Abstract
Plasmid pCXC100 from the Gram-positive bacterium Leifsonia xyli subsp. cynodontis uses a type Ib partition system that includes a centromere region, a Walker-type ATPase ParA and a centromere-binding protein ParB for stable segregation. However, ParB shows no detectable sequence homology to any DNA-binding motif. Here, we study the ParB centromere interaction by structural and biochemical approaches. The crystal structure of the C-terminal DNA-binding domain of ParB at 1.4 Å resolution reveals a dimeric ribbon–helix–helix (RHH) motif, supporting the prevalence of RHH motif in centromere binding. Using hydroxyl radical footprinting and quantitative binding assays, we show that the centromere core comprises nine uninterrupted 9-nt direct repeats that can be successively bound by ParB dimers in a cooperative manner. However, the interaction of ParB with a single subsite requires 18 base pairs covering one immediate repeat as well as two halves of flanking repeats. Through mutagenesis, sequence specificity was determined for each position of an 18-bp subsite. These data suggest an unique centromere recognition mechanism by which the repeat sequence is jointly specified by adjacent ParB dimers bound to an overlapped region.
INTRODUCTION
Stable transmission of duplicated genetic material to daughter cells is essential for all life forms. The active partitioning of low-copy-number plasmids has served as a simple model system for DNA segregation studies (1–5). Plasmid partition systems are composed of only three elements: a cis-acting centromere-like site called parS; a filament-forming NTPase, ParA; and a centromere-binding protein (CBP), ParB. These elements are normally arranged as a par operon, with parB located downstream of parA. The centromere region typically consists of multiple direct or inverted repeats near the par operon. Multiple ParB proteins assemble on the centromere repeats to form a high-order nucleoprotein complex, the partition complex. The partition complex recruits ParA to form the segrosome. ParA can polymerize into filaments in an NTP dependent manner. The dynamics of partition filaments is important to separate the attached plasmids into the opposite poles of dividing cells, but the underlying molecular mechanism remains unclear (1–5).
There are two main plasmid partitioning systems, distinguished from each other on the basis of whether the NTPase is a Walker-type (type I) or actin-type (type II) protein (6). The type I partition system is subdivided into types Ia and Ib according to different structure of ParA and ParB, and the relative position of the centromere (6). The centromere region is located downstream of the par loci in the type Ia system but is located upstream in the type Ib and II systems. Transcription of the par operon is under autoregulation by the Par proteins. Type Ia ParA is larger than type Ib ParA because it has an additional N-terminal helix–turn–helix (HTH) domain that binds the promoter region of the par locus to repress transcription. In type Ib and II systems, ParB associates with the centromere sequence located upstream of the par operon to repress transcription. Type III partition systems were recently characterized that are composed of a tubulin-like GTPase TubZ and a CBP TubR with a dimeric winged HTH motif (7–11).
Type Ia CBPs have a complex multidomain organization, but they share a conserved HTH DNA-binding motif (12–14). In contrast, type Ib and II CBPs are extremely divergent in their primary sequences, obscuring recognition of their DNA binding domain. The structures of two type Ib CBPs—omega and ParG—have been determined (15–17). Each contains a dimeric ribbon–helix–helix (RHH) DNA-binding domain and a flexible N-terminal domain. The RHH motif was also found in the structures of type II CBPs pB171 ParR and pSK41 ParR (18,19). Although type Ib and II CBPs specifically interact with Walker- and actin-type NTPase, respectively, they appear to share a common DNA-binding structure. CBPs need to function in the context of the partition complex. A type Ib omega–centromere complex shows an extended structure (17), whereas a type II pSK41 ParR–centromere complex forms a superhelical structure (18). The different higher order structures of the partition complex may impact the recruitment of specific ParA filament.
pCXC100 is a 51-kb plasmid in Gram-positive bacterium Leifsonia. xyli subsp. cynodontis (20). We previously identified a partition cassette in pCXC100, in which ParA has 317 residues and is homologous to type Ib Walker-type NTPase, but its downstream orf4 is not homologous to any known CBP (20). We recently showed that orf4 is essential for plasmid stability and its protein product specifically binds to a ∼90-bp region upstream of the parA gene (21). We concluded that orf4 encodes a bona fide CBP ParB and its binding target constitutes the centromere parS site. The Walker-type ParA and the upstream location of parS in the par locus suggest that the partition cassette in pCXC100 belongs to type Ib. However, the DNA-binding motif of pCXC100 ParB is unknown, raising the question of how it is related to the previously characterized type Ib ParB.
In this study, we applied X-ray crystallography to determine the structure of the C-terminal DNA-binding domain of pCXC100 ParB, hereafter referred to as ParB, and revealed that it forms a dimeric RHH fold. We also extensively characterized the interaction of ParB with the centromere by hydroxyl radical footprinting, electrophoretic mobility shift assays (EMSA) and isothermal titration calorimetry (ITC). We have mapped the binding sites of ParB on the centromere and divided the structure of the centromere into nine direct repeats. We also characterized in detail how ParB recognizes single subsites and binds cooperatively to tandem subsites. Interestingly, a ParB dimer binds to an extended 18-bp region of single subsites and recognizes the sequence of the immediate repeat as well as flanking repeats.
MATERIALS AND METHODS
Gene expression and protein purification
The ParB gene (residues 1–139) was PCR-amplified from plasmid YB411 (21) with primers WTF1 (5′-ATATACCATGGCTGATCGCACGGTTGC-3′; the restriction site is underlined) and WTR139 (5′-CCGCAAGCTTGCTTCCCAGTGGGCGCCCG-3′) and cloned into pET28a using NcoI and HindIII sites. The full-length ParB protein contains a non-cleavable six-His-tag at the C-terminus encoded in the plasmid. ParB fragments spanning residues 60–139, 65–139 and 65–135 were generated with primers PF60 (5′-GCCCGAAGCTTCGGAGCCCCGAGGGGCGCG-3′) and PR139 (5′-CCGGAATTCTTAGCTTCCCAGTGGGCGCCCGC-3′), PF65 (5′-GCCCGAAGCTTCCGCGCGTTCTGAGGTCAAGAT-3′) and PR139 and PF65 and PR135 (5′-CCGGAATTCTTAGCGCCCGCGAGTAACGCCTCG-3′), respectively. The fragments were cloned into a modified pET-Duet1 plasmid (Novagen) using HindIII and EcoRI sites, in which ParB was fused to an upstream six-His-tagged DsbA with a PreScission-cleavable linker.
