Abstract
Coactivator-associated arginine methyltransferase 1 (CARM1), the histone arginine methyltransferase and coactivator for many transcription factors, is subject to multiple post-translational modifications (PTMs). To unbiasedly investigate novel CARM1 PTMs we employed high-resolution top-down mass spectrometry. Surprisingly, mouse CARM1 expressed in insect and mammalian expression systems was completely dimethylated at a single site in the C-terminal domain (CTD). We demonstrate that dimethylation of CARM1 occurs both in vivo and in vitro and proceeds via an automethylation mechanism. To probe function of automethylation, we mutated arginine 551 to lysine to create an automethylation-deficient CARM1. Although mutation of CARM1's automethylation site did not affect its enzymatic activity, it did impair both CARM1-activated transcription and pre-mRNA splicing. These results strongly imply that automethylation of CARM1 provides a direct link to couple transcription and pre-mRNA splicing in a manner differing from the other steroid receptor coactivators. Furthermore, our study identifies a self-regulatory signaling mechanism from CARM1's catalytic domain to its CTD.
INTRODUCTION
Protein arginine methyltransferases (PRMTs) are a family of enzymes that mono-and di-methylate arginine residues on protein substrates. The PRMTs are generally classified as either type I or II enzymes. While both types catalyze the formation of a monomethylated arginine intermediate, type I PRMTs further catalyze the production of an asymmetrical dimethylarginine, and type II PRMTs catalyze the formation of a symmetrical dimethylarginine (1). PRMTs play multiple roles in cellular function, including the regulation of transcription, pre-mRNA splicing, ribosome biogenesis, cytokine signaling and DNA repair (2).
Coactivator-associated arginine methyltransferase 1 (CARM1), also known as PRMT4, is a type I PRMT which was originally identified as an associated protein for glucocorticoid receptor-interacting protein 1 (GRIP1), the p160 family steroid receptor coactivator (3). CARM1 is best characterized as a coactivator for a number of transcription factors, either relying on GRIP1 as a tether or binding directly to these factors (3–5). CARM1 activates transcription via multiple mechanisms, including histone H3 methylation at R17 (6,7), methylation of other key coactivators (8,9), and recruitment of chromatin remodeling proteins (10). Two functional domains have been identified in CARM1: a central catalytic core and the C-terminal domain (CTD). The central catalytic core contains both S-adenosyl-methionine (AdoMet) binding and substrate binding domains, a structural characteristic of PRMT family proteins (11,12). The methyltransferase activity of CARM1 plays a pivotal role in its ability to regulate transcription (3) with one exception (13). The enzyme-dead CARM1 knockin mice have defects similar to those seen in their knockout counterparts (14). These observations suggest that the enzymatic activity of CARM1 is essential for most CARM1-regulated processes. In contrast to the catalytic core, the CTD of CARM1 is unique among PRMTs. While it is dispensable for methylation of known CARM1 substrates (11,12), deletion of the CTD of CARM1 greatly impairs transcriptional coactivation by CARM1 (15).
Multiple steroid receptor coactivators regulate alternative splicing. PGC-1α, CoAA and CAPER harbor RNA-recognition motifs (RRMs) and/or arginine–serine-rich regions characteristic of SR splicing factors, consistent with their involvement in mRNA processing (16–18). CARM1 does not possess any structural features implicating it in splicing. However, recent studies showed that RNA-binding proteins HuR and HuD (19,20) and several splicing and transcription elongation factors (SmB, SAP49, U1C and CA150) (21) are bona fide substrates for CARM1. CA150, a molecule that links transcription to splicing, only interacts with SMN, a spliceosome component, when it is methylated by CARM1 (21). Furthermore, the enzymatic activity of CARM1 is required to promote exon skipping on CD44 (21). Thus, CARM1′s enzymatic activity appears to regulate both transcription and pre-mRNA splicing events, although the mechanism of CARM1's regulation of these seemingly coupled events remains unknown.
CARM1 is unique among PRMTs in that various identified post-translational modifications (PTMs) on CARM1 are emerging as alternative mechanisms to regulate its function. Phosphorylation of CARM1 occurs on at least three serine residues. Phosphorylation of two CARM1 residues appears to regulate the enzymatic activity of CARM1 during mitosis (22,23). We previously observed that phosphorylation of serine 228 blocks CARM1 dimerization, interrupting proper binding of the dimerization ‘arm’ and thus inhibiting its methylation activity (23). Phosphorylation at serine 217 appears to blocks AdoMet binding to the catalytic site of CARM1 (22). More recently, serine 448 on CARM1 was found to be phosphorylated by PKA, which facilitates a direct interaction between CARM1 and estrogen receptor α (ERα) (24). Besides phosphorylation, CARM1 is modified by O-linked acetylglucosamine (25), however the location and function of this modification remains to be determined.
Top-down mass spectrometry (MS) is a powerful approach for mapping PTMs on a protein of interest (26–31). In top-down MS, the intact protein ions are directly introduced into the gas phase, which allows highly accurate determination of the molecular mass of proteins and a global view of all detectable modification forms. The specific modified forms can be isolated and subsequently fragmented in the mass spectrometer by tandem MS (MS/MS) such as collisionally activated dissociation (CAD) and electron capture dissociation (ECD) for highly reliable PTMs mapping (26–31). Furthermore, this method allows relative quantitation of each protein ion population to provide accurate measure of the percent of protein being modified (32–35).
Using high-resolution top-down combined with middle-down (limited digestion) MS, we precisely mapped a single mouse CARM1 automethylation site to R551, which is conserved among all vertebrate CARM1 proteins. Mutation of the automethylation site from arginine to lysine does not alter the enzymatic activity of CARM1. However, mutation of the automethylation site on CARM1 impairs both CARM1-activated transcription and pre-mRNA splicing, strongly implying that regulation of transcription and splicing events are jointly regulated by the PTM of CARM1.
MATERIALS AND METHODS
Protein expression and purification
FLAG-CARM1 was expressed from Sf9 cells as described previously (10). Full-length mouse CARM1 was PCR cloned into pFC14K Flexi vector (Promega, Madison, WI, USA). The CARM1 R551K mutant was obtained by site-directed mutagenesis of CARM1 in the pFC14K vector (CARM1R551K forward primer: 5′ gTCAATCACACCCACTCCAAgATgggCTCCATAATgAg 3′ and reverse primer: 5′ TCATTATggAgCCCATCTTggAgTgggTgTgATTgAC 3′).
The CARM1 construct contains a C-terminal HaloTag. The construct was expressed in transiently transfected HEK293T cells, affinity purified using HaloLink resin (Promega), and eluted by TEV protease. The purified CARM1 does not contain Halo tag, which remains covalently coupled to the resin. The exact details of expression and purification of full-length mouse CARM1 using the HaloTag are described in a separate publication (36).
Mass spectrometry
Top-down MS: high-resolution FT-ICR MS analysis of FLAG-CARM1 from Sf9 and CARM1 derived from HEK293T was performed under similar conditions as described previously (35). The intact mass spectra were collected under broad ion-trap isolation of three charge states. Fragmentation was performed by narrow isolation of a single charge state and then dissociated by CAD using 15% collision energy.
