Abstract
High affinity and specificity RNA–RNA binding interfaces can be constructed by combining pairs of GNRA loop/loop–receptor interaction motifs. These interactions can be fused using flexible four-way junction motifs to create divalent, self-assembling scaffolding units (‘tecto-RNA’) that have favorable properties for nanomedicine and other applications. We describe the design and directed assembly of tecto-RNA units ranging from closed, cooperatively assembling ring-shaped complexes of programmable stoichiometries (dimers, trimers and tetramers) to open multimeric structures. The novelty of this work is that tuning of the stoichiometries of self-assembled complexes is achieved by precise positioning of the interaction motifs in the monomer units rather than changing their binding specificities. Structure-probing and transmission electron microscopy studies as well as thermodynamic analysis support formation of closed cooperative complexes that are highly resistant to nuclease digestion. The present designs provide two helical arms per RNA monomer for further functionalization aims.
INTRODUCTION
Structured RNA molecules form complex structures by hierarchical folding and comprise different types of recurrent modular motifs that can be utilized to design artificial self-assembling RNA supra-molecular systems of diverse architectures and novel functionalities (1–4). The repertoire of natural RNA motifs is augmented by new motifs obtained by efficient in vitro selection methods and having novel binding selectivities or enzymatic capabilities (5,6). The programmability of RNA secondary structure, its hierarchical folding, and the modularity of recurrent motifs make RNA an attractive medium for diverse nanotechnological applications.
Modular RNA 3D motifs suitable for nano-fabrication can be broadly categorized according to their functional properties: interaction motifs, architectural motifs and ligand-binding motifs. (i) ‘Interaction’ motifs make possible specific intermolecular assembly of smaller molecular building units to form larger nanoassemblies. Examples include kissing hairpin loops and other pseudoknots, cognate hairpin loop/loop–receptor pairs, T-loops and paranemic motifs (7–14). (ii) ‘Architectural’ motifs define the topological and geometric relationships of the folded structure and include the canonical A-form double helix, kink-turns, C-loops and most importantly, multi-helix junctions (15). RNA helices provide highly regular, variable-length struts for positioning other motifs in 3D space. Kink-turn and related motifs create sharp bends in helical elements in which they are embedded (16), while C-loops and related motifs change the helical twist of RNA helices (17). Multi-helix junctions (3-way, 4-way, etc.) increase the topological complexity of RNA by introducing branch points (18,19). Up to 10-way junctions have been found in RNA (20). Some RNA junctions are flexible while others stabilize the stacking of helices at the branch point in a specific manner that can be used in molecular design (19,21). (iii) ‘Ligand-binding’ motifs are found in naturally occurring RNAs such as riboswitches or can be generated by in vitro selection (so-called ‘aptamers’) (6,22).
A variety of RNA nano-objects have been constructed by combining modular RNA using rational, computer-aided design. For example, square-shaped RNA complexes were constructed by combining the kissing hairpin loop interactions motifs with the tRNA multi-helix junction (23,24). More elaborate constructions including design of cubic and octahedron RNA scaffolds were recently reported (25,26). Pairs of loop/loop–receptor interaction motifs were combined to construct efficient platforms for in vitro selection of novel receptor motifs (5). The malachite green (MG) RNA aptamer was coupled to RNA paranemic binding motifs to design label-free biosensors which target internal loop structures in pre-folded RNA analytes in a programmable and sequence-specific manner (27).
RNA has a number of additional properties such as low immunogenicity in the human body and amenability to in vitro selection (SELEX) that make it an ideal material for constructing multivalent nano-particles for drug delivery. Additionally, RNA can be readily derivatized using a variety of technologies to incorporate therapeutic or imaging functionalities, including RNA sequence elements that can act as silencing RNAs inside the targeted cell to suppress specific genes.
These technologies have been combined by Guo and co-workers (28–31) to engineer multivalent RNA-based nanoparticles for specific delivery of therapeutic agents into cancer cells and virus-infected cells by modifying the RNA component (‘pRNA’) of the bacteriophage Phi29 packaging motor. Multimer formation of pRNA units is accomplished by Watson–Crick base-pairing of complementary hairpin loops.
Multimeric RNA self-assembly can also be achieved using specific GNRA loop/loop–receptor interaction motifs that can be combined in pairs for high affinity self-assembly of a variety of nanoscale RNA objects (32,33). Pairs of GNRA loop/loop–receptor interaction motifs were fused using the four-way junction from the hairpin ribozyme to engineer RNA monomers that assembled to form straight, micrometer-long filaments (34). Using two orthogonal loop/loop–receptor interaction motifs, it was possible to design RNA monomers capable of directional assembly in either the parallel (‘up–up’) or anti-parallel (‘up–down’) assembly modes (34). In anti-parallel assembly of interacting molecules, as exemplified by molecule 9 (Figure 1A), each incorporated monomer switches the directionality of the growing chain and thus compensates for its intrinsic bending, producing long, relatively straight multi-unit chains (fibrils).
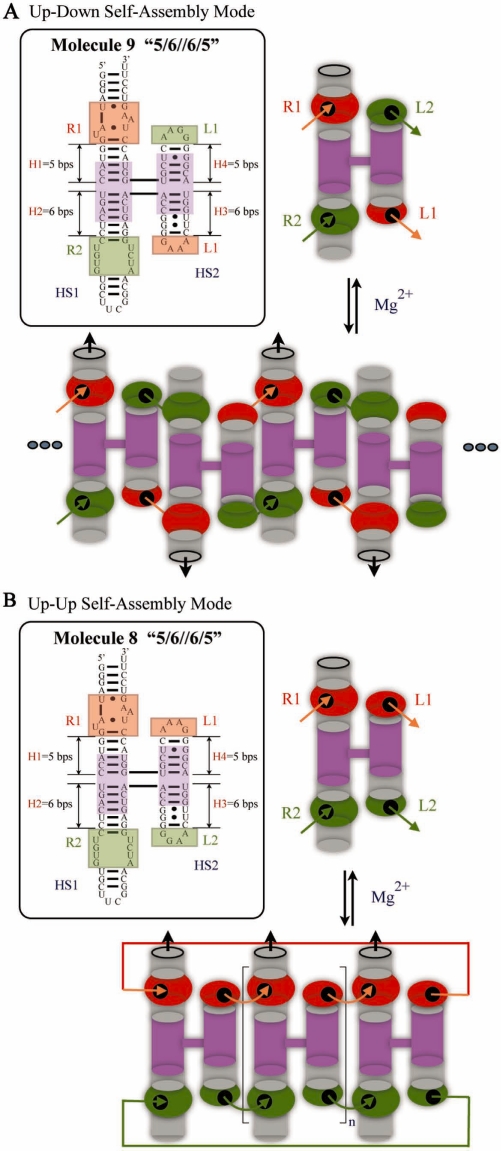
Figure 1.
Molecules 9 and 8, examples of anti-parallel ‘up–down’ and parallel ‘up–up’ self-assembly modes, respectively. (A) Left panel shows the detailed sequence design of molecule 9, where the central core of the molecule consists of the 4WJ from the hairpin ribozyme (shown in purple) and various motifs placed on its helical arms. GAAA-receptor (R1) is positioned in H1 and its cognate GAAA loop (L1) at the end of H3, while the GGAA-receptor (R2) is placed in H2 and its GGAA loop (L2) at the end of H4. R1 and L1 are shown in red, R2 and L2 in green. Right panel provides a schematic diagram of the molecule 9 monomer. Orange arrows indicate the GAAA-loop/GAAA-receptor motifs and green arrows indicate the GGAA-loop/GGAA-receptor motifs to show the orientation of interaction interfaces. Molecule 9 can bind to its partner molecules via HS1 (which carries receptors) or HS2 (which carries loops) with a subsequent formation of one pair of receptor–loop motifs on each side. Upon addition of magnesium, lower panel, molecule 9 self-associates so that each incorporated monomer switches the directionality of the molecular unit (shown by black arrows). (B) Left panel shows the detailed sequence design of molecule 8. The positions of loops L1 and L2 are switched, compared to molecule 9. Molecule 8 binds to its partner molecules via HS1 or HS2, without changes in directionality, thus forming bands of defined stoichiometry on electrophoresis gels.
In this work, we turn our attention to H-shaped tecto-RNA molecules designed to assemble with uncompensated curvature to form closed ring-shaped complexes of defined and, in principle, programmable stoichiometries, exemplified by molecule 8 in Figure 1B. We have carried out a systematic study to define the rules of uncompensated self-assembly of molecule 8 and molecules derived from it, with the goal learning how to tune the stoichiometry of this process.
MATERIALS AND METHODS
RNA design
We used Mfold (www.mfold.bioinfo.rpi.edu) to check the secondary structures of each RNA sequence prior to synthesis. If the calculated lowest free-energy secondary structure corresponded to the desired RNA conformation, and no other secondary structure was closer than 15% in energy to the lowest energy structure, the sequence was used as is (35). Otherwise, minor changes were made to Watson–Crick base-paired positions to destabilize competing conformations. The detailed designs and sequences of RNA molecules used in this study are provided in the Supplementary Data section.
