Abstract
Several methods have been proposed for detecting insertion/deletions (indels) from chromatograms generated by Sanger sequencing. However, most such methods are unsuitable when the mutated and normal variants occur at unequal ratios, such as is expected to be the case in cancer, with organellar DNA or with alternatively spliced RNAs. In addition, the current methods do not provide robust estimates of the statistical confidence of their results, and the sensitivity of this approach has not been rigorously evaluated. Here, we present CHILD, a tool specifically designed for indel detection in mixtures where one variant is rare. CHILD makes use of standard sequence alignment statistics to evaluate the significance of the results. The sensitivity of CHILD was tested by sequencing controlled mixtures of deleted and undeleted plasmids at various ratios. Our results indicate that CHILD can identify deleted molecules present as just 5% of the mixture. Notably, the results were plasmid/primer-specific; for some primers and/or plasmids, the deleted molecule was only detected when it comprised 10% or more of the mixture. The false positive rate was estimated to be lower than 0.4%. CHILD was implemented as a user-oriented web site, providing a sensitive and experimentally validated method for the detection of rare indel-carrying molecules in common Sanger sequence reads.
INTRODUCTION
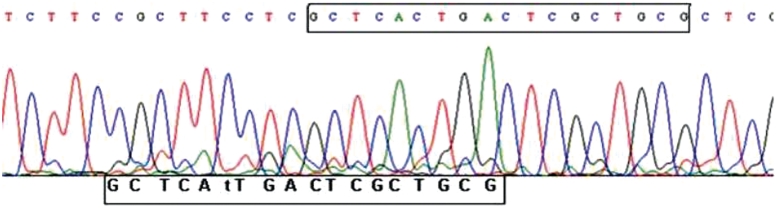
Heritable short insertion and deletions (indels) account for as many as 2.5 M polymorphic sites in the human genome, representing more than 25% of the entire human variation repertoire (1). Short indels have also been associated with diseases (2), underlining their importance to human health. In diploids, germ line-inherited indels are expected to encode either 50% or 100% of the molecules in a given genome. However, in many cases, indels are expected to encode different fractions of molecules as a result of splice variants (3–5), de novo mutations in cancer (6–8) and mutations found in organelles that carry multiple genomes (i.e. mitochondria, chloroplasts) (9–12), for example. An attractive approach for identifying novel indels in specific genomic regions is to amplify these regions via the polymerase chain reaction (PCR) and directly sequence products with the standard Sanger chain termination sequencing method. In this method, sequencing of mixed molecular species will give rise to a ‘double trace’, in which chromatograms that would have resulted from the sequencing of each molecular species separately are superimposed. In the case of an indel, the two traces following the inserted/deletion region should be identical but shifted. In other words, the traces comprising the double trace should self-align (Figure 1). Alternative approaches for de novo indel detection are more laborious or costly, such as cloning and sequencing of multiple molecules. Massive parallel sequencing for this purpose is currently only cost-efficient if used on large genomic regions (10,13–15).
Figure 1.
Representative fragment of an ABI trace file with mixed intact and 9-bp-deleted templates. The framed bases on the top panel represent the strongest calls, while bottom sequences represent the second-best calls harboring the 9-bp deletion and hence, the sequence frame-shift.
Resolving chromatograms for rare indel detection is complicated by the minor contribution of the rare variant to the superimposed chromatogram, a contribution that may be close to noise levels. A number of tools have been developed that can be used for the purpose of detecting relatively rare indels. Such tools are all based on calling not only the best base for every position in the trace but also the base defined by the second highest intensity peak in the chromatogram. Two sequences are thus generated from each chromatogram (or, in some cases, a single degenerate sequence is formed), which can then be aligned to each other (Figure 1), with or without comparison to a reference sequence (16–21). Interestingly, all of the published methods presented thus far rely on specialized sequence alignment algorithms specifically designed to search for near-perfect shifted alignment, rather than standard alignment algorithms, such as the Smith–Waterman local alignment algorithm (22). Consequently, none of these tools provides a robust statistical significance estimate of the results. ShiftDetector, for example, compares two sequences derived from primary and secondary peak calls using a window-based comparison and provides an estimate of the likelihood to observe by chance a given number of matches in a window, based on the binomial distribution. However, ShiftDetector does not account for the multiple hypothesis testing involved in considering many windows and many possible gap sizes, and does not account for sequence composition biases (20). Other methods, such as Indelligent, offer no statistical test.