Middle-down MS: CARM1 was digested with endoprotease AspN for FT-ICR MS analysis. Fifty microgram of CARM1 from HEK293T cells was incubated with 250 ng of AspN for 20 min at 37°C in 2.5 mM ZnSO4 and 50 mM Tris pH 7.5. The resulting reaction was desalted on a 10 kDa NMWL Amicon Ultra (Millipore). Nano-electrospray ionization was performed in a 50% MeOH, 1% acetic acid spray solution on a Triversa Nanomate injector (Advion). Fragmentation was performed by narrow isolation of a single charge state and then dissociated by CAD using 12–16% collision energy or ECD using 3% activation energy for 50 ms.
32P-labeled PCR
The forward primer for the CT/CGRP mini-gene was labeled by incubation with polynucleotide kinase (PNK) in PNK buffer (New England Biolabs) with γ-labeled 32P-ATP at a final concentration of 1 µM per primer. Reactions were held at 37°C for 30 min. PCR was performed with Taq polymerase (Promega), 2 µl of 1:100 diluted cDNA per 10 µl reaction and a final primer concentration of 600 nM. Twenty cycles of PCR were performed, with 20 s annealing step at 59°C and 30 s extension step at 72°C. Reactions were run on 5% non-denaturing TBE-acrylamide gels, vacuum-dried and stored at −80°C prior to exposure to film.
In vitro methylation assays
Enzymes and substrates were incubated in 15 µl of 5 mM MgCl2, 20 mM Hepes pH 7.9, 1 mM EDTA, 1 mM DTT, 10% glycerol containing 1 µl of 3H–S-adenosylmethionine (78 Ci/mmol, GE Healthcare) for 5 min–1 h. The reaction mix was then separated by SDS–PAGE and fixed in methanol: acetic acid (50%:5%) containing Coomassie brilliant blue for 2 h followed by destaining in Coomassie-free methanol: acetic acid for 1 h. Fixed gels were incubated in ‘Amplify’ (Amersham Biosciences) scintillation fluid for 30 min, stored at −80°C overnight and exposed to a film.
Transient transfections
For mini-gene reporter assays
For mini-gene reporter assays, HEK293T cells were seeded into 6-well culture dishes with high glucose DMEM containing 10 % FBS. Twenty-four hours later each well was transfected with 2 µg ERE-CT/CGRP reporter, 5 ng CMX-ERα and 1 µg of pSG5-vector, pSG5-CARM1WT or pSG5-CARM1R551K with 8 µl of TurboFect (Fermentas). Media was replaced after 6 h and cells were grown in the presence of 10 nM 17β-estradiol (E2) or DMSO alone for 48 h.
For qRT-PCR of endogenous gene expression
For qRT-PCR of endogenous gene expression, ER18 cells (HEK293 cells with stable expression of ERα) or CARM1 null MEF cells were plated in 6-well culture dishes with 5% charcoal stripped FBS in high glucose-DMEM. Twenty-four hours later, ER18 cells were transfected with 1 µg of pSG5-vector, pSG5-CARM1WT or pSG5-CARM1R551K with 2.3 µl of TurboFect for 6 h. CARM1-null MEF cells were transfected with 2 µg of pSG5-vector, pSG5-CARM1WT or pSG5-CARM1R551K and 6 µl Fugene HD (Roche). At 24 h, media was changed fresh. At 48 h, transfected cells were treated with 10 nM E2 or DMSO for 3 h before harvesting.
Gal4-luciferase assays
These were performed in HEK293T cells by cotransfecting 1 µg of pSG5-vector, pSG5-CARM1WT or pSG5-CARM1R551K with CMX-ERα (5 ng), renilla-luciferase expression vector (100 ng), Gal4DBD-fusion protein (200 ng) and a UAS-Luc reporter (200 ng). Cells were then treated with DMSO or 10 nM E2 for 48 h prior to lysis and detection of luciferase. Firefly luciferase from the UAS reporter was normalized with internal control renilla luciferase.
RNA isolation, reverse transcription and qPCR
RNA was isolated using RNeasy kit (Qiagen) following the manufacturers’ instructions. DNA contamination was digested using DNAfree (Ambion). RNA was quantified and 2 µg was added to Superscript II (Invitrogen) reverse transcription reactions with oligo-d (T) as a primer. Completed reactions were diluted 100-fold prior to use as a template in qPCR and 32P-labeled reactions. qPCR was performed using 2× Sybr Green MasterMix (Invitrogen). Four microliters of 1:100 diluted cDNA was added to each 20 µl reaction and four replicates were performed per condition. Ct levels were compared to a relative standard curve of similarly prepared cDNA from the same cell type.
RESULTS
Mouse CARM1 is methylated at arginine 551 in vitro and in vivo
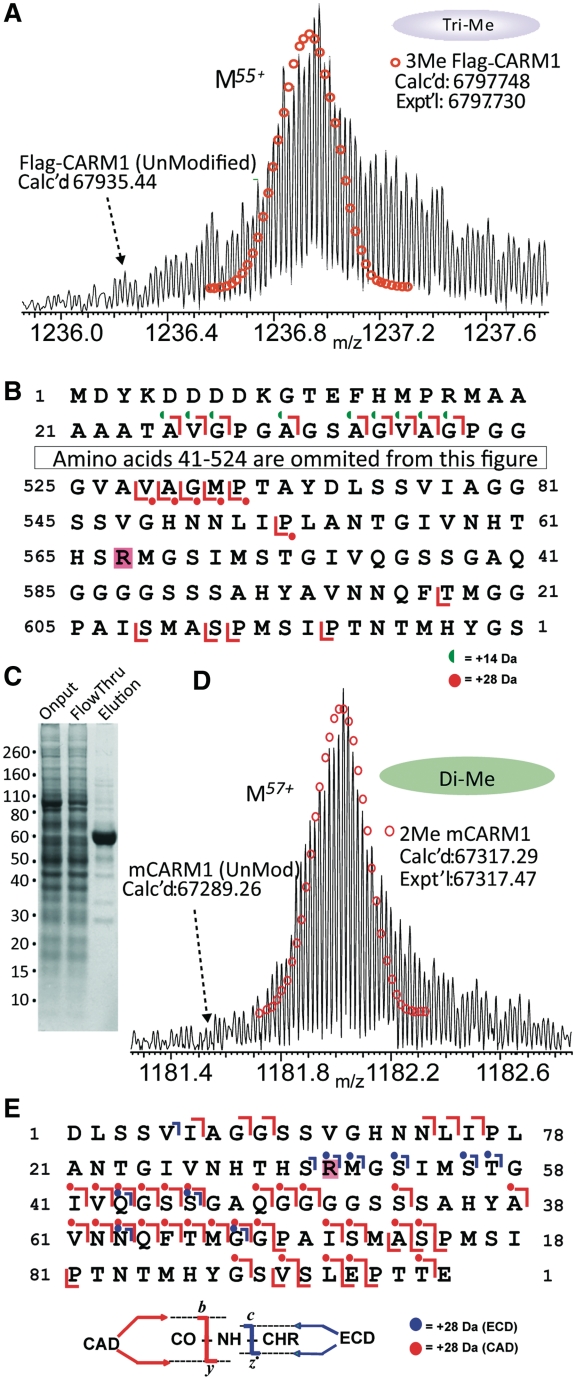
To map and determine the relative quantity of mouse CARM1 PTMs, we employed high-resolution top-down MS. We first attempted to analyze PTMs on full-length FLAG-CARM1 expressed in Sf9 insect cells using a Linear Trap Quadrupole (LTQ) Fourier Transform-Ion Cyclotron Resonance (FT-ICR) mass spectrometer. No ion peak was found at the predicted unmodified mass of FLAG-CARM1 (calculated mass 67 935.44, Figure 1A). In contrast, the major peak was 42 Da larger than the unmodified mass (Figure 1A). The experimentally determined mass was 67 977.30, which is very close to calculated mass 67 977.48 if CARM1 is trimethylated. To map the location of this additional mass, the 55+ charge state of the 67 977.3 Da mass peak was isolated in the LTQ and fragmented by CAD. MS/MS of FLAG-CARM1 identified an additional mass of 14 Da (consistent with monomethylation) on all N-terminal fragments of FLAG-CARM1. The minimal N-terminal sequence that is modified was primarily the FLAG-tag and linker sequence, which encompasses commonly methylated residues (lysines and arginines). Further site-mapping was not pursued because this modification would not occur on endogenous CARM1. An additional mass of 28 Da (consistent with dimethylation) was observed on C-terminal fragments that included the sequence from amino acids 537–582 (amino acids 555–600 of the FLAG-CARM1 fusion protein). This region only contains one likely dimethylation site, arginine (R) 551 (Figure 1B).
