Abstract
In recent years, the field of epigenetics has grown dramatically and has become one of the most dynamic and fast-growing branches of molecular biology. The amount of diseases suspected of being influenced by DNA methylation is rising steadily and includes common diseases such as schizophrenia, bipolar disorder, Alzheimer’s disease, diabetes, atherosclerosis, cancer, major psychosis, lupus and Parkinson’s disease. Due to cellular heterogeneity of methylation patterns, epigenetic analyses of single cells become a necessity. One rationale is that DNA methylation profiles are highly variable across individual cells, even in the same organ, dependent on the function of the gene, disease state, exposure to environmental factors (e.g. radiation, drugs or nutrition), stochastic fluctuations and various other causes. Using a polymerase chain reaction (PCR)-slide microreaction system, we present here a methylation-sensitive PCR analysis, the restriction enzyme-based single-cell methylation assay (RSMA), in the analysis of DNA methylation patterns in single cells. This method addresses the problems of cell heterogeneity in epigenetics research; it is comparably affordable, avoids complicated microfluidic systems and offers the opportunity for high-throughput screening, as many single cells can be screened in parallel. In addition to this study, critical principles and caveats of single cell methylation analyses are discussed.
INTRODUCTION
Despite significant effort, understanding the causes and mechanisms of complex non-Mendelian diseases such as schizophrenia, bipolar disorder, diabetes, Alzheimer’s disease, Parkinson’s disease and various cancers remains a major challenge. In recent years, it became evident that the study of epigenetic mechanisms, which are consistent with various non-Mendelian irregularities of complex diseases, may hold the key to the understanding of some of the characteristics of these disorders (1). One of the central obstacles hampering progress in the burgeoning field of epigenetics related to human disease is the inherent cell heterogeneity of a given tissue. Standard population measurement techniques merely describe average behavior and are insufficient to investigate variability among cells (2). Epigenetic analyses traditionally probe cell ensembles, thereby completely averaging over relevant individual cell responses, such as differences in cell proliferation, lack of synchrony of cells in a culture, responses to external stimuli and disease onset or stochastic events (3). As a consequence, up to now it is impossible to understand whether a small increase in the methylation level measured in the ensemble results from a small, homogeneous increase across all cells or a large increase in a subset of cells. In cancer studies, methylation patterns in CpG islands that are important in gene regulation could be different from cell to cell even in a single tumor tissue. Cells with distinct epigenetic profiles eventually display distinct phenotypic behavior and drug response (4,5). For example, variability in adipogenesis activity among preadipocytes presents significant challenges to drug intervention (6). To improve the efficacy of drug treatment, the source of cell-to-cell variability should be identified and targeted. Thus, the ability to detect methylation patterns of regulatory sequences from a single cell is an essential factor for understanding the mechanism of tumor initiation, the screening of molecular markers and early diagnosis of cancers. This holds true for the molecular characteristics of most diseased tissues (especially highly compartmented tissues such as brain or pancreas) where current analyses are extremely limited by examination of pooled cell lysates. However, in contrast to one-cell transcriptome analyses, the interrogation of DNA methylation patterns in single cells is lagging behind, primarily because of more complex methodologies. As a consequence, epigenetic cause of cell-to-cell variability cannot be readily investigated in an average laboratory setup. For example, the study of methylation patterns in single cells using standard equipment such as polymerase chain reaction (PCR) reaction tubes with 0.2–1.5 ml volume is usually unsuccessful due to numerous reasons, including partial recovery of cell nuclei, loss of DNA during various experimental steps, incomplete chemical or enzymatic modification and insufficient amplification of the DNA, among other causes. Indeed, previous attempts to establish protocols for single cell analyses of methylation pattern suffered from low success rate and low throughput (7). In contrast to gene expression analyses, successful methylation studies using microfluidic devices are still lacking, presumably because microfluidic protocols are expensive, usually have limited multiplexing capabilities, are laborious or cannot handle the necessary larger number of chemical or enzymatic steps involved. One of the main restrictions of microfluidic devices is the inefficient transfer of the analyte (i.e. DNA) between each processing step and elimination of waste products (8,9). To circumvent these limitations, we developed a protocol that is comparably affordable and contains all the necessary elements for single-cell DNA methylation profiling of CpG sites integrated in a single device. The protocol consists of cell placement and cell lysis, enzymatic DNA fragmentation, cleavage of DNA with methylation-sensitive restriction enzymes (MSREs) and parallelized multi-well DNA amplification using 48-well slides.
To demonstrate the usefulness of the restriction enzyme-based single-cell methylation assay (RSMA) technology in a clinical setup, we chose several CpG island sequences of genes such as CDKN2A/INK4a (p16) and COL1A2, which are known to be aberrantly methylated in various cancers. Interrogation of DNA methylation patterns in regulatory regions such as CpG islands has become an important tool for diagnostic purposes and understanding of tissue-specific gene regulation in both normal land pathological conditions. Ultimately, the basic methodology described in this report may be adapted to include real-time methylation protocols and even whole-genome ‘epigenetic’ microarrays (1,10).
MATERIALS AND METHODS
Cell lines
As an epigenetically well-characterized test system, the human SW480 cancer cell line derived from a colorectal adenocarcinoma, grade III–IV, was used (11). To study tissue-specific effects, human lymphocytes (Pachmann SIMFO, Dr. Med. Ulrich Pachmann, Bayreuth/Germany) were analyzed. SW480 cells were cultured under standard conditions (humidified incubator, at 37°C and 5% CO2) in Dulbecco’s modified eagle medium (DMEM) containing 10% fetal bovine serum (FBS), 50 mg/l gentamycin (Biochrom, Berlin/Germany) and 100 mg/l kanamycin (Biochrom). Human lymphocytes were used as delivered. Cells were stained with Bisbenzimide H33342 (Fluka) in a 10 µg/ml final concentration and propidium iodide (Sigma) with a 50 µg/ml final concentration. Living single cells (propidium iodide negative) in 1 × PBS were deposited on the reaction sites of AmpliGrid slides (Beckman Coulter Biomedical GmbH, Advalytix Products, Munich/Germany, BCB) according to their physical properties (size, granularity) with a fluorescence-activated cell sorter (FACS), fitted with an AmpliGrid slide holder (BCG). Cell deposition was validated with a fluorescence microscope fitted with a DAPI filter. Slides with deposited cells were stored at 4°C.