Here, we describe CHILD, a new short indel detection algorithm based on double-trace resolution which utilizes standard alignment algorithms and the robust statistics implemented in the SSEARCH algorithm (20,23). CHILD was specifically designed to consider rare indels and offers a user-friendly web interface. We experimentally evaluated CHILD performance in terms of both sensitivity and specificity using a set of controlled mixes of cloned molecules containing indels. We show that indels could be detected with a sensitivity as low as 5% and a specificity as high as 98%, although these values may vary, depending on the different primers and templates used.
MATERIALS AND METHODS
The CHILD algorithm
The algorithm involves three consecutive steps: (i) inference of primary and a secondary DNA sequences from trace files (Sanger sequencing) by parsing the output of the PHRED algorithm, (ii) local alignment of the primary and secondary sequences using the Smith–Waterman algorithm and (iii) indel detection within the resulting alignment.
Inference of primary and a secondary DNA sequences from trace files. Trace files are processed with PHRED (24). Using the detailed output option (i.e. ‘–d’), the amplitudes of all four traces at every sequence position are written to a ‘*.poly’ file. This file is parsed to extract a primary sequence, representing those bases with the highest intensity at each position, and a secondary sequence, containing those bases with the second-best amplitude at every position. In positions where the secondary peak intensity was lower than 2.3% of the primary peak intensity, the secondary sequence is assigned the same base as the primary base. This threshold was chosen based on the distribution of the intensity ratio between primary and secondary peaks in 47 cloned samples (totaling 22 363 chromatogram positions), which was found to reach its maximal frequency at this value. When two identical second-best amplitude values are found to have passed the ratio threshold, one base is randomly chosen. To reduce noise, only positions 20–700 of the chromatogram are considered, thus trimming the beginning and ends of the sequenced fragments.
Local alignment of the primary and secondary sequences. The primary and secondary sequences are aligned using the SSEARCH program (22,23) from the FASTA package (version 3.5), which implements the Smith–Waterman local alignment algorithm with shuffling-based significance estimation. Default parameters are used with the following modifications: gap penalty score (the ‘-f’ flag) is set to −80, maximal expectation value (the ‘-E’ flag) is set to 10−4 and the number of shuffling tests is set to 1000.
Indel detection in the resulting sequence alignment. The results are parsed to extract the best alignment significance level and coordinates (i.e. the beginnings and ends of the primary and secondary sequences). An indel is reported only for statistically significant alignments (E-value <10−4). If a gapped alignment is detected, only the 5′ terminal ungapped alignment is considered; if both primary and secondary gaps are detected, the longest of the 5′ terminal ungapped alignments is considered. A special warning is issued if the deduced indel size is <3-bp length. The deduced indel length is estimated from the difference in alignment ends.
Implementation and availability
CHILD is implemented in Perl (version 5). The source code and sample files are available as Supplementary Data; chromatograms can be submitted for CHILD analysis at http://bioinfo.bgu.ac.il/bsu_external/rafi/child/index.php.
Plasmid constructs
The program sensitivity and accuracy were validated by analyzing plasmid constructs harboring human mitochondrial DNA (mtDNA) fragments encompassing naturally occurring deletions. Specifically, two sets of constructs were generated carrying deletions of different sizes (pMitA and pMitB). For pMitA, PCR products of human mtDNA fragments corresponding to positions 1–722 (Revised Cambridge Reference Sequence (rCRS) NC_012920), with or without a 51-bp deletion corresponding to positions 297–348, (pMitA and pMitAΔ51, respectively) were amplified and cloned into the pUC18 vector, as described previously (25). Similarly, pMitB and pMitBΔ9 were created by cloning the PCR fragments corresponding to rCRS positions 8123–8388, with or without a 9-bp deletion corresponding to positions 8271–8281, using the pGEM-T vector, as described previously (26). A series of dilutions varying the ratios of the deleted and intact constructs were prepared in triplicate for all of the above-described constructs.
Sequencing
The construct mixtures were sequenced using an Applied Biosystems 3130 Genetic Analyzer DNA sequencer. For the pUC18 vector, the M13 reverse standard primer was used. For the pGEM-T vector, the standard T7 and SP6 primers were used.
Two types of calibration chromatograms were generated for examining the shadow effect (see text) relying on: (i) the primers and template provided with the BigDye Terminator v1.1 sequencing kit (Applied Biosystems) and an ABI 3130 Genetic Analyzer [Sequencing Facility, National Institute for Biotechnology in the Negev, Ben Gurion University (BGU)], and (ii) the pGEM ezf+ plasmid (Applied Biosystems) with an in-house synthesized, column-purified T7 primer (Biological Services, Weizmann Institute of Science) and the ABI 3730 DNA analyzer.
