Abstract
Sld2 is essential for the initiation of DNA replication, but the mechanism underlying its role in replication is not fully understood. The S-phase cyclin dependent kinase (S-CDK) triggers the association of Sld2 with Dpb11, and a phosphomimetic mutation of Sld2, Sld2T84D, functionally mimics the S-CDK phosphorylated state of Sld2. We report that Sld2T84D binds directly to the single-stranded (ss) DNA of two different origins of replication, and S-CDK phosphorylation of Sld2 stimulates the binding of Sld2 to origin ssDNA. Sld2T84D binds to a thymine-rich ssDNA region of the origin ARS1, and substitution of ARS1 thymines with adenines completely disrupts binding of Sld2T84D. Sld2T84D enhances the ability of origin ssDNA to pulldown Dpb11, and Sld2 binding to origin ssDNA may be important to allow Sld2 and Dpb11 to associate with origin DNA. We also report that Sld2T84D anneals ssDNA of an origin sequence. Dpb11 anneals ssDNA to low levels, and the addition of Sld2T84D with Dpb11 results in higher annealing activity than that of either protein alone. Sld2-stimulated annealing may be important for maintaining genome stability during the initiation of DNA replication.
INTRODUCTION
Sld2 is required for the initiation of DNA replication in budding yeast cells (1,2). Temperature sensitive mutations of SLD2 are defective for the initiation of DNA replication and S-phase progression at the restrictive temperature (1,2). High copy SLD2 suppresses the growth defect caused by a mutation of dpb11 (dpb11-1), and high copy DPB11 suppresses the growth defect of a mutation of sld2 (sld2-6) (1). These mutations of dpb11 (dpb11-1) and sld2 (sld2-6) are also defective in DNA replication (1). Yeast two-hybrid and immunoprecipitation assays show that Dpb11 interacts physically with Sld2 specifically during S phase (1,3). These results suggest that Sld2 and Dpb11 form a complex in vivo that is necessary for the initiation of DNA replication. Furthermore, a recent study demonstrates that Sld2 forms a transient complex with Dpb11, GINS and polε called the pre-loading complex (4). However, the function of Sld2 at origins is not well understood.
Dpb11 and its functional homologs Cut5, Mus101 and TopBP1 are essential for DNA replication in eukaryotic cells (5–8). Dpb11 was first cloned as a multi-copy suppressor of a temperature-sensitive mutant in a polymerase ε subunit, Dpb2, and was also found to suppress mutants in the catalytic subunit (Pol2) of Polε (7). Dpb11 associates preferentially with DNA fragments containing autonomously replicating sequences during S phase at the same time as polε, suggesting that Dpb11 co-localizes with polε at replication origins (9). There are also data suggesting that Dpb11 may interact with GINS (10), and that Xenopus TopBP1 (Mus101) is required for Cdc45 recruitment to origins (6). However, unlike polε, chromatin immunoprecipitation experiments show that Dpb11 does not migrate with the replication fork (9). Dpb11 also has a cell-cycle checkpoint function (11–13), and dpb11 mutants exhibit sensitivity to replication stress and DNA-damaging agents (2,7).
At the onset of S phase, the S-phase cyclin-dependent kinase (Clb5, 6-Cdc28 in budding yeast) is essential for chromosomal DNA replication (14,15). S-phase cyclin dependent kinase (S-CDK) phosphorylates Sld2 in yeast cells, and S-CDK phosphorylation of Sld2 is critical for the interaction of Sld2 with Dpb11 and for DNA replication initiation (3). S-CDK phosphorylation of one residue of Sld2, Sld2T84, is critical for the interaction between Sld2 and Dpb11, and the phosphomimetic mutant Sld2T84D can productively bind to Dpb11 in vivo 18). Interestingly, when the Sld2T84D mutation is combined with a fusion of Sld3 with Dpb11 (17), the cellular requirement for S-CDK activity is bypassed. Thus, the mutant Sld2T84D mimics S-CDK phosphorylation of Sld2, and Sld2T84D can help bypass the cellular requirement for S-CDK activity.
The N-terminal region of Sld2 is homologous to the N-terminal region of Xenopus and human RecQ4 (19,20). In Xenopus egg extracts, RecQ4 is essential for the initiation of DNA replication, in particular for chromatin binding of DNA polymerase-α (19). It is currently not clear if RecQ4 is a functional homolog of Sld2, since amino conservation is limited to the N-terminal region. Sld2 does not have any ATPase or helicase motifs, unlike RecQ4.
We report in this article that Sld2T84D binds directly to single-stranded (ss) DNA found at the ARS1 origin, and it also binds to ssDNA from the origin ARS305. The T84D phosphomimetic mutation stimulates the binding of Sld2 to ssDNA, and S-CDK phosphorylation of Sld2 stimulates the binding of Sld2 to ssDNA as well. ssARS1-1 is thymine rich, and substitution of the ssARS1-1 thymines with adenines completely disrupts binding to Sld2T84D. Replication origins in budding yeast contain a thymine-rich strand (21). Sld2T84D stimulates the ability of origin ssDNA to pulldown Dpb11; thus, Sld2 binding to origin DNA may be important for the association of Sld2 and Dpb11 with origin DNA. We also find that Sld2T84D anneals ssDNA containing an origin sequence. Dpb11 alone also anneals ssDNA to low levels, and the combination of Sld2T84D and Dpb11 results in higher dsDNA formation than that achieved by either Sld2T84D or Dpb11 alone. Sld2-annealing activity at an origin may limit the accumulation of ssDNA at an origin of replication, thereby preserving genomic stability.
MATERIALS AND METHODS
Cloning and purification of proteins
Cloning and purification of GST–Sld2 and GST–Sld2T84D
The full-length open reading frame of Saccharomyces cerevisiae Sld2 was cloned into a pET-41a vector. The Sld2-pET vector was transformed into Escherichia coli BL-21 DE3 Codon Plus cells and a colony from the transformation was inoculated into 12 liters of auto-induction media (22) containing 100 µg/ml kanamycin. When the cells reached an OD600 of 0.6, the temperature was lowered to 14°C. Cells were harvested 16 h later, and lysed by French Press in 200 ml of solution containing 500 mM NaCl, 10 mM imidazole and 50 mM Na2HPO4, pH 8.0. The lysate was applied to a 10 ml chelating sepharose fast flow resin (GE Healthcare) pre-charged with nickel sulfate. The column was washed with solution containing 500 mM NaCl, 50 mM imidazole and 50 mM Na2HPO4, pH 8.0. The GST–Sld2 protein was eluted in solution containing 10% glycerol, 500 mM NaCl, 250 mM imidazole and 50 mM Na2HPO4, pH 8.0, and incubated 1 h with gentle shaking at 4°C with 2 ml glutathione agarose (Sigma). The glutathione agarose was collected in a micro bio-spin chromatography column (Bio-Rad) and the column was washed with a solution containing 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 0.1 mg/ml BSA and 20 mM Tris–HCl, pH 7.5. The GST–Sld2 protein was eluted in solution containing 10% glycerol, 500 mM NaCl, 0.1 mM EDTA, 1 mM DTT, 500 mM glutathione and 20 mM Tris–HCl, pH 7.5, and flash frozen.
