Abstract
The analysis of the mitochondrial DNA of Isoetes engelmannii as a first representative of the lycophytes recently revealed very small introns and indications for extremely frequent RNA editing. To analyze functionality of intron splicing and the extent of RNA editing in I. engelmannii, we performed a comprehensive analysis of its mitochondrial transcriptome. All 30 groups I and II introns were found to be correctly removed, showing that intron size reduction does not impede splicing. We find that mRNA editing affects 1782 sites, which lead to a total of 1406 changes in codon meanings. This includes the removal of stop codons from 23 of the 25 mitochondrial protein encoding genes. Comprehensive sequence analysis of multiple cDNAs per locus allowed classification of partially edited sites as either inefficiently edited but relevant or as non-specifically edited at mostly low frequencies. Abundant RNA editing was also found to affect tRNAs in hitherto unseen frequency, taking place at 41 positions in tRNA-precursors, including the first identification of U-to-C exchanges in two tRNA species. We finally investigated the four group II introns of the nad7 gene and could identify 27 sites of editing, most of which improve base pairing for proper secondary structure formation.
INTRODUCTION
Even 20 years after the original discovery of RNA editing that exchanges cytidine and uridine nucleotides in plant mitochondrial and chloroplast transcripts, many aspects of the phenomenon remain enigmatic (1–4). The reasons why RNA editing came into being during land plant evolution remain unclear. No obvious functional gain or evolutionary adaptation can be connected with RNA editing in plant organelles. Similarly, strong evidence that plant RNA editing may functionally modulate gene activity is lacking, although several publications have reported variability of RNA editing among different environments, ecotypes or plant tissues (5–13). RNA editing in plant organelles largely seems to act as a correction mechanism which reinstalls codons conserved during evolution for proper protein function. In other words: RNA editing mainly re-establishes sequences on RNA level that could directly be encoded as such in the DNA (and in fact often are so in related species). Significant progress, however, has come lately with the identification of specific RNA-binding PPR (pentatricopeptide repeat) proteins providing sequence recognition specificities to determine nucleotide positions for editing in organelle transcripts (14–17).
It seems well supported that RNA editing, which is absent in algae, arose with the emergence of the earliest land plants. Now it is universally distributed among plants with the unique exception of the marchantiid liverworts where it appears to be secondarily lost (18,19). Correspondingly, RNA editing varies widely in appearance and frequency, ranging from zero sites in the marchantiid liverworts, over only 11 in the mitochondrial transcriptome of the model moss Physcomitrella patens (20), to some 200–500 sites in flowering plant mitochondria (21–27).
A further mystery in the evolution of RNA editing during 500 million years of plant diversification concerns the direction of pyrimidine exchanges. The initially discovered type of cytidine-to-uridine editing, most likely a simple deamination (28–30), is the dominating or even exclusive form of editing in angiosperms, mosses and also in the jungermanniid liverworts (which show editing in contrast to their marchantiid sister group). RNA editing in the reverse direction, which converts uridines into cytidines, has been discovered early (31,32) but is obviously very rare among the flowering plants. Other plant clades such as the hornworts and the ferns, however, show substantial additional ‘reverse’ U-to-C editing, incompatible with a simple biochemical deamination step (18,33–36).
We have recently analyzed the mitochondrial DNA sequence of a lycophyte, the quillwort Isoetes engelmannii (37). Extant lycophytes represent the most ancient lineage of vascular plants and are the sister group to all other tracheophytes. Among other peculiarities, the I. engelmannii mtDNA sequences seemed to require a substantial amount of RNA editing, possibly even exceeding 1500 sites, to correct sequences of its 24 encoded proteins (plus one intron-encoded maturase). First cDNA sequence analyses supported this hypothesis. In particular, I. engelmannii mt sequences seemed to require substantial amounts of reverse U-to-C exchanges in addition to numerous C-to-U edits. Furthermore, editing appeared also to be required for generating intact tRNAs. Additionally, the I. engelmannii mtDNA is characterized by particularly small intron sequences.
Here, we describe the results of an exhaustive analysis of mitochondrial transcripts in I. engelmannii which shows the hitherto most extensive degree of RNA editing observed among plants affecting mRNAs, tRNAs and intron sequences. Moreover, the data strongly support that (i) RNA editing mainly acts before other forms of RNA maturation such as splicing or tRNA processing; (ii) partial editing at moderate to high-frequency reflects inefficient editing; and (iii) partial editing at particularly low frequency indicates mis-operation of the RNA editing machinery. Also, we verifed splicing of the particularly small introns in the I. engelmannii mtDNA and discuss the concomitant secondary loss of introns and neighboring RNA editing sites by retro-processing.
MATERIALS AND METHODS
Plant material and RNA isolation
Isoetes engelmannii plant material originally collected in South Central Indiana (USA) by Jerry Gastony, and subsequently greenhouse cultivated, was kindly made available through Jeff Palmer and Erin Badenhop (Bloomington, IN, USA) and further cultivated at the Botanical Garden of the University of Bonn. Total RNA was isolated from whole tissue using the TRI-Reagent (Sigma-Aldrich, Steinheim, Germany) or the NucleoSpin RNA Plant Kit (Macherey Nagel, Düren, Germany) and treated with DNase I (Fermentas, Burlington, ON, USA) for 30 min at 37°C to remove contaminating genomic DNA.
Molecular work
cDNA was synthesized with RevertAid First Strand cDNA Synthesis Kit (Fermentas) in the presence of random hexamer primers as specified by the manufacturer. Oligonucleotide pairs (all sequence information available from the authors upon request) were designed to anneal to 5′- and 3′-untranslated gene regions (UTRs). Gene specific cDNA products were amplified by PCR according to the GoTaq protocol (Promega, Madison, WI, USA). A GeneAmp PCR System 2700 (Applied Biosystems, Foster City, CA, USA) with annealing temperatures between 50°C and 55°C was used. Amplicons were recovered from agarose gels using the NucleBond Xtra Midi EF Kit (Macherey Nagel) and cloned into pGEM T Easy vector (Promega). For analysis of processed tRNA species, a transcript end mapping protocol was used (38). Total I. engelmannii RNA was ligated by T4 RNA ligase (New England Biolabs, Ipswich, MA, USA) and cDNAs were synthesized with RevertAid First Strand cDNA Synthesis Kit (Fermentas), in presence of 200 pmol of a reverse oriented primer. RT–PCR across the RNA-ligation site was done with the same reverse primer in combination with an upstream forward-oriented primer according to the BD Advantage 2 protocol (BD Bioscience, Franklin Lakes, NJ, USA) with annealing temperatures at 47°C.
Sequence handling and analyses
On average 16 independent cDNA clones were sequenced for each locus to estimate the degree of partial editing. Partial editing was considered authentic when found in at least two independent cDNA clones. For cross-validation, selected RT–PCR amplicons were sequenced directly (Supplementary Figure S1). All cDNA sequences were deposited in the database under accession numbers HQ616410-HQ616434 with editing sites annotated using the recently proposed nomenclature (20,39). Sequence handling and sequence alignment were essentially done using the alignment explorer of the MEGA software (40). Prediction, analysis and graphic display of RNA editing was done with the PREPACT software (39) and display and shading of sequence alignments was done with GeneDoc (http://www.nrbsc.org/gfx/genedoc/).
RESULTS
Massive mRNA editing characterizes the I. engelmannii mitochondrial transcriptome
To allow amplification of complete coding sequences, RT–PCR primers were designed to anneal in the respective 5′- and 3′-flanking UTRs. This has the added benefit of reducing the risk of introducing a bias for differentially edited transcripts by accidentally targeting edited sequence regions (25). To exclude potential DNA contamination, we strived for amplification across introns to select for spliced RNAs whenever possible, and performed control PCR assays without reverse transcriptase.
RNA editing was identified for all I. engelmannii mitochondrial protein-coding genes without exception (Table 1). In total, 1782 sites of pyrimidine exchanges in both directions were identified in messenger RNAs, which change 1406 codon meanings—the highest number of editing sites reported so far for a plant mitochondrial or chloroplast transcriptome. About 1/7th of these events (222 of 1782) are U-to-C changes. Interestingly, the fraction of silent RNA editing events leaving the encoded amino acid unchanged is far lower for the reverse U-to-C edits (six of 222, 2.7%) when compared to silent events among the C-to-U editing events (297 of 1560, 19%). A detailed listing of all editing sites following a recently proposed nomenclature (20,39) is given in Supplementary Table S1. This nomenclature is composed of the name of the respective gene, followed by an ‘e’ (for editing), the respective nucleotide introduced by the editing event (U or C), the nucleotide position in the transcript (with position 1 corresponding to the A of the AUG start codon) and finally the resulting amino acid change e.g. nad5eU22PS. Further qualifiers may be added such as ‘p’ for partial editing as we will discuss below.
Table 1.