RESULTS
We have developed CHILD, a computer program for the detection of small sub-populations of molecules carrying indels using ABI trace files. The program compares the sequence of the strongest base calls at each position with the sequence of the second-best calls (Figure 1). Alignment of the two sequences and shuffling tests are then used to test whether the sequences generated from the secondary peaks (namely the secondary sequence) are not random, i.e. represent a shifted version of the primary sequence.
Evaluation of CHILD performance
The data were experimentally generated to test the performance of CHILD, evaluating (i) sensitivity, i.e. the minimal fraction of the molecules carrying an indel that can be detected; (ii) specificity, i.e. the fraction of falsely reported indels in pure samples; and (iii) size accuracy, i.e. the fraction of correctly determined indel sizes. Two sets of plasmid constructs were used to evaluate CHILD performance, with the first carrying a 51-bp deletion and the second a 9-bp deletion in the human mitochondrial genome. The tests were conducted separately for each mutation, including both intact and deleted constructs of the relevant mutation (see ‘Materials and Methods’ section). Mixtures of each construct, with and without the deletions, were prepared in triplicate, sequenced at the BGU sequencing facility and analyzed with CHILD (Table 1). The sensitivity of CHILD varied, depending on the construct and sequencing primers used. CHILD successfully detected the deleted molecule while analyzing chromatograms that originated from mixtures containing 5% or more of the plasmid pMitAΔ51 construct. For the plasmid pMitBΔ9 construct, on the other hand, only mixtures containing 10% or more of the deleted molecule were reproducibly detected, especially when the sequencing reactions were conducted using the T7 primer. Furthermore, when the SP6 primer was used to sequence mixtures containing this construct, 20% or more of the deleted molecules were required for detection.
Table 1.
Experimental evaluation of CHILD sensitivity and reproducibility
| % Δ construct | Predicted |
% Δ construct | Predicted |
||||
|---|---|---|---|---|---|---|---|
| P-value | Loc | Size | P-value | Loc | Size | ||
| Plasmid pMitA | |||||||
| 0 | 1E-111 | 421–423 | 2a | 15 | 8E-168 | 315–366 | 51 |
| NA | NA | NA | 2E-29 | 301–352 | 51 | ||
| 1E-99 | 183–184 | 1a | 1E-99 | 23–24 | 1a | ||
| 2.5 | NA | NA | NA | 20 | 1E-29 | 321–372 | 51 |
| 2E-68 | 354–405 | 51 | 6E-36 | 321–372 | 51 | ||
| 1E-99 | 105–106 | 1a | 1E-99 | 292–344 | 52 | ||
| 5 | 4E-129 | 319–370 | 51 | 25 | 2E-33 | 313–364 | 51 |
| 7E-140 | 323–374 | 51 | 7E-179 | 321–372 | 51 | ||
| 2E-10 | 312–363 | 51 | 1E-99 | 308–360 | 52 | ||
| 7.5 | 4E-19 | 317–368 | 51 | 30 | 1E-158 | 320–371 | 51 |
| 4E-19 | 310–361 | 51 | 4E-161 | 323–374 | 51 | ||
| 4E-27 | 320–371 | 51 | 1E-99 | 293–345 | 52 | ||
| 10 | 3E-25 | 321–372 | 51 | 50 | 2E-29 | 323–374 | 51 |
| 2E-23 | 323–374 | 51 | 6E-47 | 389–440 | 51 | ||
| 6E-129 | 304–356 | 52 | 3E-29 | 410–461 | 51 | ||
| % Δ construct | primer used: T7 | % Δ construct | primer used: SP6 | ||||
|---|---|---|---|---|---|---|---|
| Predicted |
Predicted |
||||||
| P-value | Loc | Size | P-value | Loc | Size | ||
| Plasmid pMitB | |||||||
| 5 | 1E-99 | 78–80 | 2a | 5 | 1E-99 | 44–46 | 2a |
| 1E-99 | 100–102 | 2a | 1E-99 | 22–24 | 2a | ||
| 9E-149 | 105–107 | 2a | 1E-99 | 21–23 | 2a | ||
| 10 | 1E-99 | 550–559 | 9 | 10 | 1E-99 | 22–24 | 2a |
| 1E-99 | 550–559 | 9 | 1E-99 | 31–33 | 2a | ||
| 1E-99 | 185–194 | 9 | 1E-99 | 22–24 | 2a | ||
| 15 | 1E-99 | 540–549 | 9 | 15 | 1E-99 | 32–34 | 2a |
| 1E-99 | 547–556 | 9 | 1E-99 | 46–48 | 2a | ||
| 1E-99 | 547–556 | 9 | 1E-99 | 23–25 | 2a | ||
| 20 | 1E-99 | 542–551 | 9 | 20 | 1E-99 | 423–432 | 9 |
| 1E-99 | 497–506 | 9 | 1E-99 | 206–215 | 9 | ||
| 1E-99 | 191–200 | 9 | 1E-99 | 195–204 | 9 | ||
The table shows an analysis of the 51- and 9-bp deletion constructs with CHILD.