Figure 1.
High-resolution top- and middle-down MS reveal dimethylation at R551 of the C-terminal domain (CTD) of CARM1. (A) Mass of the 55+ precursor ion (intact protein) of an N-terminally tagged FLAG-CARM1 from Sf9 cells. The isolated spectra represents a single population of FLAG-CARM1 at isotopic resolution. LTQ/FT-ICR MS was used to determine the intact mass of recombinant CARM1 protein and map the site of methylation. The calculated (Calc’d) mass represents the mass predicted based on amino acid composition and putative PTMs. The theoretical peak distribution is represented graphically by a series of open (methylated) circles. The experimental (Expt’l) mass represents the mass determined from the MS spectra. (B) CAD fragmentation of full-length FLAG-CARM1 maps dimethylation site to the sequence between amino acids 555 and 600. Green half circles represent a fragment that is ∼14 Da larger than predicted (indicating monomethylation), while a red circle represents a fragment that is ∼28 Da larger than the predicted fragment (indicating dimethylation). Sequence map shows the coverage and placement of CAD fragments of FLAG-CARM1. (C) Coomassie staining of CARM1 (with Halo tag removed) purified from HEK293T cells. (D) CARM1 from HEK293T cells is dimethylated. Mass of the 57+ precursor ion of full-length CARM1 purified from HEK293T cells is shown, no peak is observed at the expected mass of unmodified CARM1. (E) CAD and ECD fragmentation map of an AspN proteolytic peptide (D520–E616) of CARM1. Blue marks represent ECD fragments while red marks represent CAD fragments as indicated in the figure graphic at the bottom.
The presence of monomethylation on the FLAG-tag potentially complicated the analysis of the role of methylation in CARM1 function, thus we switched to a mammalian expression system to overexpress CARM1 fused to a cleavable HaloTag® purification tag (Promega). Halo-CARM1 was expressed in HEK293T cells, purified on Halo-link resin, and cleaved with TEV protease to obtain full-length CARM1. Surprisingly, this approach produced a very pure protein (Figure 1C) and a significantly higher yield than FLAG-CARM1 purified from Sf9 cells. Top-down MS analysis of CARM1 from HEK293T matched the calculated mass of full-length CARM1 with a dimethyl modification (calculated mass is 67317.29 and experimentally determined mass is 67317.47), while unmodified CARM1 was undetectable (Figure 1D).
MS/MS of full-length CARM1 exhibited limited fragmentation data due to the poor stability of CARM1 during electrospray ionization. Thus, we took a middle-down MS approach by partially digesting CARM1 with the endoproteinase AspN prior to analysis on the LTQ/FT-ICR to determine the site(s) of dimethylation. Multiple charge states (M7+–M10+) in the FT-ICR fragmentation spectrum corresponding to an Asp-N proteolytic peptide (D521–E617) were detected to be dimethylated, with no observable peak matching the mass of the unmodified peptide (Supplementary Figure S1). Isolation of the M9+ charge state of the dimethylated D521–E617 peptide and subsequent CAD or ECD fragmentation unequivocally localized R551 as the single dimethylation site (Figure 1E).
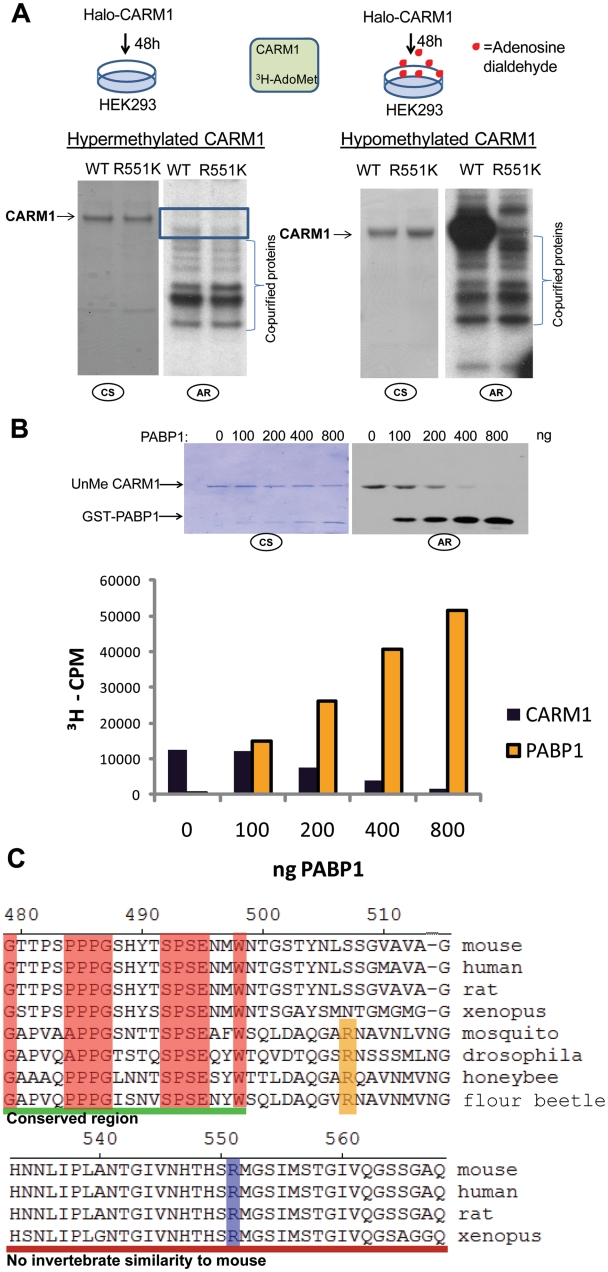
We created an arginine (R) to lysine (K) mutation in the HaloTag expression vector to preclude CARM1 methylation (CARM1R551K) and subsequently purified CARM1R551K from HEK293T cells. To monitor CARM1 automethylation, purified CARM1WT or CARM1R551K was incubated with a radiolabeled methyl donor S-adenosyl-methionine (3H-AdoMet) in vitro. Comparison of the two proteins in the absence of additional substrate indicated that neither protein was significantly methylated in vitro; however, extended exposure indicates in vitro methylation of co-eluted proteins (Figure 2A). To determine whether this lack of in vitro methylation was due to saturating levels of methylation in vivo, hypomethylated proteins were purified by pre-treating Halo-CARM1 transfected HEK293T cells with a methylation inhibitor, adenosine dialdehyde (Adox). Adox treatment for 48 h at 20 µM in the culture media significantly reduced basal level of endogenous asymmetric, di-methylated proteins as detected by a di-methyl H3R17 antibody (Supplementary Figure S2) as this antibody was shown to recognize histone H3R17me2 and other CARM1 substrates (37). When CARM1 proteins from these Adox-treated cells were compared in vitro, CARM1WT exhibited strong incorporation of the 3H-methyl groups, while CARM1R551K did not (Figure 2A). These biochemical data corroborate our MS data and indicate that R551 is the primary methylation site on CARM1 both in vivo and in vitro.