The threonine to aspartate substitution at position 84 was introduced in full-length open reading frame of Sld2 by PCR stitching, and then cloned into the pET vector. Purification of GST–Sld2T84D was similar to the purification of GST–Sld2 described above.
Cloning and purification of GST–Dpb11
The full-length open reading of S. cerevisiae Dpb11 was cloned into a pET-41a vector. The Dpb11-pET vector was transformed into E. coli BL-21 DE3 Codon Plus cells, and GST–Dpb11 was purified as described above for GST–Sld2.
Cloning and purification of native Sld2 and Sld2T84D
The full-length open reading frame of S. cerevisiae Sld2 was cloned into the NdeI/HindIII sites of the pET-33b vector. The Sld2–pET-33b vector was transformed into E. coli BL-21 DE3 Codon Plus cells and a colony from the transformation was inoculated into 12 liters of auto-induction media (22) containing 100 µg/ml kanamycin. When the cells reached an OD600 of 0.6, the temperature was lowered to 14°C. Cells were harvested 16 h later, and lysed by French Press in 200 ml of solution containing 500 mM NaCl, 10 mM imidazole and 50 mM Na2HPO4, pH 8. The lysate was applied to a 10 ml chelating sepharose fast flow resin (GE Healthcare) pre-charged with nickel sulfate. The column was washed with solution containing 500 mM NaCl, 50 mM imidazole and 50 mM Na2HPO4, pH 8. The Sld2 protein was eluted in solution containing 10% glycerol, 500 mM NaCl, 250 mM imidazole and 50 mM Na2HPO4, pH 8. The his-tag was cleaved, and the sample buffer was then exchanged via Amicon Ultra Centrifugal Units (Millipore) into 2 ml buffer containing 100 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol and then the sample was subjected to FPLC and applied to 124 ml sephadex 6 resin. Size exclusion chromatography was carried out in buffer containing 250 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol. Following size exclusion chromatography, the sample containing Sld2 was diluted in 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol until conductivity matched that of buffer containing 50 mM NaCl. The sample was then applied to 2 ml fast flow Q sepharose (Pharmacia) and washed with solution containing 50 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol. Sld2 was then eluted in solution containing 1 M NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol. The sample was then dialyzed in buffer containing 100 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol.
The threonine to aspartate substitution at position 84 was introduced in full-length open reading frame of Sld2 by PCR stitching, and then cloned into the NdeI/HindIII sites of the pET-33b vector. Purification of Sld2T84D was identical to the purification of Sld2 described above.
Cloning and purification of native Dpb11
The full-length open reading frame of S. cerevisiae Dpb11 was cloned into the NdeI/XhoI site of the pET-33b vector. The Dpb11–pET-33b vector was transformed into E. coli BL-21 DE3 Codon Plus cells and a colony from the transformation was inoculated into 12 liters of auto-induction media (Studier, 2005 #721) containing 100 µg/ml kanamycin. When the cells reached an OD600 of 0.6, the temperature was lowered to 14°C. Cells were harvested 16 h later, and lysed by French Press in 200 ml of solution containing 500 mM NaCl, 10 mM imidazole and 50 mM Na2HPO4, pH 8. The lysate was applied to a 10 ml chelating sepharose fast flow resin (GE Healthcare) pre-charged with nickel sulfate. The column was washed with solution containing 500 mM NaCl, 50 mM imidazole and 50 mM Na2HPO4, pH 8. The Dpb11 protein was eluted in solution containing 10% glycerol, 500 mM NaCl, 250 mM imidazole and 50 mM Na2HPO4, pH 8. The his-tag was cleaved, and the sample buffer was then exchanged via Amicon Ultra Centrifugal Units (Millipore) into 2 ml buffer containing 100 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol and then the sample was subjected to FPLC and applied to 124 ml sephadex 6 resin. Size exclusion chromatography was carried out in buffer containing 250 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol. Following size exclusion chromatography, the sample containing Dpb11 was diluted in 0 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol until the conductivity matched that of buffer containing 50 mM NaCl. The sample was then applied to 0.5 ml phospho-cellulose resin and washed with solution containing 50 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol. Dpb11 was then eluted in solution containing 200 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol. The sample was then dialyzed in buffer containing 100 mM NaCl, 20 mM Tris pH 7.5, 1 mM DTT and 10% glycerol.
S-CDK was purified as previously described (16).
Radiolabeling DNA
DNA was end labeled with T4 polynucleotide kinase (NEB) according to the manufacturer’s instructions. For double stranded (ds) DNA substrates requiring annealing, 500 nM radiolabeled DNA was incubated overnight at 37°C with 1 µM complementary DNA in 20 mM Tris–HCl, 4% glycerol, 0.1 mM EDTA, 40 µg/ml BSA, 5 mM DTT and 5 mM magnesium acetate in a final volume of 12 µl. Following the overnight incubation, the reaction was diluted to a final concentration of 50 nM (concentration of radiolabeled DNA) with 20 mM Tris–HCl, 0.1 mM EDTA.
GST–Sld2 pulldown of radiolabeled DNA
GST (13 pmol), GST–Sld2 or GST–Sld2T84D attached to glutathione agarose beads were incubated 5 min at 30°C with increasing amounts of radiolabeled DNA in a solution containing 0.1 mM EDTA, 0.2 mM DTT, 10 mM magnesium acetate, 10% glycerol, 40 µg/ml BSA and 20 mM Tris–HCl pH 7.5 in a final volume of 10 µl. Following the 5 min incubation the reactions were shifted to room temperature, the glutathione agarose beads were allowed to settle and the supernatant was removed. The beads were washed twice with a solution containing 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 0.1 mg/ml BSA and 20 mM Tris–HCl, pH 7.5. The beads were allowed to settle, the supernatant was removed and the beads were boiled at 95°C for 10 min in a solution containing 2% SDS, 2 mM DTT, 4% glycerol, 4 mM Tris–HCl and 0.01% bromophenol blue. The reactions were analyzed by SDS/PAGE. The gel was dried for 1 h at 80°C and exposed to a phosphorimaging screen for 1 h. Results shown are the mean plus/minus SEM for three or more experiments, and data were combined if the substrate analyzed is the same in more than one figure (e.g. ssARS1-1 is analyzed in Figures 1 and 5).
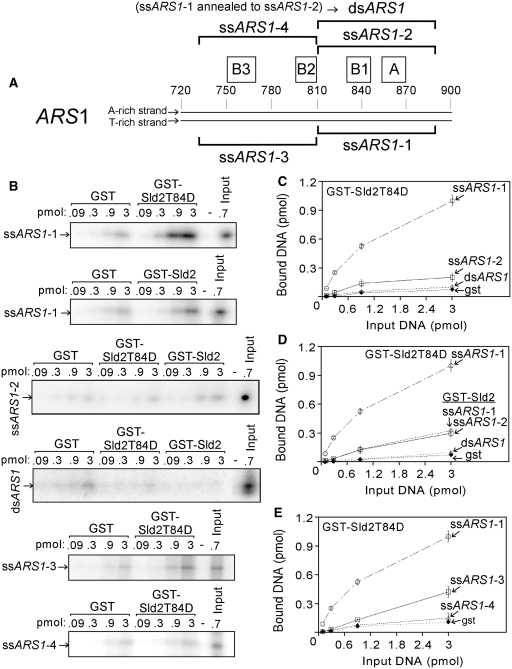
Figure 1.