(Upper part) Results for pMitA and pMitAΔ51.
Analysis was conducted for chromatograms generated with increasing concentration of the deletion-containing construct (%Δconstruct) and with two different sequencing primers (T7 and SP6).
The confidence that the indel is not the result of noise (P-value), the indel positions in the corresponding ABI trace file (LOC) and the inferred length of the indel (size) are given as provided by CHILD, analyzing each biological replication separately.
(lower part) The same analysis as in upper part, using pMitB and pMitBΔ9.
NA: no indel was found in the corresponding ABI trace file (the alignment was statistically insignificant or otherwise incompatible with an indel).
aIndels of length 1–2 bp are likely to be artificial (see text) and are ignored in subsequent analysis.
CHILD was found to be highly specific. When shadow-induced short (1–2 bp) indels were ignored, false positive were not found in nine pure samples (three of plasmid pMitA and six of plasmid pMITB, with either the T7 or the SP6 primers). To further investigate the specificity of the program, we analyzed 3024 traces (available at the NCBI Trace Archive: http://nsdl.org/resource/2200/test.20061004111541306T, from chromosome 20 of the ENCODE project). As these traces originated from cloned molecules, they were not expected to include indel mixtures. Less than 1% of these samples (N = 28) were reported by CHILD to carry indels of a length >2.
CHILD also identified the indel size with high accuracy. For 85% of the mixtures involving plasmid pMitAΔ51, the deletion size was accurately determined, while for the remaining four samples, it was overestimated by 1 bp. For plasmid pMitBΔ9, all of the reported indel sizes were precise.
Unlike the size determination, position determination with CHILD was inaccurate, with indel start position varying by as much as 200 bp (Table 1). Since we think that these errors may very likely be the result of an intrinsic property of the sequencing reaction, we conclude that CHILD can offer only a rough estimate of indel position.
The impact of double peaks on indel detection from chromatograms
Manual inspection of the alignment between the primary and secondary sequences of the 2.5% plasmid pMitAΔ51 mixture revealed a statistically significant alignment involving a 1-bp indel (Figure 2A). In fact, a high-quality ‘shadow’ alignment was frequently found 5′ to the deletion, with a shift of 1 or 2 bp. This phenomenon is documented in the ABI troubleshooting guide (Applied Biosystems, Carlsbad CA, USA), where it is suggested to be influenced by the primer used for the PCR or by the sequencing steps. Suggested causes of such shadow alignment include length variation introduced during primer synthesis, homopolymer runs in the primer sequence or homopolymer runs embedded within the beginning of the template. Surprisingly, the primer–plasmid combination used for standard calibration of the sequencing kits (Figure 2B) also produced shadow alignments, suggesting that a low level of shifted molecules is commonly found in sequencing reactions, regardless of the presence of homopolymers in the template or the primer sequences.
Figure 2.
Examples of n + 1-shifted chromatograms (‘shadow sequences’). Alignment of secondary (upper) and primary (lower) sequences generated using (A) a highly diluted mixture of plasmids pMitA and pMitAΔ51 (2.5%) or (B) a pure sample of the pGEM ezf+ plasmid and a column-purified T7 primer. The primary and secondary sequences from each chromatogram were aligned using SSEARCH (see ‘Materials and Methods’ section).
A comparison of CHILD performance versus that of Indellignet and ShiftDetector
The chromatograms generated from controlled indel/wild-type mixtures were analyzed with two other algorithms, namely Indelligent and ShiftDetector (Table 2). VarDetect was not tested since it repeatedly failed to analyze these chromatograms, either due to technical problems or due to the increase of the maximal indel size parameter to 51 bp. The results suggest that CHILD is both more accurate and more sensitive in detecting rare variants. CHILD was the only algorithm that repeatedly detected indels involving 5% of plasmid pMitΔA. ShiftDetector did report a 51-bp indel for mixtures involving higher concentrations of the deletion-carrying molecule but often reported other deletions as well. For example, given a 30% mixture sequenced with the M13 primer, ShiftDetector reported two to three indels for each replica, correctly reporting a 51-bp indel for two out of the three repeats (compared to three out of three for CHILD, which reports only the correct indel). Indelligent performed very poorly in this analysis, never detecting the correct indel for plasmid pMitΔA, and detecting plamsid pMitΔB with a sensitivity of ≥20% (as compared to 10% with CHILD). We note that using Indelligent requires an extra step of secondary peaks calling with Sequencher, and that we did not optimize this step. Nevertheless, our results indicate that a naïve user can submit chromatograms to CHILD without any optimization or manual editing, and that rare indel variants are best detected with CHILD.