All proteins were expressed in Escherichia coli BL21-Gold (DE3) induced with 0.2 mM isopropyl β-d-1-thiogalactopyranoside at 16°C. Harvested cells were resuspended in buffer P300 (300 mM KCl and 50 mM phosphate, pH 7.6) and lysed by sonication. Cell lysate was clarified by centrifugation and loaded onto a HisTrap column (GE Healthcare). After a wash with 25 mM imidazole in P300, bound protein was eluted with 500 mM imidazole in P300 and pooled. The six-His-DsbA tag was cleaved from the ParB fragments by PreScission protease. ParB was loaded onto a heparin column (GE Healthcare) and eluted at ∼300 mM KCl in a gradient from 50 to 1000 mM KCl in buffer H (20 mM HEPES, pH 7.6). The protein was further purified with a Superdex 200 gel filtration column in a buffer containing 5 mM HEPES-K, pH 7.6, and 100 mM KCl. The protein monomer concentration was measured by its absorbance at 280 nm using a molar extinction coefficient of 2980 M−1 cm−1 for all ParB constructs. This value was calculated on the basis of amino acid composition. The protein was concentrated to 20 mg/ml in buffer containing 5 mM HEPES-K, pH 7.6, and 100 mM KCl and stored at −80°C as aliquots. The molar concentration of ParB protein is expressed for its dimeric form.
Limited proteolysis
The limited digestion reactions were set up with 24 μg of the full-length ParB protein and 0, 0.004, 0.016 or 0.064 μg trypsin (Sigma) in a 20 μl volume containing 5 mM HEPES-K, pH 7.6, 5 mM MgCl2 and 100 mM KCl. For the DNA protection experiment, ParB was preassembled with 10 μg R4–18 DNA for 30 min before digestion. The digestion reactions proceeded at 20°C for 24 h. Half of each sample was separated on a 4–20% gradient SDS denaturing gel and stained with Coomassie blue. Half of each of the DNA-bound sample was also analyzed on a 5% native polyacrylamide gel and strained with ethidium bromide. For mass spectroscopy identification, the band corresponding to the major cleavage product was excised, in-gel digested by trypsin and analyzed with Q-Star (ABI).
Crystallization and structure determination
The crystal of the free ParB RHH domain was obtained during crystallization of ParB–DNA complexes. ParB 65–135 was assembled with DNA R4–18 in a 1:1 molar ratio and concentrated to 10 mg/ml in 5 mM HEPES-K (pH 7.6), 5 mM MgCl2 and 100 mM KCl. The DNA complex was incubated with 0.1% (w/w) trypsin on ice for 2 h prior to crystallization to trim disorder regions (22). Crystals were grown by vapor diffusion at 20°C in a hanging drop containing 1 μl of the ParB–DNA complex and 1 μl of mother solution containing 0.01 M MgCl2, 2.4 M (NH4)2SO4, 0.05 M MES, pH 5.6. The crystal was cryoprotected with 30% glycerol in the mother solution and flash-frozen in liquid nitrogen. To make a gold derivative, the crystal was soaked in the mother liquor supplemented with 10 mM KAu(CN)2 for 12 h. This structure was initially solved using the single isomorphous replacement with anomalous scattering method (SIRAS) based on a 1.7 Å native dataset and a 3.0 Å derivative dataset collected at an in-house generator (Rigaku), and revealed only the free protein. We later obtained a better crystal of free protein during the screen of ParB 65–139 and DNA complex. This crystal grew from 1.8 M Li2SO4, 0.01 M MgSO4, 0.05 M cacodylate-Na and pH 6.0, diffracted to 1.4 Å at Shanghai Synchrotron Radiation Facility (SSRF) beamline BL17U1, and provided a dataset for final refinement.
Diffraction data were reduced using Denzo and Scalepack in-house or HKL2000 at the synchrotron (23). Heavy atoms were detected by SHELXD (24). The phases were calculated, refined and solvent modified in SHARP (25). ARP/wARP was used for automated model building (26). The model was built in COOT (27) and refined in Refmac (28). The current model contains two ParB chains containing residues 69–128 and 70–128, respectively, 10 sulfate ions and 162 water molecules. The Ramachandran plot indicates that 100% of the residues are in a favored region.
Hydroxyl radical footprinting assay
Hydroxyl radical footprinting was performed as previously described (29). For 3′-labeling of the forward strand, a 170-bp centromere DNA sequence was amplified by PCR from a plasmid containing the whole par operon (20,21) with primers ParS3141-F (5′-CTAGAGCAAAGGACCCACCGACCG-3′) and ParS3310EcoRI-R (5′-GGAATTCTCTGGAATTTCCAGCCCCGC-3′). The PCR product was cleaved by EcoRI at one end. The resultant sticky end was filled in by a Klenow fragment in the presence of [α-32P] dATP (Furui Biotech, Beijing) and other unlabeled dNTPs, resulting in 3′-end labeling of the forward strand. The sequencing ladders were prepared using the Sanger dideoxy method with the 5′-labeled primer ParS3310-5R (5′-TCGAGTCTGGAATTTCCAGCCCCGC-3′). The sequence reads have been converted to denote complementary nucleotides. The reverse strand of the centromere DNA was similarly 3′ labeled using primers ParS3141EcoRI-F (5′-GGAATTCGCAAAGGACCCACCGACCG-3′) and ParS3310-R (5′-TCTGGAATTTCCAGCCCCGC-3′). The corresponding sequencing ladders were prepared using the 5′-labeled primer ParS3141-5F (5′-CTAGAGCAAAGGACCCACCGACCG-3′).