Sld2T84D binds to ssARS1. (A) Schematic of ARS1. The complete nucleotide sequences are found in Supplementary Table S1. (B) Purified GST–Sld2T84D, GST–Sld2 or GST were studied for interaction with radiolabeled DNA as indicated to the left of the gel. GST–Sld2T84D, GST–Sld2 or GST (13 pmol) and varying amounts of DNA, as indicated in the figure, were mixed with 1.3 mg glutathione agarose beads and incubated for 5 min at 30°C. The beads were washed and analyzed as described in ‘Materials and Methods’ section. The results from experiments similar to (B) were quantified and plotted as pmol of DNA bound versus pmol of input DNA (C, D and E). Sld2T84D binds tighter to ssARS1-1 than to either ssARS1-2 or dsARS1. Sld2T84D binds tighter than Sld2 to ssARS1-1.
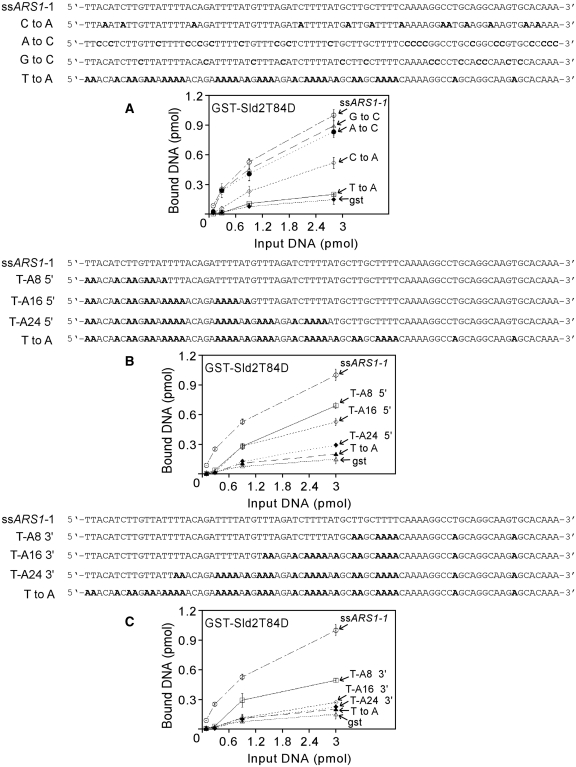
Figure 5.
Sld2T84D binding to ssARS1-1 is disrupted by substituting the thymines with adenines. (A–C) Sequence of ssARS1-1 and modifications of ssARS1-1. Purified GST–Sld2T84D or GST was studied for interaction with radiolabeled DNA as indicated on the graph. GST-Sld2T84D or GST (13 pmol) and varying amounts of DNA, as indicated in the figure, were mixed with 1.3 mg glutathione agarose beads and incubated for 5 min at 30°C. The beads were washed and analyzed as described in ‘Materials and Methods’ section. The results were quantified and plotted as pmol of DNA bound versus pmol of input DNA. (A) A substitution of T-to-A completely abolishes binding of ssARS1-1 to Sld2T84D. (B, C) Sld2T84D binds to multiple regions of ssARS1-1.
GST–Dpb11 pulldown of radiolabled Sld2T84D
GST or GST–Dpb11 (40 pmol) attached to glutathione agarose beads were incubated 5 min at 30°C with increasing amounts of radiolabeled Sld2T84D in a solution containing 0.1 mM EDTA, 0.2 mM DTT, 10 mM magnesium acetate, 10% glycerol, 40 µg/ml BSA and 20 mM Tris–HCl pH 7.5 in a final volume of 10 µl. Following the 5 min incubation the reactions were shifted to room temperature, the glutathione agarose beads were allowed to settle and the supernatant was removed. The beads were washed twice with a solution containing 100 mM NaCl, 0.1 mM EDTA, 10% glycerol, 0.1% Triton X-100, 1 mM DTT, 0.1 mg/ml BSA and 20 mM Tris–HCl, pH 7.5. The beads were allowed to settle, the supernatant was removed and the beads were boiled at 95°C for 10 min in a solution containing 2% SDS, 2 mM DTT, 4% glycerol, 4 mM Tris–HCl and 0.01% bromophenol blue. The reactions were analyzed by SDS/PAGE. The gel was dried for 1 h at 80°C and exposed to a phosphorimaging screen for 1 h. For reactions containing ssARS1-1, the experimental procedures are the same as described above except varying amounts of unlabeled ssARS1-1 were added at the same time as radiolabeled Sld2T84D.
Biotin-pulldown assay
Biotinylated DNA (7 pmoles) conjugated to streptavidin–agarose magnetic beads (Dynal) were incubated 5 min at 30°C with increasing concentrations of radiolabeled Sld2 or Sld2T84D in a solution containing 0.1 mM EDTA, 0.2 mM DTT, 10 mM magnesium acetate, 10% glycerol, 40 µg/ml BSA and 20 mM Tris–HCl pH 7.5 in a final volume of 25 µl. Following the 5 min incubation the beads were collected at room temperature using a magnet (Dynal). The supernatant was removed and the beads were washed twice with a solution containing 0.1 mM EDTA, 0.2 mM DTT, 10 mM magnesium acetate, 10% glycerol, 40 µg/ml BSA and 20 mM Tris–HCl pH 7.5. The beads were collected with a magnet, the supernatant was removed and the beads were boiled at 95°C for 10 min in a solution containing 2% SDS, 2 mM DTT, 4% glycerol, 4 mM Tris–HCl and 0.01% bromophenol blue. The reactions were analyzed by SDS/PAGE. The gel was dried for 1 h at 80°C and exposed to a phosphorimaging screen for 1 h.
Kinase treatment of Sld2, Sld2T84D and Dpb11
Sld2 or Sld2T84D (1 mg/ml) was incubated 1 h at 30°C with 0.13 mg/ml CDK in a solution containing 0.10 mM magnesium chloride, 1 mM DTT, 150 µCi γ32P-ATP, 50 mM Tris–HCl pH 7.5 in a final volume of 100 µl. The reaction was stopped with the addition of 2 µl of 0.5 M EDTA and kept on ice until it could be used in subsequent reactions.
For reactions containing protein kinase A (PKA), 1 mg/ml Sld2 or Sld2T84D or Dpb11 was incubated with 0.017 mg/ml PKA 1 h at 30°C in a solution containing 0.10 mM magnesium chloride, 1 mM DTT, 150 µCi γ32P-ATP, 50 mM Tris–HCl pH 7.5 in a final volume of 100 µl. The reaction was stopped with the addition of 2 µl of 0.5 M EDTA and kept on ice until it could be used in subsequent reactions.
