Abstract
Structured RNA regions are important gene control elements in prokaryotes and eukaryotes. Here, we show that the mRNA of a cyanobacterial heat shock gene contains a built-in thermosensor critical for photosynthetic activity under stress conditions. The exceptionally short 5′-untranslated region is comprised of a single hairpin with an internal asymmetric loop. It inhibits translation of the Synechocystis hsp17 transcript at normal growth conditions, permits translation initiation under stress conditions and shuts down Hsp17 production in the recovery phase. Point mutations that stabilized or destabilized the RNA structure deregulated reporter gene expression in vivo and ribosome binding in vitro. Introduction of such point mutations into the Synechocystis genome produced severe phenotypic defects. Reversible formation of the open and closed structure was beneficial for viability, integrity of the photosystem and oxygen evolution. Continuous production of Hsp17 was detrimental when the stress declined indicating that shutting-off heat shock protein production is an important, previously unrecognized function of RNA thermometers. We discovered a simple biosensor that strictly adjusts the cellular level of a molecular chaperone to the physiological need.
INTRODUCTION
Cyanobacteria are ubiquitiously distributed on earth and—together with plants—provide the foundation of aerobic life by the photosynthetic generation of oxygen. The integrity of the photosynthesis machinery is challenged by highly fluctuating environmental conditions. In particular, heat, high light intensities, reactive oxygen species, salt and metal stress are known to cause defects of the thylakoid membrane-associated photosystems (1,2).
The small heat shock protein Hsp17 (also known as Hsp16.6 or HspA) is essential for stress tolerance in the model cyanobacterium Synechocystis sp. PCC 6803 (3,4). Hsp17 belongs to the ubiquitous family of α-crystallin-type ATP-independent chaperones (5). Small heat shock proteins (sHsps) capture unfolded proteins to prevent formation of irreversible aggregates (6). Synechocystis Hsp17 not only possesses protein-protective activity but also stabilizes the lipid phase of membranes, thus maintaining thylakoid membrane integrity under stress conditions (7).
The exposure of Synechocystis to a sudden increase in temperature or light intensity triggers expression of the heat shock regulon including hsp17 (3,8). Shifting Synechocystis cells from 34°C to 44°C results in a >60-fold induction of hsp17 mRNA (9). Global gene expression profiling revealed a 20-fold induction of the hsp17 transcript under light stress (8). Transcription of heat shock genes, including hsp17, was shown to rely on the alternative sigma factors SigB and SigE (10,11). Furthermore, hsp17 transcription is strongly regulated by changes in the physical order of membranes (12). A combined transcriptomics and proteomics approach suggested that regulation of heat shock gene expression in Synechocystis is governed by transcriptional and yet unknown translational regulation (9,11,13,14).
In recent years, the universal importance of regulatory RNAs as posttranscriptional gene control elements has been recognized (15,16). In bacteria, small regulatory RNAs (sRNAs) are very abundant regulators that often act through base pairing with target mRNAs, thereby modulating translation efficiency and mRNA stability (17,18). Biocomputational predictions and experimental strategies have revealed several hundred sRNAs in Synechocystis; some acting as trans-encoded regulatory RNAs with short and discontinuous complementarity to their targets, others as cis-encoded perfectly complementary antisense RNAs (19–21).
In contrast to numerous sRNAs, mRNA-inherent riboregulators like riboswitches and RNA thermometers have received little attention in cyanobacteria. Riboswitches are mRNA leader sequences that fold into a complex structure whose conformation changes upon ligand binding (22). RNA thermometers are translational control elements built-into the 5′-untranslated region (5′-UTR) of bacterial heat shock or virulence genes (23). Typically, they fold into a complex structure that traps the Shine–Dalgarno (SD) sequence at low temperatures. An increase in temperature to 37°C (virulence genes) or higher (heat shock genes) destabilizes the structure, liberates the SD sequence and permits formation of the translation initiation complex. Functionality of RNA thermometers in the relevant temperature range of mesophilic bacteria requires a structure that is stable enough to resist opening at temperatures below 30°C but sufficiently unstable to melt as the temperature increases. This can be achieved by a delicate balance of Watson–Crick base pairs, internal bulges and loops and non-canonical base pairing (24–27).
An RNA thermometer-like structure is located in the 5′-UTR of the Synechocystis hsp17 transcript. The hairpin engages the SD sequence and part of the AUG start codon in a secondary structure, contains an internal loop and might thus act as RNA thermometer (Figure 1A). With only 44 nucleotides in length, the hsp17 5′-UTR is the smallest natural thermometer candidate discovered yet. In this work, we provide genetic and biochemical proof that it acts as bona fide RNA thermometer that has important not previously described physiological functions.
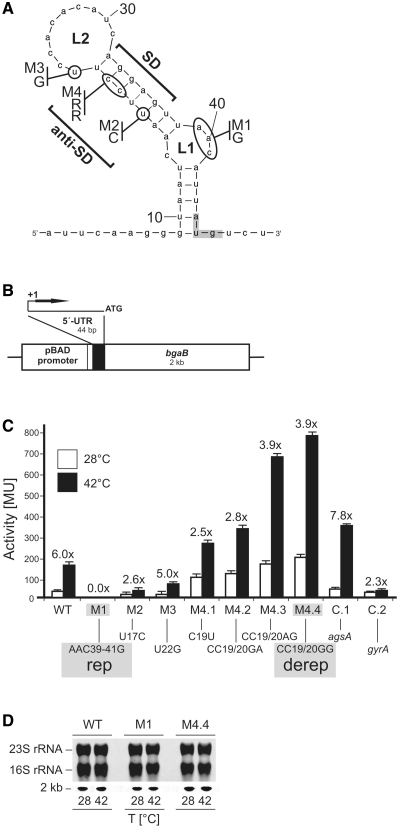
Figure 1.
Translational control by the hsp17 UTR element in E. coli. (A) The secondary structure as predicted by the mfold program (40) of the entire hsp17 5′-UTR is shown. The start codon (AUG, marked by gray box) is located 45 nt downstream of the transcription start site. The SD and anti-SD sequences, loop1 (L1) and loop2 (L2) are labeled. Site-directed mutations M1–M4 and the exchanged nucleotides are indicated; RR, variable nucleotides derived from random mutagenesis (primer: hsp17therm-M4-fw + hsp17therm-M4-rv, Supplementary Table S1). (B) Schematic representation of the reporter gene fusion on plasmid pBAD-bgaB. Additional nucleotides inserted due to the position of the NheI cloning site relative to the pBAD promoter are indicated by a box upstream of the 5′-UTR. The artifical nucleotides at the 5′-end of the hsp17 transcript do not influence RNA folding and expression of the gene (data not shown). (C) Expression of the translational bgaB reporter fusions (Miller Units, MU) to various hsp17 5′-UTRs. Escherichia coli DH5α cells containing the corresponding plasmids were grown in LB medium at 28°C and either kept at this temperature (white columns) or transferred to 42°C (black columns) for 30 min before β-galactosidase activity was measured. All experiments were repeated at least in triplicate. Induction rates are shown above each fusion. A Salmonella agsA-bgaB fusion [fourU element; (27)] was used as a positive control (C.1), while E. coli gyrA-bgaB (27) served as a negative control (C.2). Absolute β-galactosidase levels are listed in Supplementary Table S2. (D) mRNA levels of hsp17-bgaB fusions before and after heat shock. Total RNA was extracted from E. coli cultures incubated at either 28 or 42°C. Equal amounts were separated on a 1.2% denaturing agarose gel and northern blot experiments were carried out using digoxygenin-labeled RNA probes to detect bgaB transcripts. Ethidum–bromide stained rRNAs from the gel before blotting are shown as loading control. The bgaB fusion transcript runs at 2 kb.
MATERIALS AND METHODS
Strains and growth conditions
Escherichia coli cells (DH5α and DH5αZ1) were grown at 28 or 37°C in Luria–Bertani (LB) medium supplemented with ampicillin (Ap, 150 µg/ml) or chloramphenicol (Cm, 50 µg/ml). For induction of the pBAD promoter in strains carrying translational bgaB fusions, 0.01% (w/v) l-arabinose was added. Expression of translational gfp fusions was induced via inactivation of the Tet repressor with 50 ng/ml doxycycline.
Synechocystis cells were grown under low light conditions (30 µmol photons m−2 s−1) at 28°C on BG11/agar (28) plates or in BG11 liquid media as described (29). Liquid media was supplemented with 5 mM glucose when appropriate.
For the measurement of chlorophyll concentrations, cells were sedimented by centrifugation and extracted with 100% methanol. The concentration of chlorophyll was calculated from the absorbance values of the extract at 666 and 750 nm (30).
Plasmid construction
The hsp17-bgaB fusion (pBO1293) was constructed by transfer of the hsp17 5′-UTR from pBO1292 upon NheI/EcoRI digestion into the corresponding site in pBO415 (Supplementary Table S1). To obtain the translational gfp fusion (pBO1325), the hsp17 5′-UTR was transferred from pBO1292 via NheI and an introduced PstI site into pXG-10 (31). Site-directed mutagenesis to generate pBO1312, 1310, 1311, 1316, 1314, 1315, 1313, 1801 and 1802 was performed according to the instruction manual of the QuikChange® mutagenesis kit (Stratagene, La Jolla, USA). Plasmid pBO1292 served as a template for PCR with mutagenic primers (Supplementary Table S1). The inserts containing mutated hsp17 UTRs were isolated upon NheI/EcoRI or PstI/NheI digestion and cloned into the corresponding site upstream the bgaB or gfp gene, respectively. The entire hsp17 coding region including its 5′-UTR was amplified and cloned into pUC18 (pBO1347) for subsequent site directed mutagenesis (Supplementary Table S1). Using unique restriction sites BamHI/CpoI, hsp17 thermometer variants rep, derep and wild-type (WT) were cloned into pNaive.16 (29) resulting in pBO1834, 1806 and 1807. The correct nucleotide sequence was confirmed by automated sequencing (eurofins, Martinsried, Germany). pNaive16 (pAZ877) was provided by Prof. Dr Elisabeth Vierling (University of Arizona).