Table 2.
The performance of indel detection algorithms with controlled mixtures of indel constructs
| % Δ construct | primer | CHILD | ShiftDetector | Indelligent |
|---|---|---|---|---|
| Plasmid pMitA (Δ = 51 bp) | ||||
| 2.5 | M13 | 51 | – | 1 |
| 1 | – | 1 | ||
| – | ||||
| 51 | – | 1 | ||
| 5 | M13 | 51 | – | 1 |
| 51 | – | 1 | ||
| 51 | – | 1 | ||
| 7.5 | M13 | 51 | 51 | 1 |
| 51 | 1 | |||
| 51 | – | 1 | ||
| 10 | M13 | 51 | – | 1 |
| 51 | 1 | 1 | ||
| 51 | 2,51,53 | 1 | ||
| 15 | M13 | 51 | – | 1 |
| 1 | 1,29 | 1 | ||
| 51 | 2,51,53 | 1 | ||
| 20 | M13 | 51 | 51 | 1 |
| 51 | 1,13,32,52 | 1,2 | ||
| 51 | 51 | 1,2 | ||
| 25 | M13 | 51 | 51 | 1,2,3,4 |
| 51 | 1,13,32 | 1,2,3,4,5,6 | ||
| 51 | 28,51 | 1,2,3,4 | ||
| 30 | M13 | 51 | 2,51,53 | 1,2,3,4 |
| 51 | 1,32,50 | 1,4,52 | ||
| 52 | 51 | 1,4,51 | ||
| 50 | M13 | 51 | 51 | 1,2,3,4 |
| 51 | 51 | 51 | ||
| Plasmid pMitB (Δ=9 bp) | ||||
| 5 | T7 | – | – | 1 |
| – | 1 | 1 | ||
| – | – | 1 | ||
| SP6 | – | 2 | 1,2 | |
| – | 2 | 1,2 | ||
| – | 2 | 1,2 | ||
| 10 | T7 | 9 | 7 | 1 |
| 9 | 2 | 1 | ||
| 9 | 9 | 1,2 | ||
| SP6 | – | 2 | 1,2 | |
| – | 2 | 1,2 | ||
| – | 2 | 1,2 | ||
| 15 | T7 | 9 | 9 | 1.2 |
| 9 | 9 | 1 | ||
| 9 | 1 | 1 | ||
| SP6 | – | 2 | 1,2 | |
| – | 2 | 1,2 | ||
| – | 2 | 1,2 | ||
| 20 | T7 | 9 | – | 1,2,9 |
| 9 | 1 | 1,9 | ||
| 9 | 9 | 1,9 | ||
| SP6 | 9 | 2 | 1,2,7,9 | |
| 9 | 2 | 1,8,9 | ||
| 9 | 2 | 1,9 |
The performance of CHILD, ShiftDetector and Indelligent in resolving traces resulting from mixtures of indel-carrying and wild-type molecules.
The plasmids pMitA and pMitB (see text) were used to generate chromatograms with increasing concentration of the deletion-containing construct (%Δ construct), using the M13, T7 or SP6 primers, and repeating the entire process three times.
Each chromatogram was analyzed with all three algorithms and the results are summarized for each replicate (‘-’ indicates no result was reported).
Correct indel size assignments are indicated in bold.
For Indelligent, chromatograms were converted using the Sequencher package (GeneCodes, Ann Arbor MI), using default parameters.
For ShiftDetector, the significance cutoff was set to 0.0001 to match the default stringency of CHILD.
Where applicable, multiple indel size predictions are provided as a comma-separated list.
DISCUSSION
We present here CHILD, a dynamic programming-based software for the identification of indels from ABI sequencing traces. CHILD infers primary and secondary DNA sequences from sequence traces using PHRED and aligns the two using SSEARCH. A statistically significant similarity between the two reads is used to rule out the possibility that the secondary sequence is a random sequence. The beginning of the alignment indicates the position of the indel and the shift between the reads reveals the size of the indel.