Summary of editing sites detected in the mRNAs of the 25 protein encoding genes of the I. engelmannii chondrome
| Gene | Size [bp] | Codon changes by C > U editing |
C-to-U |
Codon changes by U > C editing |
U-to-C |
Total AA changes | Total sites | Creation of |
|||||||||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| A>V | H>Y | L>F | P>F | P>L | P>S | Q>* | R>* | R>C | R>W | S>F | S>L | T>I | T>M | Total | Silent | V>A | Y>H | F>L | F>P | L>P | S>P | *>Q | *>R | C>R | W>R | F>S | L>S | I>T | M>T | Total | silent | Start | Stop | ||||
| atp1 | 1545 | 5 | 9 | 1 | 3 | 18 | 12 | 0 | 0 | 5 | 0 | 5 | 20 | 8 | 4 | 104 | 10 | 3 | 0 | 0 | 0 | 0 | 1 | 12 | 3 | 5 | 0 | 0 | 1 | 2 | 0 | 27 | 0 | 117 | 131 | Yes | No |
| atp4 | 588 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 6 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 2 | 6 | No | No |
| atp6 | 762 | 3 | 4 | 1 | 7 | 12 | 5 | 0 | 1 | 3 | 1 | 9 | 15 | 3 | 3 | 83 | 9 | 1 | 1 | 0 | 0 | 1 | 0 | 3 | 2 | 2 | 0 | 0 | 1 | 1 | 0 | 12 | 0 | 79 | 95 | Yes | Yes |
| atp8 | 489 | 0 | 3 | 0 | 1 | 6 | 1 | 0 | 0 | 1 | 1 | 0 | 6 | 1 | 1 | 31 | 9 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 25 | 35 | Yes | No |
| atp9 | 225 | 1 | 0 | 1 | 2 | 4 | 1 | 1 | 0 | 0 | 0 | 4 | 4 | 2 | 2 | 30 | 6 | 3 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 26 | 34 | Yes | Yes |
| cob | 1170 | 4 | 11 | 6 | 4 | 17 | 1 | 0 | 0 | 1 | 6 | 8 | 13 | 2 | 1 | 99 | 20 | 3 | 3 | 0 | 0 | 4 | 3 | 4 | 1 | 1 | 0 | 0 | 2 | 1 | 0 | 22 | 0 | 96 | 121 | No | No |
| cox1 | 1632 | 0 | 5 | 5 | 2 | 16 | 4 | 0 | 0 | 2 | 6 | 11 | 11 | 2 | 3 | 88 | 18 | 4 | 4 | 1 | 0 | 0 | 2 | 2 | 1 | 2 | 0 | 2 | 1 | 1 | 0 | 22 | 1 | 87 | 110 | Yes | No |
| cox2 | 753 | 0 | 0 | 2 | 0 | 1 | 0 | 0 | 0 | 1 | 2 | 0 | 1 | 0 | 1 | 10 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 12 | 14 | Yes | No |
| cox3 | 792 | 3 | 3 | 1 | 8 | 17 | 6 | 1 | 0 | 1 | 9 | 10 | 5 | 2 | 6 | 98 | 18 | 0 | 1 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 0 | 75 | 101 | Yes | Yes |
| nad1 | 987 | 1 | 1 | 1 | 2 | 8 | 5 | 1 | 0 | 1 | 3 | 1 | 7 | 1 | 5 | 48 | 9 | 0 | 0 | 0 | 0 | 1 | 2 | 3 | 1 | 1 | 0 | 1 | 0 | 0 | 0 | 9 | 0 | 46 | 57 | Yes | No |
| nad2 | 1464 | 0 | 8 | 5 | 2 | 13 | 8 | 1 | 0 | 8 | 1 | 8 | 12 | 1 | 4 | 94 | 20 | 0 | 0 | 0 | 0 | 1 | 2 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 6 | 0 | 77 | 100 | No | Yes |
| nad3 | 357 | 0 | 2 | 0 | 1 | 6 | 3 | 1 | 0 | 2 | 1 | 12 | 8 | 3 | 0 | 53 | 13 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 4 | 0 | 42 | 57 | No | Yes |
| nad4 | 1488 | 2 | 8 | 1 | 9 | 23 | 6 | 1 | 0 | 5 | 5 | 13 | 17 | 3 | 6 | 140 | 30 | 0 | 2 | 1 | 0 | 2 | 3 | 2 | 2 | 0 | 0 | 1 | 2 | 0 | 0 | 16 | 0 | 114 | 156 | Yes | Yes |
| nad4L | 303 | 0 | 0 | 0 | 0 | 2 | 4 | 1 | 0 | 0 | 0 | 3 | 6 | 2 | 4 | 26 | 4 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 23 | 27 | Yes | Yes |
| nad5 | 1986 | 2 | 7 | 6 | 7 | 15 | 13 | 0 | 0 | 8 | 8 | 19 | 25 | 4 | 7 | 152 | 22 | 1 | 1 | 0 | 0 | 0 | 0 | 3 | 3 | 0 | 0 | 0 | 3 | 2 | 0 | 14 | 1 | 134 | 166 | No | No |
| nad6 | 627 | 2 | 1 | 1 | 4 | 11 | 2 | 0 | 0 | 1 | 3 | 1 | 8 | 2 | 5 | 58 | 12 | 0 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 6 | 1 | 46 | 64 | Yes | No |
| nad7 | 1191 | 5 | 7 | 0 | 2 | 16 | 3 | 1 | 0 | 5 | 2 | 8 | 13 | 5 | 8 | 95 | 18 | 0 | 4 | 1 | 0 | 2 | 0 | 5 | 5 | 1 | 0 | 0 | 0 | 1 | 0 | 20 | 0 | 94 | 115 | No | Yes |
| nad9 | 558 | 1 | 1 | 1 | 3 | 2 | 2 | 0 | 0 | 1 | 5 | 4 | 8 | 1 | 3 | 40 | 5 | 1 | 0 | 0 | 0 | 2 | 0 | 3 | 2 | 0 | 0 | 0 | 2 | 0 | 0 | 10 | 0 | 42 | 50 | Yes | No |
| rpl5 | 549 | 2 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 1 | 0 | 1 | 3 | 1 | 1 | 23 | 10 | 1 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 1 | 1 | 0 | 0 | 7 | 2 | 18 | 30 | No | No |
| rps2 | 699 | 0 | 0 | 0 | 0 | 2 | 2 | 0 | 0 | 3 | 0 | 0 | 8 | 0 | 0 | 23 | 8 | 0 | 0 | 0 | 0 | 1 | 1 | 1 | 0 | 0 | 0 | 1 | 0 | 1 | 0 | 5 | 0 | 20 | 28 | No | No |
| rps3 | 1200 | 2 | 4 | 0 | 1 | 5 | 4 | 0 | 0 | 3 | 2 | 6 | 8 | 1 | 0 | 56 | 19 | 0 | 0 | 0 | 0 | 0 | 0 | 4 | 2 | 0 | 0 | 0 | 0 | 0 | 0 | 6 | 0 | 42 | 62 | No | No |
| rps4 | 765 | 2 | 4 | 0 | 0 | 12 | 7 | 0 | 0 | 1 | 1 | 5 | 5 | 3 | 0 | 48 | 8 | 0 | 0 | 0 | 0 | 0 | 0 | 3 | 2 | 0 | 0 | 0 | 0 | 1 | 0 | 7 | 1 | 46 | 55 | No | No |
| sdh3 | 297 | 0 | 2 | 1 | 0 | 4 | 4 | 1 | 0 | 1 | 0 | 1 | 2 | 1 | 0 | 21 | 4 | 0 | 0 | 0 | 0 | 0 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 0 | 0 | 2 | 0 | 19 | 23 | No | Yes |
| tatC | 756 | 0 | 4 | 3 | 2 | 11 | 8 | 0 | 0 | 4 | 3 | 8 | 3 | 4 | 0 | 62 | 10 | 0 | 0 | 0 | 0 | 1 | 1 | 2 | 0 | 1 | 0 | 0 | 1 | 0 | 0 | 6 | 0 | 56 | 68 | No | No |
| mat* | 2019 | 1 | 8 | 0 | 0 | 24 | 8 | 0 | 0 | 3 | 3 | 3 | 6 | 5 | 2 | 72 | 9 | 1 | 0 | 0 | 0 | 0 | 0 | 3 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 5 | 0 | 68 | 77 | Yes | No |
| Partial | 1 | 7 | 4 | 0 | 16 | 9 | 2 | 0 | 3 | 1 | 12 | 15 | 10 | 3 | 282 | 198 | 2 | 0 | 2 | 0 | 1 | 0 | 16 | 6 | 0 | 0 | 1 | 2 | 2 | 0 | 38 | 6 | |||||
| Full | 35 | 85 | 32 | 60 | 232 | 101 | 8 | 1 | 58 | 61 | 129 | 199 | 47 | 63 | 1278 | 99 | 16 | 17 | 2 | 0 | 16 | 16 | 45 | 22 | 15 | 1 | 7 | 14 | 9 | 0 | 184 | 0 | |||||
| Total | 36 | 92 | 36 | 60 | 248 | 110 | 10 | 1 | 61 | 62 | 141 | 214 | 57 | 66 | 1560 | 297 | 18 | 17 | 4 | 0 | 17 | 16 | 61 | 28 | 15 | 1 | 8 | 16 | 11 | 0 | 222 | 6 | 1406 | 1782 | 12 | 9 | |
Codon conversions resulting from C-to-U and U-to-C editing that were observed in the complete coding sequences are listed separately from the respective silent editing events. Additionally indicated are editing events causing the creation of start and stop codons, and the fraction of sites for which either full or partial editing was observed in the respective cDNA pools. The total editing counts contain number of amino acid changes, silent editing and multiple editings affecting single codons (S>L [3], L>S [9], L>P [1], P>F [60]). The atp9i87 maturase (mat) is translated in frame with the two upstream atp9 exons and the asterisk indicates that editing sites in these exons were not included here but only counted once for the line labelled atp9.