RNA and protein detection
Preparation of total RNA from E. coli and Northern analysis followed previously published protocols (32). Preparation of total RNA from Synechocystis was performed with a Qiagen RNeasy® kit (Qiagen, Hilden, Germany) according to the instruction manual. RNAs were detected with a DIG-labeled bgaB or hsp17 RNA probe, respectively. The DIG-HIGH prime labeling kit (Roche Applied Science, Mannheim, Germany) was used for preparation of probes (see Supplementary Table S1 for details). For E. coli protein extraction, cell pellets were resuspended in lysis buffer (10 mM sodiumphosphate, pH 7.0, 20% glycerol, 0.5 mM DTT) according to their cell density (100 μl buffer per OD580 of 1.0). SDS sample buffer was added, cells were boiled for 5 min and equal volumes were subjected to SDS–PAGE. Synechocystis whole cell lysates were prepared according to Klinkert et al. (33). Soluble fractions corresponding to 20 µg of chlorophyll were separated on 12.5% polyacrylamide gels. SDS–PAGE and western blot was performed as described (34). GFP was detected on a western blot using a polyclonal α-GFP antibody (abcam209; Abcam, Cambridge, USA). Hsp17 antigen was revealed by α-Hsp17 antibody, kindly provided by Prof. Dr Elisabeth Vierling (University of Arizona). Signals were detected with a ChemiImager™ Ready (Alpha Innotech, Biozym, Wien, Austria).
In vitro transcription
RNAs were synthesized in vitro by runoff transcription with T7 RNA polymerase from linearized plasmid templates. Plasmids pBO1301, 1304 and 1302 (linearized with MlsI) were used to generate RNA for enzymatic structure probing and plasmids pBO1305, 1349 and 1348 (linearized with HpyCH4V) to generate RNA for toeprinting experiments (Supplementary Table S1).
Structure probing experiments
RNA structure formation at either 28 and 42°C was compared. RNA was 5′-end labeled as described (35). Partial digestion of 5′-end-labeled RNAs with ribonucleases T1 (0.004U and 0.01U), V (0.008U and 0.02U) was conducted according to Waldminghaus et al. (32). For chemical probing, labeled RNA was mixed with 2 µl 5× lead buffer (250 mM tris–acetate pH 7.5, 25 mM Mg–acetate; 250 mM Na–acetate) and 1 µg tRNA in a total volume of 10 µl. Lead(II) probing assay was conducted as described (36). RNA fragments were separated on denaturing 8% polyacrylamide gels. Alkaline ladders were generated as described previously (35).
Toeprinting analysis
Primer extension inhibition experiments were carried out using 30 S ribosomal subunits, target mRNA and tRNAfMet basically according to (37). The 5′-[32P]-labeled hsp17-specific oligonucleotide hsp17therm-runoff-toeprint (Supplementary Table S1) was used as a primer for cDNA synthesis. An aliquot of 0.08 pmol mRNA (5′-UTR plus 63 nt of the hsp17 coding region) annealed to the oligonucleotide was incubated for 10 min at 28 or 42°C, respectively, together with 16 pmol of uncharged tRNAfMet (Sigma-Aldrich, St Louis, MO, USA). Six picomole of 30S subunits or water (negative control) was added and incubation for another 10 min followed before 2 µl of MMLV-Mix [VD+Mg2+-buffer, BSA, dNTPs and MMLV reverse transcriptase (USB, Cleveland, OH, USA)] was added. cDNA synthesis was performed at 28°C. Reactions were stopped after 10 min by adding formamide loading dye and aliquots were separated on a denaturing 8% polyacrylamide gel.
CD spectroscopy
Circular dichroism (CD) unfolding and refolding curves were recorded with a JASCO spectropolarimeter J-810. RNA concentration was adjusted to 25 µM. Buffer conditions: 15 mM KxHy(PO4), 25 mM KCl, pH 6.5. CD unfolding curves were recorded with a temperature slope of 1°C/min at a wavelength of 263 nm between 5 and 90°C. CD refolding curves were recorded with a temperature slope of −1°C/min starting from 90 to 5°C. The CD melting curves were normalized to determine the fraction of the unfolded RNA α(T) according to Equation (1).
| (1) |
In Equation (1) T is the temperature, θ(T) is the ellipticity in [mdeg], θunfolded(T) is the ellipticity of the unfolded RNA, θfolded(T) is the ellipticity of the folded RNA and α(T) the temperature dependent fraction of unfolded RNA (RNAunfolded) with respect to the total amount of RNA (RNAtotal). At the melting temperature Tm the fraction of unfolded RNA is α(Tm) = 0.5.
β-Galactosidase assays
β-Galactosidase activities of E. coli strains carrying bgaB-fusions (pBO1312, 1310, 1311, 1316, 1314, 1315, 1313, 602 and 1056; Supplementary Table S1) were measured as described previously (38), except that enzyme activity was measured at 55°C.
GFP-fluorescence assays
Twenty-five mililiter of LB-Cm medium was inoculated with 2 ml of E. coli DH5αZ1 overnight cultures, carrying translational gfp-fusions (pBO1325, 1801 and 1802, Supplementary Table S1) of the respective hsp17 thermometer variants. Cultures were grown at 28°C to a final OD600 of 0.5–0.6. Ten milliliter of the culture was transferred to a prewarmed flask at 42°C. Subsequent to heat shock; cells were shifted to 28°C for 90 min to allow full maturation of GFP-fusion proteins. Aliquots were taken for Western analysis. Two times 2.5 ml of cells were spun down (13 000 rpm, 1 min) and resuspended in 500 µl 1 × PBS buffer. Ten microliter of cell suspension was pipetted on object slides and fixed with 10 µl 0.5% low-melting agarose. The samples were analyzed under a fluorescence microscope (BX51; Olympus, Hamburg, Germany).
Engineered Synechocystis strains
The Synechocystis strains used in this work were generated by transforming pNaive- hsp17 derivates (pBO1834, 1806 and 1807) into the kanamycin-resistant hsp17 deletion strain HK1-1 (Kosaka and Fukuzawa, Kyoto University, Japan) kindly provided by Prof. Dr Elisabeth Vierling (University of Arizona). Transformations were performed according to Klinkert et al. (33), prior to selecting for increasing spectinomycin resistance, at concentrations up to 250 µg/ml spectinomycin dihydrochloride as described (29). Homologous recombination of the hsp17 variants (WT, rep and derep) into Synechocystis genome was confirmed by PCR, northern and western analysis.
Synechocystis heat shock assays
Heat shock treatments (42°C) for subsequent analysis were conducted by shifting cells from 28°C to prewarmed flasks at 42°C for various time lengths as indicated in the figure legends.
Synechocystis high-light stress assays
High-light experiments were performed by exposing cells to 600 µmol of photons m−2 s−1 at 28°C for various time periods as indicated in the figure legends. Light intensities were measured outside the flasks. Potential light-induced heat generation was monitored in a reference flask. We measured a maximal increase of 2.5°C over a 360-min period.
Chlorophyll determination
Chlorophyll determinations of Synechocystis cells was based on the method described by Porra et al. (39). Five hundred microliter of each culture was pelleted and resuspended in 1 ml MeOH prior to sonification for 15 min. After centrifugation, absorbance of the supernatant was read at 652, 665.2 and 750 nm on a Novaspec spectrophotometer (Pharmacia Biotech, Freiburg, Germany). Chlorophyll a (Chl a) concentration was calculated using the following formula:
Concentration (Chl a) [mg/ml] = 18.22 × (OD665.2−OD750)−9.55 × (OD652−OD750).
Thylakoid stability assays
In vivo examinations of thylakoid thermostability were conducted with a luminescence spectrometer (Aminco Bowman II, Thermo Fisher Scientific, Langenselbold, Germany) as described (12). The temperature of respective Synechocystis cultures (‘WT’, Rep and Derep) was increased at 2°C/min. Thylakoid photostability was determined by exposing cells to high light at 28°C for 380 min. Changes in chlorophyll a (Chl a) fluorescence were measured every 20 min.
Photosynthetic assays
A water-cooled Clark-type electrode (Bachofer, Reutlingen, Germany) was used to measure the photosynthetic rates of Synechocystis strains carrying different hsp17 mutations. Samples of heat shocked or high-light stressed cells were immediately transferred to an oxygen electrode chamber. To prevent CO2 depletion, 1 mM NaHCO3 was added to the culture as described (3). For analysis of cellular recovery, cells were transferred to physiological conditions (28°C and low light).
In silico RNA structure prediction
RNA secondary structures were predicted by using the mfold server (http://frontend.bioinfo.rpi.edu/applications/mfold/cgi-bin/rna-form1.cgi) running version 3.2 (40).
RESULTS
Characterization of the first cyanobacterial RNA thermometer
In contrast to other RNA thermometers, the 5′-UTR of Synechocystis hsp17 exhibits a rather minimalistic architecture. Based on the previously determined transcription start site (41,42), the calculated structure of the 5′-UTR consists of a single hairpin of 44 nt, in which the SD sequence site is flanked by two loops, the internal asymmetric L1 loop and the large L2 loop at the top (Figure 1A). Nearly perfect canonical base pairing exists between the SD (AGGAG) and the anti-SD (UCCUU) sequence. The AUG triplet is only partially embedded in the structure at the base of the hairpin. The calculated free energy of the entire structure is −5.5 kcal mol−1 [mfold; (40)].