RNA synthesis, purification and labeling
RNA molecules were synthesized by run-off transcription of PCR-amplified DNA templates. DNA templates were designed from the desired RNA sequences using the ‘RNA to DNA’ web applet (http://rna.bgsu.edu/rnatodna) written by J. Stombaugh. DNA was purchased from Integrated DNA Technology (www.idtdna.com) and amplified using PCR primers, one of which is complementary to the 3′-end of the template and the other to the 5′-end of the template. The forward primer also carries the T7 promoter, as described previously (34). PCR-amplified DNA molecules were purified using the QiaQuick PCR purification kit (Qiagen Sciences) and transcribed in vitro using T7 RNA polymerase purchased from Takara Bio Inc. (www.takara-bio.com). The synthesized RNA were purified by gel electrophoresis (10% acrylamide, 29:1 acrylamide:bisacrylamide, 8 M Urea) and extracted from excised gel slices using 0.5 ml of ‘crush and soak’ buffer (10 mM Tris–HCl, pH 7.9, 1 mM EDTA, 0.3 M of sodium acetate) with overnight shaking at 4°C. RNA molecules were ethanol precipitated (2.5:1 volume ratio), rinsed twice with cold 80% ethanol, dried and redissolved in water.
Radiolabeling of RNA molecules was achieved using T4 RNA ligase (New England Biolabs Inc.) and 5′-[32-P]-pCp (MP Biomedicals). Typically, 40 pmol of RNA in 4 µl water was mixed with 1 µl of 10× T4 RNA ligase buffer (0.5 M Tris–HCl, pH 7.8, 0.1 M MgCl2, 0.1 M DTT, 10 mM ATP), 1 µl of T4 RNA ligase (20U), 1 µl of DMSO and 3 µl of 250 µCi 5′[32-P]-pCp. The mixture was incubated overnight at 4°C and labeled RNA products were purified on denaturing acrylamide gels (10% acrylamide, 29:1 acrylamide:bisacrylamide, 8 M Urea).
Assembly experiments
For assembly assays, one RNA component was radiolabeled. RNA samples included trace amounts (∼0.5 nM) of the 3′-radiolabeled RNA supplemented by sufficient unlabeled RNA to achieve the desired RNA concentration. All RNA samples were heated to 94°C in de-ionized water for 1–2 min and snap-cooled on ice. Tris–borate/Mg2+ buffer (89 mM Tris–borate, pH 8.3, 1 mM Mg(CH3COO)2) was then added and the resulting mixtures incubated at 30°C for 5 min (annealing step). The final concentration of Mg2+ was increased to 15 mM and incubation was continued for additional 30 min (assembly step). An equal amount of gel loading buffer (89 mM Tris-borate, 15 mM Mg(CH3COO)2, 0.01% bromophenol blue (BPB), 0.01% xylene cyanol blue (XCB), 50% glycerol) was added to each sample prior to loading on native gels. Samples were run at 4–6°C on 7% native polyacrylamide gels (29:1 acrylamide: bisacrylamide) containing 15 mM magnesium acetate with constant recycling of the running buffer [89 mM Tris–borate, pH 8.3, 15 mM Mg(CH3COO)2] to maintain constant pH. The gels were then dried under vacuum and imaged using a phosphor storage screen with the Storm phosphoimager (Amersham, Storm 860).
Dissociation constant measurements
Small amounts of radiolabeled RNA (<5 fmol) were incubated with unlabeled cognate partner molecules ranging in concentration from 1 to 1000 nM, as described above in the ‘Assembly experiments’ section. RNA samples were run on 7% native polyacrylamide gel to separate monomers from multimers, and reaction products were quantified using the ImageQuant software. The percentage of dimer or trimer was plotted as a function of unlabeled partner RNA concentration and analyzed using the ORIGIN software (www.originlab.com) by non-linear regression curve fitting. Detailed description of Kd determination is provided in the Supplementary Data section of this work.
Structure probing of RNA by lead cleavage
Labeled RNA (1 µl, ∼2 pmol, 50 000–100 000 c.p.m.), either alone or mixed with an unlabeled partner RNA molecule, was brought to 5 µl total volume. For example, in probing the dimer 1bDA–1bDB, 1 µl of labeled 1bDA (∼2 pmol) was mixed with sufficient unlabeled 1bDB to give a final concentration of 1µM. RNA samples in pure water were then denatured at 94°C for 2 min, snap-cooled on ice for 2–3 min and folded at 30°C for additional 3 min. After addition of 2 µl of 5× Assembly buffer to a final concentration of 25 mM HEPES, pH 7.5, 15 mM Mg(CH3COO)2 and 25 mM CH3COOK, and 1 µg of yeast RNA, the mixture was incubated for 30 min at 30°C to ensure proper folding of RNA prior to cleavage reactions. Next, the mixture was adjusted with freshly prepared Pb(CH3COO)2 to a final concentration of 20 mM of Pb2+ and a total volume of 10 µl. RNAs were incubated in the presence of Pb2+ for 8 min at 30°C and the cleavage reactions were terminated by the addition of 10 µl of 0.1M EDTA. RNA molecules were precipitated with 2.5 volumes of ethanol on dry ice, pelleted and washed with 80% EtOH, dried and re-suspended in 10 µl of gel loading buffer. All samples were heated at 94°C for 5 min prior to loading on the gel. The analysis of the digestion products was performed on thin (0.5 mm) 8–10% polyacrylamide gels (19:1 acrylamide/bisacrylamide ratio) containing 8 M UREA. All sequencing gels were run for about 4–6 hours in TBE running buffer. At the end of the run, all gels were rinsed and fixed in 10% ethanol/6% acetic acid mixture and transferred to Whatman paper (Grade 3) for drying under vacuum. The RNA products were visualized using a phosphor imaging screen and the Storm phosphoimager (Amersham, Storm 860).
Sequencing ladders for locating positions of lead cleavage were generated by digestion of control samples with alkali (pH 9.2, cleavage at all positions) or with RNase T1 (cleavage of unpaired G’s). The reagents for these two controls were purchased as the RNase T1 toolkit from AMBION (ambion.com). For RNase T1 cleavage, 1 µl of labeled RNA (∼2 pmol, 50 000–100 000 c.p.m.) was denatured in pure water (7 µl total volume) at 94°C for 2 min then cooled on ice for 2–3 min. Then 1 µl of 10× RNA structure buffer and 1 µg of yeast RNA were added, and the sample was incubated for 20 min at 25°C. This step was followed by the addition of 1 µl of RNase T1 to the mixture, and an additional incubation for a 15-min period. RNA digestion products were precipitated with 20 µl of inactivation/precipitation buffer, placed on dry ice for 15 min and centrifuged at 13 000g. The pellet was washed with 80% ethanol, dried and resuspended in gel loading buffer. For the alkaline ladder, 1 µl of labeled RNA (∼2 pmol, 50 000–100 000 c.p.m.) was mixed with 1 µg of yeast RNA and brought with an alkaline hydrolysis buffer to a 5 µl total volume. RNA samples were heated at 94°C for 10 min and snap cooled on ice. At the end, the gel loading buffer (5 µl) was added, and the samples were directly loaded on the gel.
3D modeling
Computer models of RNA monomers were assembled using modular 3D motifs extracted from atomic resolution structures from the PDB databank (www.pdb.org) with the help of SWISS-PDB viewer (www.spdbv.vital-it.ch). For example, molecule 8 monomer was built by extracting the 4WJ modular unit from hairpin ribozyme (nucleotides: A, 3–11: B, 11–19, 64–72, 81–89, PDB ID: 1M5O), and the GAAA-receptor–loop motif from group I intron (nucleotides: 145–158, 215–231, 243–254, PDB ID: 1HR2), (36–38). Motifs were assembled by overlapping helical domain extensions of motifs to position them at the correct angle and distance using the ‘autofit’ tool in Swiss-PDB Viewer and manual adjustment (39). The backbones of the assembled structures were connected using the NanoTiler software (40).
RESULTS
RNA design and nomenclature
The central core of each H-shaped tecto-RNA molecule derived from molecule 8 consists of the 4-way junction (4WJ) from the hairpin ribozyme (shown in purple in Figure 1B), which preferentially folds in an anti-parallel configuration with helix 1 (H1) stacked on helix 2 (H2) and helix 3 (H3) stacked on helix 4 (H4). We note that the crystallographic structure of the hairpin ribozyme shows that the coaxial helical stacks of the 4WJ cross at an angle of about 60° (36). In each tecto-RNA unit, two receptors and two loops are positioned in the helical arms of the 4WJ, one interaction motif (loop or receptor) per arm at specific distances relative to the 4WJ branchpoint center. In molecule 8, which is designed to assemble in parallel mode (‘up–up’), the GAAA-receptor (R1) is positioned in H1 and its cognate GAAA loop (L1) at the end of H4, while the GGAA-receptor (R2) and its cognate GGAA-loop (L2) are placed at the ends of H2 and H3 (Figure 1B). For anti-parallel assembly (‘up–down’), as in molecule 9, the positions of loops L1 and L2 are switched, as shown in Figure 1A. Helical stack 1 (‘HS1’) comprises H1 and H2, and helical stack 2 (‘HS2’) comprises H3 and H4.