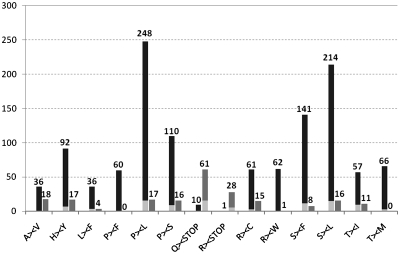
All predictable cases for reconstitution of appropriate AUG start codons from ACG threonine codons (12 cases) and of stop codons introduced through conversion of glutamine or arginine codons (nine cases) were confirmed (Table 1). Both ends of the reading frame are actually introduced by editing in five genes: atp6, atp9, cox1, nad4 and nad4L. Conversely, a total of 89 U-to-C editings are necessary to remove genomically encoded stop-codons within reading frames to recover 61 conserved glutamine and 28 arginine codons, respectively (Figure 1). In fact, only three of the 25 protein coding sequences—the small atp4, nad3 and nad4L genes—are free of in-frame stop codons whereas between one and up to 15 (in atp1) have to be removed at the transcript level in all others (Table 1). These conversions constitute a major share (42%) of the U-to-C type of editing events. In contrast, the recreation of leucine codons is the major effect of C-to-U editing with 462 codon changes (38.7%) altering either genomic proline or serine codons (Figure 1).
Figure 1.
Bar chart displaying the numbers of observed codon changes introduced through RNA editing in the I. engelmannii mitochondrial transcriptome. Dark gray bars display C-to-U and light gray bars represent the ‘reverse’ U-to-C exchanges. The numbers of conversions in the C-to-U direction is significantly higher for the 12 possible codon sense changes introduced by pyrimidine transitions with ratios ranging from 2:1 for A–V to 62:1 for R–W codon sense changes. Reverse U-to-C editing in contrast dominates in conversions involving stop codons. Lighter colours indicate the fraction of codon conversions in each type, for which partial editing was observed.
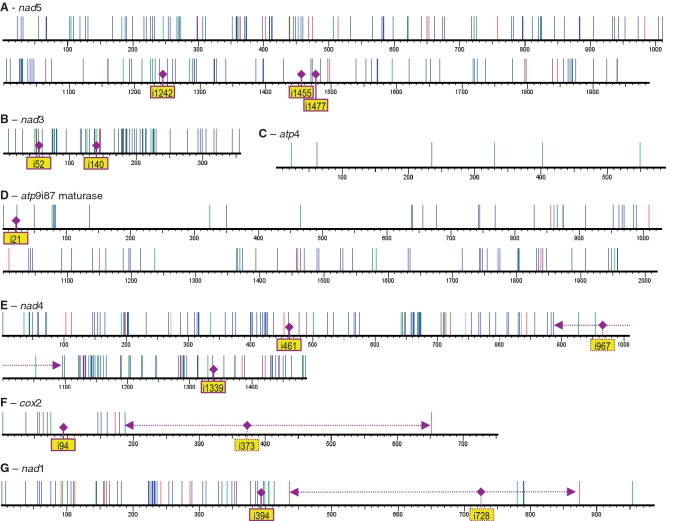
The numbers of editing sites per particular transcript varies widely. Most affected by RNA editing is the (large) nad5 mRNA, where 152 pyrimidine changes effect 133 codon changes (Figure 2A). The highest density of editing sites, however, is reached in the (small) nad3 mRNA, in which 57 out of 357 nucleotides are edited (Figure 2B). On the other end of the scale, only six edited sites were identified in atp4 mRNAs (Figure 2C). Among these, only one site (atp4eU62SF) is efficiently edited, four sites are partially edited silent sites (see below) and one partial editing event erroneously introduces a stop codon (atp4eU235Q*). This exceptionally deviant pattern of RNA editing in atp4 may suggest that the mitochondrial gene copy is evolving into a pseudogene to be replaced by a functional nuclear copy.
Figure 2.
RNA editing sites in I. engelmannii mitochondrial genes displayed with the default graphic tool options of PREPACT (39). Non-silent C-to-U and U-to-C editings are indicated by blue or red lines, respectively, and green lines indicate silent editings. RNA-editing patterns are exemplarily shown for selected genes nad5 (A), nad3 (B), atp4 (C), the maturase in atp9i87g2 (D), nad4 (E), cox2 (F) and nad1 (G). Group II intron insertion sites are indicated, those shown with stippled lines are absent in Isoetes, coinciding with regions lacking RNA-editing sites (horizontal lines with arrowheads). The atp9i87 maturase reading frame is in frame with the first 29 codons of atp9 which contain the upstream group II intron atp9i21.
RNA editing of the single intron-encoded ORF in Isoetes mtDNA
Introns in the I. engelmannii mtDNA are generally very small, irrespective of the class they belong to (groups I or II) and do not carry intron-encoded open reading frames (ORFs) for endonucleases or maturases, respectively. The unique exception is group II intron atp9i87g2, which carries maturase sequence similarities with its Marchantia polymorpha homolog. Two overlapping intron regions were RT–PCR-amplified to gain complete sequence insights on potential functional significance of this maturase homology on cDNA level. Indeed, a conserved reading frame with a size of 2019 bp (672 amino acids) was found to be reconstituted by RNA editing. Altogether, 81 codons in the maturase sequence are affected, including conversion of four stop codons into one arginine and three glutamine codons (Figure 2D). The majority of editing events in the I. engelmannii atp9i87g2 maturase is located in conserved maturase domains and significantly increases sequence similarity with the Marchantia homolog (Figure 3). Similar to the situation in Marchantia, the maturase ORF in atp9i87g2 is translated in-frame with upstream atp9 exon(s). Hence, apart from RNA editing, splicing of the upstream atp9 intron atp9i21g2 not present in Marchantia (Figures 2D and 3) would be an additional prerequisite for proper maturase translation in I. engelmannii. In addition to the editing sites within the atp9i87g2 maturase reading frame, we could identify two more editing sites in the downstream domains V and VI of the group II intron secondary structure (see below).
Figure 3.
Sequence alignment of the I. engelmannii maturase protein sequence in intron atp9i87g2 and its homolog from M. polymorpha. The Isoetes cDNA-derived protein sequence results from RNA editing of 81 DNA-encoded codons as shown below the alignment. Background shading colours indicate C-to-U (cyan), U-to-C (magenta) and silent edits (green). Codon changes increasing sequence similarity with Marchantia are underlined and are mostly located within conserved maturase domains 0–7 and ‘X’ (56) as indicated. The atp9i87 maturase is translated in frame with the small upstream exon(s) of atp9 across the splice donor site (arrowhead). Upstream intron atp9i21g2 (vertical line) is present in Isoetes, but not in Marchantia.
Partial RNA editing at 320 sites in the I. engelmannii mitochondrial transcriptome
Although operating very efficiently at most sites in plant organelles, RNA editing is not a yes/no process. The analyses of independent cDNA clones or of sequence data generated by direct sequencing of RT–PCR amplicons can lead to the discovery of partially edited transcripts. To account for recognized partial editing in the recently proposed RNA editing nomenclature (39) we have introduced the additional qualifier ‘p’. With sufficient cDNA data available the ‘p’ qualifier may be followed by the percentage of cDNA clones reflecting a given editing event. To estimate the extent of partial editing we generally sequenced several independent cDNA clones (on average 16 per locus). Partial editing sites were considered as verified only when deviant pyrimidines were determined at least twice independently in the respective cDNA population (i.e. above the threshold of occasional PCR-derived sequence errors). Independent cDNA clone sequencing versus RT–PCR product bulk sequencing has the benefits of higher sequence qualities and, more importantly, allows to detect editing site interdependence among the diverse cDNA-editing patterns (as discussed below for the nad4 case). However, independent cDNA clone sequencing comes at the risk of cloning bias. For comparison, we sequenced several selected RT–PCR amplicons (for nad1, nad3, nad4 and nad7) directly without observing significant discrepancies in comparison to populations of cDNA clones, as exemplarily outlined for nad3 (Supplementary Figure S1). All events of full (100%) editing were equally detected as such both in direct RT–PCR product and cDNA-clone sequencing. Similarly, no further evidence for any editing was found by direct amplicon sequencing that was not equally reflected in cDNA pool sequencing. In contrast, however, only some of the partial editing events of particularly low (<10%) or high (>90%) efficiency identified in the cDNA populations were adequately reflected in direct RT–PCR sequences, whereas others would have been missed in the latter approach. Given that such sites are equally detected in spliced and unspliced cDNA clones (see below for comparison of mature versus immature nad7 transcripts), we consider the cDNA clone sequences to faithfully and better reflect the RNA-editing status of transcripts. Certainly the P-value percentages may suggest higher precision of determining partial editing efficiencies than actually given before some 100 independent cDNA clones are sequenced. As an alternative for such pseudo-precise values five stages of editing efficiency could alternatively be distinguished, e.g. pVH, very high (>80%); pHI, high (>60%); pME, medium (40–60%); pLO, low (<40%) and pVL, very low (<20%).