Figure 2.
The CTD of CARM1 is automethylated in vivo and in vitro. (A) CARM1WT and CARM1R551K expressed in the absence or presence of a methylation inhibitor (Adenosine dialdehyde, Adox) prior to in vitro methylation without any additional substrates. In vitro methylation reactions were performed in the presence of 3H-AdoMet, separated by SDS–PAGE, stained with coomassie and exposed to film. Autoradiograph shown represents a long exposure to detect low-level methyation. CS = coomassie staining, AR = autoradiograph. (B) Poly (A) binding Protein 1 (PABP1) can efficiently compete with hypomethylated, CARM1WT from automethylation in vitro. Hypomethlated (Adox treated) CARM1WT (1 µg) was incubated alone or in the presence of increasing amounts of PABP1 (0–800 ng). After exposure to a film, bands were excised and quantitiated for 3H incorporation. (C) Partial CLUSTALW alignment of known and putative CARM1 sequences indicates high conservation of the automethylation site within vertebrates (purple).
The strong in vitro methylation of purified, hypomethylated CARM1WT expressed in HEK293T implies an automethylation mechanism. However, the presence of minor co-eluted proteins as indicated in the in vitro methylation assay (Figure 2A) raised the possibility that one of these co-eluted proteins may methylate CARM1. To rule out this possibility, we incubated hypomethylated CARM1WT with increasing amounts of the CARM1 specific substrate poly(A) binding protein 1 (PABP1) (38). The presence of PABP1 effectively inhibits methylation of CARM1WT in a dose-dependent manner, and this inhibition is highly effective even at low concentrations of PABP1 (Figure 2B). This result strongly suggests that methylation of CARM1 is in fact automethylation, as other PRMTs do not methylate PABP1 (38,39). Sequence comparison of the CTD of CARM1 in other annotated species indicates a high degree of conservation among vertebrate species near the CARM1 automethylation site (Figure 2C).
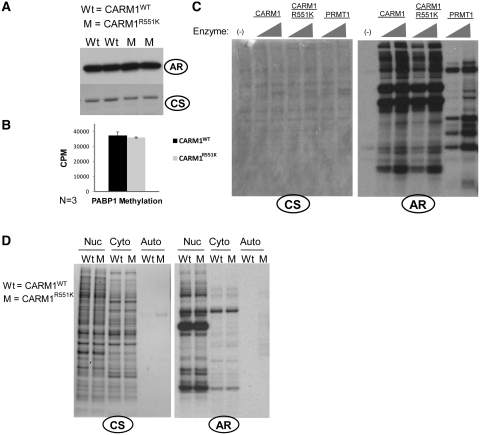
The main cellular functions of CARM1 are currently attributed to its methyltransferase activity. To determine the effect of mutation of the automethylation site on CARM1′s enzymatic activity, we performed in vitro methylation assays with the CARM1 specific substrate PABP1. Both CARM1WT and CARM1R551K were able to methylate PABP1 to a similar degree (Figure 3A and B), and exhibited similar activity and specificity when incubated with nuclear or cytoplasmic extracts from MCF7 in a dose-dependent manner (Figure 3C and D). Separation of CARM1 substrates in two-dimentional SDS–PAGE also revealed no quantitative differences between CARM1WT and CARM1R551K methylation profiles (Supplementary Figure S3). Thus, we conclude that mutation of the automethylation site of CARM1 has no effect on its methyltransferase activity.
Figure 3.
CARM1 enzymatic activity and substrate specificity are not affected by mutation of automethylation site R551→ K. (A) In vitro methylation of PABP1 by CARM1WT (WT) or CARM1R551K (Mut) (n = 2). AR represents autoradiograph, CS represents coomassie staining. (B) In vitro methylation of PABP1 by CARM1WT (WT) or CARM1R551K (Mut) for 5 min, followed by excision of the PABP1 bands and quantification by scintillation counting (n = 3). Error bars represent standard deviations from the mean. (C) In vitro methylation of 7.5 µg nuclear lysates from CARM1-silenced MCF7 cells by 0.25 and 1.25 µg of recombinant CARM1WT, CARM1R551K or GST-PRMT1. (D) In vitro methylation of control buffer (Auto), 15 µg nuclear lysates (Nuc) or 15 µg cytoplasmic lysates (Cyto) from CARM1−/− MEF cells by 1 µg CARM1WT or CARM1R551K.
Figure 4.
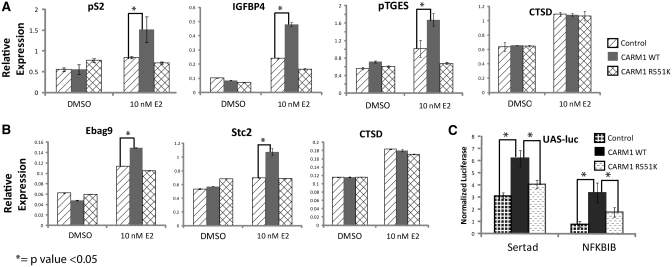
Loss of automethylation impairs transcriptional activation activity of CARM1. (A and B) ER18 cells (A) are HEK293T cells stably expressing ERα, and CARM1 null MEF cells (B) were cotransfected with pSG5-vector: pSG5-CARM1WT or pSG5-CARM1R551K. After 48 h transfection, cells were treated with DMSO or 10 nM E2 for 3 h, followed by RNA isolation and qRT-PCR analysis of endogenous gene products. A mean (±SD, n = 3) quantification is graphically displayed. (C) CARM1R551K displayed low transcriptional activity as compared with CARM1WT to activate Sertad and NFκBIB in Gal4-reporter assays. A mean (±SD, n = 3) quantification is graphically displayed.