For consistency with our previous work (34), we use the same numbers (#8 and #9) for the tecto–RNAs shown in Figure 1 and also designate the number of basepairs in each helix as before. Thus, molecule 8 is designated as 8 ‘5/6//6/5’ to indicate the number of basepairs between the interacting motifs in each helix and the crossover point of the 4WJ. Specifically, ‘5/6//6/5’ indicates that the 4WJ crossover point is 5 bp from the R1 motif, 6 bps from R2, 6 bp from L2 and 5 bp from L1. A single forward slash ‘/’ separates two helices belonging to the same helical stack and a double forward slash ‘//’ separates HS1 from HS2.
In all designs, we maintained a distance of 11 basepairs (one helical turn) between the interacting modules in each helical stack (HS1 and HS2), whether loops or loop-receptors, to maintain the optimal orientation of interacting motifs for self-assembly. Extending or reducing the length of the helical region between interacting modules by one basepair to 12 or 10 bp weakens the intermolecular interactions significantly, whereas an additional basepair increment or decrement, yielding a total of 13 or 9 bp distance between motifs, completely abolishes self-assembly (33).
Determination of the stoichiometry and cooperativity of molecule 8 self-association
Several observations led us to suspect that molecule 8 self-assembles cooperatively to form a complex comprising at least three monomers (34). First, on native gels, molecule 8 migrates as a sharp discrete band with lower mobility than that expected of a dimer. Second, monomer bands disappear at low nanomolar concentrations. Third, no intermediate dimer species are observed on native gels at any concentrations.
In this work, we establish the cooperativity and the stoichiometry of molecule 8 association with the following experiments: first, we performed dilution experiments of pre-assembled, radiolabeled molecule 8 over the low nM to picomolar range, and failed to observe any intermediate species, even at very low dilution, as shown in Supplementary Figure S3. Upon dilution to picomolar range, an abrupt transition from the complex to the monomer units is observed. This suggests that the complex assembles cooperatively, indicating that each molecule 8 monomer interacts with two other monomers to form a closed complex, with no monomers found on the ends of a linear array, and thus less tightly bound.
Next, we performed an experiment to measure the rate of incorporation of radiolabeled molecule 8 into pre-associated non-radioactive complexes (Supplementary Figure S4). The half-time for reaching exchange equilibrium was 20 min, which suggests that these complexes are kinetically stable. This result provides further support for the cooperative assembly of molecule 8.
To determine the stoichiometry of molecule 8 complexes, we designed new sets of molecules derived from molecule 8 that have the potential to form closed hetero-oligomeric complexes with stoichiometry ranging from n = 2 to n = 6 units by combining three sets of high-affinity, selective and orthogonal loop/loop–receptor interaction motifs, as shown in Supplementary Figure S5. To allow for the formation of closed complexes, the interaction interface of HS2 of the n-th subunit was designed to complement the interaction interface of HS1 of the first subunit. The monomers were designed so that the interacting motifs on each monomer avoid self-assembly and can only bind to one other subunit, in a directional ‘up–up’ manner. Thus, a given set of molecules would only be able to form closed cooperative complex if it happened to contain the same number of units as the original molecule 8.
For hetero-dimer design 8DA–8DB, large amounts of monomers are observed on native gels even at 1 μM, Supplementary Figure S6A. ‘D’ refers to the design that was optimized to form a dimer, and A, B refer to the individual monomers, designed to bind in the order A to B and B to A. The appearance of small amounts of complexes with mobility similar to the molecule 8 complex suggests the formation of non-cooperative but ‘saturated’ 8DA–8DB*–8DA or 8DB–8DA*–8DB systems (star indicates radiolabeling).
Full cooperativity was only achieved with the heterotrimeric set of molecules, 8TA–8TB–8TC (Supplementary Figure S6C, ‘T’ stands for trimer-optimized design). These molecules were designed for unique binding of the interaction interface HS2 of 8TA to HS1 of 8TB, HS2 of 8TB to HS1 of 8TC and HS2 of 8TC to HS1 of 8TA to potentially form a closed complex (Supplementary Figure S5). For this design, each molecule was assayed individually on native gels and none showed any self-assembly. 8TA was found bound to 8TC in the absence of 8TB, however, 8TB failed to bind to either 8TA or 8TC in the absence of the third molecule. When all three molecules were combined, a sharp band appeared with mobility identical to that of the complex of the original molecule 8. Thus, the behavior of 8TB serves as a good indicator of the cooperativity of trimer formation in the 8TA–8TB–8TC system. One interaction interface of 8TB contains the GUAA loop and the other contains the GUAA loop–receptor, which have significantly weaker affinities than the GAAA and GGAA binding motifs. Only in the presence of both 8TA and 8TC, when 8TB can bind with both interfaces, does it associate to form a complex of any type.
Furthermore, in the tetrameric system 8TA–8TB–8TetC–8TetD Supplementary Figure S6B, 8TB remains a monomer in the presence of the other three subunits. Individual radiolabeling of each of the other subunits 8TA, 8TetC or 8TetD shows that they associate non-cooperatively to form smeared bands. These results indicate molecule 8 does not form tetramers. Pentameric and hexameric sets also failed to show any traces of cooperative assembly, Supplementary Figure S6B. Again, small amounts of complexes with mobility no slower than that of the molecule 8 complex were obtained for some combinations of subunits in the pentamer and hexamer series.
Structure probing of hetero-trimer derived from molecule 8
Overall, the stoichiometry determination experiments fully support molecule 8 forming cooperative trimers. However, to obtain definitive proof of its cooperative association and to assess the formation of all loop/loop–receptor interactions in the molecule 8 trimer, we used the hetero-trimeric system, 8TA–8TB–8TC, for structure probing experiments. Use of the hetero-trimer design allows chemical probing of individual units in the monomer, dimer and trimer states. The design of these molecules is shown in Supplementary Figure S2 and Figure 2C. Individual 8TA and 8TC molecules were radiolabeled and probed with lead ions to monitor changes in solvent accessibility of the interaction motifs between monomer, dimer and trimer states. Alkaline hydrolysis (lane 1) and RNase T1 cleavage (lane 2) were performed to generate sequencing ladders for reference (Figure 2).
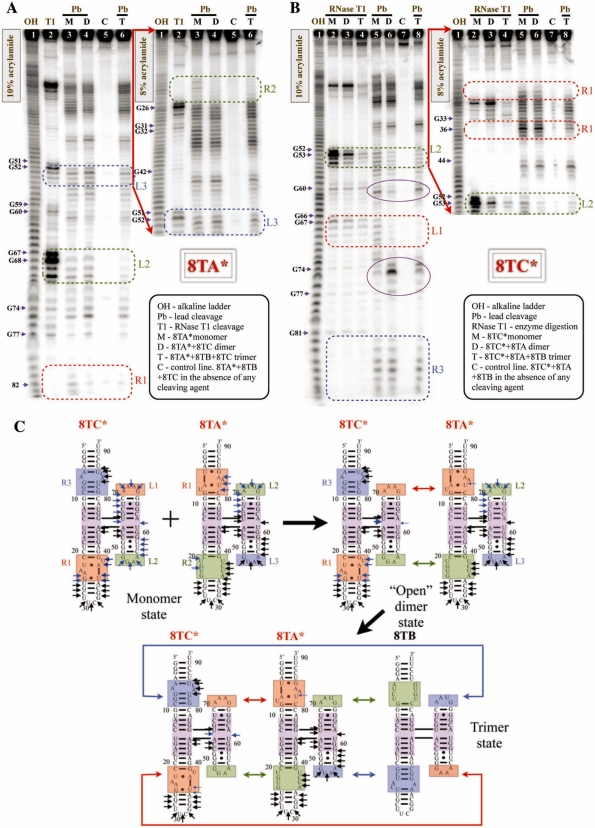
Figure 2.
Lead cleavage of hetero-trimer 8TA–8TB–8TC. (A) Denaturing gels of 8TA* (asterisk indicates radiolabeled RNA) cleavage in monomer, dimer and trimer complexes. Regions of primary interest are labeled and outlined with dotted squares. Cuts induced in the 4WJ region by complex formation are circled in purple. (B) Structure probing experiments of 8TC* in monomer, dimer and trimer complexes. (C) Summary of lead-induced cleavage obtained for 8TA* and 8TC*. Lead-induced cuts that remain unchanged between monomer, dimer and trimer states are indicated with black arrows, while lead-induced cuts that change their intensity over the course of self-assembly are noted with cyan blue.