Fluorescence anisotropy
Sld2T84D binding to ssARS1-1 or ssARS1-2 was measured by the increase in anisotropy as varying concentrations of Sld2T84D were incubated with 50 nM fluorescent DNA (5-FAM) for 5 min in a solution containing 0.1 mM EDTA, 0.2 mM DTT, 10 mM magnesium acetate, 10% glycerol, 40 µg/ml BSA and 20 mM Tris–HCl pH 7.5 in a final volume of 40 µl. Polarized fluorescence intensities were measured at excitation and emission wavelengths of 495 and 538 nm, respectively. The data were plotted as fraction bound versus protein concentration, and dissociation constants (Kd) were derived by fitting the data with the equation y = m0 / (m0 + m1), where y = fraction bound, m0 = concentration of Sld2T84D and m1 = Kd. Sld2T84D and Sld2 binding to ssARS1-1 is the same as above except 25 nM fluorescent DNA was used.
DNA annealing
Each annealing reaction was prepared on ice and contained 10 mM magnesium acetate, 10% glycerol, 5 mM DTT, 40 µg/ml BSA, 20 mM Tris–HCl pH 8.0, 1 nM radiolabeled DNA and proteins as detailed in each figure legend, in a final volume of 10 µl. Then the reactions were shifted to 30°C for the times indicated in the figure legends. At the end of the reaction 1 µl proteinase K (10 mg/ml) was added, and the sample was incubated an additional 1 min at 37°C. Stop solution (5µl; 2% SDS, 80 mM EDTA) was added, followed by 5 µl 6× loading dye (30% glycerol + 0.25% bromophenol blue) and finally the reactions were snap frozen on dry ice. Reactions were analyzed by an 8% native polyacrylamide gel containing 1 × TBE (90 mM Tris–HCl–borate, 2 mM EDTA) at 175 V. The gel was dried for 1 h at 80°C and exposed to a phosphorimaging screen overnight.
RESULTS
Sld2T84D binds to origin ssDNA
Sld2 is found at origins of replication in budding yeast cells. To investigate if Sld2 has the ability to bind directly to DNA at origins of replication, we assayed the ability of Sld2 to bind directly to DNA found at an origin of replication. We initially studied the ARS1 origin because it is a well-characterized yeast origin (23). ARS1 has an adenine-rich strand (top strand), and a thymine-rich strand (bottom strand), a characteristic of yeast replication origins (21). ARS1 also contains four elements that are important for DNA replication: the A, B1, B2 and B3 elements (23). The A element contains the ARS consensus sequence (ACS), while the B region, containing the B1, B2 and B3 elements, is an easily-unwound sequence positioned adjacent to the A site (24). We initially studied the ability of Sld2 to bind directly to the DNA sequence containing the A and B1 elements, encompassing nucleotides 810–889 of ARS1, using the GST-pulldown assay (Figure 1B and Supplementary Table S1).
Purified GST, GST–Sld2T84D or GST–Sld2 attached to glutathione beads was used to pull-down increasing amounts of radiolabeled DNA (Figure 1B). After washing, the bound DNA was analyzed by gel electrophoresis followed by phosphorimaging to quantify the amount of DNA bound. The DNA examined included ssARS1-1, ssARS1-2 or dsARS1. The fraction of DNA bound was quantified and plotted as a function of input DNA concentration (Figure 1C and D). When different concentrations of ssARS1-1 were added to GST–Sld2T84D, GST–Sld2T84D pulled-down a substantial fraction of ssARS1-1 (Figure 1B, top gel, and C). In contrast, GST–Sld2T84D bound very weakly to the top strand of ARS1 (ssARS1-2), or to dsARS1 (Figure 1B and C). GST–Sld2 pulled-down a lower fraction of ssARS1-1 compared to GST–Sld2T84D (Figure 1B and D). These data suggest that the T84D mutation activates binding of Sld2 to ssARS1-1. Weak interactions were also detected for Sld2 binding to ARS1-2 and dsARS1 (Figure 1B and D), similar to that observed for Sld2T84D (Figure 1B and C). These data suggest that the T84D mutation does not activate binding of Sld2 to ssARS1-2 or dsARS1.
We next examined if GST–Sld2T84D binds to the ss region of ARS1 containing the B2 and B3 elements (ssARS1-3 or ssARS1-4, Figure 1A and B). GST–Sld2T84D bound weakly to these two sequences (Figure 1B and E). These data suggest that Sld2T84D binds more tightly to ssARS1-1 than to other regions of ARS1 tested in this assay. The thymidine content of ssARS1-1 is 41%, and we therefore next examined if Sld2T84D binds to poly-thymidine ssDNA. We found that GST–Sld2T84D pulls down a low fraction of 80-dT compared to ssARS1-1 (Supplementary Figure S1), suggesting that Sld2T84D interaction with ssARS1-1 is not simply the result of nonspecific poly-thymidine binding.
We next investigated if Sld2T84D binds to a different yeast origin, ARS305 (Figure 2). GST, GST–Sld2T84D or GST–Sld2 was incubated with the single-strands of ARS305 (ssARS305-1 or ssARS305-2), or ds ARS305. GST–Sld2T84D bound tightly to ARS305-1, and weakly to ARS305-2 or dsARS305 (Figure 2A and B). Sld2 bound modestly to ARS305-1 and ARS305-2, but weakly to dsARS305 (Figure 2C). Sld2T84D bound more tightly than Sld2 to ARS305-1 (Figure 2C), suggesting that the phosphomimetic mutation activates binding of Sld2 to the single-strand of at least two different origins of replication.
Figure 2.
Sld2T84D binds to ssARS305 bottom strand (ssARS305-1). (A) Purified GST–Sld2T84D, GST–Sld2 or GST were studied for interaction with radiolabeled ssARS305-1 (top panel), ssARS305-2 (middle panel) or dsARS305 (bottom panel). GST–Sld2T84D, GST–Sld2 or GST (13 pmol) and varying amounts of DNA, as indicated in the figure, were mixed with 1.3 mg glutathione agarose beads and incubated for 5 min at 30°C. The beads were washed and analyzed as described in ‘Materials and Methods’ section. The results from experiments similar to (A) were quantified and plotted as pmol of DNA bound versus pmol of input DNA (B and C). Sld2T84D binds tighter to ssARS305-1 than to either ssARS305-2 or dsARS305. Sld2T84D binds tighter than Sld2 to ssARS305-1.