Labeled DNA probes (20 nM) were incubated in a buffer containing 5 mM sodium cacodylate, pH 7.0, 5 mM MgCl2, 100 mM KCl, 0.02% NP-40, 0.05 mg/ml poly(dI–dC) with ParB 60–139 or full-length ParB in a final volume of 10 μl at 25°C for 10 min. The concentrations of ParB 60–139 and full-length ParB were both 0.5 and 1 μM in the forward-strand reaction. In the reverse-strand reaction, ParB 60–139 was used at 0.25, 0.5 and 1 μM, and full-length ParB was used at 0.1, 0.2, 0.25, 0.33, 0.5, 0.66, 1 and 2 μM. Three 1 μl drops of 10 mM sodium ascorbate, 0.6% H2O2 and 1 mM FeCl2/2 mM EDTA were added separately on the wall of tube, mixed on the wall by pipetting and spun down to the sample. The reactions were incubated for 4 min and quenched by adding 1 μl of 100 mM thiourea. The reactions were then diluted with 300 mM sodium acetate, pH 5.2, to 200 μl and extracted with phenol–chloroform. The upper phase was collected and 1 μl of GlycoBlue (15 mg/ml; Ambion), and 500 μl of ethanol were added. DNA was precipitated in liquid nitrogen for 2 min and harvested by centrifugation. The pellets were washed with 70% ethanol, briefly dried, resuspended in 8 μl of loading buffer containing 95% (v/v) deionized formamide, 0.5 mM EDTA, 0.025% (w/v) SDS, 0.05% (w/v) xylene cyanol, 0.05% (w/v) bromophenol blue, and separated on an 8% polyacrylamide/8 M urea sequencing gel. The gel was visualized by autoradiography. Software SAFA (semi-automated footprinting analysis) was used to extract the volumes of resolved bands (30). The volume data were imported into OriginPro 8 (OriginLab) and fit to a sine function:
| (1) |
where y is the band volume, x is the nucleotide number, w is the period, y0 is the baseline, x0 is the phase and A is the amplitude of protection.
EMSA
DNA oligos were synthesized and gel-purified by Invitrogen (Supplementary Table S1). Complementary strands (500 μM) were annealed in 5 mM HEPES-K, pH 7.6, at 95°C for 1 min. DNA duplexes were radiolabeled at the 5′ end with [γ-32P]ATP (5000 Ci/mmol; Furui Biotech, Beijing) by T4 polynucleotide kinase (NEB). The 170-bp centromere DNA was PCR amplified as described above followed by dephosphorylation and phosphorylation. Labeled DNAs (∼0.2 nM) were incubated in binding buffer (5 mM MgCl2, 100 mM KCl, 0.01% NP-40, 5 mM HEPES-K and pH 7.6) with various concentrations of ParB 65–139 in a final volume of 10 μl at 4°C for 30 min. An equal volume of loading buffer containing 10% glycerol, 0.05% (w/v) xylene cyanol FF and 0.05% (w/v) bromophenol blue was added to the reactions, which were then resolved on a 5% native polyacrylamide gel run in 1× Tris-glycine buffer at 20 V/cm at 4°C for ∼1 h. Gels were dried, exposed to phosphorimager screens and read by a Typhoon 9400 scanner (GE Healthcare). The volumes of the DNA bands were integrated in Quantity One (BioRad). For stoichiometic assembly, 5 μM DNA was mixed with 0, 2.5, 5, 7.5, 10, 12.5, 15 and 20 μM ParB 65–139 in a 10 μl volume containing the same binding buffer indicated above. The gel was stained by ethidium bromide.
Analysis of two-ParB binding data
We consider the case of ParB binding to a DNA probe with two identical binding sites. The fraction of DNA bound to i ParB molecules Θi (i = 0,1,2) is given by (31):
| (2) |
| (3) |
| (4) |
where K is the intrinsic microscopic binding constant of ligand to a single binding site, ω is the cooperativity factor between adjacent interacting ligands, L is the concentration of free ligand, and Z is the binding polynomial:
| (5) |
Parameters K and ω were determined by non-linear global fit of experimentally measured values Θi (i = 0,1,2) to Equations (2)–(5) as a function of L. Because the labeled DNA has a concentration of ∼0.2 nM in our experimental conditions and the amount of bound ligand is negligible, the concentration of free ligand L is approximated by the total concentration of ligand. The global non-linear fit analysis of the 2R3′-27 and 2R6′-27 data was carried out in MATLAB 6.1 with home-written scripts. The intrinsic dissociation constant Kd,int is equal to 1/K.
ITC
The titrations were performed at 25°C using an ITC-200 microcalorimeter (MicroCal Inc.). DNA solutions (5 or 10 μM) were prepared by diluting concentrated stocks into the binding buffer containing 5 mM HEPES-K, pH 7.6, 5 mM MgCl2 and 100 mM KCl. ParB 65–139 was prepared in the same binding buffer with a concentration of 50 or 100 μM. Solutions were degassed for 2–5 min before loading. The sample cell was filled with 200 μl of DNA. ParB 65–139 was injected in a volume of 0.4 μl for the first injection and 2 μl for the next 19 injections using a computer-controlled 40-μl microsyringe with an injection interval of 150 s. Titration of protein into the binding buffer or titration of the binding buffer into the DNA solution produced negligible heat. Integrated heat data were analyzed using a one-set-of-sites model in MicroCal Origin following the manufacturer's instructions. The first data point was excluded in analysis. The binding parameters ΔH (reaction enthalpy change in cal mol−1), K (binding constant in M−1) and n (bound ParB dimer per DNA) were floating in the fit. The binding free energy ΔG and reaction entropy ΔS were calculated using the relationships ΔG = −RT ln K (R = 1.9872 cal mol−1 K−1, T = 298 K) and ΔG = ΔH − TΔS. The dissociation constant Kd was calculated as 1/K. The change of the ParB binding free energy of a subsite relative to R3–18 was calculated using the equation ΔΔG = ΔGsubsite − ΔGR3–18.
RESULTS
The C-terminal half of ParB forms a DNA-binding domain resistant to proteolysis
Sequence analysis of pCXC100 ParB did not reveal any known domains. Searching for ParB homologs by Phi-Blast returned ∼10 significant hits, but none of them has been biochemically characterized. To experimentally probe the domain organization of ParB, full-length ParB protein was subjected to limited proteolysis by increasing amounts of trypsin and was analyzed by SDS–PAGE (Figure 1A). Full-length ParB protein was digested into a major species with a molecular weight of ∼6 kDa. Mass spectroscopy analysis showed that the species includes residues 67–129, suggesting that this region folds into a structural core of ParB.
Figure 1.