DNA preparation
A volume of 0.5 µl of a 2 % solution of CEK enzyme (BCG) in 1 × reaction buffer was pipetted to the reaction site of an AmpliGrid slide containing a single cell and covered immediately with 5 µl of sealing solution (BCB). After complete loading, the AmpliGrid slide was incubated for 2 h at 60°C on an AmpliSpeed slide cycler (BCB). Subsequently the sealing solution was removed by dipping the slide into a staining jar filled with hexane for 30 s, followed by drying of the remaining aqueous phase at 37°C on the AmpliSpeed slide cycler.
Methylation-sensitive enzymatic cleavage of DNA
A double digestion solution was prepared containing 0.25 U/µl NlaIV (BspLI) and 0.25 U/µl MspI, HpaII or Hin6I in 0.5 × Tango buffer. All restriction enzymes were purchased from Fermentas, St. Leon-Rot/Germany. For the PCR positive control a single digestion solution was prepared containing 0.25 U/µl NlaIV in 0.5 × Tango buffer. For each digestion, 0.5 µl of reaction mix was pipetted to an AmpliGrid reaction site containing a single cell and immediately covered with 5 µl of sealing solution. The slide was incubated for 6 h at 37°C on an AmpliSpeed slide cycler followed by a 30-min heat denaturation of the enzymes at 65°C. Subsequently, the sealing solution was removed by dipping the slide into a staining jar filled with hexane for 30 s followed by drying of the remaining aqueous phase at 45°C on the AmpliSpeed slide cycler.
Single-cell PCR
Primers were designed by the Beckman Coulter Biomedical GmbH primer design service (Advalytix Products, Munich, Germany) and purchased from MetaBion, Martinsried/Germany as 100 µM solutions in water. Primer mixes for the COL1A2 promoter: 0.8 µM long forward (5′-TTCGGCTAAGTTGGAGGTACTG-3′), 0.3 µM short forward (5′-CTAGACATGCTCAGCTTTGTGG-3′) and 0.3 µM reverse (5′-CTTACATTGGCATGTTGCTAGG-3′). CDKN2A gene: each 0.5 µM for long forward (5′-CTCTGGAGGACGAAGTTTGC-3′), short forward (5′-CTTCCTGGGGAGTTTTCAGA-3′) and reverse (5′-ATTCCTCTTCCTTGGCTTCC-3′). One microliter of each primer mix was pipetted to a reaction site with restriction-enzyme-treated cells and dried at 45°C on an AmpliSpeed slide cycler. Subsequently, 1.65 µl of PCR mix containing 1 × Multiplex Master Mix (Qiagen, Hilden/Germany) that contains HotStarTaq DNA Polymerase and 0.9x Q-solution were pipetted onto AmpliGrid’s reaction sites and covered immediately with 5 µl sealing solution. PCR program: initial denaturation at 95°C for 10 min, followed by 60 cycles of (30 s at 94°C, 1:15 min annealing at 60°C and 1:15 min elongation at 72°C), followed by a final 10-min elongation step at 72°C. After the amplification, 4 µl of loading dye (1.25×) was added to each reaction site of the AmpliGrid slide. After joining of aqueous phase and loading dye, 4 µl of each PCR product was transferred to an 8% PAGE gel (Anamed, Gross-Bieberau/Germany) and separated for 30 min at 250 V. Silver staining was performed with 0.1% AgNO3-solution for 5 min, followed by a 10 s washing step in DI water and development in 0.4 M NaOH/0.1% formaldehyde for 10 min.
Bisulfite sequencing
DNA from SW480 colorectal cancer cells and from whole blood was purified using the QIAamp mini kit and QIAamp DNA blood mini kit (Qiagen), respectively, according to the manufacturer's standard protocols. Bisulfite treatment was performed using a modified standard protocol after Clark et al. (12). Briefly, 500 ng genomic DNA was split into four reactions and separately fragmented by EcoRI, PvuII and HindIII (Fermentas). In the fourth reaction, DNA was sheared briefly (1–2 s) with an ultrasound sonicator. After heat inactivation of the enzymes, all reactions were combined and then purified using a MiniElute reaction cleanup kit (Qiagen) and eluted in 35 µl H2O. Bisulfite conversion was performed using an EpiTect Bisulfite Kit (Qiagen) according to the manufacturer’s manual and finally eluted in 20 µl EB buffer. Bisulfite-treated DNA was stored at −80°C until needed. 10 µl of each sample was used for bisulfite hot-start PCR in 25 μl volume reactions containing 1 μM primers, and a master mix containing TrueStart Taq polymerase (Fermentas). PCR program: Initial denaturation at 95°C for 2 min, followed by 35 cycles of (20 s at 94°C, 1:10 min annealing at 61.5°C and 1 min elongation at 72°C), followed by a final 10-min extension step at 72°C. CDKN2A bisulfite primer: F: 5′-GTAGTATGGAGTTTTYGGTTGATTG-3′; R: 5′-AAAAACTAAATAATCCCAACACATCTT-3′ (product size 498 bp). COL1A2 bisulfite primer: F: 5′-GGAGGTATTTTAGGGTTAGGGAAAT-3′; R: 5′-CAAACAAACTAAAAACACTTACATTAA-3′ (product size 295 bp). All PCR reactions were checked on a 1.0% agarose gel to ensure successful amplification and specificity before proceeding with sequencing. The amplification product of the expected size were cut from the gel and purified by QIAquick gel extraction kit (Qiagen). PCR products were diluted to 2 ng/µl and subjected to direct sequencing (Eurofins MWG Operon, Ebersberg/Germany).