To determine the solution affinity of Sld2T84D for ssARS1-1, we incubated fluorescently-labeled ARS1-1 DNA with various concentrations of native Sld2T84D, and measured the anisotropy change (Figure 3A). The data were fit to a single-binding site equation, and the apparent Kd was calculated to be 179 nM + 43 nM. In contrast, the Kd of Sld2T84D binding to ssARS1-2 is greater than 1 µM (Figure 3A). GST–Sld2T84D may weakly pull down ssARS1-2 (Figure 1B and C) because this assay will show weakly positive results if there is a transient interaction between the protein and DNA. Fluorescence anisotropy is not as sensitive to weak protein–DNA interactions at the low levels of DNA used (50 nM). To determine the effect of the phosphomimetic mutation on ssARS1-1 binding affinity, we next compared the Kd of Sld2T84D binding to ssARS1-1 to Sld2 binding to ssARS1-1 (Figure 3B). The apparent Kd of Sld2 binding to ssARS1-1 is 694 nM + 50 nM, which is 3.5-fold higher than that calculated for Sld2T84D (Figure 3B). These data further support that the T84D phosphomimetic mutation modestly increases the affinity of Sld2 for ssARS1-1.
Figure 3.
Sld2T84D binds to ssARS1-1. (A) Fluorescently labeled ssARS1-1 or ssARS1-2 (50 nM) was mixed with varying concentrations of native Sld2T84D, as indicated in the figure, and incubated for 20 min at room temperature. Polarized fluorescence intensities were measured at excitation and emission wavelengths of 495 and 538 nm, respectively, at room temperature. The data were plotted as the change in anisotropy versus protein concentration, and the curve was fit with the equation y = m0/(m0 + m1). The table to the right of the graph shows the apparent Kd for Sld2T84D interaction with ssARS1-1 or ssARS1-2. Sld2T84D binds tighter to ssARS1-1 than ssARS1-2. (B) Fluorescently labeled ssARS1-1 (25 nM) was mixed with varying concentrations of either Sld2T84D or Sld2, as indicated in the figure, and incubated for 20 min at room temperature. Polarized fluorescence intensities were measured at excitation and emission wavelengths of 495 and 538 nm, respectively, at room temperature. The data were plotted as the change in anisotropy versus protein concentration in nM, and the curve was fit with the equation y = m0/(m0 + m1). The table to the right of the graph shows the apparent Kd for Sld2T84D and Sld2 interaction with ssARS1-1. Sld2T84D binds tighter than Sld2 to ssARS1-1.
To confirm the interaction between Sld2T84D and ssARS1-1, we engineered Sld2T84D with a PKA sequence at the N-terminus, and radiolabeled Sld2T84D with PKA. PKA is not physiologically relevant here—the PKA is used solely to radiolabel the protein. Biotinylated dsARS1, ssARS1-2 or ssARS1-1, or biotin alone was conjugated to streptavidin beads, and the mixtures were then incubated with radiolabeled Sld2T84D. After washing, the bound protein was analyzed by SDS/PAGE and phosphorimaging (Figure 4A), followed by quantitation (Figure 4B). Biotinylated-ssARS1-1 pulled-down a significant fraction of Sld2T84D at input quantities of 4.8 and 13 pmoles (Figure 4B), while a lower fraction of Sld2T84D was pulled down by either biotinylated-ssARS1-2 or biotinylated-dsARS1. The trend follows that of the GST-pulldown and fluorescence anisotropy assays, further supporting the conclusion that Sld2T84D binds tightly to ssARS1-1.
Figure 4.
Sld2T84D binds to ssARS1-1. (A) Biotinylated dsARS1, ssARS1-2 or ssARS1-1 (7 pmol) was conjugated to streptavidin agarose and then mixed with varying amounts of radiolabeled Sld2T84D, as indicated in the figure, for 5 min at 30°C. As a control, beads without DNA were incubated with Sld2T84D. The beads were washed and analyzed as described in ‘Materials and Methods’ section. The results from experiments similar to (A) were quantified and plotted as the percent Sld2T84D bound versus the input protein concentration (B). Sld2T84D binds more tightly to ssARS1-1 than either ssARS1-2 or dsARS1. (C) Biotinylated ssARS1-1 was tested for binding to CDK-phosphorylated Sld2 and PKA-phosphorylated Sld2. The results from experiments similar to (C) were quantified and plotted as the fraction of Sld2 bound versus the input protein concentration (D). These results suggest CDK phosphorylation of Sld2 stimulates binding to ssARS1-1.
S-CDK activates Sld2 binding to origin ssDNA
The observation that Sld2T84D binds tighter than Sld2 to ARS1-1 suggests that S-CDK phosphorylation of Sld2 increases the affinity of Sld2 for DNA. To test this hypothesis, we radiolabeled Sld2 with either PKA or S-CDK, and then matched the levels of radioactive counts and total Sld2 protein (Figure 4C). We incubated PKA-labeled Sld2 or S-CDK-labeled Sld2 with biotinylated ssARS1-1 conjugated to streptavidin beads, and then pulled-down Sld2 protein (Figure 4C). We found that a greater fraction of S-CDK-labeled Sld2 was pulled-down compared to PKA-labeled Sld2 (Figure 4C and D). These data suggest that S-CDK phosphorylation of Sld2 increases the affinity of this protein for ssARS1-1. Since S-CDK phosphorylation of Sld2 will add negative charge to the Sld2 protein, and negative charge is generally repulsive for DNA, the data also suggest that CDK-activation of Sld2 binding to DNA is not the result of additional non-specific attractive charge. Furthermore, the data lead us to speculate that CDK-phosphorylation of Sld2 induces a conformational change in Sld2 that activates DNA binding activity.
Replacing the origin thymines with adenines completely disrupts Sld2T84D binding
ssARS1-1 has a thymine content of 41%, and replication origins in budding yeast have a thymine-rich strand (21). We speculated that Sld2T84D may be recognizing the thymines of ssARS1-1. To determine if Sld2T84D recognizes the thymines of ssARS1-1, we substituted all of the ssARS1-1 thymines with adenines (T-to-A, Figure 5A). Binding was reduced to GST-background levels as a result of this T-to-A substitution of ssARS1-1. These data suggest that Sld2T84D recognizes the thymines of ssARS1-1. As a control, we substituted all of the guanines with cytosines (G-to-C), all of adenines with cytosines (A-to-C) or all of the cytosines with adenines (C-to-A). The G-to-C or A-to-C substitution had little effect on Sld2T84D binding, while the C-to-A substitution resulted in modest inhibition of Sld2T84D binding. The C-to-A substitution may inhibit Sld2T84D binding because cytosine is a pyrimidine, like thymine.
To determine if GST–Sld2T84D recognizes the thymines within a region in ssARS1-1, we replaced regions of ssARS1-1 with T-to-A substitutions (Figure 5B and C). When eight thymines of ssARS1-1 were substituted with adenines from either end, there was a modest reduction in DNA bound by the Sld2T84D (Figure 5B and C). When 16 thymines of ARS1-1 were substituted from the 5′ end, there was also a modest decrease in DNA binding (Figure 5B and C); however, when 16 thymines of ssARS1-1 were substituted from the 3′-end, DNA binding was substantially decreased (Figure 5B and C). When 24 thymines were substituted with adenines, DNA bound by Sld2T84D was reduced to nearly background (GST) levels (Figure 5B and C). These data suggest that Sld2T84D is recognizing the thymines in multiple regions within ssARS1-1.