ParB contains a C-terminal DNA-binding domain. (A) SDS–PAGE of limited digestion products of ParB. Full-length ParB protein (24 μg) in the absence and presence of 10 μg R4–18 DNA was digested by 0, 0.004, 0.016 and 0.064 μg trypsin for 24 h at 20°C. The SDS–PAGE was stained with Coomassie blue. The structure core resistant to digestion covers residues ∼67–129. (B) Native gel analysis of DNA-protected ParB digestions. The gel was stained by ethidium bromide. Free DNA and DNA complexes with full-length ParB and its structural core are indicated.
We also digested ParB in the presence of an 18-bp DNA (R4–18) derived from the centromere. The DNA-bound ParB degraded into a stable species of a size similar to that of the free ParB. Native gel analysis showed that the stable species remained bound by DNA (Figure 1B). In the absence of DNA, the ∼6 kDa species could be completely degraded within 24 h at the highest concentration of trypsin tested whereas the DNA-bound species remained highly resistant to digestion, indicating that DNA interaction stabilizes the structure core. Hence, these results suggest that the C-terminal half of ParB forms the DNA-binding domain.
Crystallization and structure determination
Given the distinct sequence of ParB, determination of its three-dimensional structure would be essential for resolving its DNA-binding motif. We obtained the crystal of the ParB DNA-binding domain in the free-state during crystallization of ParB–DNA complexes. The structure was solved by the SIRAS method based on a gold derivative. Although the derivative dataset has a high Rmerge value of 0.183, the experimental electron density map is of excellent quality (Supplementary Figure S1). The structure revealed only the DNA-binding domain of ParB but no bound DNA in the crystal. The asymmetric unit of the crystal contains two chains composed of residues 69–128 and 70–128 that fold into an intertwined dimeric structure. The structure has been refined to 1.4 Å resolution with an R factor of 0.198 and a free R-value of 0.212 (Table 1).
Table 1.
Data collection and refinement statistics
| Crystal form | KAu(CN)2 | Native |
|---|---|---|
| Data collection | ||
| Space group | P3221 | P3221 |
| Cell dimensions | ||
| a, b, c (Å) | 73.9, 73.9, 52.9 | 74.2, 74.2, 52.6 |
| α, β, γ (°) | 90, 90, 120 | 90, 90, 120 |
| Wavelength (Å) | 1.5418 | 0.9796 |
| X-ray source | CuKα | SSRF, BL17U1 |
| Resolution range (Å)a | 20–3.0 (3.05–3.00) | 50–1.4 (1.42–1.40) |
| Unique reflections | 3472 | 33 276 |
| Redundancy | 5.6 (4.6) | 6.2 (4.1) |
| I/σ | 9.5 (2.8) | 28.0 (2.3) |
| Completeness (%) | 99.3 (93.9) | 99.9 (99.9) |
| Rmergeb | 0.183 (0.376) | 0.113 (0.627) |
| Structure refinement | ||
| Resolution range (Å) | 20–1.4 (1.44–1.40) | |
| No. reflections | 31 419 | |
| No. atoms | 1187 | |
| Rworkc | 0.198 (0.239) | |
| Rfreed | 0.212 (0.248) | |
| Rmsd bond length (Å) | 0.007 | |
| Rmsd bond angles (°) | 1.029 |
aThe values for the data in the highest resolution shell are shown in parentheses.
bRmerge = ∑|Ii − Im|/∑Ii, where Ii is the intensity of the measured reflection and Im is the mean intensity of all symmetry-related reflections.
cRwork = ∑hkl|Fo − Fc|/∑hklFo, where Fo and Fc are the observed and calculated structure factor amplitudes of reflection hkl.
dRfree is the same as Rwork, but calculated on 5% reflections not used in refinement.
Dimeric RHH structure of the ParB DNA-binding domain
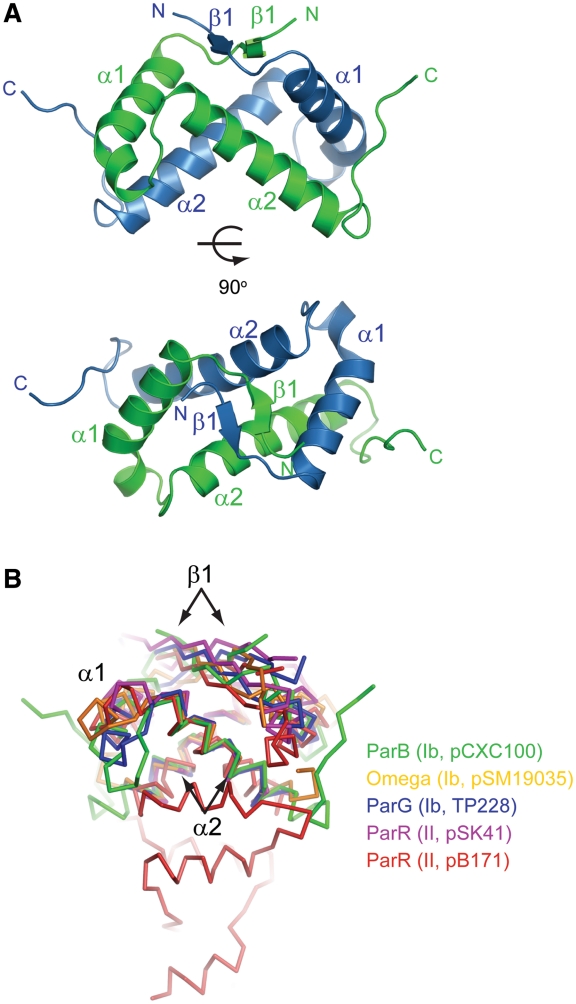
The DNA-binding domain of ParB adopts a dimeric RHH structure (Figure 2A). Each subunit consists of a short β-strand, β1 (residues 72–74), followed by two helices, α1 (residues 77–94) and α2 (residues 99–118). Two monomer subunits are tightly intertwined to form a symmetric homodimer. The N-terminal β1 strands pair with each other, forming an antiparallel β-ribbon. The symmetry-related α2 helices cross over by 107° and occupy the center of the structure. The α1 helices are located at the peripheral region with a bend in the last helical turn. The dimerization interface is extensive and buries a total of 1808 Å2 of solvent accessible surface area. Residues V73, V75, L83, A86, Y87, L89, F99, F102, I103, L107, V111, L114 and F124 from both subunits constitute the hydrophobic core of the structure, and most of them are conserved among homologs of ParB (Figure 3).