A well-established Escherichia coli reporter gene system (27) was used to test whether the cyanobacterial RNA element is able to confer temperature-dependent translational control. The hsp17 5′-UTR element was cloned between the l-arabinose-inducible pBAD promoter and the bgaB gene coding for a heat-stable β-galactosidase (43) (Figure 1B). At 28°C the WT fusion allowed a basal β-galactosidase activity of 30 MU that increased 6-fold to 180 MU after a shift to 42°C (Figure 1C). The induction by the hsp17 5′-UTR is comparable to the Salmonella agsA fourU thermometer in our control experiments and clearly exceeds induction of a control fusion to the 5′-UTR of the E. coli housekeeping gene gyrA coding for the DNA gyrase.
As the efficiency of translation initiation correlates with the accessibility of the ribosome binding site (44,45), stabilizing and destabilizing point mutations should have an impact on RNA thermometer function. Selected sites for oligonucleotide-directed mutagenesis are outlined in Figure 1A. The first set of mutations (M1–M4) was aimed at stabilizing the overall thermometer structure. Indeed, M1, M2 and M3 significantly lowered expression both at 28 and 42°C (Figure 1C). Introduction of a G–C pair instead of the kinked internal loop in M1 (AAC39–41G; henceforth called rep for ‘repressed’) resulted in complete loss of reporter activity at both low and high temperatures. Mutation M2 (U17C) substituted a G–U basepair by a more stable G–C basepair resulting in tighter repression at both temperatures. M3 (U22G) was constructed to reduce the size of loop L2 (Figure 1A). The extra G–C pair provided additional stability to the stem and reduced β-galactosidase activity at 28 and 42°C but retained a 5-fold induction (Figure 1C).
A collection of destabilizing mutations was obtained by partially randomized oligonucleotides that introduced various nucleotides at the positions C19 and C20 (see ‘Material and Methods’ section and Supplementary Table S1). Four different hsp17 5′-UTR variants (M4.1–M4.4) that were predicted to reduce the thermodynamic stability of the SD/anti-SD interaction resulted in elevated expression at 28 and 42°C (Figure 1C). As an example, the M4.4 variant (henceforth called derep for ‘derepressed’) produced 200 and 790 MU (as compared to 30 and 180 MU, respectively, in the WT). Elevated reporter gene activity at 28°C resulted in reduced heat induction (2.5- to 3.9-fold) by all four constructs.
To demonstrate that the changes in expression levels at different temperatures were due to translational control rather than to different transcript levels, we conducted northern blot experiments (Figure 1D). The amounts of WT, rep and derep mRNAs were monitored at 28 and 42°C using a probe directed against the coding region of the bgaB reporter gene. Temperature changes had no influence on the steady-state level of the transcripts.
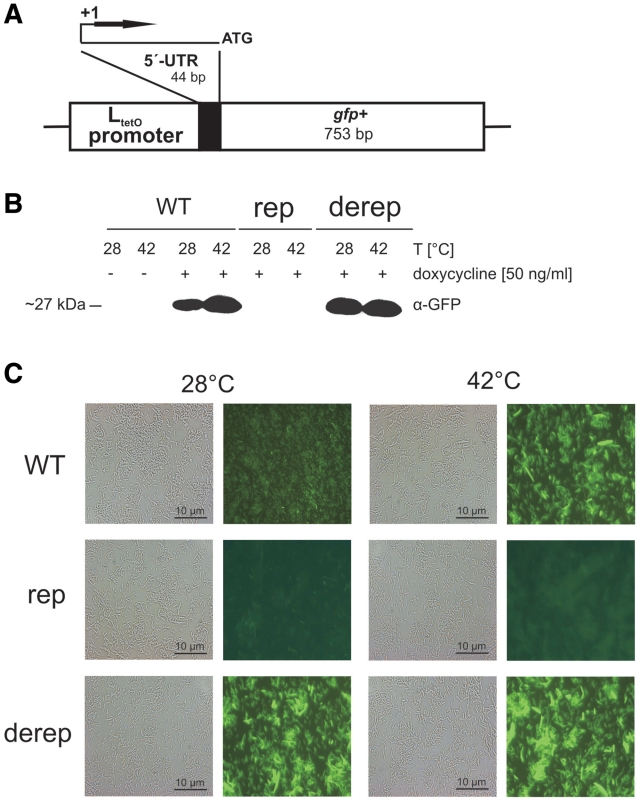
Translational control by the hsp17 5′-UTR was further assessed by a newly developed green-fluorescent protein (gfp)-based reporter system (Figure 2A). In contrast to the bgaB system, it does not add any vector-derived nucleotides to the 5′-end of the transcript. Transcription is driven by the PLtetO-1 promoter and initiates at the natural +1 site immediately at the 5′-end of the inserted fragment (46). In E. coli DH5αZ1, this promoter is tightly repressed by the Tet repressor and can be regulated by supplying 50 ng/ml doxycycline to the culture. Western analysis demonstrated doxycycline-dependent and temperature-controlled expression of the translational WT fusion (Figure 2B). In case of the rep fusion, expression occurred neither at 28°C nor at 42°C confirming the importance of loop L1 for thermometer function. The derep fusion produced equal amounts of GFP protein at both 28 and 42°C. The determined protein levels were fully reflected by GFP fluorescence in vivo (Figure 2C). Fluorescence microscopy revealed weak basal GFP activity at 28°C from the WT fusion that was enhanced when cells were shifted to 42°C. The rep variant produced no fluorescence at all, whereas cells containing the derep fusion emitted fluorescence at both temperatures.
Figure 2.
The hsp17 5′-UTR controls gfp expression in E. coli. (A) Schematic representation of the reporter gene fusions on the low-copy vector pXG10 (31). (B) Detection of GFP fusion proteins. Escherichia coli DH5αZ1 cells (46) carrying reporter plasmids with the WT, rep and derep hsp17 UTR element were grown in LB medium at 28°C. Following heat shock as described in the legend to Figure 1C, cells were incubated at room temperature for 90 min to allow fully maturation of GFP fusion proteins. Samples were subjected to western blot analysis using monoclonal α-GFP antibodies. Transcription of the gfp fusions was induced by application of doxycycline (50 ng/ml), which inactivates the Tet repressor. (C) Aliquots from the same samples were inspected by phase contrast and fluorescence microscopy. GFP fluorescence was excited at 460 nm, and light emission was recorded using a 510 nm filter. Representative examples are shown from three independent experiments, all of which gave similar results.
Taken together, the 5′-UTR of the Synechocystis hsp17 gene displays all features characteristic of a typical RNA thermometer when tested in E. coli reporter systems. The rep and derep versions are presumably locked into ‘closed’ and ‘open’ states, respectively.
Temperature-mediated structural changes in the hsp17 leader sequence
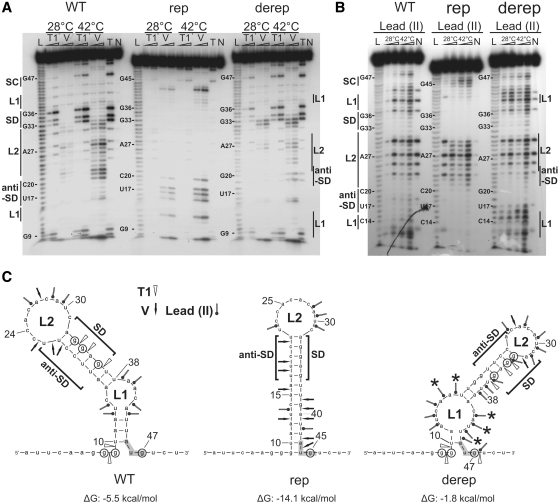
To map the structure and temperature responsiveness of the hsp17 5′-UTR in vitro, we synthesized WT, rep and derep fragments by T7 RNA polymerase-mediated in vitro transcription and performed comparative enzymatic and chemical probing experiments. Cleavages introduced at 28 and 42°C by lowest enzyme or lead(II) concentrations, respectively, were used for structural interpretations. The cleavage pattern of the WT UTR after limited digestion with various RNases indicated that the computer-predicted structure (Figure 1A) is almost correct (Figure 3A). Nucleotide G9 proposed to mask the uridine of the AUG start codon appeared to be partially single stranded at 28°C and became highly susceptible to RNase T1, which cuts 3′ of single-stranded nucleotides, at 42°C. Temperature-mediated disengagement of the AUG start codon was also suggested by enhanced T1 cleavage of G47 at 42°C. The SD sequence-forming nucleotides G33, G34 and G36 were moderately cleaved by RNase T1 at 28°C. The susceptibility of this region was significantly increased at 42°C, providing evidence for temperature-induced liberation of the SD region. G36 was most sensitive toward RNase T1 suggesting that melting initiates from L1 toward G36.
Figure 3.
Enzymatic and chemical probing of hsp17 UTR variants. (A and B) Enzymatic hydrolysis and lead(II)-induced cleavages performed on 5′-end-labeled hsp17 5′-UTR at 28 or 42°C. The conditions for RNase and lead(II) concentrations were as follows: (A) RNase T1 (0.004 and 0.01U), RNase V (0.008 and 0.02 U); (B) lead(II) (10 and 20 mM). RNA fragments were separated on 8% polyacrylamide gels. Lanes N, controls without RNase or modifying agents; lanes T, RNase T1 cleavages under denaturing conditions; lane L, alkaline ladder. Start codon (SC), internal loop1 (L1), Shine–Dalgarno (SD), loop2 (L2) and anti-SD regions are indicated. (C) Computer-predicted secondary structures and enzymatic and lead(II)-induced cleavage sites of WT and mutated hsp17 UTRs. Cleavage sites introduced at 28°C by lowest enzyme or lead(II) concentrations are depicted by arrows as indicated. Circled nucleotides are highly susceptible to RNase T1 at 42°C. Enhanced lead(II) cleavage at 28°C occurred at nucleotides marked by asterisks.