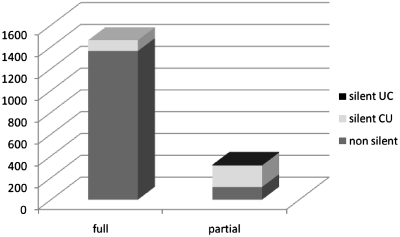
In total, 320 out of the 1782 editing sites in mRNAs were identified as partially edited. A full 63.8% of these partial editing events (204/320) affect silent editing sites, leaving codon identities unchanged (Figure 4). In contrast, only 6.8% of the fully edited sites (99/1462) affect silent positions. Partial editing at silent sites is particularly ineffective: 111 of the 206 partially edited silent sites are edited in <25% of sequenced cDNA clones (Supplementary Table S1).
Figure 4.
Bar chart displaying the fractions of silent editings for both directions of pyrimidine conversions among fully (left) versus partially (right) edited sites. All six silent U-to-C editing sites are partially edited (right).
The remaining, non-silent partial editing sites affect 115 codon changes (Figures 1 and 4). On average, partial editing affects 8.7% of codon conversions of a particular type. Individual exceptions with higher degrees of partial editing are the C-to-U type threonine into isoleucine codon conversions (10/57 = 18%) and the U-to-C editings converting phenylalanine into leucine (2/4 = 50%), stop into glutamine (16/61 = 26%) and stop into arginine (6/28 = 21%) codons (Figure 1). In some instances partial editing may (to generally low degrees) lead to mis-conversions of evolutionarily conserved codons on DNA level or erroneously introduce stop codons: atp6eU182SFp13, atp6eU430HYp13, nad1e U790Q*p13, atp4eU235Q*p32. The latter case, in particular may indicate an ongoing degeneration of the I. engelmannii atp4 gene into a pseudogene as mentioned above. Such events with low efficiencies of partial editing and the high proportion of silent sites among the partial editing events support the idea that a majority of those reflect collateral misfiring of the editing machinery targeting other sites. In other cases, however, partial editing of relevant sites may simply reflect inefficiencies of the particular editing factors.
Different editing patterns among independent nad4 cDNA clones
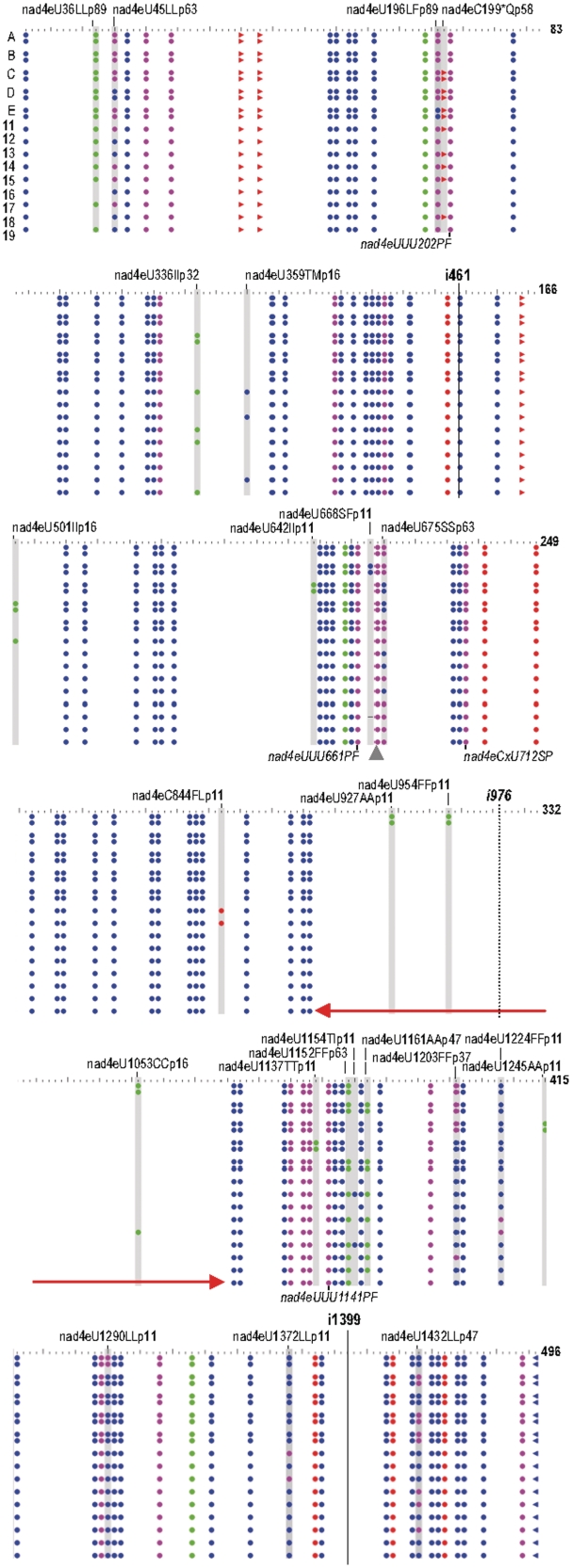
We present the nad4 gene as a typical model case for frequent RNA editing including numerous reverse, silent and partial RNA-editing events. With only five pairs of identical sequences, 14 different editing patterns were revealed in a pool of 19 nad4 cDNA clones (Figure 5). In the case of the nad4 gene, 140 C-to-U and 16 U-to-C editing sites lead to 114 codon changes (Figure 1E). Both the start and the stop codon have to be introduced by RNA editing to define the correct nad4 reading frame ends and four genomically encoded stop-codons have to be eliminated from the reading frame to avoid early termination of translation (Table 1). Additionally, 30 silent editing sites are located at first or third triplet positions not influencing the encoded protein (Supplementary Table S1). The majority of partial editing sites (19 of 25) affects silent sites to varying degrees (between 2 and 17 out of 19 clones): nad4eU36LLp89, nad4eU45LLp63, nad4e U675SSp63, nad4eU1152p63, nad4eU672FFp58, nad4e U1161AAp47, nad4eU1432LLp47, nad4eU1203FFp37, nad4eU336IIp32, nad4eU501IIp16, nad4eU1053CCp16, nad4eU642IIp11, nad4eU927AAp11, nad4eU954FFp11, nad4eU1137TTp11, nad4eU1224FFp11, nad4eU1245 AAp11, nad4eU1290LLp11 and nad4eU1372LLp11 in descending order of editing frequencies. Three additional partial editing events are expected as they reinstate evolutionarily conserved amino acid codons: nad4eU196LFp89, nad4eC199*Qp58 and nad4eU359 TMp16. Finally, three further sites of partial editing are unexpected as they introduce evolutionarily non-conserved amino acid codons. Notably, all three occur at low frequencies in only two out of 19 clones each: nad4eU668 SFp11, nad4eC844FLp11 and nad4eU1154TIp11. Considering the above, only two of 19 clones (cDNAs #13 and #18) actually reflect ‘proper’ editing as predicted with editing at the three expected but not at the three unexpected partial sites (Figure 5). These cDNAs are also non-edited at most silent partial editing sites. On the other side of the spectrum, cDNA pair #A lacks several important codon conversions but in contrast shows silent editing at the infrequently, partially edited positions (Figure 5).
Figure 5.
RNA-editing status in a population of 19 nad4 cDNA clones. The graphic is based on the cDNA variants overview of PREPACT (39) with default settings: blue and red circles indicate codon sense changes derived from C-to-U or U-to-C editings, respectively, green circles indicate silent codon changes and purple circles reflect more than one editing in a codon. Forward arrows indicate stop codon removal in codons 35, 38, 67, 164 and 496 while reverse arrows indicate stop-codon creation at the end of the reading frame. Positions of partial editing are highlighted with gray columns and the labeling for the respective editing events are indicated. With five pairs of identical cDNA sequences (A–E) each showing identical partial editing patterns, a total of 14 different editing patterns is recognized. Group II intron insertion sites nad4i461 and nad4i1399 (solid lines) in I. engelmannii mtDNA are indicated, intron nad4i976 (dotted line) is absent but conserved in other taxa. The intron loss coincides with absence of editing sites over an extended nad4 region (red arrows) which may be the result of cDNA retroprocessing.
Concomitant loss of introns and RNA-editing sites
A total of 27 group II introns and three group I introns were predicted on the basis of the I. engelmannii mitochondrial DNA sequence (37). The cDNA analyses performed here showed that all 30 introns were correctly spliced as predicted in spite of their generally small sizes. As common for plant mitochondrial introns, many of them are shared with other land plant groups whereas some Isoetes introns were novel discoveries. In contrast, some ancient introns conserved in other plant clades (including non-liverwort bryophytes and angiosperms) were absent in I. engelmannii: cox2i373, nad1i728 and nad4i967.