In addition to the development of this new tool, we present here, for the first time, a controlled experimental evaluation of the sensitivity and specificity of this approach. In previous reports, the ability to detect indels was evaluated with DNA extracted from individuals shown to carry an indel with other methods, either in a heterozygous situation (see for example ref. 20) or at unknown ratios (1). Our results suggest that the sensitivity of this approach depends on template and primer choice. While variation in DNA purity could contribute to this difference, a 2-fold decrease in sensitivity was observed when the same template was sequenced using different primers (Table 1). The sensitivity of chromatogram-based indel detection most likely depends on signal and noise levels. One source of noise known to be template- and primer-dependent is the occurrence of n + 1/n–1 ‘shadow’ peaks in the chromatogram. Sufficiently high levels of shadow peaks will confound shift detection algorithms. Additionally, random noise may also be template- and primer-dependent, and could overshadow the secondary peaks truly originating from indels. Furthermore, ‘shadow’ sequences are expected to be detrimental to indel localization by CHILD. The observed position at which the indel-related alignment begins is determined by the relative strength of the secondary sequence and the shadow sequence. As a result, CHILD provides only a rough estimate of the indel position, as indicated in the output of the program.
Our controlled analysis suggests that CHILD frequently fails to resolve traces resulting from equal amounts of the two molecular species (Table 1). When considering the algorithm, the occasional successes are more surprising than the failures. In the case of equimolar species, the peaks originating from the two molecules should be equal in intensity. As a result, their assignment as ‘primary’ and ‘secondary’ should be random. The resulting sequences are thus expected to align very poorly. We note that this should hold true for other algorithms that are based on primary/secondary sequence calling. This stands in contrast with the reported successes of other algorithms in resolving traces originated from heterozygous indels, as well as the success of ShiftDetector reported here (Table 2) and the occasional success of CHILD in resolving 50% mixtures. This contradiction can be explained by (i) minute differences in the original concentration of the two molecules; (ii) differences in the amplification efficiency of the two species in PCR; or (iii) differences in the efficiency of the sequencing reactions in reading the two molecules. Consistent differences between the intensities of the peaks derived from the two molecules through either of these mechanisms would result in proper assignment of peaks to primary and secondary sequences, and successful peak detection. Further research is required to understand why and when CHILD and other secondary-sequence-based algorithms succeed in detecting heterozygous mutations. We note that Indelligent is not expected, even in theory, to have difficulties resolving traces from equimolar mixtures as it calls a degenerate sequence without trying to assign the secondary peaks into a separate sequence (see below).
Due to design considerations, CHILD will always report a single indel and may successfully report only one indel in a molecule carrying multiple indels. In fact, we present results in which a very similar scenario is successfully resolved: shadow sequences imitate the presence of two indels co-occurring in the same chromatogram, involving a short (1–2 bp) indel and a longer (9- or 51-bp long) indel. In most of the above-reported cases, one indel was successfully identified in each chromatogram and the other was ignored. Nevertheless, it is difficult to predict the behavior of CHILD with real multiple indels: it is expected to depend on the length of the indels, their position, the quality of the alignment before and after each gap and the presence of a shadow sequence. The windows-based approach of ShiftDetector may be more suitable for chromatograms originating from mixtures involving rare multi-indel variants, if adjustments are made to make it more suitable for the detection of rare variants.
Some modifications to the algorithm could be considered that would further improve its ability to detect very rare variants. Chromatogram positions in which the primary and secondary bases are accidentally identical are currently identified based on the ratio of intensity between the secondary and primary bases passing some threshold. However, as sequence quality increases at the very beginning of the sequence and drops toward the ends, a more dynamic threshold could be developed that better resolves such identities. A simpler improvement is adding some warning when simple repeats flank the indel, such as implemented in ShiftDetector, as such repeats are expected to lead to inaccuracy in indel positioning (20). However, such an improvement will have impact on CHILD performance only after the impact of shadow sequences is eliminated, as CHILD currently reports only rough estimates of indel location (as discussed earlier).
Our analysis of controlled indel mixtures revealed that in some cases, CHILD can resolve indel mixtures even when one of the deleted species accounts for only 5% of the mixture. Such sensitivity is sufficient to detect rare tumor-related indels, splice variants and heteroplasmic mitochondrial deletions. This level of sensitivity or better can be achieved, in principle, in next-generation massive parallel sequencing efforts (6,10,15). However, standard Sanger sequencing will most likely continue to be more cost effective than massive parallel sequencing for targeted analysis of specific genomic regions/transcripts. Thus, increasing indel detection sensitivity and accuracy provides benefits to those researchers not conducting genome-scale analysis.