The structure of the rep RNA clearly deviates from the WT structure (Figure 3A). Absent T1 cuts supported the predicted tight structure. Nucleotides U42 and A43 were cleaved by RNase V at low and high temperature suggesting permanent sequestration of the start codon. RNase V primarily cleaves double stranded and stacked regions. The anti-SD nucleotides were also cleaved by RNase V at 28 and 42°C. Most importantly, the SD sequence did not become accessible to RNase T1 at 42°C indicating a ‘closed’ state, in which the SD sequence is not accessible to the 16S rRNA.
In the derep RNA, the introduction of two Gs instead of two Cs at positions 19 and 20 (pairing with the two Gs of the SD sequence in the WT RNA, Figure 1A) resulted in an altered and less stable structure with a shifted L2 loop and a wider L1 loop (Figure 3C). The instable structure at low temperature was most evident by chemical probing (Figure 3B). Lead was used as a single-strand-specific probe to detect subtle conformational changes in rather flexible regions, e.g. internal loops and bulged nucleotides (47). Nine nucleotides (A12, U13, C14, U44, and in particular A15, A16, C41, A42, U43) in or flanking the internal loop L1 of the derep RNA were attacked by lead (II) at 28°C. Loop L1 of the WT structure was much less sensitive toward lead(II). Lead probing confirmed the predicted existence of the L2 loop in the WT and rep RNAs. The SD sites of all RNAs remained unaffected by lead(II) attack probably due to strong base stacking of the purine nucleotides.
Collectively, these results support the model that the 5′-UTR of Synechocystis hsp17 blocks access to the SD sequence at low temperatures and responds to a temperature upshift by melting that liberates the ribosome binding site.
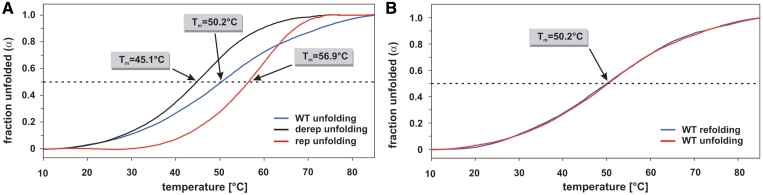
To provide further evidence for the melting process and the differences between the three RNA species, commercially synthesized RNA was subjected to temperature-gradient circular dichroism (CD) spectrometry (Figure 4A). The unfolding curves exhibit Tm values of 45.1°C for the derep RNA, 50.2°C for the WT RNA and 56.9°C for the rep RNA. Unfolding and refolding curves of the WT RNA are nearly identical (Figure 4B) indicating that unfolding is a completely reversible process.
Figure 4.
Melting studies of hsp17 thermometer element by CD spectroscopy. (A) Temperature-dependent fraction of unfolded RNA α(T) of the hsp17 WT (blue line), derep (black line) and rep (red line) RNA as derived from CD unfolding curves recorded at a wavelength of 263 nm. (B) Comparison of unfolding and refolding α(T) curves of the hsp17 WT RNA as derived from CD unfolding and refolding curves recorded at a wavelength of 263 nm.
Temperature-controlled ribosome binding to the hsp17 thermometer
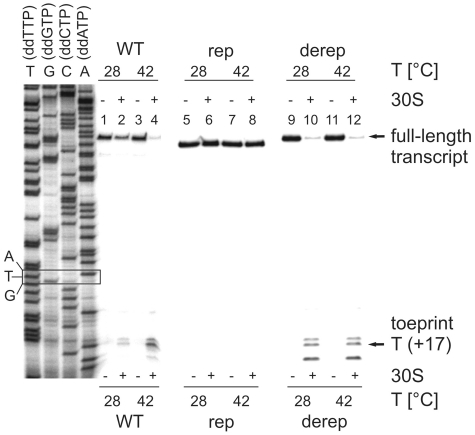
In order to directly examine binding of the ribosome to the Synechocystis hsp17 5′-UTR, we performed toeprinting (primer extension inhibition) assays. Complex formation between the radio-labeled primer, 30S ribosome and initiator tRNA and the hsp17 UTRs (WT and derep: 104 nucleotides; rep: 102 nt; see full-length products in Figure 5) was allowed at 28 or 42°C. The extent of ternary complex formation was determined by primer extension. Prematurely terminated products (toeprints) corresponding to the +17 position downstream of the AUG start codon were expected if ribosome was bound to the RNA. At 28°C, the majority of reverse transcripts were extended to the 5′-end of the template (Figure 5). Only small amounts of truncated cDNAs were obtained at the characteristic toeprint position. In contrast, prominent toeprint signals accompanied by a reduced yield of full-length cDNA occurred at 42°C indicating that formation of a translation initiation complex was greatly stimulated at heat shock temperatures.
Figure 5.
Temperature-dependent ribosome binding to the hsp17 5′-UTR. Toeprinting was carried out on 2 pmol of WT, rep and derep RNAs as described in ‘Materials and Methods’ section. The absence (−) or presence (+) of 30S subunits is indicated. Terminated reverse transcription products (toeprints) at position +17 relative to the A of the translation start codon and full-length products are pointed out by arrows. TGCA refers to a sequencing ladder generated with the same oligonucleotide as in the toeprint experiments. The position of the start codon is boxed.
The complete absence of a toeprint signal when the rep RNA was used and constitutive, temperature-independent formation of a ternary complex with the derep RNA confirmed the importance of the RNA architecture for ribosome binding.
Introduction of hsp17 thermometer variants into the Synechocystis background
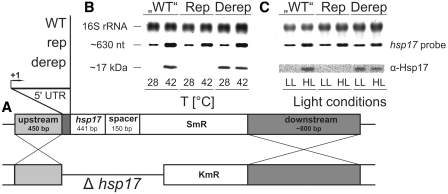
As reported previously, inactivation of the Synechocystis hsp17 gene results in reduced thermotolerance and thylakoid stability under heat stress conditions (3,48). Conversely, constitutive expression of hsp17 confers thermotolerance and protection of the photosynthetic apparatus (49). This offers the unique opportunity to test for the physiological importance of the hsp17 thermometer element. We made use of a Δhsp17 mutant and engineered stable Synechocystis strains carrying chromosomally integrated WT, rep and derep 5′-UTRs upstream of hsp17 (henceforth called ‘WT’, Rep and Derep strains). The Synechocystis HK-1 strain, which lacks the hsp17 open reading frame sll1514 (50) was used for homologous recombination via flanking sequences enabling selection for increasing spectinomycin resistance (Figure 6A). Correct insertion into the Synechocystis genome was confirmed by PCR (data not shown).
Figure 6.
Effect of chromosomally integrated hsp17 UTR variants on Hsp17 production in Synechocystis. (A) Outline of the strategy for construction of hsp17 mutants. The Synechocystis HK-1 (Δhsp17) strain was used for integration of hsp17 and its 5′-UTR via up and downstream flanking sequences. SmR, spectinomycin resistance; KmR, kanamycin resistance. Homologous recombination resulted in generation of the so-called ‘WT’, Rep and Derep strains. (B and C) Determination of hsp17 mRNA and Hsp17 protein levels by northern and western analysis, respectively. Total RNA and protein was extracted from Synechocystis cells incubated at either 28 or 42°C. Ethidium bromide stained 16 s rRNA served as a loading control in northern blot experiments. Equal amounts of total protein were checked by Coomassie-stained SDS–PAGE gels (data not shown). A digoxygenin-labeled RNA probe was used to detect hsp17 transcripts, Hsp17 protein was detected via monoclonal α-Hsp17 antibody. (C) Synechocystis cells were incubated under low light (LL: 30 µmol photons m−2 s−1) or high light (HL: 600 µmol photons m−2 s−1) conditions at 28°C for 30 min. Temperature-stability of the culture was monitored. Subsequent northern and western analysis was conducted as described above.
Expression of hsp17 in the recombinant strains was assayed by northern and western blot analyses. All three strains produced comparable amounts of hsp17 mRNA (Figure 6B and C). Consistent with previous results (42), the level of hsp17 transcript was low at 28°C and increased after a shift to 42°C for 1 h (Figure 6B). Despite the presence of hsp17 mRNA, the small heat shock protein was not detectable at low temperature suggesting translation inhibition. Hsp17 protein accumulated at 42°C. The Rep strain was incapable of translating hsp17 mRNA whereas the Derep mutant synthesized equal amounts of Hsp17 at 28 and at 42°C.
Induction of hsp17 in Synechocystis occurs not only after heat-shock but also after exposure to strong visible light (8). As Synechocystis cells exposed to 300 µmol photons m−2 s−1 transcribe only little hsp17 mRNA (3), we choose higher light intensities of 600 µmol photons m−2 s−1 for 30 min. Light stress-dependent synthesis of Hsp17 in ‘WT’, Rep and Derep cells followed the same pattern as in heat-shocked cells; i.e. it was induced, absent or constitutively high, respectively (Figure 6C). Comparative western blot analysis showed that heat shock is a stronger inductor of hsp17 expression than high light stress (Supplementary Figure S2). Moreover, Hsp17 protein is a very stable protein as it was still traceable in the late recovery phase (60 min post-shock) of stressed ‘WT’, Rep and Derep strains.
Open and closed hsp17 UTRs confer growth defects under stress conditions
In order to determine their stress resistance, Synechocystis ‘WT’, Rep and Derep strains were challenged with different stress conditions. First, cells were exposed to a 6 h heat shock at 42°C and subsequently incubated at 28°C under low light conditions (30 µmol photons m−2 s−1) for 5 days (Figure 7A, top panel). The pale phenotype showed that the Δhsp17 strain was sensitive to this treatment. The complemented ‘WT’ strain was protected, and the Rep and Derep mutants behaved like the Δhsp17 and ‘WT’ strain, respectively.