CARM1 is best characterized as a transcriptional coactivator. Furthermore, the CTD of CARM1 is required to maintain its full transcriptional activity (15). To assess whether mutation of the automethylation site of CARM1 affects its transcriptional activity, we examined expression of several endogenous ERα target gene by overexpressing CARM1. CARM1 was transiently overexpressed in ER18 cells, a HEK293T cell line stably expressing ERα (40), and treated with or without 17-β-estradiol (E2) for 3 h. mRNA levels of CARM1WT and CARM1R551K were present at similar levels in these transfected cells (Supplementary Figure S4). Known CARM1-responsive, ERα-target genes were tested for expression by qRT-PCR. pS2, IGFPB4 and pTGES exhibited increased expression in the presence of overexpressed CARM1WT in a ligand-dependent manner (Figure 4A). The expression of these genes in CARM1R551K transfected cells was identical to that of cells transfected with vector control, suggesting that mutation of the automethylation site impaired CARM1′s ability to activate transcription. The mRNA levels of cathepsin D, an E2-responsive but CARM1-independent gene, showed no enhancement of transcription by transfecting either CARM1WT or CARM1R551K. A similar pattern was observed for Ebag9 and Stc2 in CARM1−/− MEF cells, using cathepsin D as a negative control (Figure 4B). Ebag9 and Stc2 were analyzed in CARM1−/− MEFs because those genes were shown to be CARM1-dependent in MEF cells (23,37) while pS2, IGFPB4 and pTGES were found not expressed in MEFs. To test whether the lack of transcriptional activity by the automethylation deficient mutant is specific to ERα target genes, we determined transcription of two other transcription factors known to be activated by CARM1 in a Gal4 reporter assay. The plasmid encoding Gal4-Sertad or Gal4-NFκBIB was co-transfected with vector, CARM1WT or CARM1R551K along with a UAS-reporter, into HEK293T cells. With these two transcription factors, the automethylation-deficient CARM1R551K also exhibited compromised activity in transcriptional activation (Figure 4C), suggesting that automethylation-dependent CARM1 activation of transcription is broadly significant.
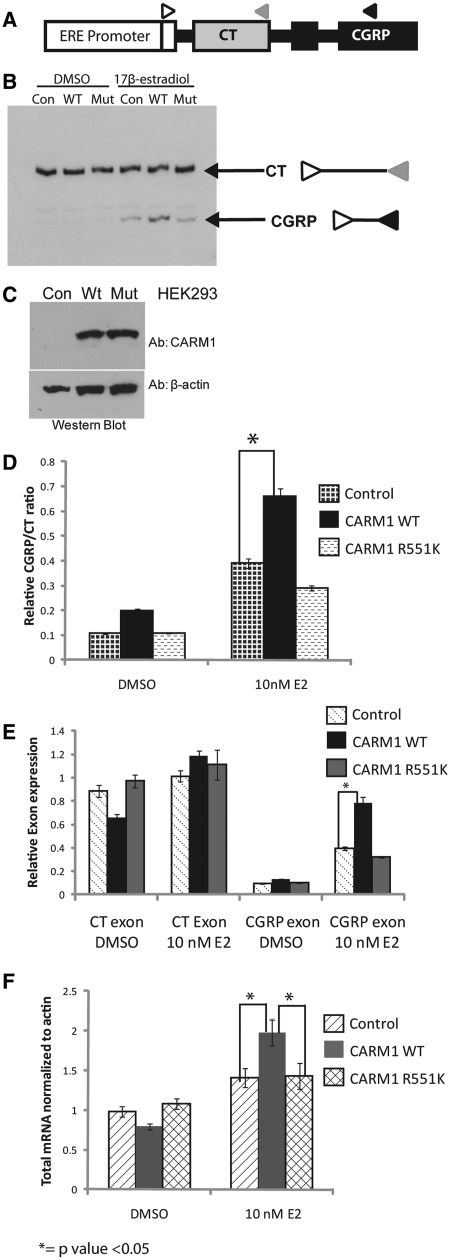
Recently CARM1 was shown to methylate a number of splicing factors and regulate alternative pre-mRNA splicing (21). To assess the role of CARM1 automethylation in its regulation of alternative splicing, we performed a splicing assay using a mini-gene reporter. HEK293T cells were transfected with a CT/CGRP reporter gene under the control of an ERE promoter (17) (illustrated in Figure 5A). Cells were additionally transfected with an ERα expression vector and CARM1R551K, CARM1WT or vector control prior to treatment with E2 or DMSO. The mRNA levels of each alternative exon were quantified by 32P-labeled RT-PCR (Figure 5B) and Sybr Green qRT-PCR using splicing form-specific primers (Figure 5D–F). The ratio of inclusion of the CGRP exon over inclusion of the CT exon increased with E2 treatment, and this increase was enhanced by the overexpression of CARM1WT, but not CARM1R551K (Figure 5B and D). We confirmed that the protein levels (Figure 5C) of CARM1WT and CARM1R551K are similar. CARM1 appears to mediate change in the mini-gene splicing ratio by increasing expression of the CGRP exon without affecting the expression of the CT exon (Figure 5E). This observation is consistent with a model where CARM1 co-regulates transcription and mRNA splicing in an interdependent manner. To further examine this possibility, the total mRNA expressed from the mini-gene reporter was normalized to β-actin and compared between the three transfection conditions. Interestingly, the total mRNA of both alternative transcripts increased with overexpression of CARM1WT but not CARM1R551K in a ligand-dependent manner (Figure 5F). Thus, the CARM1 automethylation-defective mutant exhibited impaired activity in mRNA splicing as well as transcriptional activation of the same transcriptional target.
Figure 5.
Loss of automethylation affects CARM1-mediated alternative splicing. (A) Cartoon representation of the ERE-CT/CGRP mini-gene reporter with primers indicated; non-specific forward primer (open triangle), CT exon-specific reverse primer (gray triangle) and CGRP exon-specific reverse primer (black triangle). (B) Overexpression of CARM1WT but not CARM1R551K promotes exon skipping in E2-dependent manner. 32P-labeled forward primer was used in 20-cycle PCR of cDNA from mini-gene reporter cDNAs, separated by non-denaturing PAGE and exposed to a film. (C) CARM1WT or CARM1R551K were expressed at a similar level as determined by western blot. Whole cell lysates from HEK293T cells transfected with control, CARM1WT or CARM1R551K expression vector were probed with a rabbit polyclonal CARM1 antibody. (D) E2-dependent exon-skipping of mini-gene splicing reporter (A) by CARM1WT but not CARM1R551K. Two splicing products was quantitated using qRT-PCR and presented as CGRP/CT ratio. qRT-PCR was performed on cDNA from mini-gene reporter assays. (E) The expression level of each alternative exon from the splicing reporter was normalized to β-actin. (F) CARM1WT increases total mRNA of CT/CGRP exons as compared with CARM1R551K. The total mRNA levels of CT and CGRP exons from the mini-gene reporter were normalized to endogenous β-actin levels to determine total mRNA expression from the ERE-mini-gene reporter.
DISCUSSION
Using high-resolution MS, we identified a single automethylation site at the CTD of mouse CARM1, which is conserved among vertebrate species. Surprisingly, unmethylated CARM1 is undetectable when it is expressed in either insect or mammalian expression systems. Mutation of the automethylation site of CARM1 impairs both its regulation of transcription and alternative mRNA splicing, highlighting an interesting paradigm that transcription and splicing are co-regulated by PTM of CARM1. Our study also enhances our understanding of the poorly defined CARM1 CTD function in transcriptional regulation and presents a self-regulatory mechanism to signal from CARM1's catalytic domain to its CTD.
Mapping CARM1 PTMs using top- and middle-down MS
Here our high-resolution top-down MS/MS data unambiguously revealed a 28 Da increase in the mass of C-terminal fragments containing the R551 site, which indicated that CARM1 is modified near its C-terminus. However, the poor stability of CARM1 during electrospray ionization compromised the MS/MS fragmentation process and produced a limited number of fragmentation ions. Therefore, we used a combination of top- and middle-down (limited digestion) MS to map the site of dimethylation to a single amino acid at R551. All detectable CARM1 was found already automethylated. The absence of unmethylated CARM1 explains previous observations of extremely weak automethylation of purified CARM1 in vitro (8,41) because CARM1 automethylation is nearly complete in vivo (Figures 1A and D, and 2A). Among the dimethylated CARM1 population from 293T cells, ∼50% was also found to have an O-linked acetylglucosamine (O-GlcNAc) modification (data not shown). We are currently mapping the site of the O-GlcNAc modification and are investigating its function. As expected, we did not observe phosphorylation of CARM1 at the characterized locations because phosphorylation on these three sites only occurs during mitosis (22,23) or under specific stimuli (24). The cells we used to purify CARM1 were unsynchronized, thus the abundance of phosphorylated CARM1 would have been very low. This low abundance would make detection of a modified intact protein ion difficult by FT-ICR MS.