Sld2T84D enhances the ability of ssARS1-1 to pulldown Dpb11
Since Dpb11 and ssARS1-1 bind independently to Sld2T84D, we next investigated if ssARS1-1 competes with Dpb11 for Sld2T84D binding. Before assaying for competition, it is first critical to demonstrate a binary interaction between Sld2T84D and Dpb11. Sld2T84D has been previously shown to bind Dpb11 in vivo (16), but an in vitro assay with purified proteins has not been accomplished. We incubated radiolabeled Sld2T84D with GST–Dpb11 or GST and then pulled-down the radiolabeled Sld2T84D (Figure 6A). We detected an in vitro binary interaction between Dpb11 and Sld2T84D (Figure 6A and B), as expected based upon previous in vivo studies.
Figure 6.
Sld2T84D enhances the ability of ssARS1-1 to pulldown Dpb11. (A) A direct interaction between Sld2T84D and Dpb11 was studied. GST–Dpb11 or GST (40 pmol) and varying amounts of Sld2T84D, as indicated in the figure, were mixed with 1.3 mg glutathione agarose beads and incubated for 5 min at 30°C. The beads were washed and analyzed as described in ‘Materials and Methods’ section. The results from experiments similar to (A) were quantified and plotted as the fraction of Sld2T84D bound versus the pmol input of Sld2T84D (B). (C) The interaction of Sld2T84D and Dpb11 was studied in the presence of excess ssARS1-1. The experiment was performed as described above with the addition of increasing amounts of ssARS1-1. The results from experiments similar to (C) were quantified and plotted as the fraction Sld2T84D bound versus the ratio of ssARS1-1 and Sld2T84D (D). Adding excess ssARS1-1 slightly stimulates GST-Dpb11 pulldown of Sld2T84D. (E) Dpb11 (1.27 or 4.75 pmol) and increasing amounts of Sld2T84D, as indicated in the figure, were mixed with 7 pmol biotinylated ssARS1-1 conjugated to 4.3 × 105 bead/µl streptavidin agarose and incubated for 5 min at 30°C. The beads were washed and analyzed as described in ‘Materials and Methods’ section. The results from experiments similar to (E) were quantified and plotted as the pmol of Dpb11 bound versus the pmol of Sld2T84D input (F). In the absence of Sld2T84D, Dpb11 binds weakly to ssARS1-1, and the addition of Sld2T84D enhances the amount of Dpb11 pulled down by ssARS1-1.
To determine if ssARS1-1 competes with Dpb11 for binding to Sld2T84D, varying amounts of ssARS1-1 were added to the GST–Dpb11 pulldown of Sld2T84D (Figure 6C). In the absence of ssARS1-1, 24.2% of input Sld2T84D was pulled-down by GST–Dpb11 (Figure 6D). However, when ssARS1-1 was added at a ratio of 1.2 ssARS1-1/Sld2T84D, 35.2% of the input Sld2T84D was pulled down by GST–Dpb11 (Figure 6D). Furthermore, the addition of ssARS1-1 slightly stimulated the fraction of Sld2T84D pulled down over a wide range of concentrations (Figure 6C and D). These data suggest that ssARS1-1 does not compete with Dpb11 for binding to Sld2T84D.
In budding yeast cells, Sld2–ssDNA interaction may be important for the association of Sld2 and Dpb11 with origins of replication. To determine if Sld2T84D stimulates the ability of ssARS1-1 to pulldown Dpb11, biotinylated ssARS1-1 was incubated with radiolabeled Dpb11 and different amounts of Sld2T84D, and the resulting DNA-bound protein was analyzed by SDS/PAGE followed by phosphorimaging and quantitation (Figure 6E and F). In the absence of Sld2T84D, Dpb11 bound weakly to ssARS1-1 (Figure 6E, lanes 6 and 10). However, in the presence of Sld2T84D, a substantial fraction of Dpb11 was pulled down by biotinylated ssARS1-1 (Figure 6E and F). These data suggest that Sld2T84D enhances the ability of ssARS1-1 to pulldown Dpb11.
Sld2 anneals ss origin DNA
ssDNA is likely to be formed during the initiation of DNA replication at a replication origin (24), but the accumulation of excess ssDNA at an origin might be a source of genome instability (25,26). Thus, factors that limit the accumulation of ssDNA at an origin may be important to preserve genome stability. To determine if Sld2 can anneal complementary origin ssDNA, we next examined whether Sld2 can convert origin ssDNA to dsDNA. For this assay, we examined the annealing of ssARS1-1 to modified ssARS1-2. ssARS1-2 was modified to include ssDNA on either side of the duplex DNA (ssARS1 20A-830-879-20C), thereby creating a forked DNA substrate (Figure 7A). This forked structure allows for separation of forked-product DNA from substrate ssDNA by native gel electrophoresis.
Figure 7.
Sld2T84D stimulates annealing of ssDNA to its complementary strand. (A) A schematic for annealing. Radiolabeled ssARS1-1 (1 nM) is incubated with 2 nM cold ARS1-20C-830-869-20C top strand in the presence of Sld2T84D. The annealing of these ssDNAs results in fork-shaped dsDNA. The free radiolabeled ssARS1-1 migrates faster through a native gel than the duplex containing radiolabeled ssARS1-1. (B) Sld2T84D or Sld2 (500 nM) was mixed on ice with the substrate described in (A). The mixture was then incubated at 30°C for different amounts of time, as described in the figure. Following the incubation at 30°C, the samples were analyzed as described in ‘Materials and Methods’ section. The results from (B) were quantified and plotted as the percent dsDNA versus time in minutes (C). Annealing of ssDNA in the presence of Sld2T84D or Sld2 is significantly faster than the no protein control. (D) Sld2T84D or Sld2 (50 nM) was mixed with the substrate described in (A). The results from experiments were quantified and plotted as percent dsDNA versus protein concentration. A higher fraction of DNA is annealed with Sld2T84D compared to Sld2. (E) Sld2T84D annealing ssDNA was tested with substrates containing a 5′ or 3′ overhang. The results from experiments were quantified and plotted as percent dsDNA versus time in minutes. Sld2T84D can anneal substrates with either 5′ or 3′ overhangs. (F) Dpb11, Sld2T84D or both Dpb11 and Sld2T84D (50 nM) were mixed with the substrate described in (A) for varying amounts of time, as indicated in the figure. The results from experiments similar to (F) were quantified and plotted as the percent dsDNA versus time in minutes (G). Dpb11 displays less annealing activity than Sld2T84D. Mixing Dpb11 with Sld2T84D results in an annealing activity that is greater than either protein alone.
In the absence of protein, no DNA annealing occurred over a time course of 0–16 min (Figure 7B). However, in the presence of 500 nM Sld2 or Sld2T84D, more than half of the DNA substrate was annealed after 30 s, and the fraction annealed increases over time until it exceeded 90% by 5 min (Figure 7B and C). In the presence of 50 nM Sld2T84D, nearly 50% of the DNA was annealed by 5 min (Figure 7D). Sld2 (50 nM) also stimulated DNA annealing, but higher levels of annealing are observed for Sld2T84D compared to Sld2 (Figure 7D). These data are consistent with the higher affinity for DNA of Sld2T84D compared to Sld2 (Figure 3). No annealing activity was observed when bovine serum albumin was added instead of Sld2T84D (Supplementary Figure S2), suggesting that the annealing activity observed is specific for Sld2T84D.