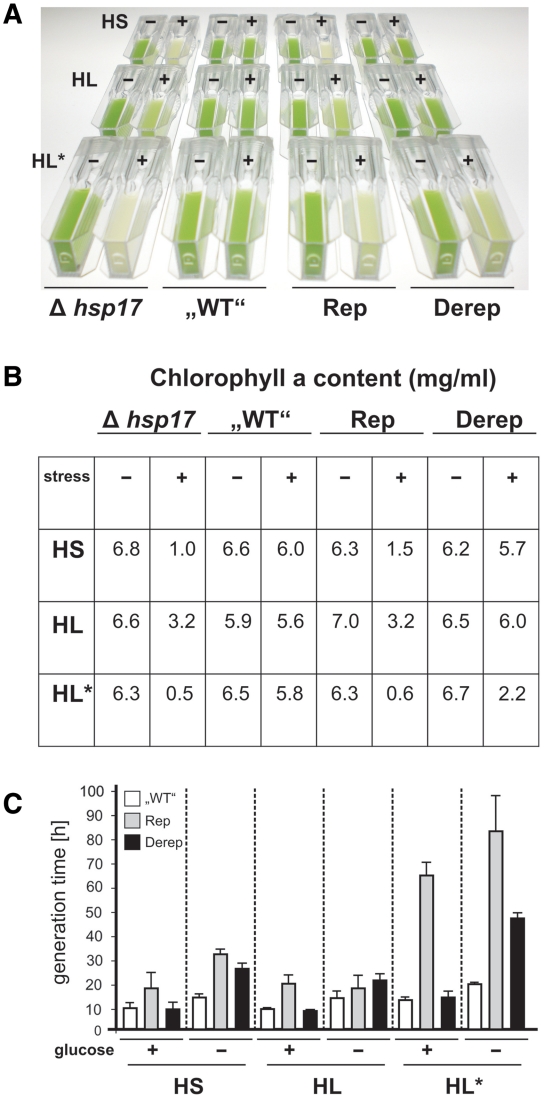
Figure 7.
Stress-induced phenotypes of Synechocystis hsp17 mutants. (A) Cells were grown to early log phase prior to stress treatment. HS, cells were incubated at 42°C for 6 h; HL, cells were exposed to 600 µmol photons m−2 s−1 for 6 h; HL*, cells were incubated at 42°C for 60 min prior to 5 h HL (600 µmol photons m−2 s−1) treatment. Following the different stress conditions, the cultures were transferred to 28°C and LL conditions for 5 days. Before documentation, cultures were diluted to an optical density (OD730) of 1.5. Representative examples of at least three experimental repetitions are shown. (B) Samples from these Synechocystis cell cultures were used for Chl a extraction. (C) Early log phase cells grown in the presence or absence of glucose were treated with HS, HL, HL* as described in (A). After treatment, cells were transferred to 28°C and LL conditions for 48 h.
Heat sensitivity of the Δhsp17 and Rep strains coincided with a reduced Chlorophyll a (Chl a) content (1.0 mg/ml in Δhsp17 and 1.5 mg/ml in Rep compared to 6.0 mg/ml in ‘WT’ cells; Figure 7B). As Chl a is functionally linked to PSII reaction centers, the reduced Chl a concentrations suggest a defective photosynthetic apparatus in the absence of Hsp17. When the four strains were exposed to high light (600 µmol photons m−2 s−1) for 6 h at 28°C, they displayed a similar phenotype as after heat stress (Figure 7A, second panel). However, light stress caused less Chl a decay in Δhsp17 and Rep cells (3- and 2-fold, respectively; Figure 7B) indicating a milder impact on cellular fitness. A defect of the Derep mutant became evident when a combined heat/light stress was applied (Figure 7A, third panel) suggesting that permanent overproduction of Hsp17 alters cell physiology. Here, a heat shock for 1 h was applied prior to 5 h high-light exposure. While the ‘WT’ strain was protected against this severe stress, the Δhsp17 and the Rep strain were equally sensitive. The Derep mutant showed an intermediate phenotype retaining one third of its original Chl a content (Figure 7B).
The importance of appropriate amounts of Hsp17 for the fitness of Synechocystis was further demonstrated by growth tests in the presence (heterotrophic growth) or absence (phototrophic growth) of glucose. The Rep mutant had an extended generation time compared to the ‘WT’ under all stress conditions tested (Figure 7C). The Derep strain grew as well as the ‘WT’ heterotrophically in the presence of glucose. Under photosynthetic growth conditions in the absence of glucose, however, the Derep strain suffered from continuous Hsp17 production. The calculated generation time was elevated to 25 h under light stress and to nearly 50 h under heat/light stress suggesting that deregulated hsp17 expression in particular affects the photosynthetic apparatus.
Open and closed hsp17 thermometer elements affect photosynthesis activity of Synechocystis
The Hsp17 protein stabilizes the lipid phase of membranes and contributes to the maintenance of the thylakoid integrity, especially under stress conditions (7). This prompted us to examine the integrity of the photosynthetic apparatus in our hsp17 mutants. Chlorophyll a (Chl a) fluorescence can be used as an intrinsic probe for the stability of PSII-protein complexes (12). Loss of thylakoid membrane stability is correlated with an increase in Chl a fluorescence. Maximal fluorescence in the ‘WT’ strain was measured at 48°C (Figure 8A). A striking reduction of thermal stability to 42°C was monitored in the Rep mutant whereas thermostability of the photosynthetic apparatus was increased to 55°C in the Derep strain. This is in good agreement with the thermostability of the corresponding 5′-UTRs (Figure 4) indicating that it is the behavior of the RNA that is responsible for the physiological outcome. Light stress led to comparable results (Figure 8B). Maximal Chl a fluorescence was measured in the ‘WT’ after 200 min of high-light exposure. Photostability was reduced to 140 min in the Rep mutant and increased to 300 min in the Derep strain.
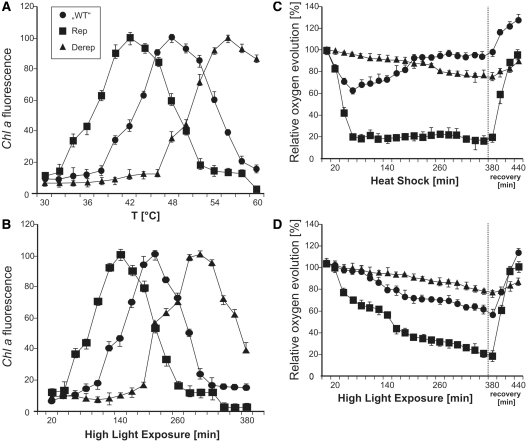
Figure 8.
Integrity of the photosynthetic machinery in Synechocystis strains with mutated hsp17 5′-UTRs. (A) The temperature effect on Chl a fluorescence was measured over a temperature range from 30°C to 60°C in ‘WT’ (circles), Rep (squares) and Derep (triangles) cells grown at 28°C under LL conditions. (B) The impact of light stress on Chl a fluorescence was measured in cells from the same pre-culture as in (A). Cultures were exposed to HL over a period of 380 min. (C) Oxygen evolution was monitored after a heat shock from 28°C to 42°C at the indicated time points. After 380 min, cultures were returned to 28°C. (D) Oxygen evolution after shift to HL conditions. For recovery, the cultures were returned to LL conditions for 60 min. Cells from the same cultures were used for the experiments in (C) and (D). Results are presented as percentage of the oxygen evolution rate measured at time 0. The photosynthetic activities before stress treatment were 3.02 ± 0.04, 2.96 ± 0.01 and 2.87 ± 0.07 µmol O2 mg Chl−1 min−1 in the ‘WT’ (circles), Rep (squares) and Derep (triangles) strains, respectively. Error bars represent standard deviations obtained from three independent experiments.
To measure photosynthetic activity of the hsp17 mutants, we determined oxygen evolution after exposure to 42°C or high-light conditions. While there was a drop in oxygen evolution of ∼30% during the first hour at 42°C in the ‘WT’, the Derep mutant was barely affected suggesting that the photosynthetic machinery was well-protected (Figure 8C). Heat sensitivity of the Rep mutant on the other hand was reflected by a sharp decrease in oxygen evolution to 20% within 60 min after heat shock. Interesting differences were observed in the recovery phase after the cultures had been returned to 28°C and low light conditions. The ‘WT’ and in particular the Rep strain, which is unable to produce Hsp17, recovered rapidly and regained full oxygen evolution capacity within 1 h. In contrast, the Derep mutant suffered most in the recovery phase and returned to pre-shock efficiency only slowly indicating that continued production of Hsp17 is counterproductive once the stress has declined.
Exposure to high-light intensities had a similar effect on oxygen evolution. Heat-preincubation of the cells increased the effect of high-light stress in all strains (data not shown). Again, intermediate damage after stress exposure occurred in the ‘WT’ background (Figure 8D). The Rep mutant was most sensitive and the Derep strain was barely affected. Under recovery conditions, the ‘WT’ and Rep strain performed best, whereas the Derep strain was delayed in full oxygen production. Apparently, well-adjusted synthesis of Hsp17 protein by the hsp17 UTR is important for stress management in Synechocystis.
DISCUSSION
A novel RNA thermometer family
RNA thermometers are widely distributed genetic control elements in the 5′-UTR of temperature controlled genes (23,51). They have been found in numerous α- and γ-proteobacteria and in the Gram-positive pathogen Listeria monocytogenes (25,27,52). Here, we document the first cyanobacterial RNA thermometer that controls translation initiation of the hsp17 gene in Synechocystis sp. PCC 6803. Consisting of only 44 nt forming a single stem–loop structure, it is by far the shortest and most simple natural RNA thermosensor known to date. Typical repression of heat shock gene expression (ROSE) elements are between 60 and 110 nt long (52). The fourU element upstream of the Salmonella agsA gene contains 58 nts (27). The E. coli rpoH thermometer that reaches far into the coding region is composed of 227 nts (24). In its simplicity, the cyanobacterial element resembles rationally designed synthetic RNA thermometers consisting of a short single stem–loop (53).