A lack of editing positions (except for the few partially edited silent sites) which are otherwise densely packed in I. engelmannii is immediately apparent in nad4 between amino acid positions 297 and 365, which corresponds to the gene region surrounding intron nad4i967g2 present in other plant species (Figures 2E and 5). In full congruence, two other extended gene regions in cox2 (Figure 2F) and nad1 (Figure 2G) lacking RNA-editing sites also perfectly coincide with the absence of introns in I. engelmannii which are present in other taxa: cox2i373 and nad1i728. Only one single U-to-C editing site (cox2eC652*Q) essential for the correction of a genomically encoded stop codon was identified in the cox2 3′-region, located >300-nt downstream of the intron cox2i373 insertion site in other species. Likewise, the closest upstream editing event, cox2eU187LF, is located in a distance of nearly 200 nt (Figure 2F). Similarly, editing sites are absent within some 300-nt upstream of the nad1i728g2 insertion site in other taxa up to editing site nad1eC436*Qp88 (Figure 2G). Interestingly, all sites downstream of the intron insertion site are irrelevant and/or weakly edited: nad1eU780SSp13, nad1eU789FFp13, nad1eU790Q*p13 and nad1eU954SSp13 with the unique exception of nad1eC874*R. Congruently only three silent sites are partially and rarely edited (nad4eU927AAp11, nad4e U954FFp11 and nad4eU1053CCp16) in proximity to the lost intron nad4i967 within the region ranging from base 888 to 1095 (Figure 2E). We conclude that simultaneous absence of RNA-editing sites and introns in these three gene regions in cox2, nad1 and nad4 is a result of recent partial retro-processing of mature mRNAs.
Editing in tRNAs
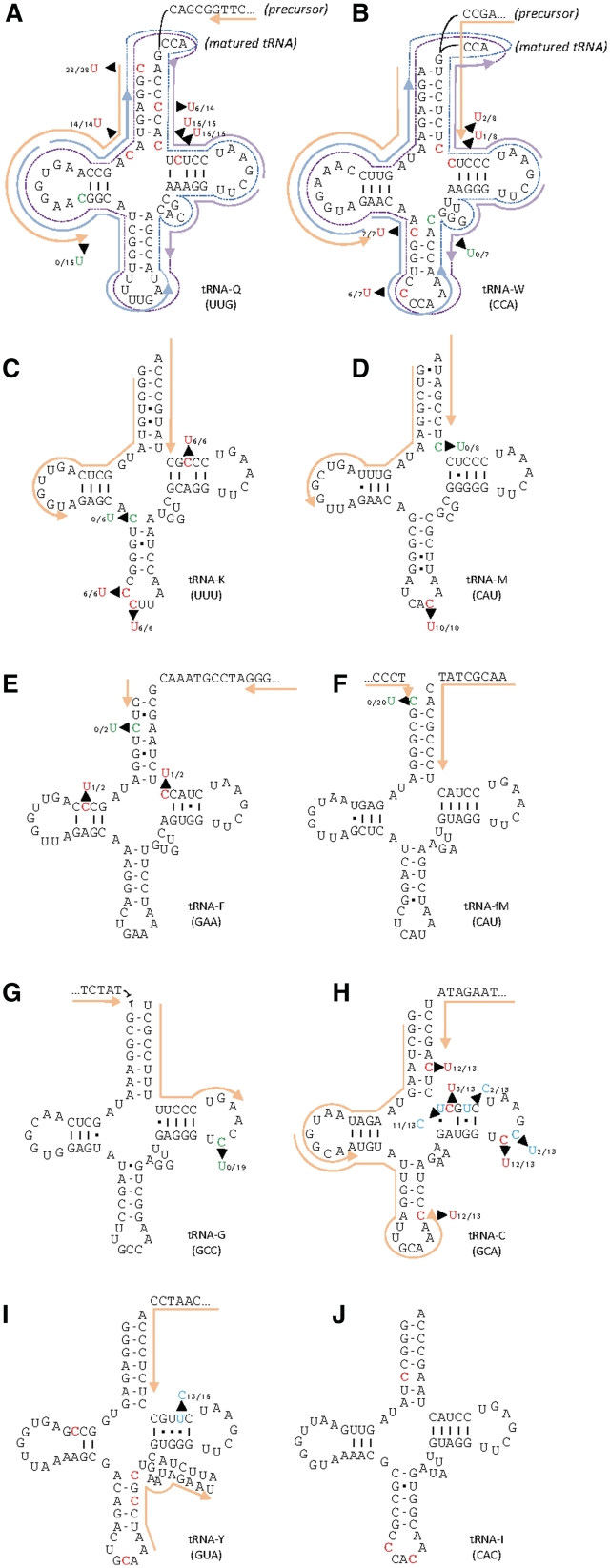
Modeling of the cloverleaf secondary structures of mitochondrially encoded tRNAs in I. engelmannii had revealed that RNA editing may also affect tRNAs to a much higher degree than previously observed in land plants (37). Altogether, 43 sites of C-to-U editings can be predicted in several crucial positions, which could reconstitute conserved uridine residues in the tRNA-consensus structure or improve base pairings in stem regions. The initial cDNA-sequence analysis of the tRNA for proline (trnP) had revealed that such editing sites could indeed be confirmed and that RNA editing takes place already in precursor transcripts with trnP still connected to the downstream sdh3 gene (37). Now, we first investigated whether the tRNA-editing status may differ between precursor versus processed tRNAs after 5′- and 3′-trimming and the addition of the CCA tail. To target the matured tRNAs, we used self-ligation across the tRNA-ends, followed by overlapping RT–PCRs. We succeeded in obtaining such fully matured, CCA-tailed tRNA sequences for tRNA–Q(UUG) and tRNA–W(CCA) in parallel to the cDNA sequences for the corresponding tRNA precursors with primers targeting sequences flanking the tRNAs (Figure 6A and B). No differences in RNA-editing status were observed, confirming that the RNA-editing machinery indeed targets tRNA precursor sequences before processing, as already suggested by the initial analysis of trnP.
Figure 6.
Outline of editing in 10 tRNAs. Nucleotides highlighted in red were found to be edited from C to U (A–J), those highlighted in blue were identified as edited from U to C in cDNA analyses with the numbers of edited and total cDNAs indicated for each site before and after the slash, respectively. Nucleotides shown in green were initially predicted to be editing sites but were found unchanged in all studied cDNAs. Primers successfully used in RT–PCR of precursor transcripts are shown in orange. No cDNA sequences could be retrieved for trnI (J) and the 5′-part of trnY (I). Overlapping primer pairs used in the parallel amplification of self-ligated, circularized trnQ (A) und trnW (B) are indicated in blue and purple with the respective amplified regions shown with stippled lines.
The analysis of the editing status of trnQ revealed five editing sites exactly as predicted: four of these re-establish A–U pairings in the acceptor stem and in the pseudouridine stem and one re-establishes the conserved uridine residue in position 8 (Figure 6A). A further potential site of editing (position 21), however, remained unedited in the trnQ dihydrouridine stem. Similarly, four of five predicted editing sites were identified in trnW (Figure 6B). Base pairing mismatches are corrected in the acceptor, pseudouridine and anticodon stems and the conserved uridine in position 33 is re-established. However, a proximal A–C mismatch in the anticodon stem remained uncorrected. Interestingly trnQ and trnW feature unmatched bases in these positions in other taxa as well (e.g. A–A in the liverwort M. polymorpha). These mispairings may in fact be relevant to tRNA functionality such as appropriate amino acid charging. Analogous observations were made for the other tRNA species as well: three predicted sites were completely edited in trnK but the corresponding base mismatch in the proximal anticodon stem position remains unaltered (Figure 6C) and again, this is a base mismatch position also present in Marchantia. Similarly, anticodon position 36 of trnM is found edited as expected whereas the proximal acceptor stem base mismatch remains unchanged (Figure 6D) and positions in the dihydrouridine and the pseudouridine stem but not in the acceptor stem are edited in trnF (Figure 6E). In two further cases of tRNAs with one potential, predicted editing each (trnfM, trnG), we did not confirm the editing events in cDNAs. This is not astonishing for trnfM where the distal acceptor stem mismatch again is also present in Marchantia (Figure 6F), but all the more in the latter case, given that the conserved GUUC motif of the pseudouridine loop is absent in trnG (Figure 6G). Moreover, the corresponding position (55 in the tRNA-consensus structure) was found to be edited both in the previously analyzed trnP and in the now investigated trnC (Figure 6H). Investigation of editing in trnC also revealed the first-documented case of reverse U-to-C editing in a tRNA. Here, the weak U–G pair in the proximal position of the pseudouridine stem is converted into a stronger C–G base pair. Two further C-to-U edits efficiently reconstitute base pairings in the acceptor and anticodon stem of trnC as predicted (Figure 6H). Interestingly, three more positions of the trnC molecule reveal inefficient partial editing and two of these are also of the reverse U-to-C type. Of these three partial editing events, two are ‘reasonable’ in the sense that they could further improve base pairing in the pseudouridine stem. However, the event affecting position 56 would destroy the GUUC consensus motif. In spite of numerous different attempts we could obtain only partial cDNA information for trnY (Figure 6I) and none for trnI (Figure 6J). Given the novel insights on U-to-C editing in trnC, we wondered whether similar events could take place in the trnY pseudouridine stem, which is particularly rich in G–U pairs. We could indeed confirm a further and efficient reverse editing event in position 62 (Figure 6I), but were unable to retrieve cDNA covering the remaining four predicted positions in trnY or the three predicted position in trnI.
