Abstract
In plants, small interfering RNAs (siRNAs) can trigger a silencing signal that may spread within a tissue to adjacent cells or even systemically to other organs. Movement of the signal is initially limited to a few cells, but in some cases the signal can be amplified and travel over larger distances. How far silencing initiated by other classes of plant small RNAs (sRNAs) than siRNAs can extend has been less clear. Using a system based on the silencing of the CH42 gene, we have tracked the mobility of silencing signals initiated in phloem companion cells by artificial microRNAs (miRNA) and trans-acting siRNA (tasiRNA) that have the same primary sequence. In this system, both the ta-siRNA and the miRNA act at a distance. Non-autonomous effects of the miRNA can be triggered by several different miRNA precursors deployed as backbones. While the tasiRNA also acts non-autonomously, it has a much greater range than the miRNA or hairpin-derived siRNAs directed against CH42, indicating that biogenesis can determine the non-autonomous effects of sRNAs. In agreement with this hypothesis, the silencing signals initiated by different sRNAs differ in their genetic requirements.
INTRODUCTION
Plants produce a variety of small RNAs (sRNAs), including microRNAs (miRNAs), small interfering RNAs (siRNAs) and trans-acting siRNAs (tasiRNAs), to regulate many different processes, such as development, stress and nutritional responses, chromatin structure and pathogen defense (1–5). A common theme in sRNA biogenesis is the processing of a double stranded RNA (dsRNA) by DICER-LIKE (DCL) enzymes into 21–24 nt long molecules. The sRNAs are then loaded onto one of several ARGONAUTE (AGO) proteins that drive transcriptional or post-transcriptional gene silencing (3,6–9).
SiRNAs are produced from perfectly-paired dsRNAs with endogenous (transposons, repetitive sequences) or exogenous (virus, transgenes) origins (3,7,8), while miRNAs originate from endogenous transcripts that include an imperfect foldback. Different from the other classes of sRNAs, a miRNA precursor often spawns just one functional sRNA. MiRNAs can trigger cleavage of target transcripts, or interfere with their translation (9). In the case of TAS targets, miRNA-initiated cleavage primes the synthesis of dsRNA by RNA DEPENDENT RNA POLYMERASE 6 (RDR6) and SUPPRESSOR OF GENE SILENCING 3 (SGS3), followed by DCL4-dependent processing of the dsRNA into 21 nt long tasiRNAs (10–15).
An important property of plant siRNAs is their non-cell autonomous activity. Even before the association of gene silencing with sRNAs was recognized, it became clear that co-suppression and post-transcriptional gene silencing (PGTS) could spread from one part of the plant to the other (16–18). Systemic silencing is transmitted via the phloem and it is dependent on RDR6 for amplification and reception of the silencing signal in other tissues (19–23). Silencing triggered by siRNAs likely moves from one cell to the other via plasmodesmata, channels that connect the cytoplasm of adjacent cells (16,20,23,24). In a first step, duplexes of 21 nt long siRNAs produced by DCL4 move 10 to 15 cells from their production site (24–26). In some cases, the primary silencing signal can spread further, relying on an RDR6- and SILENCING DEFECTIVE 3 (SDE3)-dependent amplification mechanism that supports the production of secondary siRNAs (24). Although amplification of the silencing signals is preferentially triggered by foreign RNAs, such as those derived from transgenes or from viruses (22,24), there are endogenous hairpin loci that behave very similarly (27). Furthermore, additional factors required for cell-to-cell movement of siRNA-triggered silencing include RDR2, the NRPD1a subunit of RNA polymerase IVa and CLASSY1, a SNF2 domain-containing protein (25,28,29). Grafting and deep sequencing of small RNA pools have revealed that endogenous 24 nt siRNAs can travel long distances in the plant (27,30).
While the mobility of siRNAs and its consequences are well documented, less is known about the mechanisms underlying non-autonomous effects of other classes of sRNAs, such as miRNAs and tasiRNAs. Several experiments with miRNA sensors and tissue-specific expression of natural or artificial miRNAs have indicated that the non-autonomous effects of most miRNAs do not extend very far (31–36). There are, however, several notable exceptions. MiR399 acts as a long distance signal in phosphate homeostasis (37), while miR390 accumulates in different tissues than its precursor (38). In addition, miRNAs have been detected in the phloem sap of several species (39,40). Since the phloem cells are enucleate and cannot produce RNAs, such miRNAs would need to be delivered from other cells such as phloem companion cells. Similarly, several strong lines of evidence indicate that miR165 and miR166 can move radially within the root, and thereby contribute to the patterning of root tissues (36). Finally, the precursor of tasiRNAs that regulate AUXIN RESPONSE FACTOR3 (ARF3) is transcribed in a narrow domain at the adaxial side of the leaf, but the mature tasiRNAs accumulate in a gradient that extends through much of the leaf (41,42).
While mobility of a variety of small RNAs is now accepted, their non-autonomous effects appear to differ. For example, movement of miRNAs appears to be much more limited than that of siRNAs (31–36). Because the investigated sRNAs differed in sequence in previous work, it has been difficult to disentangle the effects of sRNA sequence from the consequences of different sRNA histories due to divergent biogenesis mechanisms. We have compared sRNAs of identical sequence, but generated by either the miRNA or tasiRNA pathway. We show that similar to siRNAs, the silencing effects of miRNA can spread 10 to 15 cells from phloem companion cells to mesophyll cells, while a tasiRNA of the same sequence has much more far-reaching non-autonomous effects. Importantly, the genetic requirements for the mobile silencing signals triggered by miRNAs, tasiRNAs and siRNAs differ.
MATERIALS AND METHODS
Plant material
Arabidopsis thaliana Columbia (Col-0) is referred to as wild type. The dcl2 dcl3 dcl4 (‘dcl234′), dcl1-100, rdr6-15, rdr2-1 and nrpd1a-3 mutants and the SUC2:3xYFP transgenic lines have been previously described (43–47). Mutants expressing atasiR-SUL, amiR-SUL and siR-SUL were selected from F2 plants by PCR-based genotyping for the transgene and the mutations. F1 hybrids containing both SUC2:amiR-SUL and SUC2:3xYFP were isolated by double antibiotic selection.
Transgenic lines
The sRNA targeting the SUL homolog CH42 (At4g18480), UUAAGUGUCACGGAAAUCCCU, was designed with the WMD tool (33,48). Overlapping PCR was used to replace the mature miRNA and miRNA* in the MIR319a (AT4G23713), MIR156c (AT4G31877), MIR164b (AT2G47585) and MIR167a (AT3G22886) backbones. The same approach was used to generate the atasiR-SUL constructs, by replacing siR255 in the three members of the TAS1 family, TAS1a (AT2G27400), TAS1b (AT1G50055) and TAS1c (AT2G39675), respectively (48). For the siR-SUL construct, we cloned the same CH42 fragment (TAIR9 coordinates chromosome 4, 10, 202, 162-10, 202, 350) in both sense and antisense orientation into the pHANNIBAL vector (49). All constructs were shuttled into a modified version of the pGreen vector (50) containing the CaMV35S and SUC2 promoters (51–53). Binary constructs (Supplementary Table S3) were introduced into Agrobacterium tumefaciens strain ASE (54), which was used for floral dip transformation (55) (see Supplementary Data for additional details).