Riboswitches and RNA thermometers are simple regulatory elements that have repeatedly been suggested to originate from an ancient RNA world (23,54,55). Recently discovered cyanobacterial fossils were dated 3.5-billion-years old (56) marking the advent of oxygenic photosynthesis. Apparently the presence of a functional hsp17 5′-UTR is beneficial to Synechocystis sp. PCC 6803. This prompted us to ask whether other cyanobacteria possess similar elements. Interestingly, likely candidates were identified in the long 5′-UTRs of the Thermosynechococcus elongatus and T. vulcanus small heat shock gene hspA. At their 3′-end they exhibit significant sequence similarity to the hsp17 leader sequence (Supplementary Figure S1). According to in silico calculations, the extended 5′-end is predicted to form a long hairpin structure which might be required for overall stability of the RNA structure in a thermophilic habitat. The presence of thermometer-like sequences upstream of small heat shock genes in distantly related, thermophilic cyanobacteria suggests that this translational control element might be commonly used to withstand constantly changing temperature and light conditions.
Physiological relevance of the cyanobacterial thermometer
Our study provides first experimental evidence for the physiological importance of the on- and off-function of an RNA thermometer. Although the structure and temperature-induced conformational changes of several RNA thermometers have been studied in detail, evidence for their in vivo significance has been lacking except for the Listeria prfA thermosensor that activates virulence gene expression inducing host cell invasion (25). A major reason is that most known RNA thermometers control translation of small heat shock genes, which usually are dispensable because the cellular chaperone network is very redundant (5). In contrast, the hsp17 gene is critically important for Synechocystis because its major target, the photosynthetic machinery is very sensitive to stress-induced damage. Therefore, Hsp17 is essential for tolerance to high temperatures and light stress in several mesophilic and thermophilic cyanobacteria (3,49,57). Heterologous expression of the sHsp from Synechococcus vulcanus in Synechococcus sp. PCC 942 conferred increased thermal resistance and prevented inactivation of the photosynthetic apparatus (49). The protective effect of the amphitrophic Hsp17 protein depends on its dual role as a protein and membrane chaperone (7). In vitro, Hsp17 is able to protect model proteins from heat-induced aggregation (7). In vivo, heat stress-induced Hsp17 interacts with a large variety of proteins and protects a wide range of cellular functions (58). The second important activity is its membrane association (59). Most newly synthesized Hsp17 is associated with the thylakoid membrane (12). Here, it plays a critical role in controlling the physical order, bilayer stability and integrity of membranes (7). Further evidence for the importance of this property derives from an hsp17 mutant with increased thylakoid association, which provided elevated resistance against UV stress (60). Isolated Synechococcus elongatus PCC7942 phycocyanin interacts directly with HspA, a Hsp17 homolog (57). Thus, the severe defect of stressed Synechocystis Δhsp17 and Rep mutants (Figure 7A) might be due to detachment and degradation of accessory pigments. This assumption is well supported by the reduced Chl a levels (Figure 7B).
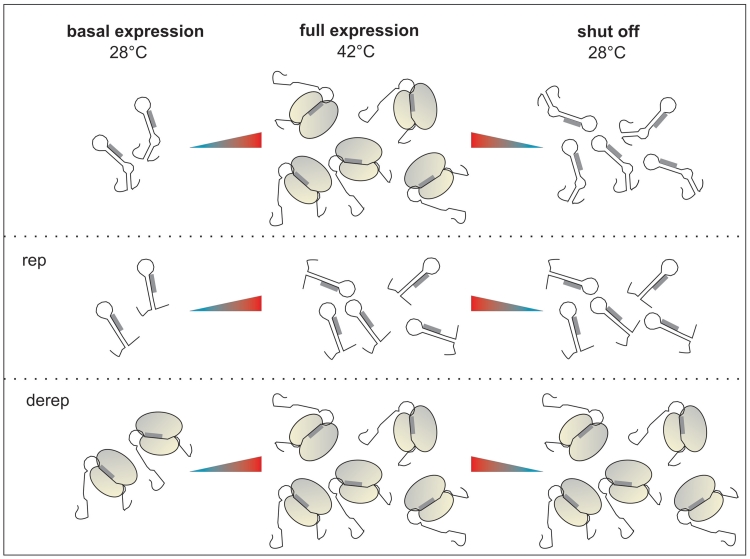
Despite the importance of Hsp17 for photosynthetic activity, the mechanisms controlling its expression were largely unexplored. Alternative sigma factors are known to be required for transcription of hsp17. However, the mRNA is not translated in the absence of stress (Figure 6). Consistent with our northern blot experiments, DNA microarray assays revealed basal transcription of the hsp17 gene even if the inducing SigB factor was missing (10,11). The absence of Hsp17 protein under normal growth conditions despite the presence of significant amounts of hsp17 mRNA (Figure 6) indicates that the 5′-UTR of hsp17 blocks translation initiation (Figure 9) when the chaperone is not required. In vitro ribosome binding assays demonstrated that translation is strictly temperature-controlled and does not require the aid of additional cellular factors. Constitutive basal transcription of hsp17 apparently provides a cellular pool of mRNA intended for instantaneous ‘translation-on-demand’, in case the conditions become unfavorable.
Figure 9.
The 5′-UTR of the Synechocystis hsp17 gene controls translation on demand. The WT RNA forms a secondary structure at physiological temperature (28°C) masking the ribosomal binding site of hsp17, thus preventing the binding of the ribosome. A heat shock induces hsp17 transcription. Concomitant melting of the hairpin structure in the 5′-UTR allows translation initiation. A downshift to physiological temperatures shuts off translation in spite of high transcript levels. Transcriptional induction of the Derep and Rep variants is unaffected. However, the blocked SD sequence in the Rep mutant prevents translation under all conditions, whereas translation is constitutively on in the Derep mutant.
Under stress conditions, the simultaneous induction of hsp17 transcription and translation allows rapid accumulation of Hsp17 protein (Figure 9). In turn, the sHsp promotes readjustment of membrane fluidity and repair of unfolded proteins in the cytoplasm. In the post-stress phase, rapid shut-down of Hsp17 synthesis is achieved by the thermometer inherent to the mRNA. The defect of the Derep mutant under phototrophic conditions and in the stress recovery phase strongly suggests that turning off hsp17 translation is as important as turning it on. Apparently, permanent accessibility of the SD sequence is deleterious in the recovery phase when hsp17 mRNA levels are still very high (Figure 9). Immediate sequestration of the SD sequence ensures that the abundant mRNA is silenced after restoration of physiological conditions.
In the course of our experiments we made the puzzling observation that translation of hsp17 mRNA was not only induced by heat stress but also, although weaker (Supplementary Figure S2), by high-light conditions (Figure 6). As we constantly monitored temperature during light exposure, we exclude that the observed induction was simply due to heating-up the growth medium. Thus, it appears that the hsp17 thermometer responds to high-light stress. At this point we can only speculate on the mechanism underlying the induction of hsp17 translation. The exposure of phototrophic organisms to light quantities above the level required for saturating photosynthetic electron flow results in irreversible damage to the D1 protein and inactivation of PSII (61). To avoid progressive photoinactivation, excess photon energy is partially dissipated as heat from non-functional PSII centres (62,63). Although this might not induce a global heat-shock response, it might be sufficient to cause partial melting of an RNA thermometer given the prolonged and intensive high-light stress (we were not able to induce Hsp17 translation with visible light intensities below 600 µmol photons m−2 s−1). In this context, it is interesting that de novo synthesis of thylakoid-associated Hsp17 at ambient temperature can be induced by altering membrane microviscosity by addition of 30 mM benzyl alcohol (12). As interaction of benzyl alcohol with ribonucleic acids has been reported, the alcohol might be able to induce partial melting of an RNA thermometer (64). Ethanol also was shown to induce sHsp biosynthesis controlled by the ROSE thermometer in Bradyrhizobium japonicum (65). In any case, alcohol was a much weaker inducer of sHsp biosynthesis than heat shock.
In γ-proteobacteria like E. coli and Salmonella, RNA thermometers modulate heat shock gene expression by adding an additional layer of control to the efficient transcriptional induction by the alternative sigma factor σ32 (27,36,66). Instead, the hsp17 thermometer is the predominant regulator of hsp17 expression in Synechocystis. Severe phenotypes were elicited by the exchange of only a few nucleotides in the chromosomally integrated thermometer. Weakening the SD/anti-SD interaction by two point mutations resulted in permanent hsp17 translation, which offered a significant protection to the photosynthetic apparatus in the first hours after stress exposure. As a drawback, cells with elevated Hsp17 levels were delayed in stress recovery and showed reduced fitness under photosynthetic growth conditions in the presence of stress. Apparently, the architecture of the hsp17 UTR is well-adjusted to supply the perfect amount of the chaperone under normal, stress and recovery conditions. It is a sensitive, efficient and rapidly responding gene control element that performs equally well in vitro, in E. coli and in Synechocystis making it an attractive candidate for applications in green biotechnology, which uses cyanobacteria or plants as biofactories (67). Owing to its simple architecture the Synechocystis element is much more suitable as versatile gene control element than all other previously described RNA thermometers.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Grant from the German Research Foundation (DFG priority program 1258) to F.N. and by a fellowship from the Studienstiftung des Deutschen Volkes to J.K. Work in the group of H.S. is funded by the SPP program 1258. H.S. is member of the DFG-funded cluster of Excellence: Macromolecular complexes. Funding for open access charge: DFG (German Research Foundation).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Elizabeth Vierling and Heather O’Neill (University of Arizona) for providing strains, plasmids and α-Hsp17 antisera. We are grateful to Matthias Rögner, Thilo Rühle, Birgit Klinkert, Nicole Frankenberg-Dinkel and Sebastian Rasche for experimental support and to Ulla Aschke-Sonnenborn for technical assistance. All members of the RNA group, Nicole Frankenberg-Dinkel, Sina Langklotz and Bernd Masepohl are acknowledged for critical comments on the manuscript and helpful discussions.