Pyrosequencing
Primers were designed with Pyrosequencing Assay Design Software v1.0.6 (Biotage, Uppsala, Sweden). The CDKN2A locus was designed to span the region containing the same HpaII and Hin6I sites that were interrogated with RSMA. A full list of primer sequences can be found in Table S1. Bisulfite conversion of genomic DNA and pyrosequencing were performed as described previously (13). All PCR reactions were checked on a 1.0% agarose gel to ensure successful amplification and specificity before proceeding with pyrosequencing.
RESULTS AND DISCUSSION
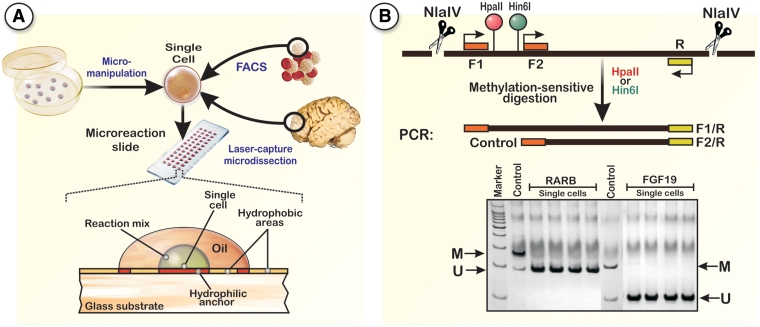
The proposed RSMA method uses a classical profiling approach with MSREs to interrogate methylation patterns at specific CpG dinucleotides. RSMA comprises four main steps (Figure 1): (i) deposition of single cells onto the microreaction sites; (ii) cell lysis and removal of DNA-bound proteins; (iii) methylation-sensitive restriction of DNA; and (iv) amplification of the interrogated DNA sequences. The final amplification is performed with two forward primers and one reverse primer (Figure 1B). The restriction sites for the MSREs are located between the two forward primers. In case the amplified DNA fragment is methylated, the restriction enzymes will not cleave, resulting in two amplification products. If the DNA is unmethylated, only the short amplification product is synthesized during the PCR. The resulting DNA products are finally separated on a gel, where the short band serves as positive PCR control. A large number of MSREs can be used with RSMA, enabling the interrogation of nearly every gene or promoter in a mammalian genome (1).
Figure 1.
Principle of the RSMA test system for the analysis of DNA methylation patterns in single cells. (A) Single cells from various sources are placed on high-throughput multi-well PCR slides (one cell per well), either by micromanipulation, laser capture microdissection or flow cytometry cell sorting. The protocol was tested with AmpliGrid microreaction slides, which are composed of a standard microscope slide sized glass surface with chemically structured, DNA free reaction centers. The reaction volume ranges from ∼500 nl up to 1.65 μl, small enough to sufficiently concentrate the reaction volume to a point where enzymatic steps proceed with high kinetics, but large enough to sufficiently dilute the cellular components (∼1 pl) of each cell. It is possible to perform 48 methylation-profiling reactions on one 75 mm × 25 mm slide. The structure of the slides allows carrying out methylation-sensitive PCRs from single cells that can be lysed directly on the reaction centers, thereby generating methylation profiles starting with DNA amounts as low as 6 pg. The hydrophilic and hydrophobic regions on the slide surface ensure that even after inaccurate pipetting the droplets containing single cells will find their way to the correct position. An important feature of this technology is the parallel analysis of multiple cells, a desirable feature that makes it possible to obtain statistically relevant methylation data on single cells in a reasonable time frame. (B) DNA is fragmented by a restriction enzyme (here NlaIV), that cleaves outside of the target region. Primer mixes contain two forward primers (F1 + F2) and one reverse primer (R). The analyzed CpG dinucleotides (here HpaII and Hin6I sites) are located between the two forward primers. If the restriction site is methylated, the restriction enzyme cannot cleave and both PCR products are synthesized, whereas for unmethylated restriction sites only the short PCR product (internal PCR positive control) is produced.
AmpliGrid slides, commonly used for single-cell PCR applications, were chosen as reaction platform (14). Each slide contains 48 independent hydrophilic reaction sites surrounded by a hydrophobic circle which holds aqueous reaction solutions, such as enzyme buffers, in place (Figure 1A). Cells were deposited on the slide surface by flow cytometry cell sorting. Before each experiment, the correct deposition of single cells on the reaction sites was verified with a microscope. The flatness of the glass substrate allows a simple optical control of the whole workflow using a microscope or a slide scanner. Lysis of single cells was performed with a protease solution (CEK) which digests proteins associated with DNA. A heat inactivation of CEK is not required as it is almost completely inactive under the conditions where most MSREs are predominantly active (37°C).The relatively low CEK reaction temperature (60°C) keeps the DNA double-stranded, a necessary prerequisite for the subsequent enzymatic cleavage. To prevent evaporation and cross-contamination, each reaction was covered with 5 µl of an oil-based sealing solution (see ‘Materials and Methods’ section). After the proteolysis, the sealing solution was removed and the aqueous droplets air-dried. The drying step is required to prevent a surplus dilution of the reaction mix in the methylation-sensitive cleavage and DNA fragmentation. Restriction enzymes should be added in 0.5 µl of 0.5 × restriction enzyme buffer, because higher buffer concentrations may result in inhibition of the following PCR reaction. Additionally, a reduced amplification efficiency using some restriction enzyme buffers was recognized (data not shown). Hence, the final PCR amplification is performed in a relatively high volume (1.65 µl) to dilute the reaction buffers of the preceding reactions as much as possible. Single-cell methylation-sensitive PCRs have to be highly efficient and sensitive as only one to two copies of the target sequence is present; hence, specific care has to be taken to avoid problems that may cause decreased amplification efficiency. One problem during amplification steps is the mixed hydrophobic–hydrophilic nature of biological macromolecules, which leads to their adsorption to a variety of surfaces; often the DNA molecules stick to reaction vessels, which may interfere with the amplification. This characteristic may result in a significant loss of sample or in the possible inaccessibility of certain DNA sequences for several initial PCR cycles and eventually becoming accessible only later during the amplification. Indeed, we found that an increase from a standard 30-cycle amplification to ∼60 cycles can improve the PCR performance significantly (data not shown). In expression studies, such cycle numbers may introduce distortions in the measured gene expression levels. However, when performing methylation analysis the high cycle number is not a disadvantage per se as the cell contains only one or two copies of each locus which is either methylated or not and no intermediate methylation levels exist. In theory, this information can be used to transform the methylation status of each CpG dinucleotide into a binary code for subsequent biostatistical analyses. Moreover, a large number of PCR cycles are essential since the amplification products are usually located within GC-rich regions that favor the formation of stable intramolecular hairpin structures interfering with the annealing of the primers. This is generally not a problem for conventional PCR with a high number of DNA templates, but can be crucial in single-cell PCR, where the amount of starting material is low. The occurrence of hairpin structures also requires special care in primer design for single-cell methylation profiling applications. To avoid primer sequences that are located in regions susceptible to hairpin structures in the chosen PCR conditions, the target regions were analyzed for their folding properties using the SirGraph package (Supplementary Data) (15). In our experience, primer annealing can also be enhanced by using a higher concentration of primers, which is often important for improving the amplification efficiency of the long PCR product. Another key component for a successful single-cell PCR is the complete denaturation of the DNA template at the beginning of the PCR. Incomplete denaturation of the DNA results in an inefficient utilization of template in the earliest amplification cycles leading to poor yield of PCR product or may interrupt amplification. Consequently, the initial denaturation should be performed over an interval of at least 5–8 min at 95°C if the GC-content of the desired DNA sequence is 50% or less. This interval should be extended up to 10 min for GC-rich templates such as gene promoters. In our experiments, the use of a hot-start DNA polymerase, i.e. HotStarTaq polymerase, which itself requires an initial denaturation step to be fully activated, resulted in good and sufficient amplification.