When 8TA* (* indicates radiolabeling) is bound to 8TC to form a stable dimer, we observe complete protection of nts 20 and 22 in the R2 receptor and significant reduction in cleavage at positions 81–85 in receptor R1 (compare lanes 3 and 4, Figure 2A). This is consistent with binding of the HS1 interaction interface comprising the R1 and R2 receptors of 8TA with the L1 and L2 loops of the HS2 interface of 8TC (Figure 2C). On the other hand, the L2 and L3 hairpin loops of HS2 of 8TA are cleaved in identical fashion in the 8TA–8TC dimer as in the monomer state, indicating that there is no interaction involving HS2 of 8TA, as intended by design. Upon addition of the third component, 8TB, to form the trimer complex, the loop–receptors R1 and R2 of 8TA* remain protected from cleavage. Moreover, a significant protection from lead cleavage is observed in HS2 of 8TA for L2 (nt 67–71, Figure 2A). However, lead cleavage in L3 of HS2 is only slightly reduced.
When 8TC* is bound to 8TA to form the stable dimer, protection from lead cleavage is observed in the L1 (GAAA) and L2 (GGAA) hairpin loops of HS2 of 8TC (compare lanes 5 and 6 in Figure 2B), consistent with the protections observed in the loop–receptors R1 and R2 of HS1 of 8TA with which they are intended to interact (Figure 2C). Also as expected, cleavage in the R3 and R1 receptors in HS1 of 8TC is unchanged in the dimer compared to the monomer format. In the 8TC*–8TA–8TB trimer complex, significant to complete protection is observed for nucleotides in R1 of 8TC, but only slight protection is observed for nucleotides in R3 which binds the GUAA loop of 8TB (Figure 2B, lane 8).
The overall lead cleavage results for 8TA and 8TC are summarized using schematic diagrams in Figure 2C, showing that in the 8TA–8TC dimer only one set of interfaces interacts—HS1 of 8TA with HS2 of 8TC. Henceforth, we will call the 8TA–8TC dimer and others like it ‘open’ dimers to emphasize that there are no interactions involving the two ‘outside’ interaction interfaces. Upon introducing the third molecule, 8TB, L2 of 8TA and R1 of 8TC exhibit enhanced protection, which supports the cooperative self-assembly mechanism. However, lead-induced structure probing experiments provide minimal evidence for interaction between the GUAA tetraloops and their loop–receptors. The GUAA-loop/GUAA-receptor interaction is known from previous work to be significantly weaker than the GAAA or GGAA loop–receptor interactions (41). To determine whether the GUAA-loop/GUAA-receptor interactions participate in the cooperative assembly of 8TA-8TB-8TC we designed 8TBmut and 8TCmut, molecules in which the GUAA-receptors are rotated by 180° so as not to be able to interact with the GUAA loops (see Supplementary Figure S2). As expected, 8TA and 8TCmut dimerize, but addition of 8TBmut to either dimer 8TA–8TC or 8TA–8TCmut did not lead to trimer formation (see Supplementary Figure S7A). Moreover, no trimers were observed when 8TCmut was added to the mixture of 8TA and 8TB. These results indicate that correctly oriented GUAA receptor is needed in both molecules 8TB and 8TC for trimer self-assembly. Thus, we can definitely conclude that formation of all possible loop–receptor interactions takes place in cooperative trimer formation of molecule 8 and its derivatives, strongly supporting our initial observations.
These data also show subtle changes in the lead cleavage pattern in the 4WJ region over the course of self-assembly. The cleavage at position 61 belonging to the 4WJ is reduced in the dimer but restored to the original intensity in the trimer, while the band at position 75 behaves in the opposite manner. This suggests that the 4WJ undergoes conformational changes sensitive to lead cleavage during assembly, consistent with the flexible nature of 4WJs such as this one.
We also used RNase T1 as a probe of accessibility of Gs in loop regions in the formed complexes. Nuclease digestion of 8TC* in monomer, dimer and trimer states shown in lanes 2–4 (Figure 2B). In summary, when comparing the monomer cleavage in lane 2 to the cleavage of dimer state, lane 3, RNase T1 digestion is reduced, especially in the GGAA loop domain. Once 8TC* is incorporated in the trimer complex, nuclease digestion is significantly minimized at all positions, demonstrating a significant protection, lane 4 in Figure 2B.
Beyond cooperative trimers—control of complex stoichiometry
In light of the previous results, we next decided to investigate whether we could change the stoichiometry of cooperative assembly by re-engineering molecule 8. The idea is that by moving the 4WJ crossover point within the interaction interfaces HS1 or HS2, while maintaining the same number of basepairs (11 bp) between the interaction motifs in each interaction interface (i.e. R1 and R2 in HS1 and L1 and L2 in HS2), the orientation of the motifs relative to the plane of the molecule can be varied systematically, while maintaining its ability to interact with a cognate interface on another molecule. Therefore, we reasoned that it should be possible, by changing one interface at a time, to design molecules that form dimers or tetramers, or, conversely, by making compensating changes in the two interfaces, to maintain trimer formation.
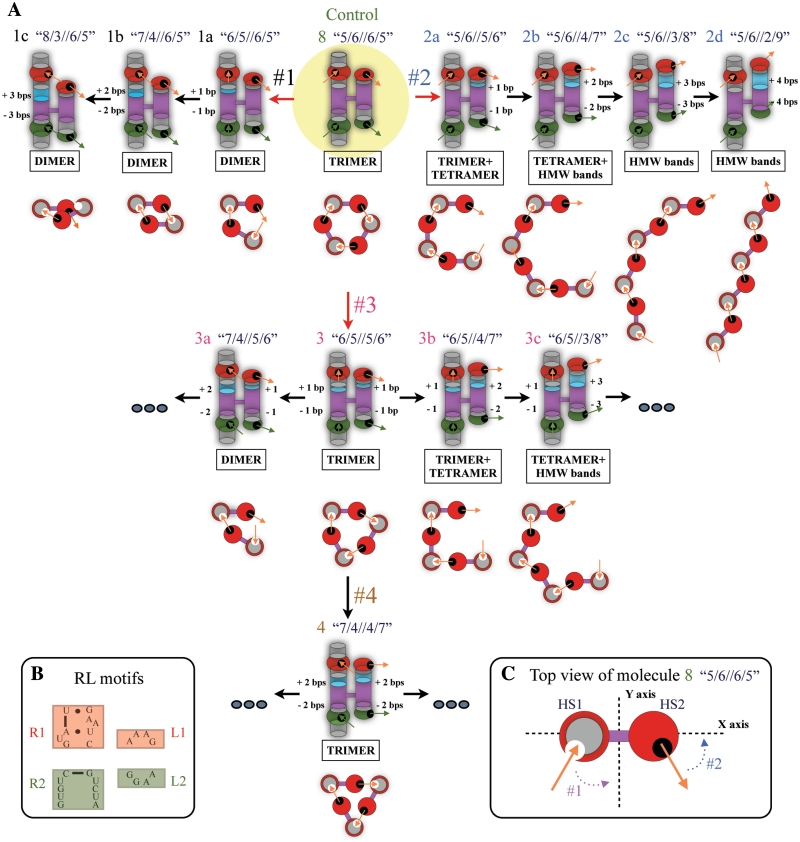
To test this hypothesis, we engineered several series of H-shaped tecto-RNA molecules derived from molecule 8. In series #1, the position of the 4WJ crossover point in HS1 was changed, while in series #2, its position in HS2 was altered (top row, Figure 3). In both series #1 and #2, up–up directionality of self-assembly was preserved, by leaving all interaction motifs in the same positions they have in molecule 8. We derived the molecules in series #1 (1a, 1b, 1c) by lengthening H1 and shortening H2 by one, two or three basepairs, which is equivalent to moving the crossover point 1 bp at a time toward the interaction motif of H2. Thus, molecule 1a ‘6/5//6/5’ has 1 bp added to H1 and 1 bp removed from H2, and retains a total separation of 11 bp between interaction motifs R1 and R2. A shift of two bps in HS1 results in molecule 1b ‘7/4//6/5’ and a shift of three bps produces molecule 1c ‘8/3//6/5’. In Figure 3, basepair insertions are shown in light blue relative to molecule 8. A similar approach was followed to engineer series #2 (2a, 2b, 2c, 2d), in which H4 is lengthened and H3 shortened by 1 bp (molecule 2a ‘5/6//5/6’), by 2 bp (2b ‘5/6//4/7’), by 3 bp (2c ‘5/6//3/8’) or by 4 bps (2d ‘5/6//2/9’), as shown in Figure 3 (upper right).
Figure 3.
Schematic diagrams of H-shaped tecto-RNAs and their self-assembly properties. (A) In the upper-central part, molecule 8 was used as a starting template (outlined in yellow). Orange and green arrows located inside receptor or loop regions show the orientation of interaction interfaces as shown in Figure 1. Basepair insertions in helical regions between interacting motifs are shown in light blue [basepair removal is not color-coded]. Note that the orientations of interaction interfaces (orange or green arrows) change with each bp insertion/deletion. Below each individual monomer unit, we provide top-view illustrations of self-assembly based on the experimental evidence and 3D modeling. (B) Detailed sequence representation of the receptor-loop motifs. (C) Top-view of molecule 8. HS1 is shown as a red circle with a grey center and HS2 is shown as a red circle corresponding to the GAAA loop. The circles are connected by the 4WJ crossover, shown as a thick purple line. The x-axis is placed to pass through HS1, HS2 and the 4WJ of the molecule.