RNA analysis
Total RNA was isolated from two-week old plants using TRIZOL (Invitrogen, Carlsbad, CA, USA). sRNA blots were prepared by resolving 10–20 µg of total RNA on a 17% PAGE gel under denaturing conditions (7 M urea) and subsequent transfer to a positively charged nylon membrane. Membranes were hybridized with DNA oligonucleotide probes that had been radioactively labeled with γ-32P-ATP and OptiKinaseTM (USB, Cleveland, OH, USA). For detection of sRNAs derived from the siR-SUL construct, we employed a DNA probe consisting of the CH42 fragment in the RNAi triggering vector, which was labeled with α-32P-dCTP using the Prime-a-genes kit (Promega, Madison, WI, USA). cDNA for RT–PCR was synthesized with the RevertAidTM First Strand cDNA Synthesis kit (Fermentas, Burlington, Canada). See Supplementary Table S4 for probes.
Small RNA sequencing
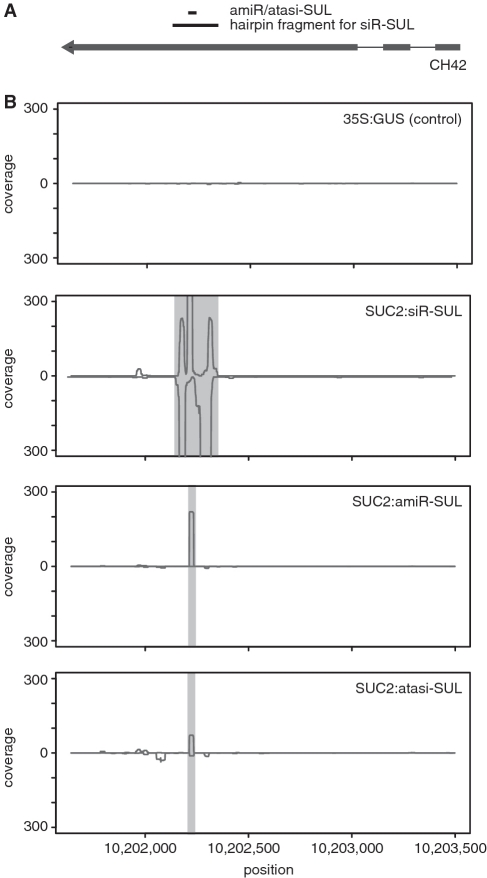
Small RNA libraries were constructed following a protocol described elsewhere (56) with modifications (Supplementary Data) and sequenced on the Illumina GAII platform (San Diego, CA, USA). Two independent libraries (biological replicates) were analyzed for the amiR-SUL and atasiR-SUL lines. The sRNA sequence tags were filtered and mapped back to the A. thaliana reference genome using SHORE (57), yielding 5.7–6.5 million aligned sRNA tags. We then calculated coverage graphs allowing or disallowing up to two mismatches to the CH42 locus. The effect of excluding repetitive matches was investigated, but found to be negligible (data not shown). We tested the significance of the secondary sRNA population observed in the SUC2:atasi-SUL line as follows. First, we defined a 500-bp region for the CH42 locus where secondary sRNAs were highly increased (Chr4:10201701.10202200, excluding the amiR/atasi-SUL region). We then determined the total number of reads for this region in both samples, which were 99 in SUC2:atasiR-SUL and 22 in SUC2:amiR-SUL. Next, starting from this region, we divided the genome in both directions, in 500-bp bins, counted the total sRNA reads in the two lines and calculated the fold change for each bin with more than 60 reads across both lines (50% of that in the CH42 bin). To avoid division by zero, we added a pseudo count of one to each bin.
Microscopy
YFP expression and natural florescence of chlorophyll were analyzed with a Leica MZ FLIII microscope (Leica Microsystems, Wetzlar, Germany) fitted with wide- and band-pass YFP filters and an AxioCam HRc (Zeiss, Jena, Germany) digital camera with Zeiss AxioVision software version 3.1.
RESULTS
Non-autonomous effects of miRNAs
To investigate movement of a silencing signal, we employed sRNAs targeting CHLORINA42 (CH42), the A. thaliana homolog of tobacco SULPHUR (SUL). Inactivation of CH42 causes bleaching of green plant tissues (58), resulting in an easily-scorable phenotype. We targeted CH42 with an artificial miRNA, amiR-SUL (33). We compared the effects of amiR-SUL with those of siRNAs spawned from a transcribed inverted repeat of CH42 sequences (siR-SUL) (49). Both constructs were introduced into A. thaliana plants under the control of the SUCROSE-PROTON SYMPORTER 2 (SUC2) promoter, which confers strong expression in phloem companion cells (51,52).
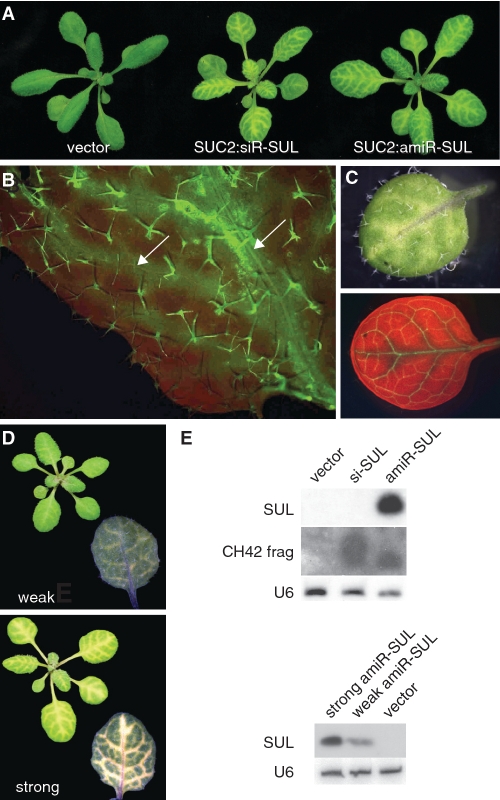
Himber and colleagues (24) have shown that the effects of siRNAs can extend 10 to 15 cells from their production site. Consistent with this report, there was prominent bleaching of green mesophyll cells along the leaf veins in SUC2:siR-SUL plants (Figure 1A). A very similar phenotype was seen in SUC2:amiR-SUL plants, suggesting that silencing initiated by miRNAs spreads over a range comparable of that of siRNAs. Closer analysis of chlorophyll autofluorescence in SUC2:amiR-SUL plants confirmed that the bleached regions extended beyond the veins (Figure 1B). To determine directly how far the silencing had spread beyond the cells in which the SUC2 promoter is active, we crossed SUC2:amiR-SUL to a plant expressing three tandem copies of yellow florescent protein in the SUC2 domain (SUC2:3xYFP) (43). The large size of 3xYFP traps it inside cells, allowing precise localization of SUC2 promoter activity. The bleached area around the veins in SUC2:amiR-SUL was indeed much larger than the SUC2 expression domain (Figure 1C). Most plants carrying the SUC2:amiR-SUL construct presented different degrees of bleaching around the vascular tissue (Figure 1D, Supplementary Figure S1). The levels of amiR-SUL were positively correlated with the severity of bleaching (Figure 1E), but the extent of the bleached area around the vasculature was similar in all lines.