Single-cell profiling in human cancer cell lines and lymphocytes
RSMA can be applied with a variety of enzymes and protocols (e.g. real-time measurements and multiplexing, among others); however, to illustrate the overall approach, we demonstrate a simple protocol, that requires two very commonly used MSREs in epigenetics research, HpaII and Hin6I. A comprehensive list of potential restriction enzymes with genomic coverage for DNA methylation profiling can be found in Schumacher & Petronis 2006 (1). One rationale in using the classical profiling approach with MSREs is the potential to use the data and principles of single-cell methylation profiling derived from this study to develop further protocols with higher genomic coverage. Indeed, the interest in MSREs is now resurging as these enzymes are the key tools for large-scale epigenomic profiling using microarrays (1) and next-generation sequencing (NGS) approaches (16), which ultimately may lead to the development of single-cell whole methylome technologies.
To validate the RSMA method, restriction sites located in the promoter region of two genes (COL1A2 and CDKN2A) were analyzed in the human colorectal cancer cell line SW480 and in human lymphocytes. In initial screenings, the FGF19 and RARB gene promoters were also analyzed; however, we detected only unmethylated CpGs in the interrogated cells (Figure 1B). Hence, we focused our analysis on the well-characterized collagen type I alpha 2 (COL1A2) and cyclin-dependent kinase inhibitor 2A (CDKN2A) genes, which are known to exhibit cancer- and tissue-specific methylation patterns. A number of reports have demonstrated the silencing of COL1A2, a candidate tumor suppressor gene, due to aberrant methylation within the promoter region in primary cancer tissues and in several cancer cell lines (17,18). Similarly, CDKN2A, a known tumor suppressor gene, is either mutated or deleted in a wide range of cancers and in addition has been found to be silenced by CpG methylation in many tumor types (19–21).
To assess the restriction enzyme efficiency in the chosen model system, DNA from single cells was also cleaved with MspI, an isoschizomer of HpaII, which cuts independent of the CpGs methylation status. If a digestion was successful, amplification of the interrogated regions should only produce the short amplification product (Figure 1B). All MSRE cleavage reactions were performed as double digests together with NlaIV (5′-GGN^NCC-3′). The main purpose of NlaIV, which cleaves independent of methylation status, is the fragmentation of the genomic DNA. Without fragmentation, we observed a significantly decreased processivity for the long PCR product of several different genes (data not shown). In our experimental setup, NlaIV does not cleave within the amplified DNA sequence, but close to the primer annealing sites outside the long PCR products (Figure 1B). As positive control for PCR efficiency, only a single digestion with NlaIV was performed.
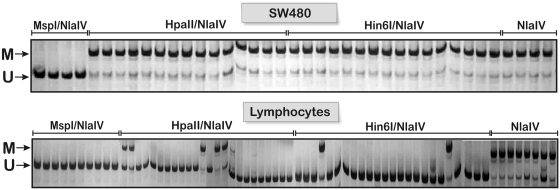
In total, we analyzed 429 single cells for COL1A and CDKN2A (Figure 2). In 1.5% of the amplification reactions using the SW480 cells, and in 5% of reactions with lymphocytes, the PCR yielded no product (on average 3% of all amplifications).
Figure 2.
Polyacrylamide gel electrophoresis of products from multiple parallel single-cell PCRs as exemplified for the CDKN2A promoter. Each lane represents a PCR product from a single cell in SW480 cells (top row) and lymphocytes (bottom row). All of the SW480 cells in the presented slide contained only fully methylated HpaII and Hin6I sites, indicated by the presence of two bands, the uncleaved (methylated) long band (191 bp, F1/R) and the short control band (157 bp, F2/R). In contrast, most of the lymphocytes contained only the short band, indicating that the analyzed CpG dinucleotides were unmethylated. Some cells were treated with MspI/NlaIV as control for the enzymatic reaction and should produce only the short band. In parallel, some cells were treated with NlaIV alone, which serves as positive control for the long PCR product. If the chosen PCR conditions are appropriate, both amplification products should be visible. Typically, the short control band is noticeable weaker compared to the long band, which may be explained by the fact that in later PCR cycles the short band may also serve to a significant extent as primer for the long PCR product. In addition, due to better hybridization efficiency, the longer PCR product may be amplified preferentially. M, methylated; U, unmethylated.