One-way, self-regulatory mechanism for two CARM1 functional domains
CARM1 has two functional domains that are essential for its full transcriptional activity (15). The enzymatic activity located in the central catalytic domain is required for most CARM1 mediated-transcription (5,8,42) and for maintaining the in vivo function of CARM1 (14). CARM1 differs from other PRMTs by carrying an extended CTD. Multiple lines of evidence suggest that CARM1′s CTD is important for CARM1's function (15,43,44). Here we present evidence that CARM1 undergoes automethylation in the CTD, and that mutation of this automethylation site inhibits CARM1's ability to regulate pre-mRNA splicing and transcription. Not only have our results highlighted the importance of CARM1's CTD but also revealed a one-way self-regulatory scheme from the CARM1 catalytic domain to the CTD. Loss of automethylation by mutation of R551 to lysine on CARM1 does not appear to affect substrate specificity or the enzymatic activity of CARM1 in vitro (Figure 3 and Supplementary Figure S3). This result is consistent with the previous observation that the CTD of CARM1 is not required for substrate methylation (15). In contrast, the central catalytic domain of CARM1 can methylate other protein substrates as well as itself, at the CTD, constituting an intriguing mode of CARM1 auto-regulation. Since methylation of the CTD is also dependent on CARM1's enzymatic activity, the functions of CARM1's CTD may have been masked in previous studies when CARM1's enzymatic activity is eliminated in vivo and in vitro.
CARM1 automethylation provides a hub for transcription and splicing co-regulation
Significant attention has been focused on the role of the methyltransferase activity in CARM1-regulated cellular functions since its discovery (3). While non-enzymatic interactions have been observed (10), the majority of CARM1 interacting proteins are also substrates (45,46), and CARM1's ability to methylate histone H3 is considered to be an integral aspect of its coactivation mechanism (6). Our results show that the mutation of the automethylation site to lysine affects neither the localization of CARM1 (Supplementary Figure S5) nor the substrate specificity or enzymatic activity in vitro (Figure 3 and Supplementary Figure S3). However, CARM1 automethylation in cells appears to play an important role in regulating multiple functions of CARM1.
Truncation of CARM1 into various domains identified the CTD as an independently important region in transcription (15); however, the mechanism for functional significance of CARM1 CTD remains completely unexplored. The only protein that was reported to interact with CTD is TIF1α, where TIF1α’s characterized role in coactivation is limited to the stabilization of interaction between CARM1 and GRIP1 (47). We surmise that the CARM1 CTD likely facilitates protein–protein interactions that are necessary for transcription, and the dimethylated arginine residue on the CTD might serve as a platform to recruit effector proteins. Our observation that CARM1 lacking the automethylation site has reduced ability to activate transcription of a GAL4 reporter (Figure 4C) as well as endogenous ERα-target genes (Figure 4B) support this notion. In fact, automethylation of a histone lysine methyltransferase G9a was found to be necessary and sufficient for mediating in vivo interaction with heterochromatin protein 1 (HP1) (48). PRMT8, a CARM1 family member, was shown to be automethylated at two sites at its N-terminus. However, the effect of the automethylation reactions on the activity of PRMT8 was not determined (41). Thus, CARM1 appears to be the first PRMT whose automethylation is demonstrated to play an important regulatory role.
Evidence of CARM1's role in regulating alternative splicing is emerging but the mechanism is still poorly defined. The current view for CARM1's regulation of pre-mRNA splicing is through direct methylation of splicing factors including CA150 (21). Methylation of CA150 by CARM1 is required for its interaction with SMN, a primary regulator of spliceosome assembly. Our findings that an automethylation-defective mutant has impaired activity in regulating alternative splicing (Figure 5) renders an alternative model that CARM1 is directly involved in splicing regulation through its CTD. Two possible models may explain how CARM1's CTD might be engaged in the regulation of alternative splicing. First, CARM1 may depend on its association with the transcriptional machinery to regulate pre-mRNA splicing, a commonly accepted theme of transcription-coupled mRNA splicing (49). Our observation that the CARM1-dependent alternative splicing of the CT/CGRP mini-gene occurs when CARM1WT also specifically activates transcription (Figure 5D and E) is consistent with a model where CARM1 co-regulates transcription and splicing. Chromatin is increasingly recognized as a scaffold for pre-mRNA splicing and the chromatin remodeling machineries have been found to be involved in splicing decisions (50). CARM1 was previously shown to associate with the SWI/SNF complex (10), a chromatin remodeling complex involved in regulation of alternative splicing (51). Second, CARM1 automethylation may regulate the direct interaction between CARM1 and the splicing factors, thus CARM1 may be an integral part of the spliceosome. Mechanistic studies are ongoing and may provide insights into the mechanism by which automethylation regulates CARM1 dual functions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
National Institutes of Health (RO1CA125387, RO3MH089442 to W.X., T32 CA009135 to P.K.); Wisconsin Partnership Fund for a Healthy Future, UW-Madison Human Proteomics Program; Promega Corporation provided financial support (to R.C.). Funding for open access charge: National Institutes of Health (RO1CA125387).
Conflict of interest statement. R.B. is required by the University of Wisconsin-Madison Conflict of Interest Committee to disclose that he has a financial interest in Promega Corp, which markets the HaloTag technology used to purify CARM1.
ACKNOWLEDGEMENTS
We thank Mark Bedford for providing the CARM1 MEF−/− cell line, Elaine Alarid for the ER18 cell line, Jiandie Lin for Gal4-Sertad and Gal4-NFκBIB plasmids and Bert O’Malley for the ERE-CT/CGRP plasmid. We would like to especially acknowledge UW-Madison Human Proteomics Program for support in acquiring mass spectrometry data. We would like to thank Promega Corporation for providing HaloTag reagents instrumental for publication of CARM1. We would like to thank Emery Bresnick, David Wassarman and Chih-hao Lee for critical reading of the manuscript. P.K., R.C., Y.G., R.B. and W.X. designed experiments. P.K., R.C. and Y.W. performed experiments. P.K., R.C. and W.X. analyzed data. P.K. Y.G. and W.X. wrote the manuscript.