To create series #3 (3, 3a, 3b, 3c) and #4 (4), we simultaneously changed both HS1 and HS2 to produce molecules 3 ‘6/5//5/6’, 3a ‘7/4//5/6’, 3b ‘6/5//4/7’, 3c ‘6/5//3/8’ and 4 ‘7/4//4/7’, as shown in Figure 3 (center and lower part). In series #3 and #4, the idea was to test the effect of compensating changes. Detailed sequence designs of all monomer units shown in Figure 3 are provided in the Supplementary data section of this work.
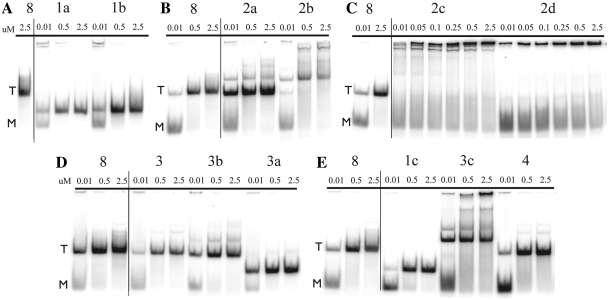
Assembly of each of these new sets of molecules was tested by native gel electrophoresis (Figure 4A–E). The primary assembly complex observed by electrophoresis is noted below each individual monomer design in Figure 3. For all the molecules in series #1 (1a,1b,1c), we observe complete disappearance of trimer bands and appearance of increased mobility bands with 100% yield (Figure 4A and E). The new bands occur about half way between the monomer and trimer bands of the trimer-forming molecule 8, and can be attributed to the formation of dimers. Self-assembly of 1a ‘6/5//6/5’ and 1b ‘7/4//6/5’ occurs to a large extent even at 10 nM concentration, which suggests cooperative self-assembly. Dimer formation of 1c ‘8/3//6/5’ requires higher concentrations, suggesting that in this complex, all potential stabilizing interactions may not be forming, possibly due to steric hindrance.
Figure 4.
Native gel data of tecto-RNA self-assembly. RNA concentrations used in this study are provided above each lane in micromolar units. Molecule 8, which self-associates into trimer complexes, is used as a mobility shift reference. ‘M’ designates monomers and ‘T’ trimers.
In contrast, quite different results are observed for the molecules in series #2, Figure 3. In molecule 2a ‘5/6//5/6’, the major product of self-assembly is still the trimer complex, but additional bands with reduced mobility are seen in a concentration-dependent manner (Figure 4B). Even at 10 nM, a discrete band, probably due to a tetramer complex, is evident, in addition to the trimer, which predominates. The self-assembly results of 2a ‘5/6//5/6’ suggest that the geometry of the 2a complex is intermediate between what is ideal for trimer or for tetramer formation. At higher concentrations, additional bands appear, possibly due to pentamer and even hexamer complexes. For molecule 2b ‘5/6//4/7’, we observe almost complete loss of the trimer band, and appearance of a discrete band, also likely due to a tetramer complex (Figure 4B). At high concentration, higher molecular weight products are also observed. For molecules 2c ‘5/6//3/8’ and 2d ‘5/6//2/9’, no discrete oligomeric products are seen, only high-molecular weight products having low mobility, which in the case of 2d are too large to even penetrate the electrophoresis gel (Figure 4C). The molecule 8 complex and complexes in series #2 were studied by TEM (Supplementary Figure S8). Their self-assembly patterns on native gels are in complete agreement with the TEM images where complexes of molecules 8, 2a, 2b can be seen as spherical-shaped objects too small to be finely resolved, while 2c and 2d self-associate to form polymeric structures.
In designing molecule 3 ‘6/5//5/6’, we made compensating 1 bp shifts in the two helical stacks, HS1 and HS2, as shown in Figure 3. We observe cooperative, homo-trimer nanoassembly, identical to that of molecule 8 (Figure 4D). Additional basepair shifts, either in HS1 to yield molecule 3a ‘7/4//5/6’, or in HS2 to produce molecules 3b ‘6/5//4/7’ and 3c ‘6/5//3/8’, result in similar distributions of assembly products as observed for 1a ‘6/5//6/5’, 2a ‘5/6//5/6’ and 2b ‘5/6//4/7’, respectively (Figures 4D and E). However, slight differences are observed for 3a and 3c. While molecule 3a ‘7/4//5/6’ resembles 1a ‘6/5//6/5’ in primarily forming dimers, a faint trimer band is observed for 3a at high concentration (2.5 µM), which was not seen for 1a (compare Figure 4D with 4A). Molecule 3c ‘6/5//3/8’, like molecule 2b ‘5/6//4/7’, forms a sharp homo-tetramer band. However, an additional pentamer complex is observed for 3c even at low nanomolar concentrations (compare Figure 4E with 4B).
To derive molecule 4 ‘7/4//4/7’, we made compensating 2 bp shifts in the helical stacks HS1 and HS2 of molecule 8, Figure 3. Molecule 4 forms a cooperative trimer complex with mobility identical to that of molecules 8 and 3 (compare Figure 4E with 4D). In conclusion, these constructions verified our hypothesis that we could systematically vary the stoichiometries of complexes by changing the positions of the 4WJ crossover points in either HS1 or HS2. Conversely, we showed with molecules 3 and 4 that we could preserve the original trimer stoichiometry by making compensating changes in HS1 and HS2.
Dimer-forming molecules in series #1: open versus closed dimers
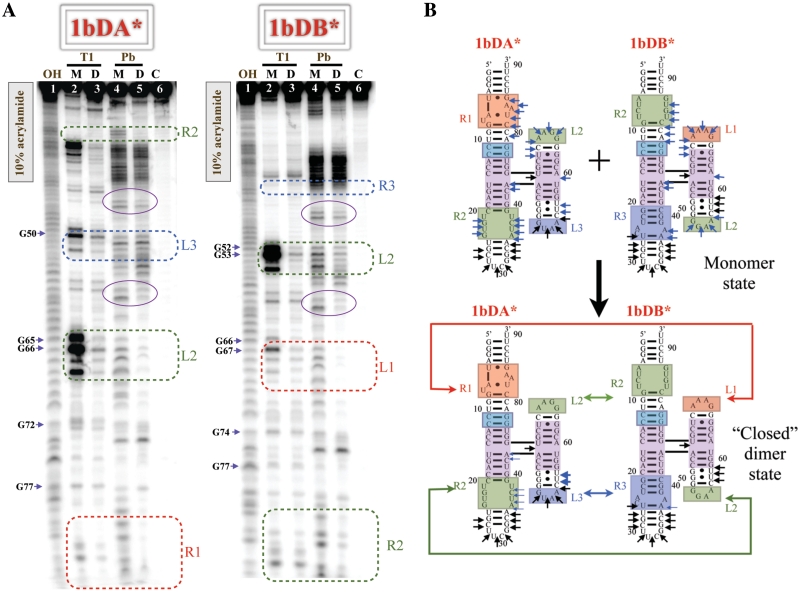
As mentioned above, native gel experiments showed molecules in series #1 forming tightly bound dimers. From the experimental evidence of cooperative molecule 8 association, we suspected that the dimer formation in series #1 might involve two interaction interfaces, resulting in the formation of ‘closed’ cooperative dimer complexes different from the open dimers formed by 8TA interacting with 8TC (see above). We decided to assess the cooperativity of the 1b complex, which potentially has an optimal geometry for very stable dimerization. Therefore, we designed the hetero-dimeric set derived from molecule 1b, 1bDA–1bDB, (see Supplementary Figure S2, ‘D’ stands for dimer-optimized design) in such a way that HS2 of 1bDA can interact with HS1 of 1bDB while HS2 of 1bDB interacts with HS1 of 1bDA with neither molecule capable of self-associating. The assembly of 1bDA–1bDB was assayed on native gels using the trimeric complex as mobility control (Supplementary Figure S7B). The obtained 1bDA–1bDB complex exhibits significantly slower mobility than that of the open 8TA–8TC dimer but runs faster than the trimer complex, supporting the idea of a distinct dimer-forming complex having a different geometry. We used chemical probing with lead ions to study the assembly.
For lead-cleavage studies, both 1bDA* and 1bDB* were radiolabeled and analyzed in their monomer and dimer forms (Figure 5A–B). The upper regions of the 1bDA and 1bDB denaturing gels were resolved using lower concentration acrylamide gels, see Supplementary Figure S9.
Figure 5.
Lead cleavage analysis of cooperative (‘closed’) hetero-dimer 1bDA–1bDB. (A) Denaturing gels of 1bDA* and 1bDB* on 10% acrylamide gels. To resolve upper regions, additional denaturing gels were performed and can be found in the Supplementary Data. (B) Lead cleavage data on 1bDA* and 1bDB*. Positions of lead cleavage are highlighted in an identical manner as in the trimer construct 8TA–8TB–8TC, see Figure 2.