In summary, out of 43 predicted sites of tRNA editing, cDNA could not be retrieved for seven. Of the remaining 36 candidate sites, 29 were confirmed to reveal C-to-U editing. Additionally, four positions of U-to-C editing were discovered of which two are partially edited to a low degree. This suggests that partial/inefficient tRNA-editing mirrors the results observed for silent or mis-editing of mRNAs.
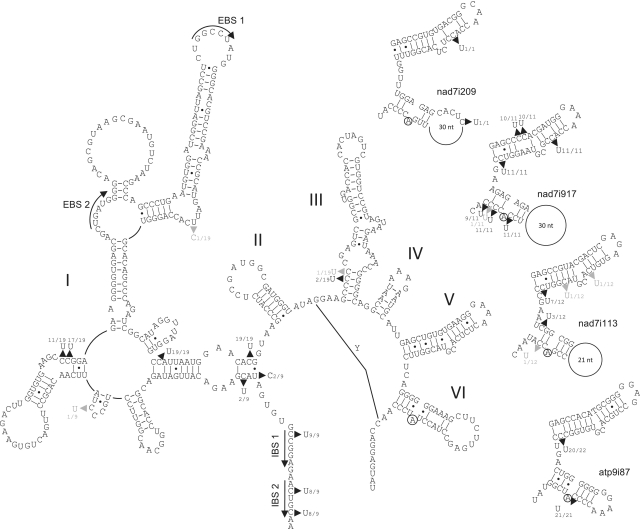
Mitochondrial intron editing in nad7
The finding of frequent tRNA editing as well as two RNA-editing events in intron atp9i87g2 downstream of its maturase prompted us to investigate more mitochondrial intron sequences systematically on cDNA level. To this end, we explored the most intron-rich nad7 gene in I. engelmannii mtDNA with its four group II introns nad7i209, nad7i676, nad7i917 and nad7i1113. Here, RT–PCR approaches could be designed to ideally select for partially matured transcripts that remained unspliced for the respective intron under investigation but were spliced for other introns of the nad7 gene. Overlapping intron amplicons were designed with one primer binding in the respective intron and one in a flanking or distant exon. The sequencing of such partially matured transcript cDNAs revealed editing sites in the flanking exons that had previously been determined in mature cDNAs as well as several editing sites in the intron regions. In total, we found 27 sites of RNA editing in the four nad7 introns reliably determined as they were identified in more than one cDNA clone each. As observed in coding regions and tRNAs, the preferential direction of editing is C-to-U, counting 26 sites. Most intron editing sites were partially edited and only seven of the 27 editing sites were edited in all sequenced cDNA clones. Mapping the sites of RNA editing onto secondary structure models suggest that many of the editing events may actually improve RNA base pairings and might be a prerequisite for splicing as exemplarily shown for nad7i676 (Figure 7). RNA editing converts five A–C mispairings in domain I of nad7i676g2 into canonical A–U base pairs. However, one obvious A–C base mismatch in domain VI is not subject to editing. Interestingly, an editing site 9-nt downstream of the 5′-splicing site has recently been found to be mandatory for splicing (41) and the corresponding position is subject to RNA editing in nad7i676g2 in addition to three more sites involved in stem formation of domain I. At the basal stem of domain I, one G–U wobble base pair is modified to a more stable G–C Watson–Crick base pair by U-to-C editing—to our knowledge the first U-to-C intron editing observed. In contrast, stem stability of domain IV is loosened by RNA editing introducing a weaker G–U wobble base pairing. No editing was observed at exon binding sites (EBS). However, editing at three positions in intron binding sites (IBS 1 and IBS 2) in the upstream nad7 exon introduce weaker wobble pairings. No editing events were seen in conserved intron domains V and VI of nad7i676. However, ten C-to-U editing sites located in intron stems and necessary to remove A–C mispairings were disclosed in the other nad7 introns nad7i209, nad7i917 and nad7i1113 and this includes several sites in their highly conserved domain V and VI structures (Figure 7). Likewise the two editing sites in atp9i87 outside of its maturase are also located in these two domains. Only a single additional editing site discovered in the loop of domain VI of intron nad7i209 is obviously not involved in intron secondary structure stability (Figure 7).
Figure 7.
Left: complete secondary structure model of I. engelmannii group II intron nad7i676g2 following the proposed consensus structure (57). Despite its reduced size the essential characteristic features in the six conserved group II intron domains I to VI are present and classify nad7i676g2 as a member of subgroup IIB. The conserved bulged adenosine for lariat formation during splicing is encircled. Selected tertiary interactions EBS-IBS and γ–γ’ are indicated. Arrow heads indicate editing sites identified in unspliced nad7 cDNAs with numbers before and after the slash indicating the amounts of edited and total sequenced cDNA clones for each site. Right: conserved domains V and VI of introns nad7i209, nad7i917, nad7i1113 and atp9i87 respectively. Editing is indicated as described above.
The nad7 dataset of immature, partially spliced cDNAs allows for a comprehensive comparison with the previously determined editing data from fully spliced cDNAs (Figure 8). The data clearly show that many non-silent sites are already edited fully or to a considerable extent in unspliced pre-mRNAs. Notable exceptions are nad7eU277RC and the stop codon removals like nad7eC82*R, which is significantly less edited in unspliced versus spliced cDNAs. Likewise, many silent edits are identified less frequently in unspliced versus spliced transcripts. This finding supports the idea that such silent editings may represent results of mis-targeting that become more apparent with transcript age in the mitochondria.
Figure 8.
Comparison of RNA-editing status for nad7 exon sites with bars indicating percentage of editing among spliced (left, dark gray) and partially spliced immature cDNAs (right, light gray). Intron-insertion sites and the overall extension of the exon region covered for partially matured transcripts (dotted line) are indicated.
DISCUSSION
Isoetes engelmannii has now surpassed all other plant taxa in frequency of reported RNA editing, including the gymnosperm Cycas taitungensis, for which close to 1000 sites were predicted (39,42) and nearly 600 confirmed among 25 of its 39 genes that were recently analyzed (43). Why and how plant organelle RNA editing has emerged in the first place still remains mysterious. General speculations on benefits of editing include a gain in gene expression variability or a compensating mechanism for mutations. However, in our opinion, strong conclusive support has never been found for any of these possible explanations. In any case, they are challenged by the striking variability of editing patterns and frequencies among land plants, which suggest an (occasionally very fast) coming and going of editing sites through the course of evolution. With this regard, we consider the editing data for the bizarre mitochondrial transcriptome of I. engelmannii a strong case in point. The maintenance of genetic machinery specifically recognizing more than 1800 editing positions, the overwhelming majority of which restores proper mRNA, tRNA and intron sequences, seems unlikely to be of regulatory benefit. It is hardly conceivable that more than 100 editing sites in a single mRNA offer important new dimensions in modulation of protein activity.
Among all hypotheses on evolutionary origins and pathways that have been put forward (44–47), we consider those most likely which simply assume merely neutral evolution of a vast molecular rococo in the genetic playground of endosymbiotic organelles. An important recent contribution in this regard highlights the convergent pathways in the evolution of peculiar phenomena including RNA editing in Euglenozoa and Dinoflagellates (48).
Much progress has recently been made to understand the underlying mechanisms of plant organelle RNA editing, notably with the identification of several protein factors specifically targeting editing sites. After the seminal discovery of a particular pentatricopetide repeat (PPR) protein necessary for an editing event in the Arabidopsis thaliana chloroplast transcriptome (14), a similar factor was discovered for mitochondrial editing (15) among several others in both organelles (16,49).
The vast amount of editing and the screening of multiple independent cDNA clones for I. engelmannii gives a conclusive picture on the issue of partial, silent, irrelevant or non-beneficial editing: while some (efficient) partial editings suggest minor inefficiency of the underlying mechanisms, other (inefficiently) partially edited sites suggest unspecific binding of PPR-proteins actually targeting other important editing sites. In one case, we assume the RNA-editing pattern to reflect an emerging pseudogene after a likely gene transfer to the nucleus. The I. engelmannii atp4 gene is characterized by low-level editing with silent sites dominating and editing event atp4eU235Q*p32 in fact de-functionalizing the gene’s transcript. Most interestingly, atp4 appears to be actually missing from the mitochondrial gene complement of Isoetes’ sister genus Selaginella (J. Hecht et al. unpublished data).
Obviously, editing sites can be gained but they can also be lost subsequently. The here reported cases of larger scale editing site losses in the environment of lost introns in three genes strongly support the idea of retro-processed mature mRNAs via reverse transcriptase mechanisms. Interestingly, the three group II introns in question which are lost from the I. engelmannii mtDNA (cox2i373, nad1i728 and nad4i967) are known to be lost independently among angiosperms, too. Yet more noteworthy, only one single maturase (in atp9i87) could possibly provide the necessary reverse transcriptase activity for retroprocessing in Isoetes mitochondria with its otherwise tiny introns. The much larger number of intron-encoded maturases in M. polymorpha (50) may actually be the cause of complete absence of RNA editing in this liverwort by providing more extensive retroprocessing.