REFERENCES
- 1.Bedford MT, Richard S. Arginine methylation an emerging regulator of protein function. Mol. Cell. 2005;18:263–272. doi: 10.1016/j.molcel.2005.04.003. [DOI] [PubMed] [Google Scholar]
- 2.Kuhn P, Xu W. Chapter 9 protein arginine methyltransferases nuclear receptor coregulators and beyond. Prog. Mol. Biol. Transl. Sci. 2009;87:299–342. doi: 10.1016/S1877-1173(09)87009-9. [DOI] [PubMed] [Google Scholar]
- 3.Chen D, Ma H, Hong H, Koh SS, Huang SM, Schurter BT, Aswad DW, Stallcup MR. Regulation of transcription by a protein methyltransferase. Science. 1999;284:2174–2177. doi: 10.1126/science.284.5423.2174. [DOI] [PubMed] [Google Scholar]
- 4.Hassa PO, Covic M, Bedford MT, Hottiger MO. Protein arginine methyltransferase 1 coactivates NF-kappaB-dependent gene expression synergistically with CARM1 and PARP1. J. Mol. Biol. 2008;377:668–678. doi: 10.1016/j.jmb.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 5.An W, Kim J, Roeder RG. Ordered cooperative functions of PRMT1, p300, and CARM1 in transcriptional activation by p53. Cell. 2004;117:735–748. doi: 10.1016/j.cell.2004.05.009. [DOI] [PubMed] [Google Scholar]
- 6.Ma H, Baumann CT, Li H, Strahl BD, Rice R, Jelinek MA, Aswad DW, Allis CD, Hager GL, Stallcup MR. Hormone-dependent, CARM1-directed, arginine-specific methylation of histone H3 on a steroid-regulated promoter. Curr. Biol. 2001;11:1981–1985. doi: 10.1016/s0960-9822(01)00600-5. [DOI] [PubMed] [Google Scholar]
- 7.Schurter BT, Koh SS, Chen D, Bunick GJ, Harp JM, Hanson BL, Henschen-Edman A, Mackay DR, Stallcup MR, Aswad DW. Methylation of histone H3 by coactivator-associated arginine methyltransferase 1. Biochemistry. 2001;40:5747–5756. doi: 10.1021/bi002631b. [DOI] [PubMed] [Google Scholar]
- 8.Xu W, Chen H, Du K, Asahara H, Tini M, Emerson BM, Montminy M, Evans RM. A transcriptional switch mediated by cofactor methylation. Science. 2001;294:2507–2511. doi: 10.1126/science.1065961. [DOI] [PubMed] [Google Scholar]
- 9.Feng Q, Yi P, Wong J, O’Malley BW. Signaling within a coactivator complex: methylation of SRC-3/AIB1 is a molecular switch for complex disassembly. Mol. Cell Biol. 2006;26:7846–7857. doi: 10.1128/MCB.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Xu W, Cho H, Kadam S, Banayo EM, Anderson S, Yates JR, III, Emerson BM, Evans RM. A methylation-mediator complex in hormone signaling. Genes Dev. 2004;18:144–156. doi: 10.1101/gad.1141704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Troffer-Charlier N, Cura V, Hassenboehler P, Moras D, Cavarelli J. Functional insights from structures of coactivator-associated arginine methyltransferase 1 domains. EMBO J. 2007;26:4391–4401. doi: 10.1038/sj.emboj.7601855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yue WW, Hassler M, Roe SM, Thompson-Vale V, Pearl LH. Insights into histone code syntax from structural and biochemical studies of CARM1 methyltransferase. EMBO J. 2007;26:4402–4412. doi: 10.1038/sj.emboj.7601856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jayne S, Rothgiesser KM, Hottiger MO. CARM1 but not its enzymatic activity is required for transcriptional coactivation of NF-kappaB-dependent gene expression. J. Mol. Biol. 2009;394:485–495. doi: 10.1016/j.jmb.2009.09.032. [DOI] [PubMed] [Google Scholar]
- 14.Kim D, Lee J, Cheng D, Li J, Carter C, Richie E, Bedford MT. Enzymatic activity is required for the in vivo functions of CARM1. J. Biol. Chem. 2010;285:1147–1152. doi: 10.1074/jbc.M109.035865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Teyssier C, Chen D, Stallcup MR. Requirement for multiple domains of the protein arginine methyltransferase CARM1 in its transcriptional coactivator function. J. Biol. Chem. 2002;277:46066–46072. doi: 10.1074/jbc.M207623200. [DOI] [PubMed] [Google Scholar]
- 16.Auboeuf D, Dowhan DH, Dutertre M, Martin N, Berget SM, O’Malley BW. A subset of nuclear receptor coregulators act as coupling proteins during synthesis and maturation of RNA transcripts. Mol. Cell Biol. 2005;25:5307–5316. doi: 10.1128/MCB.25.13.5307-5316.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Auboeuf D, Honig A, Berget SM, O’Malley BW. Coordinate regulation of transcription and splicing by steroid receptor coregulators. Science. 2002;298:416–419. doi: 10.1126/science.1073734. [DOI] [PubMed] [Google Scholar]
- 18.Monsalve M, Wu Z, Adelmant G, Puigserver P, Fan M, Spiegelman BM. Direct coupling of transcription and mRNA processing through the thermogenic coactivator PGC-1. Mol. Cell. 2000;6:307–316. doi: 10.1016/s1097-2765(00)00031-9. [DOI] [PubMed] [Google Scholar]
- 19.Fujiwara T, Mori Y, Chu DL, Koyama Y, Miyata S, Tanaka H, Yachi K, Kubo T, Yoshikawa H, Tohyama M. CARM1 regulates proliferation of PC12 cells by methylating HuD. Mol. Cell Biol. 2006;26:2273–2285. doi: 10.1128/MCB.26.6.2273-2285.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Park S, Kilburn B, Jelinek MA, Henschen-Edman A, Aswad DW, Stallcup MR, Laird-Offringa IA. Lipopolysaccharide-induced methylation of HuR, an mRNA-stabilizing protein, by CARM1. Coactivator-associated arginine methyltransferase. J. Biol. Chem. 2002;277:44623–44630. doi: 10.1074/jbc.M206187200. [DOI] [PubMed] [Google Scholar]
- 21.Cheng D, Cote J, Shaaban S, Bedford MT. The arginine methyltransferase CARM1 regulates the coupling of transcription and mRNA processing. Mol. Cell. 2007;25:71–83. doi: 10.1016/j.molcel.2006.11.019. [DOI] [PubMed] [Google Scholar]
- 22.Feng Q, He B, Jung SY, Song Y, Qin J, Tsai SY, Tsai MJ, O’Malley BW. Biochemical control of CARM1 enzymatic activity by phosphorylation. J. Biol. Chem. 2009;284:36167–36174. doi: 10.1074/jbc.M109.065524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Higashimoto K, Kuhn P, Desai D, Cheng X, Xu W. Phosphorylation-mediated inactivation of coactivator-associated arginine methyltransferase 1. Proc. Natl Acad. Sci. USA. 2007;104:12318–12323. doi: 10.1073/pnas.0610792104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Carascossa S, Dudek P, Cenni B, Briand PA, Picard D. CARM1 mediates the ligand-independent and tamoxifen-resistant activation of the estrogen receptor alpha by cAMP. Genes Dev. 2010;24:708–719. doi: 10.1101/gad.568410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheung WD, Sakabe K, Housley MP, Dias WB, Hart GW. O-linked beta-N-acetylglucosaminyltransferase substrate specificity is regulated by myosin phosphatase targeting and other interacting proteins. J. Biol. Chem. 2008;283:33935–33941. doi: 10.1074/jbc.M806199200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Siuti N, Kelleher NL. Decoding protein modifications using top-down mass spectrometry. Nat. Methods. 2007;4:817–821. doi: 10.1038/nmeth1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sze SK, Ge Y, Oh H, McLafferty FW. Top-down mass spectrometry of a 29-kDa protein for characterization of any posttranslational modification to within one residue. Proc. Natl Acad. Sci. USA. 2002;99:1774–1779. doi: 10.1073/pnas.251691898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kuhn P, Xu QG, Cline E, Zhang D, Ge Y, Xu W. Delineating Anopheles gambiae coactivator associated arginine methyltransferase 1 automethylation using top-down high resolution tandem mass spectrometry. Protein Sci. 2009;18:1272–1280. doi: 10.1002/pro.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge Y, Rybakova IN, Xu QG, Moss RL. Top-down high-resolution mass spectrometry of cardiac myosin binding protein C revealed that truncation alters protein phosphorylation state. Proc. Natl Acad. Sci. USA. 2009;106:12658–12663. doi: 10.1073/pnas.0813369106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ge Y, Lawhorn BG, ElNaggar M, Strauss E, Park JH, Begley TP, McLafferty FW. Top down characterization of larger proteins (45 kDa) by electron capture dissociation mass spectrometry. J. Am. Chem. Soc. 2002;124:672–678. doi: 10.1021/ja011335z. [DOI] [PubMed] [Google Scholar]