Figure 2.
Crystal structure of ParB DNA-binding domain. (A) Ribbon representation of the dimeric RHH structure in two orthogonal views. The two subunits are colored blue and green. Secondary-structure elements are labeled. The image on the bottom is viewed along the dyad axis of the dimer. (B) Structural superimposition of ParB (green) with other RHH CBPs: TP288 ParG (blue, 1P94), pSM19035 omega (orange, 2BNW), pSK41 ParR (magenta, 2Q2K) and pB171 ParR (red, 2JD3). These structures were aligned by their two α2 helices in PyMOL. The type and plasmid of the CBP are indicated.
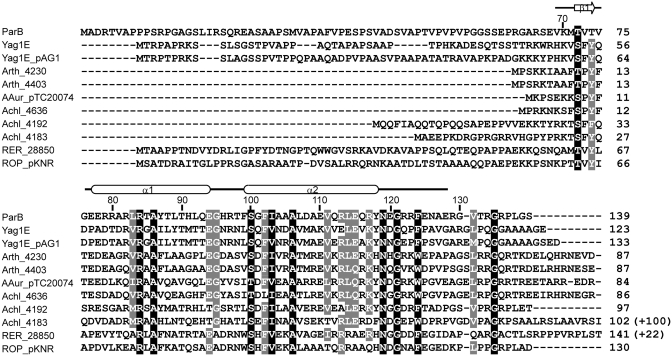
Figure 3.
Sequence alignment of ParB homologs. On the top, the residue number markers and secondary-structure elements observed in the crystal structure are indicated for ParB. The accession numbers of the 11 sequences in order are AAP69567 (ParB of pCXC100); ZP_03388950 (Yag1E in the chromosome of Propionibacterium acnes SK137); NP_052577 (Yag1E_pAG1 in plasmid pAG1 of Corynebacterium glutamicum); YP_829452 (Arth_4230 in plasmid 3 of Arthrobacter sp. FB24); YP_829155 (Arth_4403 in plasmid 1 of Arthrobacter sp. FB24); YP_950238 (AAur_pTC20074 in plasmid TC2 of A. aurescens TC1); YP_002478563 (Achl_4636 in plasmid pACHL02 of A. chlorophenolicus A6); YP_002477960 (Achl_4192 in plasmid pACHL01 of A. chlorophenolicus A6); YP_002477951 (Achl_4183 in plasmid pACHL01 of A. chlorophenolicus A6); YP_002766332 (RER_28850 in the chromosome of Rhodococcus erythropolis PR4); YP_002784447 (ROP_pKNR-00030 in plasmid pKNR of R. opacus B4). Sequence Achl_4183 is incomplete at its N-terminal region in GenBank and has been corrected here. The numbers in parentheses indicate extra C-terminal residues not shown. Residues with 100 and 80% conservation are shaded in black and gray, respectively.
The RHH motif is a well-characterized DNA-binding domain present in prokaryotic transcription repressors and plasmid partition proteins (32). In known RHH–DNA complex structures, the antiparallel β-ribbon inserts into the major groove of DNA and makes specific interactions with base edges (17,18,33–38). By analogy, ParB residues K70, T72 and T74 around strand β1 are likely responsible for sequence-specific recognition of the centromere. In addition, the conserved residue R84 in helix α1 may contact with the phosphate–sugar backbone of DNA.
Through structure determination, the RHH motif has been identified in type Ib CBPs TP288 ParG (16) and pSM19035 omega (15,17), and type II CBPs pSK41 ParR (18) and pB171 ParR (19). Structural alignment of monomer subunit to ParB yielded an rmsd of 1.4 Å over 39 Cα pairs for ParG, an rmsd of 1.9 Å over 36 Cα pairs for omega, an rmsd of 2.4 Å over 30 Cα pairs for pSK41 ParR and an rmsd of 3.2 Å over 24 Cα pairs for pB171 ParR. Hence, ParB is structurally more similar to type Ib CBP than to type II CBP. Among the five RHH CBPs including ParB, the two α2 helices display the most conserved position with an inter-helical angle of 95–113°. Alignment of the five structures by their α2 helices is shown in Figure 2B. The structural conservation of helices α2 underlies that the N-termini of α2 helices need to mediate highly conserved contacts with two phosphate groups across the DNA major groove (17,18,33–38). In the alignment, equivalent β1 strands occupy a similar position, but the conformation of helix α1 and its connecting loops to helix α2 and strand β1 are highly divergent. These regions often mediate variable interactions between adjacent RHH dimers bound to tandem repeats. In addition, the ParB RHH domain contains a unique C-terminal loop (residues 119–128) following helix α2 that makes contact with helix α1 of its dyad mate, contributing to the dimerization interface. Use of extra structural elements for dimerization was also found in the structure of pB171 ParR, which possesses an extensive C-terminal helical domain involved in dimerization (19).
ParB homologs
About 10 sequences share 48% similarity and 29% identity on average with ParB at the RHH motif region (Figure 3). Such a degree of conservation suggests that they all similarly adopt a dimeric RHH fold and likely function in binding specific DNA sequences. These sequences are encoded in seven Gram-positive actinobacteria with most in plasmid DNA and two in the genome. The function of these proteins has not been experimentally studied. Notably, Yag1E_pAG1 in plasmid pAG1 of Corynebacterium glutamicum, Yag1E in the genome of Propionibacterium acnes SK137 and ROP_pKNR in plasmid pKNR of Rhodococcus opacus B4 are located downstream of a small Walker-type ParA protein, suggesting that they belong to a type Ib partition system.
