon N Heidelberg Fly Array Consortium. Genome-wide RNAi analysis of growth and viability in Drosophila cells. Science. 2004;303:832–835. doi: 10.1126/science.1091266. [DOI] [PubMed] [Google Scholar]
- 50.Deutschbauer AM, Jaramillo DF, Proctor M, Kumm J, Hillenmeyer ME, Davis RW, Nislow C, Giaever G. Mechanisms of haploinsufficiency revealed by genome-wide profiling in yeast. Genetics. 2005;169:1915–1925. doi: 10.1534/genetics.104.036871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Copley RR, Goodstadt L, Ponting C. Eukaryotic domain evolution inferred from genome comparisons. Curr. Opin. Genet. Dev. 2003;13:623–628. doi: 10.1016/j.gde.2003.10.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.