On the one hand, when 1bDA* is pre-assembled with 1bDB and then exposed to lead (lane 5, Figure 5A), R1 and R2 from HS1 and L2 of HS2 are protected. No changes are observed upon dimerization in L3 itself, and increased cutting is observed at positions 55–57 (3′ to this loop) in 1bDA. This suggests that H3 of 1bDA, carrying the GUAA loop (L3), may in fact participate in the interaction, undergoing a conformational change that changes the cleavage pattern nearby. On the other hand, 1bDB* in the dimer state, bound to 1bDA, is significantly protected in the L1, L2 and R2 regions. For the GUAA receptor (R3), only three cuts are observed in the monomer, and two of these are partly protected upon dimerization (positions 36 and 37). The cleavage patterns of both 1bDA and 1bDB in monomer and dimer states, summarized in Figure 5B, show that all four interacting loop–receptor modules are in contact in the dimer. This is consistent with the native gel mobility shift data and supports the formation of ‘closed’ dimers that are different from the ‘open’ 8TA–8TC dimers.
Even though the cleavage of the 4WJ region is the same for all investigated molecules in their monomer state, we observe some differences in 4WJ accessibility between molecule 1b hetero-dimers and molecule 8 hetero-trimers. In contrast to the 4WJ cleavage pattern in the hetero-trimeric 8TA–8TB–8TC complex discussed above, we observe a large reduction upon dimer formation of the corresponding cuts between H3 and H4 at the 4WJ (position 59 in 1bDA and 61 in 1bDB), whereas cuts between H1 and H4 (position 73 in 1bDA and 75 in 1bDB) remain unchanged. This is consistent with the idea that the 4WJ adopts slightly different configurations to form the dimer or trimer-optimized geometry.
Both 1bDA and 1bDB were also exposed to nuclease digestion when pre-assembled in dimer complexes (see lane 3 in Figure 5A). As in the hetero-trimer system, we observe dramatic protection in the hetero-dimer, along the entire length of each of these RNAs when they are bound to each other, providing further support that closed dimers are formed.
Measurement of equilibrium constants to quantify cooperativity
To compare the affinities of interaction interfaces in different contexts and to quantify the cooperativity of association, we carried out multiple dissociation constant measurements for various homo- and hetero-systems using native gel shift analysis.
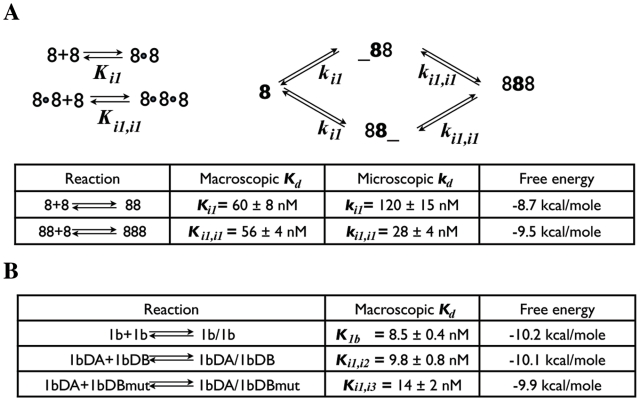
Focusing on the trimer homo-assembly of molecule 8, we know that it takes place with the formation of three identical interacting interfaces, designated as ‘i1’ in the following. In the first stage of open dimer formation, one interaction interface i1 is formed. Throughout the second stage (addition of a third molecule 8), two additional interfaces form (i1, i1) to obtain closed complexes. To quantify cooperativity correctly, the system must be characterized at the microscopic level, taking into account statistical factors reflecting the presence of multiple identical binding sites. Following standard textbooks in biophysics (42,43), the equilibria which characterize the homo-trimer formation of molecule 8 can be described in terms of two macroscopic dissociation constants Ki1 and Ki1,i1 or two microscopic dissociation constants ki1 and ki1,i1, as in Figure 6A. From the microscopic view, molecule 8 has two available interaction sites, HS1 and HS2, for binding to a second molecule. Even though the dimer complexes obtained via HS1 or HS2 are identical, it is necessary to distinguish the dimer formed by binding at HS1, which we denote with ‘88_’, from that formed by binding at HS2, denoted by ‘_88’. The macroscopic dissociation constants (Kd’s) for this process can then be expressed in terms of the microscopic dissociation constants (kd’s). Thus:
| (1) |
| (2) |
The overall dissociation constant for homo-trimer 8 formation (K8) was determined by analysis of gel-shift data (see Supplementary Figure S10) and found to be (3400±470) × 10−18 M2, as described in the Supplementary Data S2:
| (3) |
The binding of 8TA and 8TC forms an open dimer that has one interaction interface (i1), identical to the interfaces formed in molecule 8 homo-trimer. Therefore, by measuring the dissociation constant for 8TA–8TC, we can determine the microscopic dissociation constant for the i1 interface, ki1. Gel-shift analysis of the 8TA*–8TC complex, shown in Supplementary Figure S10, demonstrated that as the concentration of 8TC is increased, the mobility of the labeled 8TA gradually decreases, and at high concentrations equals the mobility of dimer. This indicates fast exchange kinetics on the timescale of the electrophoresis experiment. Analysis of the mobility shift of 8TA as a function of added 8TC allowed for determination of ki1 = 120 ± 15 nM. From ki1, we can evaluate the macroscopic Ki1 for the first step of homo-trimer formation using Equation (1):
| (4) |
Thus, from Equations (3) and (4), we can determine Ki1,i1 for the second step of trimer formation:
| (5) |
Although the macroscopic values Ki1 and Ki1,i1 are essentially the same, they should not be compared directly. One must take statistical effects into account using Equations (1) and (2) to calculate the microscopic dissociation constant, ki1,i1, corresponding to Ki1,i1 using Equation (2). The result is:
| (6) |
Comparing the microscopic kd’s, we find that ki1,i1 is at least four times smaller than ki1, indicating stronger binding for the third unit of the trimer, than expected from formation of the hetero-dimer (Figure 6A).
Figure 6.
(A) Macroscopic (left panel) and microscopic (right panel) equilibria of molecule 8 homo-trimer. (B) Table of measured equilibrium constants for closed dimers and other hetero-dimers.
To estimate the magnitude of cooperativity, we calculate the standard free energy for each step, writing each as an association reaction:
| (7) |
Thus, the difference in free energies is:
| (8) |
and the third molecule 8 binds almost 1 kcal/mol more strongly, an indication of cooperative association. However, ΔΔG is considerably less than twice the value for association at a single i1 interaction interface. This is not surprising, as each 4WJ must undergo structural rearrangements, as indicated by 3D modeling (see next section) and consistent with structure probing data. Rearrangement of the 4WJ is expected to cost energy. We estimate this energy to be about −2.7 kcal/mol per junction, obtained by dividing the apparent energy loss of binding free energy, –(2 × 8.7 – 9.5), by 3.
To compare the free energies for forming closed dimers versus open dimers, we measured Kd’s for the closed molecule 1b dimer and for the closed hetero-dimer 1bDA–1bDB (Supplementary Figure S10). The values are shown in Figure 6B. We find that the free energy of closed complex formed by 1b (−10.2 kcal/mol) is about 1.5 kcal/mole more stable than that of the corresponding open dimer, 8TA-8TC (−8.7 kcal/mol). In closed dimer-forming molecular constructs, we estimate the apparent energy loss of binding free energy to be about −7.2 kcal/mol [−(2 × 8.7 – 10.2) kcal/mol], dividing this number by two gives us −3.6 kcal/mol per monomer, which is somewhat more negative than for the trimer and may reflect increased electrostatic repulsion. Overall, these thermodynamic studies allowed us to obtain detailed information about the contribution of cooperativity to the association process, to estimate the reorganization of the 4WJ motif used in these tecto–RNA constructions and to calculate the binding affinities.