Figure 1.
Spreading of miRNA-triggered silencing from phloem companion cells. (A) SUC2:amiR-SUL and SUC2:siR-SUL plants present similar bleaching patterns. (B) UV-induced red chlorophyll autofluorescence is suppressed in bleached areas, which appear light green in a SUC2:amiR-SUL leaf. Arrows point to leaf veins. (C) SUC2:amiR-SUL SUC2:3xYFP leaf. Top, visible light; bottom, UV fluorescence. Bright green YFP signal is more restricted than the bleached areas that are dark. (D) Comparison of mild and severely bleached plants. A single leaf is shown in detail. (E) sRNA blots probed with an oligonucleotide specific for amiR-SUL (SUL) or a CH42 fragment (CH42 frag). U6 was used as loading control.
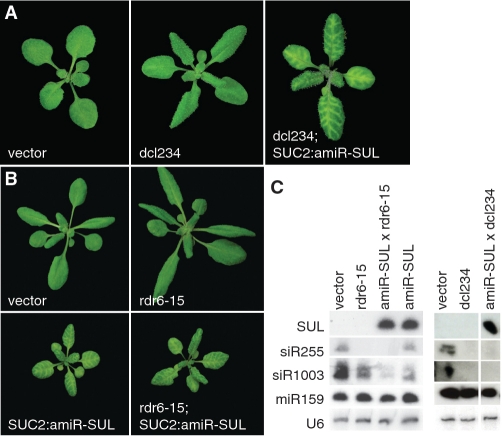
While miRNAs are produced mainly through the action of DCL1, several factors, such as secondary structure of the pre-miRNA and the tissue where the miRNA is expressed, can lead to miRNA precursors being processed by different DCLs, resulting in the production of siRNAs instead (59–61). Therefore, the non-autonomous effects in SUC2:amiR-SUL plants might be not caused by true miRNA-mediated silencing, but through siRNAs. To examine this possibility, we transformed dcl234 triple mutants with the SUC2:amiR-SUL construct. Inactivation of DCL2, DCL3 and DCL4 did not affect the bleaching phenotype (Figure 2A). As a control, we introduced the SUC2:amiR-SUL construct into dcl1 plants by crossing. In these plants, no bleaching occurred (Supplementary Figure S2).
Figure 2.
Confirmation of amiR-SUL-triggered silencing. (A) amiR-SUL production in dcl234 triple mutant background. (B) RDR6-independent spreading of amiR-SUL-triggered silencing. (C) sRNA blots. Probes are indicated on the right. siR255 production is RDR6 and DCL4-dependent, siR1003 is DCL3-dependent but RDR6-independent. MiR159 was used as an additional control. Note characteristic leaf shape of rdr6 and dcl234 mutants in (A) and (B).
In some cases, miRNA-triggered cleavage of targets can initiate transitive action of the miRNA, in which the cleaved target transcript is converted to dsRNA by RDR6 and subsequently processed into secondary siRNAs by DCLs (31,62). To test whether the cell-autonomous effect of amiR-SUL was due to transitivity, we crossed SUC2:amiR-SUL to rdr6-15 mutants, which do not generate secondary siRNAs. The SUC2:amiR-SUL-induced bleaching phenotype was unaffected by the rdr6 mutation (Figure 2B). The presence of the rdr6 mutation was confirmed both by the gross phenotype, and by the absence of tasiR255 production (Figure 2B and C). Taken together, these results suggest that the mobile silencing triggered by the SUC2:amiR-SUL is due to bona fide miRNA action.
A potential concern when using transgenes to characterize an endogenous mechanism is that expression levels much higher than those seen for endogenous miRNAs contribute to the observed effects. While commonly employed for assays of non-autonomous action of proteins or sRNAs (25,27,28,35,43,63), the SUC2 promoter is known to be strong (52,64,65). Abnormally high expression of a miRNA under control of the SUC2 promoter might saturate the processing machinery. This could in turn result in miRNA processing through pathways that are not DCL1 dependent. We therefore compared the expression levels of amiR-SUL to endogenous miRNAs by deep sequencing of the sRNA population. As reported in Supplementary Table S1, many miRNAs were expressed more strongly than amiR-SUL, with steady-state levels of some being more than two orders of magnitude higher. We conclude that our system reflects the natural action of the sRNA machinery.
Non-autonomous miRNA effects are not precursor specific
In phloem sap of the A. thaliana relative Brassica napus, a distinct subset of plant miRNAs has been identified (39,40), raising the possibility that only certain miRNA precursors can produce miRNAs that leave the cell of origin. To test the effects of the precursor, if any, on non-autonomous effects of the mature miRNA, we produced amiR-SUL from different miRNA precursors. We engineered the MIR156c, MIR164b and MIR167a precursors to produce the same mature miRNA sequence as our original amiR-SUL construct, which was in the MIR319a backbone; the corresponding constructs were named amiR-SUL_156, amiR-SUL_164 and amiR-SUL_167 (Supplementary Figure S3). We chose miR156, because it represents one of the families found in phloem sap (39,40). We chose MIR164 and MIR167, because it has been suggested that amiRNAs produced from these backbones in phloem companion cells do not have non-autonomous effects (35).
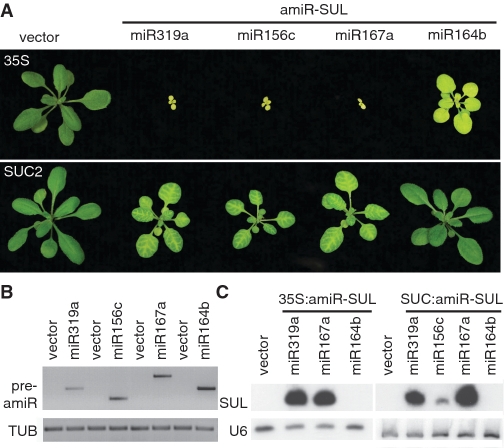
To determine the efficiency of amiRNA production from the different precursors, we first expressed these from the CaMV 35S promoter (53). Like plants that expressed amiR-SUL ubiquitously from the MIR319a precursor, 35S:amiR-SUL_156 and 35S:amiR-SUL_167 plants were very small and strongly bleached, like the original 35S:SUC2:amiR-SUL lines (Figure 3A). 35S:amiR-SUL_164 plants were larger, with variable bleaching, flowered normally and were fertile (Figure 3A). RNA blots indicated that amiR-SUL was only very inefficiently processed from the MIR164 precursor, even though it was expressed at a similar level as the other precursors (Figure 3B and C).
Figure 3.
Effect of MIRNA backbone on spreading of the silencing signal. (A) Whole-rosette phenotypes of plants expressing amiR-SUL from different precursors, with promoters indicated on the left. (B) Precursor expression monitored by RT–PCR with β-TUBULIN-2 (TUB) as control. (C) sRNA blots.