CHILD offers several advantages over existing tools. To the best of our knowledge, only three tools can utilize a single trace to detect rare variants. Indelligent (18) transforms the chromatogram into a degenerate (IUPAC) representation and applies a dynamic programming algorithm specifically designed to self-align the resulting degenerate sequences allowing for indels. This tool provides several descriptive statistics that allow users to evaluate the proposed indel but does not perform formal hypotheses testing (e.g. calculating the likelihood of a given alignment to occur by chance). VarDetect (27) uses a specialized base-calling algorithm and a matrix representation that describes both primary and secondary base calls. It then applies a search algorithm that utilizes matrix representation to detect similarity between the two. Again, no test is conducted to evaluate the statistical significance of this similarity. In addition, prior knowledge of the rare variant concentration is required for a secondary base call, and the program needs to be locally installed. For technical reasons, CHILD was not compared to VarDetect, as we were unable to use it for indel detection with out data (data not shown). ShiftDetector (20) is the algorithm most similar to CHILD. It calls primary and secondary sequences (using PHRED) from the chromatogram and uses a special running-window matching algorithm, considering a predefined range of indel sizes, reporting separately on possible alignments for each indel size. ShiftDetector does offer a statistical test, which is based on the binomial distribution, to evaluate the likelihood of fortuitous indel-like patterns. However, ShiftDetector does not account for multiple testing or for sequence composition biases. CHILD was specifically designed to address the shortcoming of these existing tools. It uses the well-tested statistical method to reject fortuitous results implemented in the SSEARCH program, in which the distribution of fortuitous alignment scores is estimated by fitting the alignment scores of shuffled sequences to the extreme value distribution. It is probably thanks to the strength of the alignment algorithm and the accuracy of the statistical test that CHILD is more accurate and sensitive in detecting rare variants than are Indelligent and ShiftDetector (Table 2). CHILD thus combines an algorithm specifically designed to handle rare variants with a user-friendly interface designed for simplicity. Only a single trace file is required. No additional tools need to be installed and no parameters need to be adjusted.
To conclude, we present a new experimentally tested bioinformatics tool for rare indel detection from single chromatograms, and offer, for the first time, an evaluation of the sensitivity of this approach. We show that even rare indels can be detected (as low as 5% of the molecules) by a friendly tool that is accessible to experimental biologists, as well as to bioinformaticians.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The National Institute for Biotechnology in the Negev and by the Israeli Cancer Association. Funding for open access charge: BGU Research budget 33591766.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank the Israel Ministry of Absorption for a scholarship granted to Ilia Zhidkov. We thank the anonymous reviewers of the manuscript for their useful comments.
REFERENCES
- 1.Bhangale TR, Stephens M, Nickerson DA. Automating resequencing-based detection of insertion–deletion polymorphisms. Nat. Genet. 2006;38:1457–1462. doi: 10.1038/ng1925. [DOI] [PubMed] [Google Scholar]
- 2.Ball EV, Stenson PD, Abeysinghe SS, Krawczak M, Cooper DN, Chuzhanova NA. Microdeletions and microinsertions causing human genetic disease: common mechanisms of mutagenesis and the role of local DNA sequence complexity. Hum. Mutat. 2005;26:205–213. doi: 10.1002/humu.20212. [DOI] [PubMed] [Google Scholar]
- 3.Breitbart RE, Andreadis A, Nadal-Ginard B. Alternative splicing: a ubiquitous mechanism for the generation of multiple protein isoforms from single genes. Annu. Rev. Biochem. 1987;56:467–495. doi: 10.1146/annurev.bi.56.070187.002343. [DOI] [PubMed] [Google Scholar]
- 4.Ner-Gaon H, Halachmi R, Savaldi-Goldstein S, Rubin E, Ophir R, Fluhr R. Intron retention is a major phenomenon in alternative splicing in Arabidopsis. Plant J. 2004;39:877–885. doi: 10.1111/j.1365-313X.2004.02172.x. [DOI] [PubMed] [Google Scholar]