On average, our analysis showed that in 91% of the SW480 cells the analyzed HpaII restriction site within the COL1A2 promoter was methylated (Supplementary Figure S2). Comparable methylation percentages were obtained for the Hin6I site, where in 87% of the cells CpG methylation was observed. The opposite pattern was found in lymphocytes, with most cells being unmethylated. Only 5% of the analyzed sites were methylated. These numbers are in agreement with published data for SW480 cells, which were reported to be hypermethylated (>80% methylation) at the +7 site in the COL1A2 promoter compared to non-cancer tissues (22). This increased methylation of the collagen gene in colorectal cancer cells was inversely correlated with collagen messenger RNA (mRNA) steady-state levels. Similarly, in our experiment, the CDKN2A promoter was found to be almost completely methylated in SW480 cells (99%), but relatively hypomethylated in lymphocytes (21%). Again, our results are in agreement with previous studies, which reported methylation levels of the CDKN2A promoter between 92% and 100% in SW480 cells and no significant methylation in blood cells (20,23–25). Additionally, using a methylation-specific polymerase chain reaction followed by analysis of the methylated cytosine content of the product by thermal denaturation, a recent study by Kovatsi and co-workers (26) also described a partial methylation of the CDKN2A promoter in peripheral blood samples from healthy individuals.
Overall, the single-cell data show the expected significant difference between the SW480 cancer cells and lymphocytes. It is important to note that relatively similar methylation levels were obtained for the closely spaced HpaII and Hin6I restriction sites within the same cell type and the same promoter region. This finding indicates that the methylation status of each CpG dinucleotide within the same promoter may be representative for the methylation status of surrounding CpG sites. This finding is significant as only a subset of CpGs in CpG islands is recognized by specific methylation sensitive restriction enzymes and therefore suitable for methylation profiling.
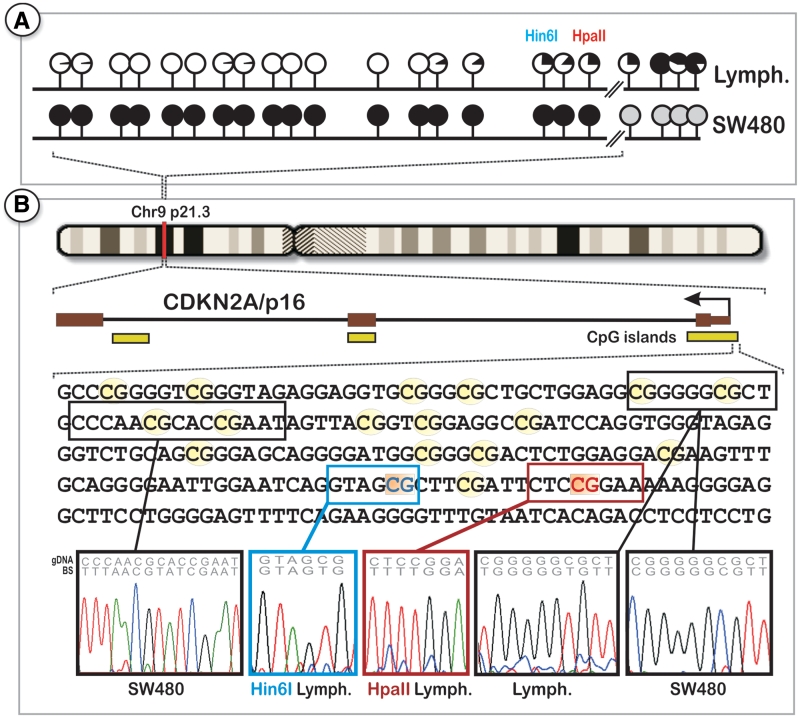
To validate our RSMA results, we performed direct bisulfite sequencing on the CDKN2A and COL1A2 promoter regions as well as quantitative pyrosequencing of the downstream CDKN2A HpaII and Hin6I sites. As exemplified for the CDKN2A promoter, direct sequencing of PCR products from bisulfite-treated DNA revealed hypermethylation (>98%) of both gene promoters in SW480 cells, whereas both CpG islands revealed to be hypomethylated in lymphocytes (Figure 3). Additionally, pyrosequencing of the 3′ border of the CDKN2A promoter in additional lymphocyte DNA samples revealed residual DNA methylation level (up to 18%), confirming our RSMA data (Supplementary Figure S3). These results underscore the power of the RSMA approach, as more detailed methylation data can be produced compared to direct sequencing. The observed variation of methylation patterns between individual cells of the same cell type illustrates that a certain stochastic variation exists, which would be masked by conventional bulk measurements. Bisulfite sequencing may not be the first choice for single-cell methylation profiling because treatment with bisulfite significantly degenerates the four nucleotide code (1), which results in the loss of specificity of a large portion of the genome and increases the chance of cross hybridization of PCR primers. Additionally, due to the harsh conditions, a certain percentage of DNA degrades during bisulfite conversion, a decisive disadvantage when working with a limited amount of DNA. As a consequence, conventional bisulfite sequencing requires several hundred or even thousands of cells in order to obtain a sufficient amount of DNA for analysis. Obtaining this number of cells of a single type is challenging, particularly when attempting to study rare stem cells such as tumor-forming cancer stem cells (CSCs) (27,28). Therapeutic innovations may emerge from a better understanding of the epigenetic mechanisms of CSCs. The high-throughput RSMA technology has the potential to transform such single-cell methylation analyses from a major technological challenge to a routine procedure for research and clinical diagnostics.
Figure 3.
Bisulfite sequencing of larger cell populations to verify the results from single-cell measurements. (A) Direct sequencing of the CDKN2A core promoter region revealed hypermethylation of all of the analyzed CpG dinucleotides within the promoter CpG island in colorectal cancer cells, whereas no significant methylation in lymphocytes could be detected. The small methylation level in the HpaII and Hin6I sites can be explained by their location at the CpG island downstream border, after which methylation levels increase. The black areas of the lollipops represent the average degree of methylation for each CpG dinucleotide, determined by semiquantitative measurement of peak height for each nucleotide from multiple bisulfite sequence electropherograms. Gray lollipops = no sequencing data. (B) Structure and analyzed sequence of the CDKN2A gene. Sample sequencing traces from different tissues are shown on the bottom. The bisulfite sequencing results confirm the methylation data derived from single-cell methylation profiling by the RSMA technology.