Similarly, SUC2:amiR-SUL_156 and SUC2:amiR-SUL_167 plants were strongly bleached, like the original SUC2:amiR-SUL lines (Figure 3A), but SUC2:amiR-SUL_164 plants were largely normal. While our results suggest that there are no specific miRNA precursor requirements for non-autonomous miRNA effects, the absence of extensive bleaching in SUC2:amiR-SUL_164 plants, apparently due to inefficient miRNA processing, indicates that expression levels are important in determining non-autonomous effects, consistent with non-selective movement of the silencing signal, similar to what appears to be the default for many proteins (66,67).
Non-autonomous effects of tasiRNAs
TasiRNAs, which like miRNAs are normally 21 nt long, are generated from TAS precursor transcripts. The phase of tasiRNA formation is determined by the miRNA cleavage event that triggers tasiRNA formation (12). This feature allows the design of artificial tasiRNAs (atasiRNAs) with specific sequences (68–70). We have previously developed a TAS1a derivative that produces an atasiRNA, atasiR-SUL_1a, with the same sequence as our amiR-SUL (48), therefore allowing a direct comparison of both sRNAs. The TAS1b and TAS1c derivatives atasiR-SUL_1b and atasiR-SUL_1c also produce the same sRNA.
SUC2:atasiR-SUL transgenic plants were much more severely affected than SUC2:amiR-SUL plants. In the most extreme cases, the phenotype of SUC2:atasiR-SUL plants began to approach that of 35S:atasiR-SUL plants, with pervasive bleaching throughout the entire leaf and much reduced stature (Figure 4A and Supplementary Figure S4). In weaker lines, which were more intensely bleached around the veins than in the remainder of the leaf, the boundary between affected and unaffected tissue was nevertheless much more diffuse than in SUC2:amiR-SUL plants (Figure 4A). The phenotypic differences between SUC2:atasiR-SUL and SUC2:amiR-SUL plants suggest that the biogenesis pathway of an sRNA, rather than its expression levels, has a major effect on its non-autonomous activity. This hypothesis was corroborated by a direct comparison of mature sRNA accumulation in the SUC2:atasiR-SUL and SUC2:amiR-SUL lines, with the first having much lower levels (Figure 6B).
Figure 4.
Non-autonomous effects of of atasiR-SUL. (A) Whole-rosette phenotype of SUC2:atasiR-SUL_1c. (B) sRNA blots. MiR173, which triggers TAS1 processing, was used as an additional control.
Figure 6.
Differential genetic requirements for non-autonomous effects of amiR-SUL and atasiR-SUL initiated silencing. (A) Whole-rosette phenotypes. (B) sRNA blots.
SiRNA-triggered silencing can spread across long distances, by means of an RDR6-dependent amplification mechanism termed transitivity (24). Unfortunately, one cannot test directly RDR6-dependence of non-cell autonomous tasiRNA effects, because tasiRNA generation itself requires RDR6 (10,11). We therefore designed an atasiR-SUL in which the two or three last nucleotides, respectively, do not pair with the target transcript (atasiR-SUL_2mm and atasiR-SUL_3mm) (Supplementary Data), based on a proposal (62) that this reduces 3′ 5′ transitivity, which depends on priming by the sRNA. Both SUC2:atasiR-SUL_2mm and SUC2:atasiR-SUL_3mm plants suffered from the same widespread bleaching as the original SUC2:atasiR-SUL plants (Supplementary Figure S5). This experiment, however, does not address the possibility of 5′ 3′ or priming-independent transitivity. We therefore sequenced the 19 to 25 nt sRNA population around the CH42 locus in these transgenic lines (Figure 5A). We found very few novel sRNAs matching the CH42 locus in SUC2:siR-SUL and SUC2:amiR-SUL plants that did not correspond to the sRNAs generated from the triggering transgene (Figure 5B and Supplementary Figure S6). Although the most abundant species was still the original atasiR-SUL trigger, the level of novel sRNAs was more than four times higher in SUC2:atasiR-SUL compared to SUC2:amiR-SUL plants (Supplementary Table S2). To confirm that this variation was not fortuitous, we compared the ratios of sRNAs reads between these two lines for different regions of the genome. We identified 1971 bins of length 500 bp in which the sum of the read counts for both samples was at least 50% of the read count for the CH42 bin. Only 60 bins (3%) had a read ratio between the two lines equal or higher than half of the ratio at the CH42 locus, and only 14 (0.7%) had a similar or higher ratio (Supplementary Figure S7). This comparison suggested that the increase in secondary sRNAs in the SUC2:atasi-SUL line is indeed significant and that transitivity has potentially a role in tasiRNA spreading.
Figure 5.
Secondary sRNAs at the CH42 locus. (A) Diagram of CH42 locus. Exons are indicated as thick lines. Regions targeted by primary sRNAs from siR-SUL and amiR/atasiR-SUL transgenes are shown. (B) Small RNA populations at the CH42 locus. About 19–26 nt sRNAs, with a maximum of two mismatches (as in amiR/atasi-SUL), are shown. See Supplementary Figure S6 for perfect-match sRNAs only. Grey regions indicate origin of primary siR-SUL and amiR/atasiR-SUL, respectively.
Genetic requirements for non-autonomous effects of different sRNAs
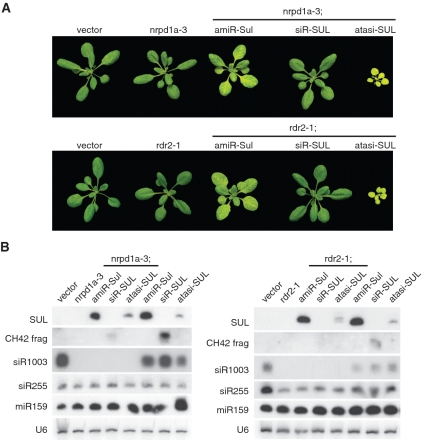
Mutations in several genes, including RDR2, NRPD1a and CLASSY compromise the non-autonomous effects of sRNAs (28,29). To determine whether the different classes of sRNAs rely on the same genetic system for spreading of the silencing signal, we crossed siR-SUL, amiR-SUL and atasiR-SUL producing lines to rdr2-1 and nrpd1a-3 mutants. As expected from previous work (28,29), the bleaching in SUC2:siR-SUL plants was completely suppressed in both mutant backgrounds. In contrast, bleaching triggered by SUC2:amiR-SUL and SUC2:atasiR-SUL was not affected in these mutants (Figure 6A), indicating that the non-autonomous effects of the different sRNAs have differential genetic requirements.
DISCUSSION
Here, we have documented that the silencing effects of an amiRNA, amiR-SUL, can extend 10 to 15 cells from the site of its production in phloem companion cells, which is in the same range as observed for transgene-derived siRNAs. In contrast to siRNAs (25,28,29), the non-autonomous effects of amiR-SUL do not depend on RDR2 and NRPD1a. While the specific precursor from which the amiRNA is processed does not seem to be essential for non-autonomous silencing, the biogenesis pathway through which a 21 nt sRNA is generated plays a crucial role, since the silencing effects of an atasiR-SUL of identical sequence as amiR-SUL extend much further.