3D modeling
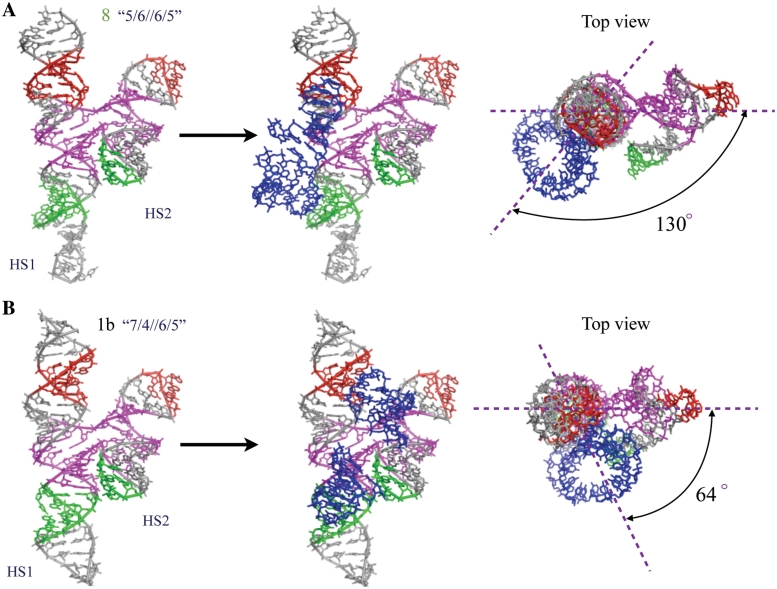
To gain insight into the trends we observed in the stoichiometries of the complexes that resulted when we moved the 4WJ crossover points along the helical stacking domains of the monomers, we constructed 3D models of trimer-forming molecule 8 and dimer-forming molecule 1b (Figure 7). Atomic resolution X-ray structures are known only for the GAAA loop bound to its receptor, therefore, all hairpin loops were changed to GAAA and all receptors to the GAAA loop receptor for modeling purposes, which should not affect the overall geometry of the molecule. We built the models by assembling motifs by superposition, as described in the ‘Materials and Methods’ section. We extracted the 3D structure of the 4-way junction from PDB file 1M5O (36), including three basepairs of each helix and the 3D structures of GAAA hairpin loops bound to their cognate receptor motifs from PDB file 1HR2 (38). At least two additional Watson–Crick base pairs were retained flanking each motif to ensure accurate superposition. The GAAA loop or receptor motifs were connected to the respective helical stems of the 4WJ junction by superposition of at least two Watson-Crick basepairs, using helical connectors, as described in ‘Materials and Methods’ section.
Figure 7.
3D models of monomer units of constructed molecules 8 (upper panel) and 1b (lower panel) by Swiss-PDB and NanoTiler. Color-coding is consistent with their schematic representations in Figure 3. The likely locations of HS2 of a neighboring molecule are denoted by blue hairpin loops docked in the loop-receptor regions.
The resulting model for molecule 8 is shown in the left panel of Figure 7A, with the nucleotides corresponding to the GAAA loop and loop–receptor of molecule 8 colored red, and those corresponding to the GGAA loop and loop–receptor colored green, as in the 2D diagrams of molecule 8 and its derivatives in other figures. In the middle panel, we added in blue the hairpin loops, correctly docked in the loop–receptors of molecule 8, to indicate the likely location of HS2 of a neighboring molecule when molecule 8 forms a complex. In the right panel, we provide a top-view of this model, with lines drawn to indicate the angle between the vertical plane through HS1 and the 4WJ of molecule 8 and the vertical plane defined by the interaction of HS1 of this molecule with HS2 of the neighboring molecule. Figure 7B shows equivalent models constructed for molecule 1b.
In the 3D model of molecule 8, the angle between the receptor interaction plane and 4WJ plane is ∼130°. We note that this value is very close to the ideal angle of 120° anticipated for trimer assembly. Assuming the flexible 4WJs of each unit rotate to align HS1 and HS2 of the same molecule, trimer formation results in a hexagon-like structure (top view in Figure 3 for 8) and consistent with TEM images. In contrast to molecule 8, the 3D model of molecule 1b, indicates that the corresponding angle is reduced by about 66°, resulting in an inter-planar angle of 64°. In this case, the motifs are oriented to favor formation of dimer having a parallelogram-like structure (top view in Figure 3 for 1b) that allows the two molecules to interact using both of their interaction interfaces. This is not possible for molecules 8TA and 8TC, which consequently form open dimers.
DISCUSSION
In this work, we have shown that H-shaped tecto–RNA units can be programmed to form cooperatively assembling closed-ring complexes of defined and predictable stoichiometries by adjusting the 4WJ crossover point within the interaction interfaces, and ensuring that ‘up–up’ association prevails between subunits.
To interpret the assembly properties of each of the molecules studied experimentally in this article, we applied the insights gained from 3D modeling of molecules 8 and 1b to generate heuristic models for the other molecules and the complexes they form. These are shown in top-view in Figure 3A, with each monomer unit represented as two circles (gray for HS1 and red for HS2) connected by a purple bridge indicating the 4WJ crossover. We emphasize these illustrations are meant as visual aids and not quantitative 3D models. We begin with series #1. Every 1 bp shift of the 4WJ in HS1 progressively reduces the inter-planar angle between the receptor–loop interaction plane of HS1 and the 4WJ plane by ∼33°. This is the helical twist per basepair in the A-form RNA helix. Even a 1 bp shift from molecule 8 reduces the available space so that no more than one molecule can bind, either by interacting with HS1 or with HS2. Thus all molecules in series #1 form dimers only (Figures 3A and 7), consistent with the assembly data.
In contrast, in series #2, the changes in the position of the 4WJ in HS2 progressively increase the inter-planar angle between the receptor–loop interaction plane of HS1 and the 4WJ plane, making it possible for these molecule to form increasingly larger nano-assemblies with more than three interacting units, as shown in Figure 3A.
In molecules 3 and 4, compensating changes were made in the positioning of the 4WJ in HS1 and HS2 so that the inter-planar angle between the receptor–loop interaction plane of HS1 and the 4WJ plane remained the same as in molecule 8. As anticipated, both these molecules form cooperative trimers, although their detailed geometries are expected to differ somewhat from that of molecule 8. Thus, molecule 4 trimer may have a more triangular structure while molecule 8 trimer probably has a hexagonal structure (Figure 3A).
It is necessary to point out that for two of the designs discussed above, molecules 1a ‘6/5//6/5’ and 2a ‘5/6//5/6’, 11 bp also separate the interacting motifs in the alternative, less-stable conformation of the 4WJ, in which H1 stacks on H4 and H2 stacks on H3. However, if these molecules did in fact change conformation before self-associating, the assembly would have to occur with change in directionality (‘up–down’), resulting in formation of long fibers, which is not observed.
Overall, we have constructed a variety of tecto–RNA molecules that form homo-complexes ranging from dimers to polymeric arrays in a highly predictable fashion. We redesigned two of these, the cooperative dimer-forming molecule 1b and the trimer-forming molecule 8, to create the cooperative hetero-complexes 1bDA–1bDB and 8TA–8TB-8TC, which have properties desirable for multi-functionality aims. The demonstrated hetero-multimerization amenability of these systems allow for independently modifying individual subunits (for example, in H1 and H2 on the other side of the loop-receptors from the 4WJ, see Figure 1B) to introduce new functionalities for biomedical applications. These systems also make it possible to probe the structures of the complexes and measure the assembly equilibria in a quantitative manner. In particular, we quantified the cooperativity of complex formation and determined the required energy for 4WJ rearrangements. The structure-probing experiments provide strong experimental evidence that the intended RNA complexes do in fact form, as designed. In addition, probing studies revealed details regarding 4WJ structural rearrangements during complex formation, consistent with modeling studies and thermodynamic measurements.
CONCLUSIONS
To advance the development of RNA nanotechnology for biomedical applications, we require deeper knowledge of RNA folding, intermolecular assembly and control of stoichiometry. This work is the first demonstration that we can rationally tune the stoichiometry of designed RNA homo-multimeric complexes.
Additionally, we have shown that the cooperatively forming closed dimers and trimers form uniquely compact structures that are resistant to nuclease degradation and thus exhibit suitable properties for use as scaffolding for diagnostic or therapeutic delivery agents. Finally, each individual building block in our systems can be derivatized at multiple sites, allowing attachment of multiple functional elements to achieve specific binding, visualization, gene silencing or other functional modalities in a single delivery complex.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Institutes of Health (2 R15GM055898-05).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Kirill Afonin, Luc Jaeger and Lorena Nasalean for useful discussions at early stages of this project and Anton Petrov for critically reading the manuscript.