Critical parameters for single-cell methylation profiling
In our experiments, we studied several other important parameters that may prove crucial in single-cell analyses. For example, in rare cases, we observed incomplete digestion of the DNA, most likely due to aged cells that were stored on the microreaction sites for longer periods. Usually, cells can be stored on the reaction sites for several months; however, we found that cells stored for more than 3 months increased the rate of failed reactions (e.g. incomplete digestion or repressed amplification) significantly. For example, control digestion with MspI, which should always produce only the small PCR product, failed in ∼7% of experiments when using old cells. Similarly, NlaIV digests failed in about 9% of old cells, whereas incomplete digestion was nearly absent in freshly prepared cells. In general, DNA methylation patterns are known to be stable in post-mortem tissues (29) and storage of cells on the slides should not affect the DNA methylation patterns as the cells are dried and no enzymatic activity is to be expected.
The main reason for the lack of technologies to study single-cell methylation patterns can be attributed to the poor yields encountered when subjecting mammalian cells to multiple enzymatic or chemical processes. In our experience, to circumvent these problems, it is advantageous to perform all experimental steps in a single reaction vessel, without transfer of the components. Ideally, to avoid loss of DNA, pumping (circulation) of solutions should be avoided. So far, researchers focusing on epigenetic profiling of a limited number of cells rely on microplates or standard reaction tubes for their assays. As a result, large dead volumes, template adsorption and no optical quality control are responsible for a considerable uncertainty and low reproducibility. We found that using microreaction slides could overcome all of these problems, whereas using standard reaction vessels, such as 0.5 ml PCR tubes resulted in inconsistent or completely failed enzymatic reactions (data not shown). For kinetic reasons, it is advantageous to perform enzymatic reactions in very small volumes, but, on the other hand, large-enough dimensions (>0.5 µl) to allow for reaction components to be easily transferred or added between reaction steps in a standard laboratory setup.
Another crucial step is the complete lysis of the cell and removal of unwanted proteins before enzymatic reactions take place, primarily to avoid protein interference. This is important because an individual cell’s total cellular protein content is quite high, averaging 8 × 109 molecules per cell, which is about equal to 700 pg (3). Many of the proteins, especially histone and transcription factors, are bound to the DNA and hence can interfere with analysis. The small absolute amount of DNA and the low DNA concentration compared to cellular proteins presents significant challenges for detection. Efficient proteolytic steps have to be applied to ensure that the minute amount of DNA can be enzymatically processed and amplified by PCR. In our experience, a 2-h treatment with CEK protease (see ‘Material and Methods’ section) was sufficient to lyse the cell and free the DNA from proteins for subsequent enzymatic treatment.
Another very crucial step in single-cell methylation profiling is the very quick transfer of the freshly prepared cell to the reaction site, followed by immediate drying or lysis of the cell. The act of placing a cultured (adherent) or microdissected cell into suspension for transfer into the analytical devices may damage the cell membrane and activates various signaling or apoptotic pathways. This in turn may disturb the epigenetic machinery of the cell, ultimately resulting in technically biased methylation patterns. For example, embryonic stem (ES) cells are easily affected by culture conditions and in vitro manipulation, which could in turn result in increased variability in the methylation and transcription patterns of the ES cell genome. Indeed, previous work on ES cells has shown that stem cell-derived tissues and embryos often fail to maintain stable epigenetic states (30,31). Often, epigenetic instability is reported in differentially methylated regions of mostly growth-related imprinted genes, but less often neuronal genes (32). The tendency for living cells to be perturbed by in vitro manipulation imposes stringent requirements in performing a successful biological experiment. Changes in the cell’s environment such as nutrients, pH, ionic strength and temperature can lead to variation in the intracellular concentrations of many molecular species involved in methylation homeostasis. Furthermore, epigenetic patterns are in a constant state of flux, e.g. cyclical methylation and demethylation of CpG dinucleotides at specific loci may occur within minutes (33). Hence, in some single-cell studies, careful consideration of cell-cycle status and transfer time to the reaction site for quick drying are warranted.
In contrast to expression studies, DNA methylation analysis of single cells are even more sensitive to contamination, as a single foreign DNA molecule could significantly alter the result, whereas a single RNA molecule would—in most cases—not dramatically alter the measured expression levels. Given the potential for contamination from even very small amounts of DNA, a useful microreaction system will likely need to be single use.
Another difficulty when studying single diploid cells comes from heterozygous DNA methylation that can be found, for instance, in imprinted genes where each allele can be differentially methylated depending on its parental origin. Depending on the applied protocol, assessing the exact methylation pattern can be challenging. For example, in our protocol, if only one allele is methylated, the resulting banding patterns in the gel will look similar to fully methylated sequences (two PCR bands), only with a higher intensity of the low-molecular PCR product. Consequently, to confirm heterozygous methylation, the resulting amount of each PCR product would have to be carefully measured, such as by quantitative real-time PCR (qPCR).
An additional caveat for single-cell approaches is that stochastic biological events or even the chance analysis of a very rare cell can create noise which may confound observations. Hence, single-cell methylation profiling must retain the capacity to perform population statistics (therefore a need for high-throughput). Although the general level of cell-to-cell epigenetic variation in mammals is still largely unknown, it can be expected that the cell’s epigenetic machinery is inherently noisy. It could be shown that significant differences in gene expression levels exist between phenotypically identical cells in vivo, and that these differences exceed any noise contribution from global mRNA amplification (34). Due to the highly parallelized methodology, RSMA also offers the opportunity to study stochastic influences in methylation dynamics, an important component of epigenetic drift (1,35,36). Indeed, it was suggested that a substantial portion of phenotypic variance in disease and aging, traditionally attributed to environmental effects, may in fact result from stochastic epigenetic events in the cell (1). By studying methylation patterns in multiple single cells (up to 48 cells per slide) under controlled environmental conditions, quantification of stochastic events becomes possible. The typical channel dimensions found in microfluidic devices (10–100 μm) and the ability to manipulate nanoliters of reagents on-chip have made these devices encouraging platforms for the analysis of single cells. However, most of these systems do not possess a high-throughput capacity or they are too expensive for the daily use in most laboratories. With our proposed system, high-throughput as well as cost efficiency can be ensured.