Recently, Tretter et al. (35) examined non-autonomous effects of sRNAs, using sRNAs targeting PHYTOENE DESATURASE (PDS), downregulation of which produces a similar phenotype as CH42/SUL knockdown. They reported that expression of siRNAs, but not amiRNAs, under indirect control of the SUC2 promoter via the LhG4 transactivator (71), resulted in bleaching beyond the veins. One reason for apparent failure to detect amiRNA non-autonomy could be relatively low sRNA expression levels due to the LhG4 system, which is known to suffer from variable efficacy (71,72). Such a scenario is in line with our finding that efficiency of amiRNA processing affects the detection of non-autonomous effects, similar to what has been reported before for siRNAs (25,29). In support of this explanation, Tretter et al. (35) did observe non-autonomous effects after simultaneous expression of two amiRNAs under direct control of the SUC2 promoter, which caused a very similar phenotype as seen in the majority of our SUC2:amiR-SUL lines.
Perhaps our most intriguing finding is that amiR-SUL and atasiR-SUL, despite having identical sequences, caused distinct silencing phenotypes in our system. Which factors could be responsible for defining how far sRNA-triggered silencing spreads? The most obvious difference is the pathway that generates the sRNA. Mallory et al. (73) have reported a case in which siRNAs produced from inverted repeats could spread systemically, while siRNAs for the same target, but derived from viral amplicons, were not able to move. It is likely that different DCLs and co-factors, which load AGO-containing RNA induced silencing complexes (RISCs), or AGOs and their co-factors, play a major role in defining the range of the silencing signal (9). This is at least indirectly supported by the different genetic requirements for miRNA- and tasiRNA-triggered silencing signals in our system.
Given several recent reports in which production and effect of sRNAs were directly examined (27,30), it seems likely that the mobile signal is the triggering sRNA itself. Differences in biogenesis could impact the production of secondary sRNAs, which in turn can mediate mobility of the silencing signal (24). We have detected more secondary sRNAs in tasiR-SUL than in amiR-SUL or siR-SUL expressing plants, showing that limited transitivity might contribute to the spreading of tasiRNA silencing.
In contrast to siR-SUL, transmission of the silencing signal triggered by an amiRNA or an atasiRNA does not rely on RDR2 and NRPD1a. Genetic screens using two different trigger loci have previously identified these two factors as being required for movement of siRNA-silencing signals (25,28,29). Some observations (28) indicate that RDR2 and NRPD1a act downstream of siRNA production, either by supporting the translocation of the silencing signal or its reception in other cells. Smith and colleagues (29), on the other hand, suggested that both proteins are involved in the amplification and/or generation of the signal. In any case, that the non-autonomous effects of amiR-SUL and atasiR-SUL are insensitive to loss of RDR2 or NRPD1a shows that not all sRNAs require these two factors for transmission of their effects to neighboring cells.
Together with previous studies (31–38), our work highlights that non-autonomous action of miRNAs is likely to be context-dependent. One of the responsible factors might be the expression level of an miRNA. Among mutants that alter cell-to-cell spreading of siRNA-mediated gene silencing, those with more extensive movement of the silencing signal also have higher siRNA levels, while one of the classes lacking non-autonomous siRNA effects no longer accumulates 21 nt siRNAs (25). The same correlation has been observed for systemic movement of siRNAs, where higher copy number of the triggering transgene may lead to more efficient systemic acquired silencing (20). As discussed above, this could be one of the reasons why different levels of non-autonomy have been detected for the same amiRNAs (35).
Plants expressing higher levels of amiR-SUL present stronger bleaching, but silencing does not appear to spread further than in more weakly bleached lines. Nonetheless, expression levels still seem to be an important feature, since it affects the extent to which the neighboring cells are affected. Corroborating this idea, there are various types of published evidence for non-cell autonomy of 13 of the 19 miRNA families that are expressed more highly than amiR-SUL in SUC2:amiR-SUL plants (Supplementary Table S1) (36–40).
A second factor affecting miRNA non-autonomy could relate to time and place of expression. Both selective and non-selective intercellular mobility of molecules are affected by the tissue and developmental stage of the plant (66,74–76). In addition, trafficking of the silencing signal may depend on the cell type. RNAi initiated in epidermal cells has been shown to spread only locally, while expression of the same RNAi trigger in an entire leaf engenders systemic silencing (77). It is possible that, compared to other cell types, miRNAs expressed in phloem companion cells, as in this study, can more easily initiate non-autonomous silencing, or move themselves to adjacent cells, e.g. because phloem companion cells contain factors that promote non-autonomous behavior.
A third, less often considered possibility could be tissue- or cell type-specific processing of the precursor. Some miRNA precursors that are mainly processed by DCL1 in leaves can be processed by DCL3 in inflorescences, where they spawn a distinct class of miRNAs that are 23 to 25 nt long (61). Spreading of silencing triggered by amiR-SUL and atasiR-SUL is not due to the sRNA sequence, but more likely caused by biogenesis factors or effectors engaged in the miRNA and tasiRNA pathways. In analogy, the production of miRNAs through tissue-specific pathways could result in differential non-autonomous effects.
In summary, we propose that the question of cell-autonomy versus non-autonomy of sRNAs does not have a simple answer, but rather that it is contingent on several circumstances that include time, place and level of expression, which may interact with biogenesis and translocation pathways in a complex manner. Depending on the setting, miRNA behavior might therefore range from strictly cell-autonomous action, to local spreading that generates morphogenetic gradients, and even long-distance systemic silencing (36–38,42). The apparent behavior of tasiRNAs might be even more complex, as the non-autonomous effect of tasiRNA might depend both on tasiRNA-specific factors and on the action of the upstream triggering miRNAs.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Deutscher Akademischer Austausch Dienst (DAAD to F.F.F.); European Community (FP6 IP SIROCCO, contract LSHG-CT-2006-037900); FP7 Collaborative Project AENEAS (contract KBBE-2009-226477); Gottfried Wilhelm Leibniz Award; Max Planck Society. Funding for open access charge: Max Plank Society.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Lisa Smith and Christa Lanz for help in preparing and sequencing small RNA libraries.