REFERENCES
- 1.Castielli O, De la Cerda B, Navarro JA, Hervas M, De la Rosa MA. Proteomic analyses of the response of cyanobacteria to different stress conditions. FEBS Lett. 2009;583:1753–1758. doi: 10.1016/j.febslet.2009.03.069. [DOI] [PubMed] [Google Scholar]
- 2.Latifi A, Ruiz M, Zhang CC. Oxidative stress in cyanobacteria. FEMS Microbiol. Rev. 2009;33:258–278. doi: 10.1111/j.1574-6976.2008.00134.x. [DOI] [PubMed] [Google Scholar]
- 3.Lee S, Prochaska DJ, Fang F, Barnum SR. A 16.6-kilodalton protein in the Cyanobacterium Synechocystis sp. PCC 6803 plays a role in the heat shock response. Curr. Microbiol. 1998;37:403–407. doi: 10.1007/s002849900400. [DOI] [PubMed] [Google Scholar]
- 4.Nakamoto H, Vígh L. The small heat shock proteins and their clients. Cell. Mol. Life Sci. 2007;64:294–306. doi: 10.1007/s00018-006-6321-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Narberhaus F. Alpha-crystallin-type heat shock proteins: socializing minichaperones in the context of a multichaperone network. Microbiol. Mol. Biol. Rev. 2002;66:64–93. doi: 10.1128/MMBR.66.1.64-93.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lee GJ, Roseman AM, Saibil HR, Vierling E. A small heat shock protein stably binds heat-denatured model substrates and can maintain a substrate in a folding-competent state. EMBO J. 1997;16:659–671. doi: 10.1093/emboj/16.3.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Török Z, Goloubinoff P, Horvath I, Tsvetkova NM, Glatz A, Balogh G, Varvasovszki V, Los DA, Vierling E, Crowe JH, et al. Synechocystis HSP17 is an amphitropic protein that stabilizes heat-stressed membranes and binds denatured proteins for subsequent chaperone-mediated refolding. Proc. Natl Acad. Sci. USA. 2001;98:3098–3103. doi: 10.1073/pnas.051619498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huang L, McCluskey MP, Ni H, LaRossa RA. Global gene expression profiles of the cyanobacterium Synechocystis sp. strain PCC 6803 in response to irradiation with UV-B and white light. J. Bacteriol. 2002;184:6845–6858. doi: 10.1128/JB.184.24.6845-6858.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Suzuki I, Simon WJ, Slabas AR. The heat shock response of Synechocystis sp. PCC 6803 analysed by transcriptomics and proteomics. J. Exp. Bot. 2006;57:1573–1578. doi: 10.1093/jxb/erj148. [DOI] [PubMed] [Google Scholar]
- 10.Tuominen I, Pollari M, Tyystjarvi E, Tyystjarvi T. The SigB sigma factor mediates high-temperature responses in the cyanobacterium Synechocystis sp. PCC6803. FEBS Lett. 2006;580:319–323. doi: 10.1016/j.febslet.2005.11.082. [DOI] [PubMed] [Google Scholar]
- 11.Singh AK, Summerfield TC, Li H, Sherman LA. The heat shock response in the cyanobacterium Synechocystis sp. Strain PCC 6803 and regulation of gene expression by HrcA and SigB. Arch. Microbiol. 2006;186:273–286. doi: 10.1007/s00203-006-0138-0. [DOI] [PubMed] [Google Scholar]
- 12.Horváth I, Glatz A, Varvasovszki V, Torok Z, Pali T, Balogh G, Kovacs E, Nadasdi L, Benko S, Joo F, et al. Membrane physical state controls the signaling mechanism of the heat shock response in Synechocystis PCC 6803: identification of hsp17 as a ‘fluidity gene’. Proc. Natl Acad. Sci. USA. 1998;95:3513–3518. doi: 10.1073/pnas.95.7.3513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asadulghani, Suzuki Y, Nakamoto H. Light plays a key role in the modulation of heat shock response in the cyanobacterium Synechocystis sp PCC 6803. Biochem. Biophys. Res. Commun. 2003;306:872–879. doi: 10.1016/s0006-291x(03)01085-4. [DOI] [PubMed] [Google Scholar]
- 14.Tuominen I, Pollari M, von Wobeser EA, Tyystjarvi E, Ibelings BW, Matthijs HC, Tyystjarvi T. Sigma factor SigC is required for heat acclimation of the cyanobacterium Synechocystis sp. strain PCC 6803. FEBS Lett. 2008;582:346–350. doi: 10.1016/j.febslet.2007.12.030. [DOI] [PubMed] [Google Scholar]
- 15.Sharp PA. The centrality of RNA. Cell. 2009;136:577–580. doi: 10.1016/j.cell.2009.02.007. [DOI] [PubMed] [Google Scholar]
- 16.Waters LS, Storz G. Regulatory RNAs in bacteria. Cell. 2009;136:615–628. doi: 10.1016/j.cell.2009.01.043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Storz G, Altuvia S, Wassarman KM. An abundance of RNA regulators. Annu. Rev. Biochem. 2005;74:199–217. doi: 10.1146/annurev.biochem.74.082803.133136. [DOI] [PubMed] [Google Scholar]
- 18.Gottesman S. Micros for microbes: non-coding regulatory RNAs in bacteria. Trends Genet. 2005;21:399–404. doi: 10.1016/j.tig.2005.05.008. [DOI] [PubMed] [Google Scholar]
- 19.Dühring U, Axmann IM, Hess WR, Wilde A. An internal antisense RNA regulates expression of the photosynthesis gene isiA. Proc. Natl Acad. Sci. USA. 2006;103:7054–7058. doi: 10.1073/pnas.0600927103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Georg J, Voss B, Scholz I, Mitschke J, Wilde A, Hess WR. Evidence for a major role of antisense RNAs in cyanobacterial gene regulation. Mol. Syst. Biol. 2009;5:305. doi: 10.1038/msb.2009.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Voss B, Georg J, Schon V, Ude S, Hess WR. Biocomputational prediction of non-coding RNAs in model cyanobacteria. BMC Genomics. 2009;10:123. doi: 10.1186/1471-2164-10-123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Winkler WC, Breaker RR. Regulation of bacterial gene expression by riboswitches. Annu. Rev. Microbiol. 2005;59:487–517. doi: 10.1146/annurev.micro.59.030804.121336. [DOI] [PubMed] [Google Scholar]
- 23.Narberhaus F. Translational control of bacterial heat shock and virulence genes by temperature-sensing RNAs. RNA Biol. 2010;7:84–89. doi: 10.4161/rna.7.1.10501. [DOI] [PubMed] [Google Scholar]
- 24.Morita MT, Tanaka Y, Kodama TS, Kyogoku Y, Yanagi H, Yura T. Translational induction of heat shock transcription factor sigma32: evidence for a built-in RNA thermosensor. Genes Dev. 1999;13:655–665. doi: 10.1101/gad.13.6.655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Johansson J, Mandin P, Renzoni A, Chiaruttini C, Springer M, Cossart P. An RNA thermosensor controls expression of virulence genes in Listeria monocytogenes. Cell. 2002;110:551–561. doi: 10.1016/s0092-8674(02)00905-4. [DOI] [PubMed] [Google Scholar]
- 26.Chowdhury S, Maris C, Allain FH, Narberhaus F. Molecular basis for temperature sensing by an RNA thermometer. EMBO J. 2006;25:2487–2497. doi: 10.1038/sj.emboj.7601128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Waldminghaus T, Heidrich N, Brantl S, Narberhaus F. FourU: a novel type of RNA thermometer in Salmonella. Mol. Microbiol. 2007;65:413–424. doi: 10.1111/j.1365-2958.2007.05794.x. [DOI] [PubMed] [Google Scholar]
- 28.Rippka R. Isolation and purification of cyanobacteria. Methods Enzymol. 1988;167:3–27. doi: 10.1016/0076-6879(88)67004-2. [DOI] [PubMed] [Google Scholar]
- 29.Giese KC, Vierling E. Changes in oligomerization are essential for the chaperone activity of a small heat shock protein in vivo and in vitro. J. Biol. Chem. 2002;277:46310–46318. doi: 10.1074/jbc.M208926200. [DOI] [PubMed] [Google Scholar]
- 30.Lichtenthaler HK, Buschmann C, Rinderle U, Schmuck G. Application of chlorophyll fluorescence in ecophysiology. Radiat. Environ. Biophys. 1986;25:297–308. doi: 10.1007/BF01214643. [DOI] [PubMed] [Google Scholar]
- 31.Urban JH, Vogel J. Translational control and target recognition by Escherichia coli small RNAs in vivo. Nucleic Acids Res. 2007;35:1018–1037. doi: 10.1093/nar/gkl1040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waldminghaus T, Kortmann J, Gesing S, Narberhaus F. Generation of synthetic RNA-based thermosensors. Biol. Chem. 2008;389:1319–1326. doi: 10.1515/BC.2008.150. [DOI] [PubMed] [Google Scholar]
- 33.Klinkert B, Ossenbuhl F, Sikorski M, Berry S, Eichacker L, Nickelsen J. PratA, a periplasmic tetratricopeptide repeat protein involved in biogenesis of photosystem II in Synechocystis sp. PCC 6803. J. Biol. Chem. 2004;279:44639–44644. doi: 10.1074/jbc.M405393200. [DOI] [PubMed] [Google Scholar]