ParB binding site on the centromere
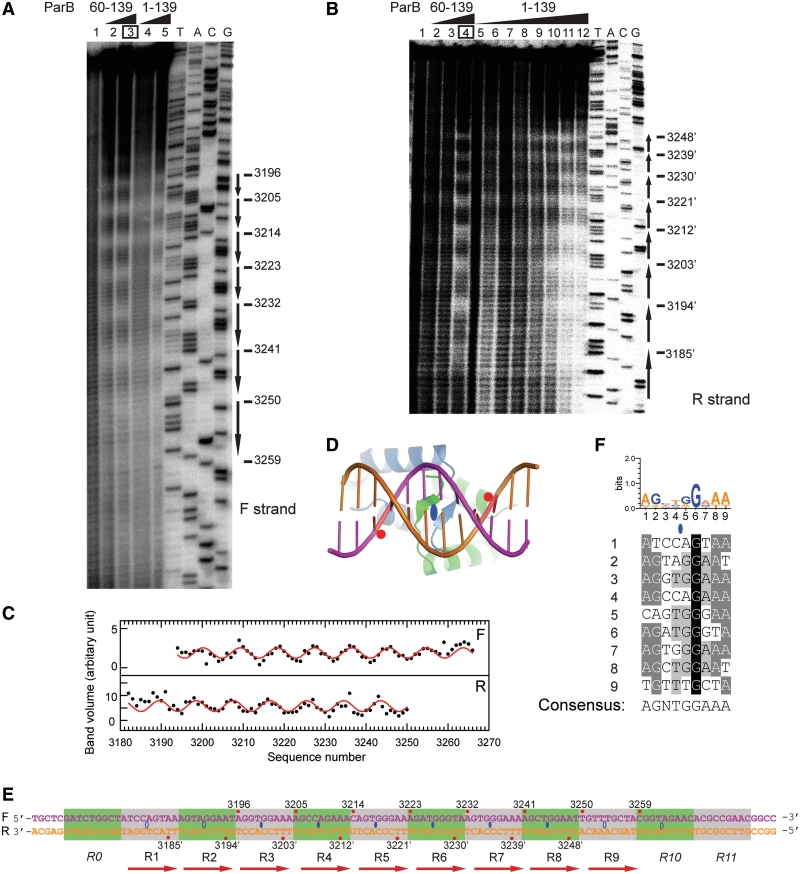
We previously showed that the binding of ParB protected a ∼90-bp centromere region upstream of the par locus from DNase I cleavage (21). To precisely locate ParB binding sites within the centromere and define the boundaries and organization of the centromere, we applied the hydroxyl radical footprinting assay to a 170-bp DNA probe covering the whole centromere region. The hydroxyl radical primarily attacks the C5′ atom of deoxyribose, resulting in the breakage of the phosphate–ribose backbone independent of the DNA sequence (29). The small size of the hydroxyl radical probe allows one to detect protein protection on the DNA backbone at a single-nucleotide resolution. The DNA was 32P-labeled at the 3′-end of either the forward or reverse strand, assembled with ParB or its RHH domain (residues 60–139), and subjected to hydroxyl radical attack. The cleaved products of either strand were resolved in sequencing gels and showed a prominent periodic pattern of protection upon ParB binding (Figure 4A and B), indicating that multiple ParB proteins successively assemble on the centromere in a specific manner. The RHH domain of ParB showed a similar protection pattern as full-length ParB, suggesting that the N-terminal region of ParB makes no significant contribution to the centromere binding specificity.
Figure 4.
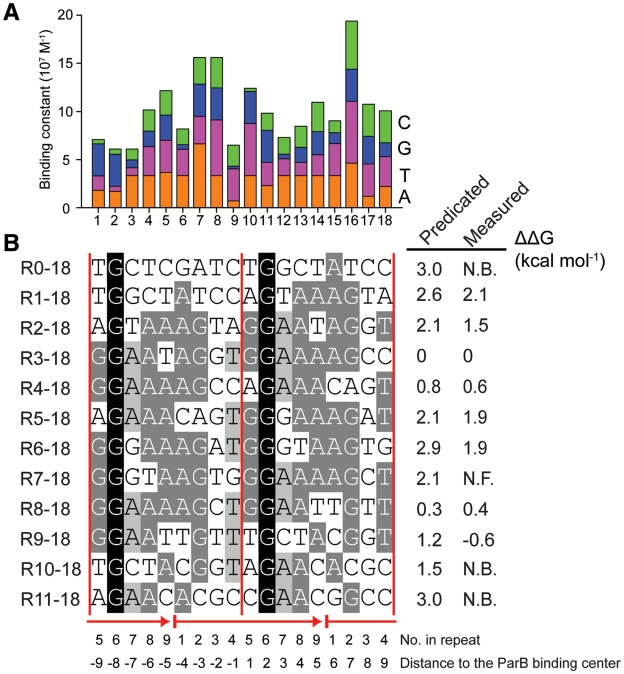
The ParB-centromere interaction probed by hydroxyl radical footprinting. (A) Footprinting of the forward strand of ParB-bound centromere. A 170-bp centromere DNA was labeled at the 3′-end of the forward strand, assembled with 0.5 and 1 μM of ParB RHH domain (residues 60–139) (lanes 2 and 3), and 0.5 and 1 μM of full-length ParB (lanes 4 and 5) and subjected to hydroxyl radical cleavage. Lane 1 contains no protein. Sequencing ladders (A, T, G, C) are indicated. The most protected nucleotides derived from curve fitting are shown on the right. (B) Footprinting on the reverse strand. The ParB 60–139 concentrations were 0.25, 0.5 and 1 μM in lanes 2–4, and the ParB 1–139 concentrations were 0.1, 0.2, 0.25, 0.33, 0.5, 0.66, 1 and 2 μM in lanes 5–12. (C) The band volumes of the cleavage products are plotted against nucleotide numbers. Lane 3 (boxed) of the forward strand and lane 4 of the reverse strand were analyzed. Smaller volume corresponds to greater protection at the respective site. The lines represent the best fit of a sine function to the protection data with a period of 9.07 ± 0.08 for the forward strand (top panel) and 8.96 ± 0.09 for the reverse strand (bottom panel). (D) Structural model of the ParB RHH domain in complex with DNA. A standard B-form DNA was positioned based on the aligned pSK41 ParR-DNA complex structure (2Q2K). Red circles refer to the DNA backbone regions contacted by the N-terminus of helix α2. (E) The structure of the centromere. The forward strand is numbered from 5′ to 3′, and nucleotides in the reverse strand are numbered as their pairing nucleotides in the forward strand and are denoted by prime. Nucleotides with maximal protection in hydroxyl radical footprinting are marked by red circles and their nucleotide numbers. Ellipses denote binding centers of ParB dimer. Hollow ellipses are extrapolated from incomplete protection data. Alternative repeats are shaded in green and gray. Repeats R0, R10 and R11 do not bind ParB by themselves and are denoted in italics. (F) Alignment of repeats R1–R9 and the consensus sequence. The sequence logo is indicated at the top. A loose criterion of 50% conservation was used to define the consensus sequence.