A further enigma concerns the occurrence of reverse U-to-C editing accompanying the classic C-to-U editing. After the early discoveries of rare U-to-C edits (31,32), similar events have been reported very rarely in flowering plants and none have been confirmed for mosses and liverworts. In contrast, significant amounts of reverse U-to-C editings can be identified in hornworts and ferns (34–36,51). Given the now well-corroborated sister-group relationship of hornworts and vascular plants (52,53), the occurrence and rise in frequency of reverse U-to-C editing in plant evolution may actually be connected to the common ancestor of hornworts and early tracheophytes after the split from liverworts and mosses. In any case, base deamination can certainly not be the biochemical mechanism behind U-to-C editing and (trans-)amination mechanisms rather need to be looked for. Hence, it will be highly exciting to see the first protein factor relevant for a U-to-C type of editing event identified. The so far ‘non-model’ taxa such as the hornworts, ferns or Isoetes will be the obvious organisms for investigation.
Finally and as conclusively shown above for I. engelmannii, it is interesting to find the same biochemical constraints defining the ratios of U-to-C versus C-to-U editing and the fractions of partial or irrelevant editings reflected in mRNAs, tRNAs and intron sequences. Similar to editings in mRNAs, which can be predicted as they reconstitute conserved codon identities, editing in the structured RNAs can be predicted as they reconstitute base pairings. In those instances where tRNA editing could have been assumed to take place in order to create base pairings but was not observed, the unedited state is most likely of functional relevance. In fact, examples have been reported, where RNA editing creates U–U mispairings in the trnC of dicots (54,55). Interestingly, we could not identify any editing sites in the mitochondrial rRNAs of I. engelmannii. In the light of ample editing seen in precursor tRNAs and unspliced mRNAs, this suggests that the RNA-editing machinery very preferentially acts on immature and single-stranded RNAs and that folding and processing of rRNAs, at least in I. engelmannii mitochondria, proceeds too fast for RNA-editing evolution to take hold in this type of RNAs as well.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutsche Forschungsgemeinschaft [DFG Kn411/6-1].
ACKNOWLEDGEMENTS
We are grateful to Dr Jeffrey Palmer and colleagues (Bloomington, IN, USA) for generously making fresh material of I. engelmannii available to us and to Dr Jeffrey Mower for comments on the atp9 maturase in Isoetes. We also wish to thank Henning Lenz for his work on the PREPACT tool and Julia Hecht for proofreading the article. Sequences obtained in this study were deposited in GenBank (accession numbers HQ616410-HQ616434).
REFERENCES
- 1.Gualberto JM, Lamattina L, Bonnard G, Weil JH, Grienenberger JM. RNA editing in wheat mitochondria results in the conservation of protein sequences. Nature. 1989;341:660–662. doi: 10.1038/341660a0. [DOI] [PubMed] [Google Scholar]
- 2.Covello PS, Gray MW. RNA editing in plant mitochondria. Nature. 1989;341:662–666. doi: 10.1038/341662a0. [DOI] [PubMed] [Google Scholar]
- 3.Hiesel R, Wissinger B, Schuster W, Brennicke A. RNA editing in plant mitochondria. Science. 1989;246:1632–1634. doi: 10.1126/science.2480644. [DOI] [PubMed] [Google Scholar]
- 4.Hoch B, Maier RM, Appel K, Igloi GL, Kössel H. Editing of a chloroplast mRNA by creation of an initiation codon. Nature. 1991;353:178–180. doi: 10.1038/353178a0. [DOI] [PubMed] [Google Scholar]
- 5.Karcher D, Bock R. Temperature sensitivity of RNA editing and intron splicing reactions in the plastid ndhB transcript. Curr. Genet. 2002;41:48–52. doi: 10.1007/s00294-002-0278-y. [DOI] [PubMed] [Google Scholar]
- 6.Bentolila S, Chateigner-Boutin AL, Hanson MR. Ecotype allelic variation in C-to-U editing extent of a mitochondrial transcript identifies RNA-editing quantitative trait loci in Arabidopsis. Plant Phys. 2005;139:2006–2016. doi: 10.1104/pp.105.069013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zehrmann A, van der Merwe JA, Verbitskiy D, Brennicke A, Takenaka M. Seven large variations in the extent of RNA editing in plant mitochondria between three ecotypes of Arabidopsis thaliana. Mitochondrion. 2008;8:319–327. doi: 10.1016/j.mito.2008.07.003. [DOI] [PubMed] [Google Scholar]
- 8.Bock R, Hagemann R, Kössel H, Kudla J. Tissue- and stage-specific modulation of RNA editing of the psbF and psbL transcript from spinach plastids - a new regulatory mechanism? Mol. Gen.Genet. 1993;240:238–244. doi: 10.1007/BF00277062. [DOI] [PubMed] [Google Scholar]
- 9.Grosskopf D, Mulligan RM. Developmental- and tissue-specificity of RNA editing in mitochondria of suspension-cultured maize cells and seedlings. Curr. Genet. 1996;29:556–563. doi: 10.1007/BF02426960. [DOI] [PubMed] [Google Scholar]
- 10.Howad W, Kempken F. Cell type-specific loss of atp6 RNA editing in cytoplasmic male sterile Sorghum bicolor. Proc. Natl Acad. Sci. USA. 1997;94:11090–11095. doi: 10.1073/pnas.94.20.11090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Miyata Y, Sugita M. Tissue- and stage-specific RNA editing of rps14 transcripts in moss (Physcomitrella patens) chloroplasts. J. Plant Physiol. 2004;161:113–115. doi: 10.1078/0176-1617-01220. [DOI] [PubMed] [Google Scholar]
- 12.Ruf S, Kössel H. Tissue-specific and differential editing of the two ycf3 editing sites in maize plastids. Curr. Genet. 1997;32:19–23. doi: 10.1007/s002940050242. [DOI] [PubMed] [Google Scholar]
- 13.Karcher D, Bock R. The amino acid sequence of a plastid protein is developmentally regulated by RNA editing. J. Biol. Chem. 2002;277:5570–5574. doi: 10.1074/jbc.M107074200. [DOI] [PubMed] [Google Scholar]
- 14.Kotera E, Tasaka M, Shikanai T. A pentatricopeptide repeat protein is essential for RNA editing in chloroplasts. Nature. 2005;433:326–330. doi: 10.1038/nature03229. [DOI] [PubMed] [Google Scholar]
- 15.Zehrmann A, Verbitskiy D, van der Merwe JA, Brennicke A, Takenaka M. A DYW domain-containing pentatricopeptide repeat protein is required for RNA editing at multiple sites in mitochondria of Arabidopsis thaliana. Plant Cell. 2009;21:558–567. doi: 10.1105/tpc.108.064535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chateigner-Boutin AL, Small I. Plant RNA editing. RNA Biol. 2010;7:213–219. doi: 10.4161/rna.7.2.11343. [DOI] [PubMed] [Google Scholar]
- 17.Tasaki E, Hattori M, Sugita M. The moss pentatricopeptide repeat protein with a DYW domain is responsible for RNA editing of mitochondrial ccmFc transcript. Plant J. 2010;62:560–570. doi: 10.1111/j.1365-313X.2010.04175.x. [DOI] [PubMed] [Google Scholar]
- 18.Steinhauser S, Beckert S, Capesius I, Malek O, Knoop V. Plant mitochondrial RNA editing: extreme in hornworts and dividing the liverworts? J. Mol. Evol. 1999;48:303–312. doi: 10.1007/pl00006473. [DOI] [PubMed] [Google Scholar]
- 19.Groth-Malonek M, Wahrmund U, Polsakiewicz M, Knoop V. Evolution of a pseudogene: Exclusive survival of a functional mitochondrial nad7 gene supports Haplomitrium as the earliest liverwort lineage and proposes a secondary loss of RNA editing in Marchantiidae. Mol. Biol. Evol. 2007;24:1068–1074. doi: 10.1093/molbev/msm026. [DOI] [PubMed] [Google Scholar]
- 20.Rüdinger M, Funk HT, Rensing SA, Maier UG, Knoop V. RNA editing: 11 sites only in the Physcomitrella patens mitochondrial transcriptome and a universal nomenclature proposal. Mol. Genet. Genom. 2009;281:473–481. doi: 10.1007/s00438-009-0424-z. [DOI] [PubMed] [Google Scholar]
- 21.Giegé P, Brennicke A. RNA editing in Arabidopsis mitochondria effects 441 C to U changes in ORFs. Proc. Natl Acad. Sci. USA. 1999;96:15324–15329. doi: 10.1073/pnas.96.26.15324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Notsu Y, Masood S, Nishikawa T, Kubo N, Akiduki G, Nakazono M, Hirai A, Kadowaki K. The complete sequence of the rice (Oryza sativa L.) mitochondrial genome: frequent DNA sequence acquisition and loss during the evolution of flowering plants. Mol. Genet. Genom. 2002;268:434–445. doi: 10.1007/s00438-002-0767-1. [DOI] [PubMed] [Google Scholar]