REFERENCES
- 1.Westhof E, Massire C. Structural biology. Evolution of RNA architecture. Science. 2004;306:62–63. doi: 10.1126/science.1104482. [DOI] [PubMed] [Google Scholar]
- 2.Jaeger L, Chworos A. The architectonics of programmable RNA and DNA nanostructures. Curr. Opin. Struct. Biol. 2006;16:531–543. doi: 10.1016/j.sbi.2006.07.001. [DOI] [PubMed] [Google Scholar]
- 3.Leontis NB, Lescoute A, Westhof E. The building blocks and motifs of RNA architecture. Curr. Opin. Struct. Biol. 2006;16:279–287. doi: 10.1016/j.sbi.2006.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Leontis NB, Westhof E. Analysis of RNA motifs. Curr. Opin. Struct. Biol. 2003;13:300–308. doi: 10.1016/s0959-440x(03)00076-9. [DOI] [PubMed] [Google Scholar]
- 5.Geary C, Baudrey S, Jaeger L. Comprehensive features of natural and in vitro selected GNRA tetraloop-binding receptors. Nucleic Acids Res. 2008;36:1138–1152. doi: 10.1093/nar/gkm1048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Klussman S, editor. The Aptamer Handbook: Functional Oligonucleotides and their Applications. Weinheim: WILEY-VCH Verlag GmbH & Co. KGaA; 2006. [Google Scholar]
- 7.Hendrix DK, Brenner SE, Holbrook SR. RNA structural motifs: Building blocks of a modular biomolecule. Q. Rev. Biophys. 2005;38:221–243. doi: 10.1017/S0033583506004215. [DOI] [PubMed] [Google Scholar]
- 8.Paillart JC, Westhof E, Ehresmann C, Ehresmann B, Marquet R. Non-canonical interactions in a kissing loop complex: the dimerization initiation site of HIV-1 genomic RNA. J. Mol. Biol. 1997;270:36–49. doi: 10.1006/jmbi.1997.1096. [DOI] [PubMed] [Google Scholar]
- 9.Oroudjev EM, Kang PC, Kohlstaedt LA. An additional dimer linkage structure in moloney murine leukemia virus RNA. J. Mol. Biol. 1999;291:603–613. doi: 10.1006/jmbi.1999.2984. [DOI] [PubMed] [Google Scholar]
- 10.Strobel SA, Doudna JA. RNA seeing double: Close-packing of helices in RNA tertiary structure. Trends Biochem. Sci. 1997;22:262–266. doi: 10.1016/s0968-0004(97)01056-6. [DOI] [PubMed] [Google Scholar]
- 11.Afonin KA, Cieply DJ, Leontis NB. Specific RNA self-assembly with minimal paranemic motifs. J. Am. Chem. Soc. 2008;130:93–102. doi: 10.1021/ja071516m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Krasilnikov AS, Mondragon A. On the occurrence of the T-loop RNA folding motif in large RNA molecules. RNA. 2003;9:640–643. doi: 10.1261/rna.2202703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Brierley I, Pennell S, Gilbert RJ. Viral RNA pseudoknots: versatile motifs in gene expression and replication. Nat. Rev. Microbiol. 2007;5:598–610. doi: 10.1038/nrmicro1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Staple DW, Butcher SE. Pseudoknots: RNA structures with diverse functions. PLoS Biol. 2005;3:e213. doi: 10.1371/journal.pbio.0030213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lescoute A, Leontis NB, Massire C, Westhof E. Recurrent structural RNA motifs, isostericity matrices and sequence alignments. Nucleic Acids Res. 2005;33:2395–2409. doi: 10.1093/nar/gki535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Klein DJ, Schmeing TM, Moore PB, Steitz TA. The kink-turn: A new RNA secondary structure motif. EMBO J. 2001;20:4214–4221. doi: 10.1093/emboj/20.15.4214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Afonin KA, Leontis NB. Generating new specific RNA interaction interfaces using C-loops. J. Am. Chem. Soc. 2006;128:16131–16137. doi: 10.1021/ja064289h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lescoute A, Westhof E. Topology of three-way junctions in folded RNAs. RNA. 2006;12:83–93. doi: 10.1261/rna.2208106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laing C, Schlick T. Analysis of four-way junctions in RNA structures. J. Mol. Biol. 2009;390:547–559. doi: 10.1016/j.jmb.2009.04.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laing C, Jung S, Iqbal A, Schlick T. Tertiary motifs revealed in analyses of higher-order RNA junctions. J. Mol. Biol. 2009;393:67–82. doi: 10.1016/j.jmb.2009.07.089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hohng S, Wilson TJ, Tan E, Clegg RM, Lilley DM, Ha T. Conformational flexibility of four-way junctions in RNA. J. Mol. Biol. 2004;336:69–79. doi: 10.1016/j.jmb.2003.12.014. [DOI] [PubMed] [Google Scholar]
- 22.Montange RK, Batey RT. Riboswitches: emerging themes in RNA structure and function. Annu. Rev. Biophys. 2008;37:117–133. doi: 10.1146/annurev.biophys.37.032807.130000. [DOI] [PubMed] [Google Scholar]
- 23.Chworos A, Severcan I, Koyfman AY, Weinkam P, Oroudjev E, Hansma HG, Jaeger L. Building programmable jigsaw puzzles with RNA. Science. 2004;306:2068–2072. doi: 10.1126/science.1104686. [DOI] [PubMed] [Google Scholar]
- 24.Severcan I, Geary C, Verzemnieks E, Chworos A, Jaeger L. Square-shaped RNA particles from different RNA folds. Nano Lett. 2009;9:1270–1277. doi: 10.1021/nl900261h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Afonin KA, Bindewald E, Yaghoubian AJ, Voss N, Jacovetty E, Shapiro BA, Jaeger L. In vitro assembly of cubic RNA-based scaffolds designed in silico. Nat. Nanotechnol. 2010;5:676–682. doi: 10.1038/nnano.2010.160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severcan I, Geary C, Chworos A, Voss N, Jacovetty E, Jaeger L. A polyhedron made of tRNAs. Nat. Chem. 2010;2:772–779. doi: 10.1038/nchem.733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Afonin KA, Danilov EO, Novikova IV, Leontis NB. TokenRNA: a new type of sequence-specific, label-free fluorescent biosensor for folded RNA molecules. Chembiochem. 2008;9:1902–1905. doi: 10.1002/cbic.200800183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shu D, Huang LP, Hoeprich S, Guo P. Construction of phi29 DNA-packaging RNA monomers, dimers, and trimers with variable sizes and shapes as potential parts for nanodevices. J. Nanosci. Nanotechnol. 2003;3:295–302. doi: 10.1166/jnn.2003.160. [DOI] [PubMed] [Google Scholar]
- 29.Khaled A, Guo S, Li F, Guo P. Controllable self-assembly of nanoparticles for specific delivery of multiple therapeutic molecules to cancer cells using RNA nanotechnology. Nano Lett. 2005;5:1797–1808. doi: 10.1021/nl051264s. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Guo S, Tschammer N, Mohammed S, Guo P. Specific delivery of therapeutic RNAs to cancer cells via the dimerization mechanism of phi29 motor pRNA. Hum. Gene Ther. 2005;16:1097–1109. doi: 10.1089/hum.2005.16.1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zhang HM, Su Y, Guo S, Yuan J, Lim T, Liu J, Guo P, Yang D. Targeted delivery of anti-coxsackievirus siRNAs using ligand-conjugated packaging RNAs. Antiviral Res. 2009;83:307–316. doi: 10.1016/j.antiviral.2009.07.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jaeger L, Leontis NB. Tecto-RNA: one-dimensional self-assembly through tertiary interactions. Angew. Chem. Int. Ed Engl. 2000;39:2521–2524. doi: 10.1002/1521-3773(20000717)39:14<2521::aid-anie2521>3.0.co;2-p. [DOI] [PubMed] [Google Scholar]
- 33.Jaeger L, Westhof E, Leontis NB. TectoRNA: Modular assembly units for the construction of RNA nano-objects. Nucleic Acids Res. 2001;29:455–463. doi: 10.1093/nar/29.2.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Nasalean L, Baudrey S, Leontis NB, Jaeger L. Controlling RNA self-assembly to form filaments. Nucleic Acids Res. 2006;34:1381–1392. doi: 10.1093/nar/gkl008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rupert PB, Massey AP, Sigurdsson ST, Ferre-D'Amare AR. Transition state stabilization by a catalytic RNA. Science. 2002;298:1421–1424. doi: 10.1126/science.1076093. [DOI] [PubMed] [Google Scholar]
- 37.Rupert PB, Ferre-D'Amare AR. Crystal structure of a hairpin ribozyme-inhibitor complex with implications for catalysis. Nature. 2001;410:780–786. doi: 10.1038/35071009. [DOI] [PubMed] [Google Scholar]
- 38.Juneau K, Podell E, Harrington DJ, Cech TR. Structural basis of the enhanced stability of a mutant ribozyme domain and a detailed view of RNA–solvent interactions. Structure. 2001;9:221–231. doi: 10.1016/s0969-2126(01)00579-2. [DOI] [PubMed] [Google Scholar]
- 39.Guex N, Peitsch MC. SWISS-MODEL and the swiss-PdbViewer: An environment for comparative protein modeling. Electrophoresis. 1997;18:2714–2723. doi: 10.1002/elps.1150181505. [DOI] [PubMed] [Google Scholar]
- 40.Bindewald E, Grunewald C, Boyle B, O'Connor M, Shapiro BA. Computational strategies for the automated design of RNA nanoscale structures from building blocks using NanoTiler. J. Mol. Graph. Model. 2008;27:299–308. doi: 10.1016/j.jmgm.2008.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Costa M, Michel F. Rules for RNA recognition of GNRA tetraloops deduced by in vitro selection: Comparison with in vivo evolution. EMBO J. 1997;16:3289–3302. doi: 10.1093/emboj/16.11.3289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Cantor C, Schimmel P. Biophysical Chemistry: Part III: The Behavior of Biological Macromolecules. San Francisco: W. H. Freeman & Co; 1980. [Google Scholar]
- 43.Klotz IM. Ligand-Receptor Energetics. A Guide for the Perplexed. New York: John Wiley & Sons, Inc; 1997. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.