CONCLUSIONS
In summary, we demonstrated an affordable technology to assess the methylation patterns of CpG dinucleotides in single cells. RSMA offers the opportunity for high-throughput screening, as many single cells can be screened in parallel. This method may prove to be especially useful for clinical studies, as the procedure is straightforward and does not require complex microfluidics devices. By coupling RSMA with micromanipulation, laser capture microdissection, or flow cytometry cell sorting, the heterogeneous nature of tumors, neurodegeneration and other complex disorders can be investigated at the single-cell level. Overall, it is argued that single-cell epigenetic strategies such as RSMA, when applied in parallel with the traditional genetic ones, may significantly advance the discovery of etiopathogenic mechanisms of complex diseases and stochastic epigenetic variance.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This work was supported by A.S. and by Beckman, which had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript. Funding for open access charge: Waived by Oxford University Press.
Conflict of interest statement. M.A.-F., P.H., M.K. and R.K. are employees of Beckman Coulter Biomedical GmbH.
ACKNOWLEDGEMENTS
We thank Gabriel Oh and Catherine Ng for the help with Pyrosequencing.
REFERENCES
- 1.Schumacher A, Petronis A. Epigenetics of complex diseases: from general theory to laboratory experiments. Curr. Top. Microbiol. Immunol. 2006;310:81–115. doi: 10.1007/3-540-31181-5_6. [DOI] [PubMed] [Google Scholar]
- 2.Le TT, Harlepp S, Guet CC, Dittmar K, Emonet T, Pan T, Cluzel P. Real-time RNA profiling within a single bacterium. Proc. Natl Acad. Sci. USA. 2005;102:9160–9164. doi: 10.1073/pnas.0503311102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Sims CE, Allbritton NL. Analysis of single mammalian cells on-chip. Lab Chip. 2007;7:423–440. doi: 10.1039/b615235j. [DOI] [PubMed] [Google Scholar]
- 4.Cluzel P, Surette M, Leibler S. An ultrasensitive bacterial motor revealed by monitoring signaling proteins in single cells. Science. 2000;287:1652–1655. doi: 10.1126/science.287.5458.1652. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson GR. Drug metabolism and variability among patients in drug response. N. Engl. J. Med. 2005;352:2211–2221. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 6.Lee YH, Chen SY, Wiesner RJ, Huang YF. Simple flow cytometric method used to assess lipid accumulation in fat cells. J. Lipid Res. 2004;45:1162–1167. doi: 10.1194/jlr.D300028-JLR200. [DOI] [PubMed] [Google Scholar]
- 7.Kerjean A, Vieillefond A, Thiounn N, Sibony M, Jeanpierre M, Jouannet P. Bisulfite genomic sequencing of microdissected cells. Nucleic Acids Res. 2001;29:E106–E106. doi: 10.1093/nar/29.21.e106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Easley CJ, Karlinsey JM, Bienvenue JM, Legendre LA, Roper MG, Feldman SH, Hughes MA, Hewlett EL, Merkel TJ, Ferrance JP, et al. A fully integrated microfluidic genetic analysis system with sample-in-answer-out capability. Proc. Natl Acad. Sci. USA. 2006;103:19272–19277. doi: 10.1073/pnas.0604663103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toriello NM, Douglas ES, Thaitrong N, Hsiao SC, Francis MB, Bertozzi CR, Mathies RA. Integrated microfluidic bioprocessor for single-cell gene expression analysis. Proc. Natl Acad. Sci. USA. 2008;105:20173–20178. doi: 10.1073/pnas.0806355106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schumacher A, Kapranov P, Kaminsky Z, Flanagan J, Assadzadeh A, Yau P, Virtanen C, Winegarden N, Cheng J, Gingeras T, et al. Microarray-based DNA methylation profiling: technology and applications. Nucleic Acids Res. 2006;34:528–542. doi: 10.1093/nar/gkj461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Leibovitz A, Stinson JC, McCombs WB, III, McCoy CE, Mazur KC, Mabry ND. Classification of human colorectal adenocarcinoma cell lines. Cancer Res. 1976;36:4562–4569. [PubMed] [Google Scholar]
- 12.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic Acids Res. 1994;22:2990–2997. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mill J, Tang T, Kaminsky Z, Khare T, Yazdanpanah S, Bouchard L, Jia P, Assadzadeh A, Flanagan J, Schumacher A, et al. Epigenomic profiling reveals DNA-methylation changes associated with major psychosis. Am. J. Hum. Genet. 2008;82:696–711. doi: 10.1016/j.ajhg.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.May A, Kirchner R, Muller H, Hartmann P, El Hajj N, Tresch A, Zechner U, Mann W, Haaf T. Multiplex rt-PCR expression analysis of developmentally important genes in individual mouse preimplantation embryos and blastomeres. Biol. Reprod. 2009;80:194–202. doi: 10.1095/biolreprod.107.064691. [DOI] [PubMed] [Google Scholar]
- 15.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31:3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lister R, Ecker JR. Finding the fifth base: genome-wide sequencing of cytosine methylation. Genome Res. 2009;19:959–966. doi: 10.1101/gr.083451.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chiba T, Yokosuka O, Fukai K, Hirasawa Y, Tada M, Mikata R, Imazeki F, Taniguchi H, Iwama A, Miyazaki M, et al. Identification and investigation of methylated genes in hepatoma. Eur. J. Cancer. 2005;41:1185–1194. doi: 10.1016/j.ejca.2005.02.014. [DOI] [PubMed] [Google Scholar]