- 23.Bentolila S, Elliott LE, Hanson MR. Genetic architecture of mitochondrial editing in Arabidopsis thaliana. Genetics. 2008;178:1693–1708. doi: 10.1534/genetics.107.073585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Handa H. The complete nucleotide sequence and RNA editing content of the mitochondrial genome of rapeseed (Brassica napus L.): comparative analysis of the mitochondrial genomes of rapeseed and Arabidopsis thaliana. Nucleic Acids Res. 2003;31:5907–5916. doi: 10.1093/nar/gkg795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mower JP, Palmer JD. Patterns of partial RNA editing in mitochondrial genes of Beta vulgaris. Mol. Genet. Genom. 2006;276:285–293. doi: 10.1007/s00438-006-0139-3. [DOI] [PubMed] [Google Scholar]
- 26.Picardi E, Horner DS, Chiara M, Schiavon R, Valle G, Pesole G. Large-scale detection and analysis of RNA editing in grape mtDNA by RNA deep-sequencing. Nucleic Acids Res. 2010;38:4755–4767. doi: 10.1093/nar/gkq202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sloan DB, MacQueen AH, Alverson AJ, Palmer JD, Taylor DR. Extensive loss of RNA editing sites in rapidly evolving silene mitochondrial genomes: selection vs. retroprocessing as the driving force. Genetics. 2010;185:1369–1380. doi: 10.1534/genetics.110.118000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rajasekhar VK, Mulligan RM. RNA editing in plant mitochondria: α-phosphate is retained during C-to-U conversion in mRNAs. Plant Cell. 1993;5:1843–1852. doi: 10.1105/tpc.5.12.1843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Blanc V, Litvak S, Araya A. RNA editing in wheat mitochondria proceeds by a deamination mechanism. FEBS Lett. 1995;373:56–60. doi: 10.1016/0014-5793(95)00991-h. [DOI] [PubMed] [Google Scholar]
- 30.Yu W, Schuster W. Evidence for a site-specific cytidine deamination reaction involved in C to U RNA editing of plant mitochondria. J. Biol. Chem. 1995;270:18227–18233. doi: 10.1074/jbc.270.31.18227. [DOI] [PubMed] [Google Scholar]
- 31.Gualberto JM, Weil JH, Grienenberger JM. Editing of the wheat coxIII transcript: evidence for twelve C to U and one U to C conversions and for sequence similarities around editing sites. Nucleic Acids Res. 1990;18:3771–3776. doi: 10.1093/nar/18.13.3771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schuster W, Hiesel R, Wissinger B, Brennicke A. RNA editing in the cytochrome b locus of the higher plant Oenothera berteriana includes a U-to-C transition. Mol. Cell Biol. 1990;10:2428–2431. doi: 10.1128/mcb.10.5.2428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Yoshinaga K, Iinuma H, Masuzawa T, Uedal K. Extensive RNA editing of U to C in addition to C to U substitution in the rbcL transcripts of hornwort chloroplasts and the origin of RNA editing in green plants. Nucleic Acids Res. 1996;24:1008–1014. doi: 10.1093/nar/24.6.1008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kugita M, Yamamoto Y, Fujikawa T, Matsumoto T, Yoshinaga K. RNA editing in hornwort chloroplasts makes more than half the genes functional. Nucleic Acids Res. 2003;31:2417–2423. doi: 10.1093/nar/gkg327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wolf PG, Rowe CA, Hasebe M. High levels of RNA editing in a vascular plant chloroplast genome: analysis of transcripts from the fern Adiantum capillus-veneris. Gene. 2004;339:89–97. doi: 10.1016/j.gene.2004.06.018. [DOI] [PubMed] [Google Scholar]
- 36.Vangerow S, Teerkorn T, Knoop V. Phylogenetic information in the mitochondrial nad5 gene of pteridophytes: RNA editing and intron sequences. Plant Biol. 1999;1:235–243. [Google Scholar]
- 37.Grewe F, Viehoever P, Weisshaar B, Knoop V. A trans-splicing group I intron and tRNA-hyperediting in the mitochondrial genome of the lycophyte Isoetes engelmannii. Nucleic Acids Res. 2009;37:5093–5104. doi: 10.1093/nar/gkp532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kempken F, Bolle N, Forner J, Binder S. Transcript end mapping and analysis of RNA editing in plant mitochondria. Methods Mol. Biol. 2007;372:177–192. doi: 10.1007/978-1-59745-365-3_13. [DOI] [PubMed] [Google Scholar]
- 39.Lenz H, Rüdinger M, Volkmar U, Fischer S, Herres S, Grewe F, Knoop V. Introducing the plant RNA editing prediction and analysis computer tool PREPACT and an update on RNA editing site nomenclature. Curr. Genet. 2009;56:189–201. doi: 10.1007/s00294-009-0283-5. [DOI] [PubMed] [Google Scholar]
- 40.Tamura K, Dudley J, Nei M, Kumar S. MEGA4: molecular evolutionary genetics analysis (MEGA) software version 4.0. Mol. Biol. Evol. 2007;24:1596–1599. doi: 10.1093/molbev/msm092. [DOI] [PubMed] [Google Scholar]
- 41.Castandet B, Choury D, Begu D, Jordana X, Araya A. Intron RNA editing is essential for splicing in plant mitochondria. Nucleic Acids Res. 2010;38:7112–7121. doi: 10.1093/nar/gkq591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaw SM, Chun-Chieh SA, Wang D, Wu YW, Liu SM, Chou TY. The mitochondrial genome of the gymnosperm Cycas taitungensis contains a novel family of short interspersed elements, Bpu sequences, and abundant RNA editing sites. Mol. Biol. Evol. 2008;25:603–615. doi: 10.1093/molbev/msn009. [DOI] [PubMed] [Google Scholar]
- 43.Salmans ML, Chaw SM, Lin CP, Shih AC, Wu YW, Mulligan RM. Editing site analysis in a gymnosperm mitochondrial genome reveals similarities with angiosperm mitochondrial genomes. Curr. Genet. 2010;56:439–446. doi: 10.1007/s00294-010-0312-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Covello PS, Gray MW. On the evolution of RNA editing. Trends Genet. 1993;9:265–268. doi: 10.1016/0168-9525(93)90011-6. [DOI] [PubMed] [Google Scholar]
- 45.Tillich M, Lehwark P, Morton BR, Maier UG. The evolution of chloroplast RNA editing. Mol. Biol. Evol. 2006;23:1912–1921. doi: 10.1093/molbev/msl054. [DOI] [PubMed] [Google Scholar]
- 46.Jobson RW, Qiu YL. Did RNA editing in plant organellar genomes originate under natural selection or through genetic drift? Biol. Direct. 2008;3:43. doi: 10.1186/1745-6150-3-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maier UG, Bozarth A, Funk HT, Zauner S, Rensing SA, Schmitz-Linneweber C, Börner T, Tillich M. Complex chloroplast RNA metabolism: just debugging the genetic programme? BMC Biol. 2008;6:36. doi: 10.1186/1741-7007-6-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Lukeš J, Leander BS, Keeling PJ. Cascades of convergent evolution: the corresponding evolutionary histories of euglenozoans and dinoflagellates. Proc. Natl Acad. Sci. USA. 2009;106(Suppl. 1):9963–9970. doi: 10.1073/pnas.0901004106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knoop V. When you can't trust the DNA: RNA editing changes transcript sequences. Cell. Mol. Life Sci. 2010 doi: 10.1007/s00018-010-0538-9. doi:10.1007/s00018-010-0538-9 (12.10.2010, last date accessed) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Ohyama K, Oda K, Ohta E, Takemura M. Gene organization and evolution of introns of a liverwort, Marchantia polymorpha, mitochondrial genome. In: Brennicke A, Kück U, editors. Plant Mitochondria. Weinheim: VCH Verlagsgesellschaft; 1993. pp. 115–129. [Google Scholar]
- 51.Malek O, Lättig K, Hiesel R, Brennicke A, Knoop V. RNA editing in bryophytes and a molecular phylogeny of land plants. EMBO J. 1996;15:1403–1411. [PMC free article] [PubMed] [Google Scholar]
- 52.Groth-Malonek M, Pruchner D, Grewe F, Knoop V. Ancestors of trans-splicing mitochondrial introns support serial sister group relationships of hornworts and mosses with vascular plants. Mol. Biol. Evol. 2005;22:117–125. doi: 10.1093/molbev/msh259. [DOI] [PubMed] [Google Scholar]
- 53.Qiu YL, Li L, Wang B, Chen Z, Knoop V, Groth-Malonek M, Dombrovska O, Lee J, Kent L, Rest J, et al. The deepest divergences in land plants inferred from phylogenomic evidence. Proc. Natl Acad. Sci. USA. 2006;103:15511–15516. doi: 10.1073/pnas.0603335103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Binder S, Marchfelder A, Brennicke A. RNA editing of tRNAPhe and tRNACys in mitochondria of Oenothera berteriana is initiated in precursor molecules. Mol. Gen. Genet. 1994;244:67–74. doi: 10.1007/BF00280188. [DOI] [PubMed] [Google Scholar]
- 55.Fey J, Tomita K, Bergdoll M, Maréchal-Drouard L. Evolutionary and functional aspects of C-to-U editing at position 28 of tRNA(Cys)(GCA) in plant mitochondria. RNA. 2000;6:470–474. doi: 10.1017/s1355838200992380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zimmerly S, Hausner G, Wu X. Phylogenetic relationships among group II intron ORFs. Nucleic Acids Res. 2001;29:1238–1250. doi: 10.1093/nar/29.5.1238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Michel F, Ferat JL. Structure and Activities of Group II Introns. Annu. Rev. Biochem. 1995;64:435–461. doi: 10.1146/annurev.bi.64.070195.002251. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.