To precisely define the phase and periodicity of the protection pattern, the band volumes of the cleavage product as a function of nucleotide number were fit to a sine function (Figure 4C). The best fitting curve yielded a protection cycle of 9.07 ± 0.08 for the forward strand and 8.96 ± 0.09 for the reverse strand. The periodicity in the protection pattern indicated that multiple ParB molecules bind at the centromere with a 9-bp interval. The best-fit curves also pinpoint the positions of maximal protection, which are otherwise difficult to locate by visual inspection.
We next used the footprinting results to deduce the binding centers of ParB on the centromere. Previous RHH-DNA complex structures show that while specific interactions between the RHH β-ribbon and DNA base are asymmetric for non-palindromic sites, non-specific interactions with DNA phosphate–sugar backbone are mostly symmetric about the dyad axis of dimeric RHH structure (17,18,33–39). By analogy, each ParB dimer is expected to interact symmetrically with the DNA backbone (Figure 4D). Hydroxyl radical footprint detects protection of DNA backbone and would generate a symmetric protection pattern on two strands of ParB-bound DNA. Indeed, the protection pattern on the two strands of the centromere DNA displays 10 symmetry centers in the middle of adjacent base pairs, each spaced 9-bp apart (Figure 4E). For example, the maximally protected nucleotides—A3196 in the forward strand and T3203′ on the reverse strand—are symmetric about a point between base pairs T3199•A3199′ and G3200•C3200′. These symmetry centers represent the binding centers for each ParB dimer. The most protected nucleotides are 4-bp from binding centers and appear to correspond to the phosphate-ribose backbone region contacted by the N-terminus of helix α2 (Figure 4D). A total of ten binding sites were identified in the centromere region, of which four peripheral sites were deduced from the protection pattern on only a single strand owing to poor resolution of the gel.
We divided the centromere region into consecutive 9-bp segments such that the binding center of ParB is located between the fourth and fifth nucleotides of a segment and each segment roughly corresponds to a binding site of ParB RHH domain. Sequence alignment of segments 1–9 reveals modest conservation with a consensus sequence ‘AGNTGGAAA’ (Figure 4F), indicating that the centromere is composed of nine unspaced direct 9-bp repeats. Repeat 10 was not included in the alignment for it does not bind ParB by itself, as shown below. The centromere is less likely to contain an inverted repeat, as none of inverted repeats match the consensus sequence better than their direct counterparts (Supplementary Figure S2).
A ParB dimer contacts an 18-bp region around its binding center
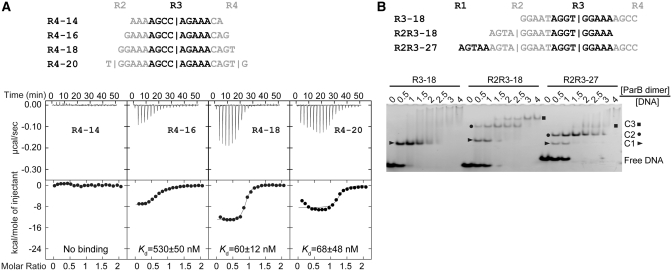
Association of a ParB dimer to a single subsite is the first step in the assembly of a full partition complex. We ask how ParB recognizes a single subsite. To find a suitable length of subsite required for ParB binding, we applied ITC to compare binding of the ParB RHH domain to four 14–20-bp probes centered around repeat R4 (Figure 5A). A 14-bp probe that fully covers repeat R4 failed to bind ParB. Elongation of the probe from 16- to 18-bp reduced Kd value from 530 to 60 nM, while further elongation to 20-bp caused no change in Kd value (68 nM). Repeat R3 also requires an 18-bp region around its binding center to bind ParB (Supplementary Table S2). We conclude that a length of 18-bp around the binding center is required for ParB to efficiently bind a single subsite.
Figure 5.
The length requirement of single subsite for ParB binding and the binding stoichiometry. (A) ITC profiles of the ParB RHH domain (65–139) and the R4 probes of different lengths. The sequences of 14, 16, −18 and 20-bp R4 probes are indicated. Repeat R4 is shown in black and its flanking sequences are shown in gray. Vertical bars denote the predicted binding centers of ParB dimer. The curves are the best fit to a one-set-of-sites model. (B) Binding stoichiometry of ParB to one-site and two-site probes. Sequences are indicated for an 18-bp one-site probe R3–18; an 18-bp two-site probe R2R3–18; and a 27-bp two-site probe R2R3–27. Alternative repeats are shaded in black and gray. ParB 65–139 was assembled with 5 μM DNA in a 10 μl volume with different protein to DNA molar ratios as indicated. The native gel was stained by ethidium bromide. C1, C2 and C3 denote complexes formed by one, two and three ParB dimers, respectively.
Binding stoichiometry of ParB with one- and two-site probes
These 18-bp one-site probes formed only a single species of complex in EMSA (Figure 6), which we interpreted as one ParB dimer-bound complex. However, the complex might actually contain a ParB dimer-of-dimers, which is the minimal functional unit of most RHH domains (32). To directly assess the binding stoichiometry, we preformed EMSA with different molar ratios of ParB 65–139 and unlabeled DNA probes (Figure 5B). We compared the assembly of an 18-bp DNA containing repeat R3 and its flanking sequences (R3–18), an 18-bp DNA containing repeats R2 and R3 (R2R3–18) and a 27-bp DNA containing repeats R2 and R3, and their flanking sequences (R2R3–27). One-site probe R3–18 was assembled almost completely into a C1 complex with equal molar amount of ParB dimer, indicating that the C1 complex contains one ParB dimer. In contrast, two-site probes R2R3–18 and R2R3–27 formed predominately a C2 complex with a ∼1.5-fold molar ratio of ParB dimer to DNA. This observation and the migration difference between the C1 and C2 complex suggest that the C2 complex contains two ParB dimers. ParB apparently associates with the two-site probes in a sequential manner as the one-ParB bound C1 complex was transitively populated.