- 18.Mori K, Enokida H, Kagara I, Kawakami K, Chiyomaru T, Tatarano S, Kawahara K, Nishiyama K, Seki N, Nakagawa M. CpG hypermethylation of collagen type I alpha 2 contributes to proliferation and migration activity of human bladder cancer. Int. J. Oncol. 2009;34:1593–1602. doi: 10.3892/ijo_00000289. [DOI] [PubMed] [Google Scholar]
- 19.Merlo A, Herman JG, Mao L, Lee DJ, Gabrielson E, Burger PC, Baylin SB, Sidransky D. 5′ CpG island methylation is associated with transcriptional silencing of the tumour suppressor p16/CDKN2/MTS1 in human cancers. Nat. Med. 1995;1:686–692. doi: 10.1038/nm0795-686. [DOI] [PubMed] [Google Scholar]
- 20.Mund C, Beier V, Bewerunge P, Dahms M, Lyko F, Hoheisel JD. Array-based analysis of genomic DNA methylation patterns of the tumour suppressor gene p16INK4A promoter in colon carcinoma cell lines. Nucleic Acids Res. 2005;33:e73. doi: 10.1093/nar/gni072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zheng S, Chen P, McMillan A, Lafuente A, Lafuente MJ, Ballesta A, Trias M, Wiencke JK. Correlations of partial and extensive methylation at the p14(ARF) locus with reduced mRNA expression in colorectal cancer cell lines and clinicopathological features in primary tumors. Carcinogenesis. 2000;21:2057–2064. doi: 10.1093/carcin/21.11.2057. [DOI] [PubMed] [Google Scholar]
- 22.Sengupta PK, Smith EM, Kim K, Murnane MJ, Smith BD. DNA hypermethylation near the transcription start site of collagen alpha2(I) gene occurs in both cancer cell lines and primary colorectal cancers. Cancer Res. 2003;63:1789–1797. [PubMed] [Google Scholar]
- 23.Herman JG, Merlo A, Mao L, Lapidus RG, Issa JP, Davidson NE, Sidransky D, Baylin SB. Inactivation of the CDKN2/p16/MTS1 gene is frequently associated with aberrant DNA methylation in all common human cancers. Cancer Res. 1995;55:4525–4530. [PubMed] [Google Scholar]
- 24.Chim CS, Fung TK, Wong KF, Lau JS, Law M, Liang R. Methylation of INK4 and CIP/KIP families of cyclin-dependent kinase inhibitor in chronic lymphocytic leukaemia in Chinese patients. J. Clin. Pathol. 2006;59:921–926. doi: 10.1136/jcp.2005.035089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nagy E, Veress G, Szarka K, Csoma E, Beck Z. Frequent methylation of p16INK4A/p14ARF promoters in tumorigenesis of Epstein–Barr virus transformed lymphoblastoid cell lines. Anticancer Res. 2005;25:2153–2160. [PubMed] [Google Scholar]
- 26.Kovatsi L, Georgiou E, Ioannou A, Haitoglou C, Tzimagiorgis G, Tsoukali H, Kouidou S. p16 promoter methylation in Pb2+-exposed individuals. Clin. Toxicol. (Phila) 2010;48:124–128. doi: 10.3109/15563650903567091. [DOI] [PubMed] [Google Scholar]
- 27.Gupta PB, Chaffer CL, Weinberg RA. Cancer stem cells: mirage or reality? Nat. Med. 2009;15:1010–1012. doi: 10.1038/nm0909-1010. [DOI] [PubMed] [Google Scholar]
- 28.Rosen JM, Jordan CT. The increasing complexity of the cancer stem cell paradigm. Science. 2009;324:1670–1673. doi: 10.1126/science.1171837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ernst C, McGowan PO, Deleva V, Meaney MJ, Szyf M, Turecki G. The effects of pH on DNA methylation state: in vitro and post-mortem brain studies. J. Neurosci. Methods. 2008;174:123–125. doi: 10.1016/j.jneumeth.2008.06.027. [DOI] [PubMed] [Google Scholar]
- 30.Humpherys D, Eggan K, Akutsu H, Hochedlinger K, Rideout WM, III, Biniszkiewicz D, Yanagimachi R, Jaenisch R. Epigenetic instability in ES cells and cloned mice. Science. 2001;293:95–97. doi: 10.1126/science.1061402. [DOI] [PubMed] [Google Scholar]
- 31.Mann MR, Chung YG, Nolen LD, Verona RI, Latham KE, Bartolomei MS. Disruption of imprinted gene methylation and expression in cloned preimplantation stage mouse embryos. Biol. Reprod. 2003;69:902–914. doi: 10.1095/biolreprod.103.017293. [DOI] [PubMed] [Google Scholar]
- 32.Schumacher A, Doerfler W. Influence of in vitro manipulation on the stability of methylation patterns in the Snurf/Snrpn-imprinting region in mouse embryonic stem cells. Nucleic Acids Res. 2004;32:1566–1576. doi: 10.1093/nar/gkh322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kangaspeska S, Stride B, Metivier R, Polycarpou-Schwarz M, Ibberson D, Carmouche RP, Benes V, Gannon F, Reid G. Transient cyclical methylation of promoter DNA. Nature. 2008;452:112–115. doi: 10.1038/nature06640. [DOI] [PubMed] [Google Scholar]
- 34.Subkhankulova T, Gilchrist MJ, Livesey FJ. Modelling and measuring single cell RNA expression levels find considerable transcriptional differences among phenotypically identical cells. BMC Genomics. 2008;9:268. doi: 10.1186/1471-2164-9-268. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fraga MF, Ballestar E, Paz MF, Ropero S, Setien F, Ballestar ML, Heine-Suner D, Cigudosa JC, Urioste M, Benitez J, et al. Epigenetic differences arise during the lifetime of monozygotic twins. Proc. Natl Acad. Sci. USA. 2005;102:10604–10609. doi: 10.1073/pnas.0500398102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang S-C, Oelze B, Schumacher A. Age-specific epigenetic drift in late-onset Alzheimer's disease. PLoS ONE. 2008;3:e2698. doi: 10.1371/journal.pone.0002698. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.