We next investigated if Sld2T84D-stimulated annealing requires a forked-shaped DNA product (Figure 7E). For these experiments, we incubated complementary DNA sequences that do not form forked-structures when annealed. The annealed products contain either 5′ or 3′ overhangs to allow for native gel separation of substrate from product. Sld2T84D (500 nM) stimulated annealing of these complementary DNA sequences, whether a 5′ or 3′ overhang is present in the product. Approximately 90% of the DNA was annealed after 5 min for both of these DNA sequence pairs. These data suggest that a forked-structured product is not a requirement for annealing.
Dpb11 and Sld2 stimulate DNA annealing
The observation that Sld2T84D stimulates the ability of ssARS1-1 to pulldown Dpb11 prompted us to investigate if Dpb11 modulates Sld2T84D-stimulated DNA annealing. We first examined whether Dpb11 alone has any DNA annealing activity using the same assay as in Figure 7A (Figure 7F and G). Dpb11 (50 nM) annealed ∼20% of the DNA after 5 min (Figure 7F and G). Thus, although the affinity of Dpb11 for ARS1-1 is low (Figure 6E and F), it is sufficient to stimulate low levels of DNA annealing. Dpb11 (50 nM) was then added to an annealing reaction that contained 50 nM Sld2T84D (Figure 7F and G). Reactions that contained both Dpb11 and Sld2T84D exhibited higher levels of ds product than Sld2T84D or Dpb11 alone (Figure 7F and G), and the level of annealing observed with both proteins approximates the sum of the annealing activities observed with single proteins.
DISCUSSION
We find that the phosphomimetic form of Sld2, Sld2T84D, binds tightly to the single-strand of two different origins of replication, ARS1 and ARS305. Furthermore, the phosphomimetic mutation T84D modestly stimulates Sld2-binding to these ss origin sequences, and S-CDK phosphorylation of Sld2 modestly stimulates Sld2-binding to ssARS1-1. ssARS1-1 is thymine rich, and substitution of the ssARS1-1 thymines with adenines completely disrupts binding to Sld2T84D. Sld2T84D binds to Dpb11 in vitro, and this interaction is slightly stimulated by ssARS1-1. Dpb11 alone interacts weakly with ARS1-1; however, in the presence of Sld2T84D, ssARS1-1 pulls down a substantial fraction of input Dpb11. These data suggest that DNA and Dpb11 do not compete for the same binding site of Sld2T84D, and that Sld2T84D enhances the ability of ARS1-1 to pulldown Dpb11. Sld2T84D anneals complementary ssDNA that contains ssARS1-1, and Dpb11 alone also anneals complementary ssDNA. Sld2T84D and Dpb11 in combination anneal a higher fraction of ssDNA compared to that achieved by either protein alone.
Sld2/Dpb11 binds to origin ssDNA
Sld2T84D, the phosphomimetic form of Sld2, binds to ARS1 ssDNA containing the A and B1 sequence elements (ARS1-1). Furthermore, Sld2T84D binds to ssDNA in ARS305 as well (ssARS305-1). Thus, Sld2T84D binds to ssDNA sequences found at more than one origin of replication. ssARS1-1 is thymine rich, and substitution of the ssARS1-1 thymines with adenines completely disrupts binding to Sld2T84D. Yeast replication origins bear a thymine-rich strand and an adenine-rich ssDNA strand (21), and our data suggest that Sld2T84D may specifically target the thymine-rich strand of yeast replication origins. Sld2 also interacts with other DNA replication initiation proteins, including Dpb11 (16), and these interactions may help target Sld2 to origin ssDNA. ss DNA will form at an origin during the initiation of DNA replication (24). Thus, Sld2/Dpb11 interaction with ssDNA formed at a replication origin may help recruit Sld2/Dpb11 to origin DNA.
In current models of initiation, S-CDK promotes the interaction between Sld2 and Dpb11 early in the initiation process. The CDK-stimulated interaction between Sld2 and Dpb11 is important for the formation of the transient pre-loading complex, composed of Sld2, Dpb11, GINS and Pol ε. In the present article, we propose that Sld2 and Dpb11 bind to origin ssDNA, and this event is modestly activated by CDK activity as well. Sld2/Dpb11 binding to ssDNA is likely modulated by other protein–protein interactions that are CDK-activated, such as Dpb11–Sld3 interaction.
Sld2 anneals ssDNA
Sld2T84D anneals ssDNA found at an origin of replication. Dpb11 alone has a weak ability to anneal ssDNA, and the combination of Sld2T84D and Dpb11 yield higher levels of annealing compared to levels achieved with either protein alone. Since Sld2 and Dpb11 bind to one another at an origin of replication, these proteins may work together to anneal complementary ssDNA at an origin of replication. Annealing DNA may be important to limit the amount of excess ssDNA that is formed at an origin, preventing the activation of the DNA damage response. This annealing activity may be particularly important in the region of low helical stability that is present in the B region of origins of replication (24). The annealing activity may also help dislodge Sld2 from origins of replication once initiation is complete. Once dsDNA is formed, the Sld2 may dissociate from the DNA, since Sld2 has weak affinity for dsDNA. While Sld2/Dpb11 annealing activity may help dislodge these proteins from origin DNA, the replisome machinery may be fully competent to dislodge these proteins from DNA as well.
A proposed timing for Sld2/Dpb11 action is as follows: at the onset of S phase, S-CDK phosphorylates Sld2, stimulating its origin ssDNA binding activity. Prior to helicase movement, the origin ssDNA will become exposed, and Sld2–Dpb11 will bind and anneal this DNA, thereby preserving genome integrity. Just prior to origin firing, this ssDNA region may be melted again, but this time Sld2/Dpb11 may fail to anneal the DNA by a mechanism that is presently unclear. This final DNA melting step may help activate the replication fork helicase.
Additional role for CDK in the cell
CDK is essential for yeast cell viability. However, the essential requirement for CDK can be bypassed if the sld2 gene bears the T84D phosphomimetic mutation, and the dpb11–sld3 genes are fused (17). The T84D mutation promotes the interaction between Sld2 and Dpb11 (16), and this biochemical activity may be mostly responsible for the CDK bypass. We now know that the T84D mutation also activates the interaction between Sld2 and ssDNA, and the T84D mutation stimulates the ability of Sld2 to anneal ssDNA. Thus, these additional biochemical functions may in part be responsible for the ability of the T84D mutation to bypass the requirement for CDK. Thus, CDK-activation of Sld2 binding to ssDNA may be an additional activity of CDK.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Vanderbilt University Discovery (grant to D.L.K.); American Cancer Society Research Scholar Grant (grant number RSG-08-124-01-CCG to D.L.K.). Funding for open access charge: American Cancer Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Hiroyuki Araki for supplying the expression construct for S-CDK, and Susan Taylor for providing purified PKA. They also thank Irina Bruck for helpful comments and purified proteins, and Leolene Jean for her helpful comments and suggestions.