REFERENCES
- 1.Mallory AC, Vaucheret H. Functions of microRNAs and related small RNAs in plants. Nat. Genet. Suppl. 2006;38:S31–S36. doi: 10.1038/ng1791. [DOI] [PubMed] [Google Scholar]
- 2.Vazquez F. Arabidopsis endogenous small RNAs: highways and byways. Trends Plant Sci. 2006;11:460–468. doi: 10.1016/j.tplants.2006.07.006. [DOI] [PubMed] [Google Scholar]
- 3.Chapman EJ, Carrington JC. Specialization and evolution of endogenous small RNA pathways. Nat. Rev. Genet. 2007;8:884–896. doi: 10.1038/nrg2179. [DOI] [PubMed] [Google Scholar]
- 4.Lu XY, Huang XL. Plant miRNAs and abiotic stress responses. Biochem. Biophys. Res. Commun. 2008;368:458–462. doi: 10.1016/j.bbrc.2008.02.007. [DOI] [PubMed] [Google Scholar]
- 5.Chuck G, Candela H, Hake S. Big impacts by small RNAs in plant development. Curr. Opin. Plant Biol. 2009;12:81–86. doi: 10.1016/j.pbi.2008.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Xie Z, Qi X. Diverse small RNA-directed silencing pathways in plants. Biochim. Biophys. Acta. 2008;1779:720–724. doi: 10.1016/j.bbagrm.2008.02.009. [DOI] [PubMed] [Google Scholar]
- 7.Carthew RW, Sontheimer EJ. Origins and mechanisms of miRNAs and siRNAs. Cell. 2009;136:642–655. doi: 10.1016/j.cell.2009.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ghildiyal M, Zamore PD. Small silencing RNAs: an expanding universe. Nat. Rev. Genet. 2009;10:94–108. doi: 10.1038/nrg2504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 10.Peragine A, Yoshikawa M, Wu G, Albrecht HL, Poethig RS. SGS3 and SGS2/SDE1/RDR6 are required for juvenile development and the production of trans-acting siRNAs in Arabidopsis. Genes Dev. 2004;18:2368–2379. doi: 10.1101/gad.1231804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vazquez F, Vaucheret H, Rajagopalan R, Lepers C, Gasciolli V, Mallory AC, Hilbert JL, Bartel DP, Crété P. Endogenous trans-acting siRNAs regulate the accumulation of Arabidopsis mRNAs. Mol. Cell. 2004;16:69–79. doi: 10.1016/j.molcel.2004.09.028. [DOI] [PubMed] [Google Scholar]
- 12.Allen E, Xie Z, Gustafson AM, Carrington JC. microRNA-directed phasing during trans-acting siRNA biogenesis in plants. Cell. 2005;121:207–221. doi: 10.1016/j.cell.2005.04.004. [DOI] [PubMed] [Google Scholar]
- 13.Gasciolli V, Mallory AC, Bartel DP, Vaucheret H. Partially redundant functions of Arabidopsis DICER-like enzymes and a role for DCL4 in producing trans-acting siRNAs. Curr. Biol. 2005;15:1494–1500. doi: 10.1016/j.cub.2005.07.024. [DOI] [PubMed] [Google Scholar]
- 14.Xie Z, Allen E, Wilken A, Carrington JC. DICER-LIKE 4 functions in trans-acting small interfering RNA biogenesis and vegetative phase change in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2005;102:12984–12989. doi: 10.1073/pnas.0506426102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Yoshikawa M, Peragine A, Park MY, Poethig RS. A pathway for the biogenesis of trans-acting siRNAs in Arabidopsis. Genes Dev. 2005;19:2164–2175. doi: 10.1101/gad.1352605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Palauqui JC, Elmayan T, De Borne FD, Crete P, Charles C, Vaucheret H. Frequencies, timing, and spatial patterns of co-suppression of nitrate reductase and nitrite reductase in transgenic tobacco plants. Plant Physiol. 1996;112:1447–1456. doi: 10.1104/pp.112.4.1447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Palauqui JC, Elmayan T, Pollien JM, Vaucheret H. Systemic acquired silencing: transgene-specific post-transcriptional silencing is transmitted by grafting from silenced stocks to non-silenced scions. EMBO J. 1997;16:4738–4745. doi: 10.1093/emboj/16.15.4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Voinnet O, Baulcombe DC. Systemic signalling in gene silencing. Nature. 1997;389:553. doi: 10.1038/39215. [DOI] [PubMed] [Google Scholar]
- 19.Brosnan CA, Mitter N, Christie M, Smith NA, Waterhouse PM, Carroll BJ. Nuclear gene silencing directs reception of long-distance mRNA silencing in Arabidopsis. Proc. Natl Acad. Sci. USA. 2007;104:14741–14746. doi: 10.1073/pnas.0706701104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palauqui JC, Balzergue S. Activation of systemic acquired silencing by localised introduction of DNA. Curr. Biol. 1999;9:59–66. doi: 10.1016/s0960-9822(99)80016-5. [DOI] [PubMed] [Google Scholar]
- 21.Schwach F, Vaistij FE, Jones L, Baulcombe DC. An RNA-dependent RNA polymerase prevents meristem invasion by potato virus X and is required for the activity but not the production of a systemic silencing signal. Plant Physiol. 2005;138:1842–1852. doi: 10.1104/pp.105.063537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Vaistij FE, Jones L, Baulcombe DC. Spreading of RNA targeting and DNA methylation in RNA silencing requires transcription of the target gene and a putative RNA-dependent RNA polymerase. Plant Cell. 2002;14:857–867. doi: 10.1105/tpc.010480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Voinnet O, Vain P, Angell S, Baulcombe DC. Systemic spread of sequence-specific transgene RNA degradation in plants is initiated by localized introduction of ectopic promoterless DNA. Cell. 1998;95:177–187. doi: 10.1016/s0092-8674(00)81749-3. [DOI] [PubMed] [Google Scholar]
- 24.Himber C, Dunoyer P, Moissiard G, Ritzenthaler C, Voinnet O. Transitivity-dependent and -independent cell-to-cell movement of RNA silencing. EMBO J. 2003;22:4523–4533. doi: 10.1093/emboj/cdg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dunoyer P, Himber C, Voinnet O. DICER-LIKE 4 is required for RNA interference and produces the 21-nucleotide small interfering RNA component of the plant cell-to-cell silencing signal. Nat. Genet. 2005;37:1356–1360. doi: 10.1038/ng1675. [DOI] [PubMed] [Google Scholar]
- 26.Dunoyer P, Schott G, Himber C, Meyer D, Takeda A, Carrington JC, Voinnet O. Small RNA duplexes function as mobile silencing signals between plant cells. Science. 2010;328:912–916. doi: 10.1126/science.1185880. [DOI] [PubMed] [Google Scholar]
- 27.Dunoyer P, Brosnan CA, Schott G, Wang Y, Jay F, Alioua A, Himber C, Voinnet O. An endogenous, systemic RNAi pathway in plants. EMBO J. 2010;29:1699–1712. doi: 10.1038/emboj.2010.65. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 28.Dunoyer P, Himber C, Ruiz-Ferrer V, Alioua A, Voinnet O. Intra- and intercellular RNA interference in Arabidopsis thaliana requires components of the microRNA and heterochromatic silencing pathways. Nat. Genet. 2007;39:848–856. doi: 10.1038/ng2081. [DOI] [PubMed] [Google Scholar]