- 5.Sammeth M, Foissac S, Guigo R. A general definition and nomenclature for alternative splicing events. PLoS Comput. Biol. 2008;4:e1000147. doi: 10.1371/journal.pcbi.1000147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell PJ, Pleasance ED, Stephens PJ, Dicks E, Rance R, Goodhead I, Follows GA, Green AR, Futreal PA, Stratton MR. Subclonal phylogenetic structures in cancer revealed by ultra-deep sequencing. Proc. Natl Acad. Sci. USA. 2008;105:13081–13086. doi: 10.1073/pnas.0801523105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Stratton MR, Campbell PJ, Futreal PA. The cancer genome. Nature. 2009;458:719–724. doi: 10.1038/nature07943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chomyn A, Attardi G. MtDNA mutations in aging and apoptosis. Biochem. Biophys. Res. Commun. 2003;304:519–529. doi: 10.1016/s0006-291x(03)00625-9. [DOI] [PubMed] [Google Scholar]
- 10.He Y, Wu J, Dressman DC, Iacobuzio-Donahue C, Markowitz SD, Velculescu VE, Diaz LA, Jr, Kinzler KW, Vogelstein B, Papadopoulos N. Heteroplasmic mitochondrial DNA mutations in normal and tumour cells. Nature. 2010;464:610–614. doi: 10.1038/nature08802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kmiec B, Woloszynska M, Janska H. Heteroplasmy as a common state of mitochondrial genetic information in plants and animals. Curr. Genet. 2006;50:149–159. doi: 10.1007/s00294-006-0082-1. [DOI] [PubMed] [Google Scholar]
- 12.Woodson JD, Chory J. Coordination of gene expression between organellar and nuclear genomes. Nat. Rev. Genet. 2008;9:383–395. doi: 10.1038/nrg2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lutz-Bonengel S, Sanger T, Pollak S, Szibor R. Different methods to determine length heteroplasmy within the mitochondrial control region. Int. J. Legal Med. 2004;118:274–281. doi: 10.1007/s00414-004-0457-0. [DOI] [PubMed] [Google Scholar]
- 14.Shendure J, Ji H. Next-generation DNA sequencing. Nat. Biotechnol. 2008;26:1135–1145. doi: 10.1038/nbt1486. [DOI] [PubMed] [Google Scholar]
- 15.Massouras A, Hens K, Carine G, Uplekar S, Decouttere F, Rougemont J, Cole ST, Deplancke B. Primer-initiated sequence synthesis to detect and assemble structural variants. Nat. Methods. 2010;7:485–486. doi: 10.1038/nmeth.f.308. [DOI] [PubMed] [Google Scholar]
- 16.Chen K, McLellan MD, Ding L, Wendl MC, Kasai Y, Wilson RK, Mardis ER. PolyScan: an automatic indel and SNP detection approach to the analysis of human resequencing data. Genome Res. 2007;17:659–666. doi: 10.1101/gr.6151507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Dicks E, Teague JW, Stephens P, Raine K, Yates A, Mattocks C, Tarpey P, Butler A, Menzies A, Richardson D, et al. AutoCSA, an algorithm for high throughput DNA sequence variant detection in cancer genomes. Bioinformatics. 2007;23:1689–1691. doi: 10.1093/bioinformatics/btm152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dmitriev DA, Rakitov RA. Decoding of superimposed traces produced by direct sequencing of heterozygous indels. PLoS Comput. Biol. 2008;4:e1000113. doi: 10.1371/journal.pcbi.1000113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Montgomery KT, Iartchouck O, Li L, Loomis S, Obourn V, Kucherlapati R. PolyPhred analysis software for mutation detection from fluorescence-based sequence data. Curr. Protoc. Hum. Genet. 2008;Chapter 7 doi: 10.1002/0471142905.hg0716s59. Unit 7 16. [DOI] [PubMed] [Google Scholar]
- 20.Seroussi E, Ron M, Kedra D. ShiftDetector: detection of shift mutations. Bioinformatics. 2002;18:1137–1138. doi: 10.1093/bioinformatics/18.8.1137. [DOI] [PubMed] [Google Scholar]
- 21.Tenney AE, Wu JQ, Langton L, Klueh P, Quatrano R, Brent MR. A tale of two templates: automatically resolving double traces has many applications, including efficient PCR-based elucidation of alternative splices. Genome Res. 2007;17:212–218. doi: 10.1101/gr.5661407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith TF, Waterman MS. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 23.Pearson WR. Searching protein sequence libraries: comparison of the sensitivity and selectivity of the Smith–Waterman and FASTA algorithms. Genomics. 1991;11:635–650. doi: 10.1016/0888-7543(91)90071-l. [DOI] [PubMed] [Google Scholar]
- 24.Ewing B, Hillier L, Wendl MC, Green P. Base-calling of automated sequencer traces using phred. I. Accuracy assessment. Genome Res. 1998;8:175–185. doi: 10.1101/gr.8.3.175. [DOI] [PubMed] [Google Scholar]
- 25.Falkenberg M, Gaspari M, Rantanen A, Trifunovic A, Larsson NG, Gustafsson CM. Mitochondrial transcription factors B1 and B2 activate transcription of human mtDNA. Nat. Genet. 2002;31:289–294. doi: 10.1038/ng909. [DOI] [PubMed] [Google Scholar]
- 26.Suissa S, Wang Z, Poole J, Wittkopp S, Feder J, Shutt TE, Wallace DC, Shadel GS, Mishmar D. Ancient mtDNA genetic variants modulate mtDNA transcription and replication. PLoS Genet. 2009;5:e1000474. doi: 10.1371/journal.pgen.1000474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ngamphiw C, Kulawonganunchai S, Assawamakin A, Jenwitheesuk E, Tongsima S. VarDetect: a nucleotide sequence variation exploratory tool. BMC Bioinformatics. 2008;9(Suppl. 12):S9. doi: 10.1186/1471-2105-9-S12-S9. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.