REFERENCES
- 1.Kamimura Y, Masumoto H, Sugino A, Araki H. Sld2, which interacts with Dpb11 in Saccharomyces cerevisiae, is required for chromosomal DNA replication. Mol. Cell Biol. 1998;18:6102–6109. doi: 10.1128/mcb.18.10.6102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wang H, Elledge S. DRC1, DNA replication and checkpoint protein 1, functions with DPB11 to control DNA replication and the S-phase checkpoint in Saccharomyces cerevisiae. Proc. Natl Acad. Sci. USA. 1999;96:3824–3829. doi: 10.1073/pnas.96.7.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Masumoto H, Muramatsu S, Kamimura Y, Araki H. S-Cdk-dependent phosphorylation of Sld2 essential for chromosomal DNA replication in budding yeast. Nature. 2002;415:651–655. doi: 10.1038/nature713. [DOI] [PubMed] [Google Scholar]
- 4.Muramatsu S, Hirai K, Tak Y, Kamimura Y, Araki H. CDK-dependent complex formation between replication proteins Dpb11, Sld2, Pol (epsilon}, and GINS in budding yeast. Genes Dev. 2010;24:602–612. doi: 10.1101/gad.1883410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Holway A, Hung C, Michael W. Systematic, RNA-interference-mediated identification of mus-101 modifier genes in Caenorhabditis elegans. Genetics. 2005;169:1451–1460. doi: 10.1534/genetics.104.036137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Van Hatten RA, Tutter AV, Holway AH, Khederian AM, Walter JC, Michael WM. The Xenopus Xmus101 protein is required for the recruitment of Cdc45 to origins of DNA replication. J Cell Biol. 2002;159:541–547. doi: 10.1083/jcb.200207090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Araki H, Leem S, Phongdara A, Sugino A. Dpb11, which interacts with DNA polymerase II(epsilon) in Saccharomyces cerevisiae, has a dual role in S-phase progression and at a cell cycle checkpoint. Proc. Natl Acad. Sci. USA. 1995;92:11791–11795. doi: 10.1073/pnas.92.25.11791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Garcia V, Furuya K, Carr A. Identification and functional analysis of TopBP1 and its homologs. DNA Repair. 2005;4:1227–1239. doi: 10.1016/j.dnarep.2005.04.001. [DOI] [PubMed] [Google Scholar]
- 9.Masumoto H, Sugino A, Araki H. Dpb11 controls the association between DNA polymerases alpha and epsilon and the autonomously replicating sequence region of budding yeast. Mol. Cell Biol. 2000;20:2809–2817. doi: 10.1128/mcb.20.8.2809-2817.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Takayama Y, Kamimura Y, Okawa M, Muramatsu S, Sugino A, Araki H. GINS, a novel multiprotein complex required for chromosomal DNA replication in budding yeast. Genes Dev. 2003;17:1153–1165. doi: 10.1101/gad.1065903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kumagai A, Lee J, Yoo H, Dunphy W. TopBP1 activates the ATR–ATRIP complex. Cell. 2006;124:943–955. doi: 10.1016/j.cell.2005.12.041. [DOI] [PubMed] [Google Scholar]
- 12.Mordes D, Nam E, Cortez D. Dpb11 activates the Mec1–Ddc2 complex. Proc. Natl Acad. Sci. USA. 2008;105:18730–18734. doi: 10.1073/pnas.0806621105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Navadgi-Patil V, Burgers P. Yeast DNA replication protein Dpb11 activates the Mec1/ATR checkpoint kinase. J. Biol. Chem. 2008;283:35853–35859. doi: 10.1074/jbc.M807435200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Hartwell L, Culotti J, Pringle J, Reid B. Genetic control of the cell division cycle in yeast. Science. 1974;183:46–51. doi: 10.1126/science.183.4120.46. [DOI] [PubMed] [Google Scholar]
- 15.Schwob E, Nasmyth K. CLB5 and CLB6, a new pair of B cyclins involved in DNA replication in Saccharomyces cerevisiae. Genes Dev. 1993;7:1160–1175. doi: 10.1101/gad.7.7a.1160. [DOI] [PubMed] [Google Scholar]
- 16.Tak Y, Tanaka Y, Endo S, Kamimura Y, Araki H. A CDK-catalysed regulatory phosphorylation for formation of the DNA replication complex Sld2–Dpb11. EMBO J. 2006;25:1987–1996. doi: 10.1038/sj.emboj.7601075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zegerman P, Diffley JF. Phosphorylation of Sld2 and Sld3 by cyclin-dependent kinases promotes DNA replication in budding yeast. Nature. 2007;445:281–285. doi: 10.1038/nature05432. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka S, Umemori T, Hirai K, Muramatsu S, Kamimura Y, Araki H. CDK-dependent phosphorylation of Sld2 and Sld3 initiates DNA replication in budding yeast. Nature. 2007;445:328–332. doi: 10.1038/nature05465. [DOI] [PubMed] [Google Scholar]
- 19.Matsuno K, Kumano M, Kubota Y, Hashimoto Y, Takisawa H. The N-terminal noncatalytic region of Xenopus RecQ4 is required for chromatin binding of DNA polymerase alpha in the initiation of DNA replication. Mol. Cell Biol. 2006;26:4843–4852. doi: 10.1128/MCB.02267-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Capp C, Wu J, Hsieh T. Drosophila RecQ4 has a 3′-5′ DNA helicase activity that is essential for viability. J. Biol. Chem. 2009;284:30845–30852. doi: 10.1074/jbc.M109.008052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Newlon CS, Theis JF. The structure and function of yeast ARS elements. Curr. Opin. Gen. Dev. 1993;3:752–758. doi: 10.1016/s0959-437x(05)80094-2. [DOI] [PubMed] [Google Scholar]
- 22.Studier FW. Protein production by auto-induction in high-density shaking cultures. Protein Expr. Purif. 2005;41:207–234. doi: 10.1016/j.pep.2005.01.016. [DOI] [PubMed] [Google Scholar]
- 23.Marahrens Y, Stillman B. A yeast chromosomal origin of DNA replication defined by multiple functional elements. Science. 1992;255:817–823. doi: 10.1126/science.1536007. [DOI] [PubMed] [Google Scholar]
- 24.Natale DA, Umek RM, Kowalski D. Ease of DNA unwinding is a conserved property of yeast replicatiion origins. Nucleic Acids Res. 1993;21:555–560. doi: 10.1093/nar/21.3.555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Segurado M, Tercero JA. The S-phase checkpoint: targeting the replication fork. Biol. Cell. 2009;101:617–627. doi: 10.1042/BC20090053. [DOI] [PubMed] [Google Scholar]
- 26.Cimprich KA, Cortez D. ATR: an essential regulator of genome integrity. Nat. Rev. Mol. Cell Biol. 2008;9:616–627. doi: 10.1038/nrm2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.