- 29.Smith LM, Pontes O, Searle I, Yelina N, Yousafzai FK, Herr AJ, Pikaard CS, Baulcombe DC. An SNF2 protein associated with nuclear RNA silencing and the spread of a silencing signal between cells in Arabidopsis. Plant Cell. 2007;19:1507–1521. doi: 10.1105/tpc.107.051540. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Molnar A, Melnyk CW, Bassett A, Hardcastle TJ, Dunn R, Baulcombe DC. Small silencing RNAs in plants are mobile and direct epigenetic modification in recipient cells. Science. 2010;328:872–875. doi: 10.1126/science.1187959. [DOI] [PubMed] [Google Scholar]
- 31.Parizotto EA, Dunoyer P, Rahm N, Himber C, Voinnet O. In vivo investigation of the transcription, processing, endonucleolytic activity, and functional relevance of the spatial distribution of a plant miRNA. Genes Dev. 2004;18:2237–2242. doi: 10.1101/gad.307804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Alvarez JP, Pekker I, Goldshmidt A, Blum E, Amsellem Z, Eshed Y. Endogenous and synthetic microRNAs stimulate simultaneous, efficient, and localized regulation of multiple targets in diverse species. Plant Cell. 2006;18:1134–1151. doi: 10.1105/tpc.105.040725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Schwab R, Ossowski S, Riester M, Warthmann N, Weigel D. Highly specific gene silencing by artificial microRNAs in Arabidopsis. Plant Cell. 2006;18:1121–1133. doi: 10.1105/tpc.105.039834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Válóczi A, Várallyay E, Kauppinen S, Burgyán J, Havelda Z. Spatio-temporal accumulation of microRNAs is highly coordinated in developing plant tissues. Plant J. 2006;47:140–151. doi: 10.1111/j.1365-313X.2006.02766.x. [DOI] [PubMed] [Google Scholar]
- 35.Tretter EM, Alvarez JP, Eshed Y, Bowman JL. Activity range of Arabidopsis small RNAs derived from different biogenesis pathways. Plant Physiol. 2008;147:58–62. doi: 10.1104/pp.108.117119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlsbecker A, Lee JY, Roberts CJ, Dettmer J, Lehesranta S, Zhou J, Lindgren O, Moreno-Risueno MA, Vaten A, Thitamadee S, et al. Cell signalling by microRNA165/6 directs gene dose-dependent root cell fate. Nature. 2010;465:316–321. doi: 10.1038/nature08977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pant BD, Buhtz A, Kehr J, Scheible WR. MicroRNA399 is a long-distance signal for the regulation of plant phosphate homeostasis. Plant J. 2007;53:731–738. doi: 10.1111/j.1365-313X.2007.03363.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nogueira FT, Chitwood DH, Madi S, Ohtsu K, Schnable PS, Scanlon MJ, Timmermans MC. Regulation of small RNA accumulation in the maize shoot apex. PLoS Genet. 2009;5:e1000320. doi: 10.1371/journal.pgen.1000320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Yoo BC, Kragler F, Varkonyi-Gasic E, Haywood V, Archer-Evans S, Lee YM, Lough TJ, Lucas WJ. A systemic small RNA signaling system in plants. Plant Cell. 2004;16:1979–2000. doi: 10.1105/tpc.104.023614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Buhtz A, Springer F, Chappell L, Baulcombe DC, Kehr J. Identification and characterization of small RNAs from the phloem of Brassica napus. Plant J. 2008;53:739–749. doi: 10.1111/j.1365-313X.2007.03368.x. [DOI] [PubMed] [Google Scholar]
- 41.Schwab R, Maizel A, Ruiz-Ferrer V, Garcia D, Bayer M, Crespi M, Voinnet O, Martienssen RA. Endogenous tasiRNAs mediate non-cell autonomous effects on gene regulation in Arabidopsis thaliana. PLoS ONE. 2009;4:e5980. doi: 10.1371/journal.pone.0005980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chitwood DH, Nogueira FT, Howell MD, Montgomery TA, Carrington JC, Timmermans MC. Pattern formation via small RNA mobility. Genes Dev. 2009;23:549–554. doi: 10.1101/gad.1770009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Mathieu J, Warthmann N, Küttner F, Schmid M. Export of FT protein from phloem companion cells is sufficient for floral induction in Arabidopsis. Curr. Biol. 2007;17:1055–1060. doi: 10.1016/j.cub.2007.05.009. [DOI] [PubMed] [Google Scholar]
- 44.Herr AJ, Jensen MB, Dalmay T, Baulcombe DC. RNA polymerase IV directs silencing of endogenous DNA. Science. 2005;308:118–120. doi: 10.1126/science.1106910. [DOI] [PubMed] [Google Scholar]
- 45.Allen E, Xie Z, Gustafson AM, Sung GH, Spatafora JW, Carrington JC. Evolution of microRNA genes by inverted duplication of target gene sequences in Arabidopsis thaliana. Nat. Genet. 2004;36:1282–1290. doi: 10.1038/ng1478. [DOI] [PubMed] [Google Scholar]
- 46.Xie Z, Johansen LK, Gustafson AM, Kasschau KD, Lellis AD, Zilberman D, Jacobsen SE, Carrington JC. Genetic and functional diversification of small RNA pathways in plants. PLoS Biol. 2004;2:E104. doi: 10.1371/journal.pbio.0020104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laubinger S, Sachsenberg T, Zeller G, Busch W, Lohmann JU, Rätsch G, Weigel D. Dual roles of the nuclear cap-binding complex and SERRATE in pre-mRNA splicing and microRNA processing in Arabidopsis thaliana. Proc. Natl Acad. Sci. USA. 2008;105:8795–8800. doi: 10.1073/pnas.0802493105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Felippes FF, Weigel D. Triggering the formation of tasiRNAs in Arabidopsis thaliana: the role of microRNA miR173. EMBO Rep. 2009;10:264–270. doi: 10.1038/embor.2008.247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wesley SV, Helliwell CA, Smith NA, Wang MB, Rouse DT, Liu Q, Gooding PS, Singh SP, Abbott D, Stoutjesdijk PA, et al. Construct design for efficient, effective and high-throughput gene silencing in plants. Plant J. 2001;27:581–590. doi: 10.1046/j.1365-313x.2001.01105.x. [DOI] [PubMed] [Google Scholar]
- 50.Hellens RP, Edwards EA, Leyland NR, Bean S, Mullineaux PM. pGreen: a versatile and flexible binary Ti vector for Agrobacterium-mediated plant transformation. Plant Mol. Biol. 2000;42:819–832. doi: 10.1023/a:1006496308160. [DOI] [PubMed] [Google Scholar]
- 51.Imlau A, Truernit E, Sauer N. Cell-to-cell and long-distance trafficking of the green fluorescent protein in the phloem and symplastic unloading of the protein into sink tissues. Plant Cell. 1999;11:309–322. doi: 10.1105/tpc.11.3.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stadler R, Sauer N. The Arabidopsis thaliana AtSUC2 gene is specifically expressed in companion cells. Bot. Acta. 1999;109:299–306. [Google Scholar]
- 53.Odell JT, Nagy F, Chua N-H. Identification of DNA-sequences required for activity of the cauliflower mosaic virus-35S promoter. Nature. 1985;313:810–812. doi: 10.1038/313810a0. [DOI] [PubMed] [Google Scholar]