- 31.Ayaz-Guner S, Zhang J, Li L, Walker JW, Ge Y. In vivo phosphorylation site mapping in mouse cardiac troponin I by high resolution top-down electron capture dissociation mass spectrometry: Ser22/23 are the only sites basally phosphorylated. Biochemistry. 2009;48:8161–8170. doi: 10.1021/bi900739f. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Pesavento JJ, Mizzen CA, Kelleher NL. Quantitative analysis of modified proteins and their positional isomers by tandem mass spectrometry: human histone H4. Anal. Chem. 2006;78:4271–4280. doi: 10.1021/ac0600050. [DOI] [PubMed] [Google Scholar]
- 33.Zabrouskov V, Han X, Welker E, Zhai H, Lin C, van Wijk KJ, Scheraga HA, McLafferty FW. Stepwise deamidation of ribonuclease A at five sites determined by top down mass spectrometry. Biochemistry. 2006;45:987–992. doi: 10.1021/bi0517584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zabrouskov V, Ge Y, Schwartz J, Walker JW. Unraveling molecular complexity of phosphorylated human cardiac troponin I by top down electron capture dissociation/electron transfer dissociation mass spectrometry. Mol. Cell Proteomics. 2008;7:1838–1849. doi: 10.1074/mcp.M700524-MCP200. [DOI] [PubMed] [Google Scholar]
- 35.Kuhn P, Xu Q, Cline E, Zhang D, Ge Y, Xu W. Delineating Anopheles gambiae coactivator associated arginine methyltransferase 1 automethylation using top-down high resolution tandem mass spectrometry. Protein Sci. 2009;18:1272–1280. doi: 10.1002/pro.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chumanov R, et al. Expression and purification of full-length mouse CARM1 from transiently transfected 60 HEK293 cells using HaloTag technology. Protein Expression and Purification. 2011 doi: 10.1016/j.pep.2010.11.010. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Yadav N, Lee J, Kim J, Shen J, Hu MC, Aldaz CM, Bedford MT. Specific protein methylation defects and gene expression perturbations in coactivator-associated arginine methyltransferase 1-deficient mice. Proc. Natl Acad. Sci. USA. 2003;100:6464–6468. doi: 10.1073/pnas.1232272100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee J, Bedford MT. PABP1 identified as an arginine methyltransferase substrate using high-density protein arrays. EMBO Rep. 2002;3:268–273. doi: 10.1093/embo-reports/kvf052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Swiercz R, Person MD, Bedford MT. Ribosomal protein S2 is a substrate for mammalian PRMT3 (protein arginine methyltransferase 3) Biochem. J. 2005;386:85–91. doi: 10.1042/BJ20041466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Valley CC, Metivier R, Solodin NM, Fowler AM, Mashek MT, Hill L, Alarid ET. Differential regulation of estrogen-inducible proteolysis and transcription by the estrogen receptor alpha N terminus. Mol. Cell Biol. 2005;25:5417–5428. doi: 10.1128/MCB.25.13.5417-5428.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Sayegh J, Webb K, Cheng D, Bedford MT, Clarke SG. Regulation of protein arginine methyltransferase 8 (PRMT8) activity by its N-terminal domain. J. Biol. Chem. 2007;282:36444–36453. doi: 10.1074/jbc.M704650200. [DOI] [PubMed] [Google Scholar]
- 42.Lee DY, Northrop JP, Kuo MH, Stallcup MR. Histone H3 lysine 9 methyltransferase G9a is a transcriptional coactivator for nuclear receptors. J. Biol. Chem. 2006;281:8476–8485. doi: 10.1074/jbc.M511093200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ohkura N, Takahashi M, Yaguchi H, Nagamura Y, Tsukada T. Coactivator-associated arginine methyltransferase 1, CARM1, affects pre-mRNA splicing in an isoform-specific manner. J. Biol. Chem. 2005;280:28927–28935. doi: 10.1074/jbc.M502173200. [DOI] [PubMed] [Google Scholar]
- 44.Matsuda H, Paul BD, Choi CY, Shi YB. Contrasting effects of two alternative splicing forms of coactivator-associated arginine methyltransferase 1 on thyroid hormone receptor-mediated transcription in Xenopus laevis. Mol. Endocrinol. 2007;21:1082–1094. doi: 10.1210/me.2006-0448. [DOI] [PubMed] [Google Scholar]
- 45.Chevillard-Briet M, Trouche D, Vandel L. Control of CBP co-activating activity by arginine methylation. EMBO J. 2002;21:5457–5466. doi: 10.1093/emboj/cdf548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lee YH, Coonrod SA, Kraus WL, Jelinek MA, Stallcup MR. Regulation of coactivator complex assembly and function by protein arginine methylation and demethylimination. Proc. Natl Acad. Sci. USA. 2005;102:3611–3616. doi: 10.1073/pnas.0407159102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Teyssier C, Ou CY, Khetchoumian K, Losson R, Stallcup MR. Transcriptional intermediary factor 1alpha mediates physical interaction and functional synergy between the coactivator-associated arginine methyltransferase 1 and glucocorticoid receptor-interacting protein 1 nuclear receptor coactivators. Mol. Endocrinol. 2006;20:1276–1286. doi: 10.1210/me.2005-0393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sampath SC, Marazzi I, Yap KL, Krutchinsky AN, Mecklenbrauker I, Viale A, Rudensky E, Zhou MM, Chait BT, Tarakhovsky A. Methylation of a histone mimic within the histone methyltransferase G9a regulates protein complex assembly. Mol. Cell. 2007;27:596–608. doi: 10.1016/j.molcel.2007.06.026. [DOI] [PubMed] [Google Scholar]
- 49.Chen M, Manley JL. Mechanisms of alternative splicing regulation: insights from molecular and genomics approaches. Nat. Rev. Mol. Cell Biol. 2009;10:741–754. doi: 10.1038/nrm2777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Allemand E, Batsche E, Muchardt C. Splicing, transcription, and chromatin: a menage a trois. Curr. Opin. Genet. Dev. 2008;18:145–151. doi: 10.1016/j.gde.2008.01.006. [DOI] [PubMed] [Google Scholar]
- 51.Batsche E, Yaniv M, Muchardt C. The human SWI/SNF subunit Brm is a regulator of alternative splicing. Nat. Struct. Mol. Biol. 2006;13:22–29. doi: 10.1038/nsmb1030. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.