- 34.Obrist M, Langklotz S, Milek S, Führer F, Narberhaus F. Region C of the Escherichia coli heat shock sigma factor RpoH (sigma 32) contains a turnover element for proteolysis by the FtsH protease. FEMS Microbiol. Lett. 2009;290:199–208. doi: 10.1111/j.1574-6968.2008.01423.x. [DOI] [PubMed] [Google Scholar]
- 35.Brantl S, Wagner EG. Antisense RNA-mediated transcriptional attenuation occurs faster than stable antisense/target RNA pairing: an in vitro study of plasmid pIP501. EMBO J. 1994;13:3599–3607. doi: 10.1002/j.1460-2075.1994.tb06667.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Waldminghaus T, Gaubig LC, Klinkert B, Narberhaus F. The Escherichia coli ibpA thermometer is comprised of stable and unstable structural elements. RNA Biol. 2009;6:455–463. doi: 10.4161/rna.6.4.9014. [DOI] [PubMed] [Google Scholar]
- 37.Hartz D, McPheeters DS, Traut R, Gold L. Extension inhibition analysis of translation initiation complexes. Methods Enzymol. 1988;164:419–425. doi: 10.1016/s0076-6879(88)64058-4. [DOI] [PubMed] [Google Scholar]
- 38.Miller JH. Experiments in Molecular Genetics. New York: Cold Spring Harbor Laboratory Press, Cold Spring Harbor; 1972. [Google Scholar]
- 39.Porra RJ, Thompson WA, Kriedmann PE. Determination of accurate extinction coefficients and simultaneous equations for assaying chlorophylls a and b extracted with four different solvents: verification of the concentration of chlorophyll standards by atomic absorption spectroscopy. Biochem. Biophys. Acta. 1989;975:384–394. [Google Scholar]
- 40.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Imamura S, Yoshihara S, Nakano S, Shiozaki N, Yamada A, Tanaka K, Takahashi H, Asayama M, Shirai M. Purification, characterization, and gene expression of all sigma factors of RNA polymerase in a cyanobacterium. J. Mol. Biol. 2003;325:857–872. doi: 10.1016/s0022-2836(02)01242-1. [DOI] [PubMed] [Google Scholar]
- 42.Fang F, Barnum SR. Expression of the heat shock gene hsp16.6 and promoter analysis in the cyanobacterium, Synechocystis sp. PCC 6803. Curr. Microbiol. 2004;49:192–198. doi: 10.1007/s00284-004-4340-5. [DOI] [PubMed] [Google Scholar]
- 43.Hirata H, Fukazawa T, Negoro S, Okada H. Structure of a beta-galactosidase gene of Bacillus stearothermophilus. J. Bacteriol. 1986;166:722–727. doi: 10.1128/jb.166.3.722-727.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Chevalier C, Geissmann T, Helfer AC, Romby P. Probing mRNA structure and sRNA-mRNA interactions in bacteria using enzymes and lead(II) Methods Mol. Biol. 2009;540:215–232. doi: 10.1007/978-1-59745-558-9_16. [DOI] [PubMed] [Google Scholar]
- 45.de Smit MH, van Duin J. Translational initiation on structured messengers. Another role for the Shine-Dalgarno interaction. J. Mol. Biol. 1994;235:173–184. doi: 10.1016/s0022-2836(05)80024-5. [DOI] [PubMed] [Google Scholar]
- 46.Lutz R, Bujard H. Independent and tight regulation of transcriptional units in Escherichia coli via the LacR/O, the TetR/O and AraC/I1-I2 regulatory elements. Nucleic Acids Res. 1997;25:1203–1210. doi: 10.1093/nar/25.6.1203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hartmann K, Bindereif A, Schön A, Westhof E. Handbook of RNA Biochemistry. 1st edn. Weinheim: Wiley-VCH; 2005. [Google Scholar]
- 48.Lee S, Owen HA, Prochaska DJ, Barnum SR. HSP16.6 is involved in the development of thermotolerance and thylakoid stability in the unicellular cyanobacterium, Synechocystis sp. PCC 6803. Curr. Microbiol. 2000;40:283–287. doi: 10.1007/s002849910056. [DOI] [PubMed] [Google Scholar]
- 49.Nakamoto H, Suzuki N, Roy SK. Constitutive expression of a small heat-shock protein confers cellular thermotolerance and thermal protection to the photosynthetic apparatus in cyanobacteria. FEBS Lett. 2000;483:169–174. doi: 10.1016/s0014-5793(00)02097-4. [DOI] [PubMed] [Google Scholar]
- 50.Kaneko T, Sato S, Kotani H, Tanaka A, Asamizu E, Nakamura Y, Miyajima N, Hirosawa M, Sugiura M, Sasamoto S, et al. Sequence analysis of the genome of the unicellular cyanobacterium Synechocystis sp. strain PCC6803. II. Sequence determination of the entire genome and assignment of potential protein-coding regions. DNA Res. 1996;3:109–136. doi: 10.1093/dnares/3.3.109. [DOI] [PubMed] [Google Scholar]
- 51.Storz G. An RNA thermometer. Genes Dev. 1999;13:633–636. doi: 10.1101/gad.13.6.633. [DOI] [PubMed] [Google Scholar]
- 52.Waldminghaus T, Fippinger A, Alfsmann J, Narberhaus F. RNA thermometers are common in alpha- and gamma-proteobacteria. Biol. Chem. 2005;386:1279–1286. doi: 10.1515/BC.2005.145. [DOI] [PubMed] [Google Scholar]
- 53.Neupert J, Karcher D, Bock R. Design of simple synthetic RNA thermometers for temperature-controlled gene expression in Escherichia coli. Nucleic Acids Res. 2008;36:e124. doi: 10.1093/nar/gkn545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Vitreschak AG, Rodionov DA, Mironov AA, Gelfand MS. Riboswitches: the oldest mechanism for the regulation of gene expression? Trends Genet. 2004;20:44–50. doi: 10.1016/j.tig.2003.11.008. [DOI] [PubMed] [Google Scholar]
- 55.Blouin S, Mulhbacher J, Penedo JC, Lafontaine DA. Riboswitches: ancient and promising genetic regulators. Chembiochem. 2009;10:400–416. doi: 10.1002/cbic.200800593. [DOI] [PubMed] [Google Scholar]
- 56.Schopf JW. Fossil evidence of Archaean life. Philos. Trans. R. Soc. Lond. B. Biol. Sci. 2006;361:869–885. doi: 10.1098/rstb.2006.1834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sakthivel K, Watanabe T, Nakamoto H. A small heat-shock protein confers stress tolerance and stabilizes thylakoid membrane proteins in cyanobacteria under oxidative stress. Arch. Microbiol. 2009;191:319–328. doi: 10.1007/s00203-009-0457-z. [DOI] [PubMed] [Google Scholar]
- 58.Basha E, Lee GJ, Breci LA, Hausrath AC, Buan NR, Giese KC, Vierling E. The identity of proteins associated with a small heat shock protein during heat stress in vivo indicates that these chaperones protect a wide range of cellular functions. J. Biol. Chem. 2004;279:7566–7575. doi: 10.1074/jbc.M310684200. [DOI] [PubMed] [Google Scholar]
- 59.Horváth I, Multhoff G, Sonnleitner A, Vigh L. Membrane-associated stress proteins: more than simply chaperones. Biochim. Biophys. Acta. 2008;1778:1653–1664. doi: 10.1016/j.bbamem.2008.02.012. [DOI] [PubMed] [Google Scholar]
- 60.Balogi Z, Cheregi O, Giese KC, Juhasz K, Vierling E, Vass I, Vígh L, Horvath I. A mutant small heat shock protein with increased thylakoid association provides an elevated resistance against UV-B damage in Synechocystis 6803. J. Biol. Chem. 2008;283:22983–22991. doi: 10.1074/jbc.M710400200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Kyle DJ, Ohad I, Arntzen CJ. Membrane protein damage and repair: Selective loss of a quinone-protein function in chloroplast membranes. Proc. Natl Acad. Sci. USA. 1984;81:4070–4074. doi: 10.1073/pnas.81.13.4070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Shikanai T, Munekage Y, Kimura K. Regulation of proton-to-electron stoichiometry in photosynthetic electron transport: physiological function in photoprotection. J. Plant Res. 2002;115:3–10. doi: 10.1007/s102650200001. [DOI] [PubMed] [Google Scholar]
- 63.Demmig-Adams B, Adams WW., 3rd Photoprotection in an ecological context: the remarkable complexity of thermal energy dissipation. New Phytol. 2006;172:11–21. doi: 10.1111/j.1469-8137.2006.01835.x. [DOI] [PubMed] [Google Scholar]
- 64.Tsukiji S, Pattnaik SB, Suga H. An alcohol dehydrogenase ribozyme. Nat. Struct. Biol. 2003;10:713–717. doi: 10.1038/nsb964. [DOI] [PubMed] [Google Scholar]
- 65.Münchbach M, Nocker A, Narberhaus F. Multiple small heat shock proteins in rhizobia. J. Bacteriol. 1999;181:83–90. doi: 10.1128/jb.181.1.83-90.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Vogel J. A rough guide to the non-coding RNA world of Salmonella. Mol. Microbiol. 2009;71:1–11. doi: 10.1111/j.1365-2958.2008.06505.x. [DOI] [PubMed] [Google Scholar]
- 67.Rittmann BE. Opportunities for renewable bioenergy using microorganisms. Biotechnol. Bioeng. 2008;100:203–212. doi: 10.1002/bit.21875. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.