- 54.Fraley RT, Rogers SG, Horsch RB, Eichholtz DA, Flick JS, Fink CL, Hoffmann NL, Sanders PR. The SEV system: a new disarmed Ti plasmid vector system for plant transformation. Biotechnology. 1985;3:629–635. [Google Scholar]
- 55.Clough SJ, Bent AF. Floral dip: a simplified method for Agrobacterium-mediated transformation of Arabidopsis thaliana. Plant J. 1998;16:735–743. doi: 10.1046/j.1365-313x.1998.00343.x. [DOI] [PubMed] [Google Scholar]
- 56.Mosher RA, Melnyk CW, Kelly KA, Dunn RM, Studholme DJ, Baulcombe DC. Uniparental expression of PolIV-dependent siRNAs in developing endosperm of Arabidopsis. Nature. 2009;460:283–286. doi: 10.1038/nature08084. [DOI] [PubMed] [Google Scholar]
- 57.Ossowski S, Schneeberger K, Clark RM, Lanz C, Warthmann N, Weigel D. Sequencing of natural strains of Arabidopsis thaliana with short reads. Genome Res. 2008;18:2024–2033. doi: 10.1101/gr.080200.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Koncz C, Mayerhofer R, Koncz-Kalman Z, Nawrath C, Reiss B, Redei GP, Schell J. Isolation of a gene encoding a novel chloroplast protein by T-DNA tagging in Arabidopsis thaliana. EMBO J. 1990;9:1337–1346. doi: 10.1002/j.1460-2075.1990.tb08248.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Rajagopalan R, Vaucheret H, Trejo J, Bartel DP. A diverse and evolutionarily fluid set of microRNAs in Arabidopsis thaliana. Genes Dev. 2006;20:3407–3425. doi: 10.1101/gad.1476406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Fahlgren N, Howell MD, Kasschau KD, Chapman EJ, Sullivan CM, Cumbie JS, Givan SA, Law TF, Grant SR, Dangl JL, et al. High-throughput sequencing of Arabidopsis microRNAs: evidence for frequent birth and death of MIRNA genes. PLoS ONE. 2007;2:e219. doi: 10.1371/journal.pone.0000219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Vazquez F, Blevins T, Ailhas J, Boller T, Meins F., Jr Evolution of Arabidopsis MIR genes generates novel microRNA classes. Nucleic Acids Res. 2008;36:6429–6438. doi: 10.1093/nar/gkn670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Moissiard G, Parizotto EA, Himber C, Voinnet O. Transitivity in Arabidopsis can be primed, requires the redundant action of the antiviral Dicer-like 4 and Dicer-like 2, and is compromised by viral-encoded suppressor proteins. RNA. 2007;13:1268–1278. doi: 10.1261/rna.541307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.An H, Roussot C, Suárez-López P, Corbesier L, Vincent C, Piñeiro M, Hepworth S, Mouradov A, Justin S, Turnbull C, et al. CONSTANS acts in the phloem to regulate a systemic signal that induces photoperiodic flowering of Arabidopsis. Development. 2004;131:3615–3626. doi: 10.1242/dev.01231. [DOI] [PubMed] [Google Scholar]
- 64.Stadler R, Lauterbach C, Sauer N. Cell-to-cell movement of green fluorescent protein reveals post-phloem transport in the outer integument and identifies symplastic domains in Arabidopsis seeds and embryos. Plant Physiol. 2005;139:701–712. doi: 10.1104/pp.105.065607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stadler R, Wright KM, Lauterbach C, Amon G, Gahrtz M, Feuerstein A, Oparka KJ, Sauer N. Expression of GFP-fusions in Arabidopsis companion cells reveals non-specific protein trafficking into sieve elements and identifies a novel post-phloem domain in roots. Plant J. 2005;41:319–331. doi: 10.1111/j.1365-313X.2004.02298.x. [DOI] [PubMed] [Google Scholar]
- 66.Kim JY. Regulation of short-distance transport of RNA and protein. Curr. Opin. Plant Biol. 2005;8:45–52. doi: 10.1016/j.pbi.2004.11.005. [DOI] [PubMed] [Google Scholar]
- 67.Wu X, Weigel D, Wigge PA. Signaling in plants by intercellular RNA and protein movement. Genes Dev. 2002;16:151–158. doi: 10.1101/gad.952002. [DOI] [PubMed] [Google Scholar]
- 68.Montgomery TA, Yoo SJ, Fahlgren N, Gilbert SD, Howell MD, Sullivan CM, Alexander A, Nguyen G, Allen E, Ahn JH, et al. AGO1-miR173 complex initiates phased siRNA formation in plants. Proc. Natl Acad. Sci. USA. 2008;105:20055–20062. doi: 10.1073/pnas.0810241105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Montgomery TA, Howell MD, Cuperus JT, Li D, Hansen JE, Alexander AL, Chapman EJ, Fahlgren N, Allen E, Carrington JC. Specificity of ARGONAUTE7-miR390 interaction and dual functionality in TAS3 trans-acting siRNA formation. Cell. 2008;133:128–141. doi: 10.1016/j.cell.2008.02.033. [DOI] [PubMed] [Google Scholar]
- 70.de la Luz Gutiérrez-Nava M, Aukerman MJ, Sakai H, Tingey SV, Williams RW. Artificial trans-acting siRNAs confer consistent and effective gene silencing. Plant Physiol. 2008;147:543–551. doi: 10.1104/pp.108.118307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Moore I, Gälweiler L, Grosskopf D, Schell J, Palme K. A transcription activation system for regulated gene expression in transgenic plants. Proc. Natl Acad. Sci. USA. 1998;95:376–381. doi: 10.1073/pnas.95.1.376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Rutherford S, Brandizzi F, Townley H, Craft J, Wang Y, Jepson I, Martinez A, Moore I. Improved transcriptional activators and their use in mis-expression traps in Arabidopsis. Plant J. 2005;43:769–788. doi: 10.1111/j.1365-313X.2005.02486.x. [DOI] [PubMed] [Google Scholar]
- 73.Mallory AC, Mlotshwa S, Bowman LH, Vance VB. The capacity of transgenic tobacco to send a systemic RNA silencing signal depends on the nature of the inducing transgene locus. Plant J. 2003;35:82–92. doi: 10.1046/j.1365-313x.2003.01785.x. [DOI] [PubMed] [Google Scholar]
- 74.Kim I, Cho E, Crawford K, Hempel FD, Zambryski PC. Cell-to-cell movement of GFP during embryogenesis and early seedling development in Arabidopsis. Proc. Natl Acad. Sci. USA. 2005;102:2227–2231. doi: 10.1073/pnas.0409193102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kim I, Kobayashi K, Cho E, Zambryski PC. Subdomains for transport via plasmodesmata corresponding to the apical-basal axis are established during Arabidopsis embryogenesis. Proc. Natl Acad. Sci. USA. 2005;102:11945–11950. doi: 10.1073/pnas.0505622102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kobayashi K, Zambryski P. RNA silencing and its cell-to-cell spread during Arabidopsis embryogenesis. Plant J. 2007;50:597–604. doi: 10.1111/j.1365-313X.2007.03073.x. [DOI] [PubMed] [Google Scholar]
- 77.Ryabov EV, van Wezel R, Walsh J, Hong Y. Cell-to-Cell, but not long-distance, spread of RNA silencing that is induced in individual epidermal cells. J. Virol. 2004;78:3149–3154. doi: 10.1128/JVI.78.6.3149-3154.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.