Figure 6.
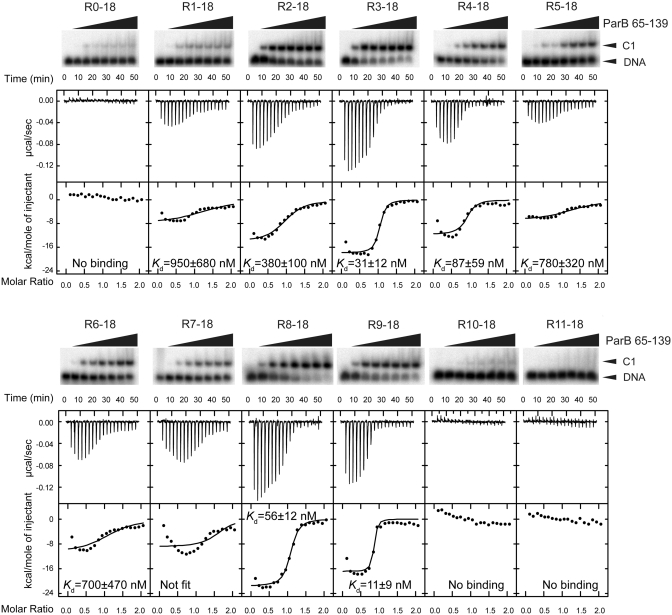
Binding affinity of individual sites. Twelve 18-bp probes covering repeats R0-R11 were assessed for binding of ParB RHH domain (residues 65–139) by EMSA (top panel) and ITC (bottom panel). The concentrations of ParB dimer in EMSA were 0, 2, 8, 16, 32, 64, 128 and 512 nM. The curves in the ITC panels are the best fit to a one-set-of-sites model that dose not account for possible additional binding events. Fit of the R7-18 data was unsuccessful.
The above EMSA experiments were preformed with protein and DNA concentrations in the micromolar range. Under these conditions, excess protein was found to lead to larger complexes that are likely caused by non-specific binding. These complexes appeared as smeared bands for R3–18 and R2R3–27. Notably, a discrete C3 complex is evident for R2R3–18, and less so for longer R2R3–27. The C3 complex likely contains 3 ParB dimers as judged from its migration rate, but its assembly mode is unclear. The non-specific complexes were not observed in EMSA using low concentrations (∼0.2 nM) of 32P-labeled DNA.
Affinity variation among individual subsites
The 10 centromere repeats are relatively divergent in sequence, raising the question of whether they have a similar affinity toward ParB. To this end, we characterized the binding of ParB RHH domain (residues 65–139) to individual subsites. We used EMSA and ITC to evaluate the binding affinity of 12 18-bp DNA probes that center around each of 10 repeats (R1–R10) as well as the putative repeat R0, which is upstream of R1, and the putative repeat R11, which is downstream of R10 (Figure 6). The ITC data were analyzed with a one-set-of-sites model that assumes one ParB dimer binding to one DNA. Some fit curves show systemic deviation from the data, which is probably caused by additional binding events, as shown in Figure 5B. The Kd values derived from ITC are indicated in Figure 6, and the full sets of binding parameters are listed in Supplementary Table S2.
Subsites around R0, R10 and R11 display virtually no binding in both EMSA and ITC experiments, indicating that the centromere core contains only repeats R1–R9. Although binding of repeat R10 was detected in the footprinting experiment, this result is likely due to the high affinity of subsite R9 and binding cooperativity between adjacent ParB molecules (see below). ITC measurements showed that subsites R9, R3, R8 and R4 have the highest binding affinities with Kd values of 11–87 nM, whereas the rest subsites show Kd values in the submicromolar range (380–950 nM). The two orders of magnitude variation in binding affinity among individual subsites is probably caused by their divergent sequences.
Sequence recognition of ParB
To assess the contribution of individual nucleotide to ParB recognition, we replaced each of 18 base pairs in the one-site probe R3–18 to other three alternatives and measured the ParB binding affinity by ITC (Table 2, Supplementary Table S2, and Figure 7A, Supplementary Figure S3). Sequences in positions 4, 5, 7, 8, 11, 14 and 16 are not recognized by ParB, as corresponding point mutants retain the similar binding affinity. The most significant mutational effect occur at positions 9 and 10, which are immediately adjacent to the ParB binding center. Mutation of T9 to adenine and guanine reduced the binding affinity by 5- and 12-fold, respectively, while replacement of G10 by cytosine yielded an ITC profile lacking two-state transition (Supplementary Figure S3). Moreover, guanine substitution of A6 or A12 decreased the binding affinity by 7-fold. Since steps 6–12 are within 4-bp of the ParB binding center, these sequences may be recognized by the β-ribbon structure of ParB dimer, according to the known RHH–DNA interaction mode.
Table 2.
Dissociation constants Kd (nM) of R3–18 point mutants and ParB 65–139 measured by ITC
| No. | WT | Replacement |
|||
|---|---|---|---|---|---|
| A | T | G | C | ||
| 1 | G | 56 ± 12 | 67 ± 6 | 30 ± 10 | 216 ± 22 |
| 2 | G | 59 ± 29 | 187 ± 20 | 30 ± 10 | 189 ± 22 |
| 3 | A | 30 ± 10 | 123 ± 12 | 121 ± 31 | 89 ± 13 |
| 4 | A | 30 ± 10 | 33 ± 8 | 62 ± 15 | 45 ± 12 |
| 5 | T | 27 ± 11 | 30 ± 10 | 38 ± 9 | 39 ± 13 |
| 6 | A | 30 ± 10 | 37 ± 5 | 209 ± 77 | 60 ± 28 |
| 7 | G | 15 ± 5 | 35 ± 5 | 30 ± 10 | 36 ± 9 |
| 8 | G | 30 ± 7 | 17 ± 6 | 30 ± 10 | 32 ± 8 |
| 9 | T | 143 ± 31 | 30 ± 10 | 365 ± 57 | 46 ± 11 |
| 10 | G | 30 ± 10 | 18 ± 2 | 30 ± 10 | Not fit |
| 11 | G | 44 ± 21 | 41 ± 9 | 30 ± 10 | 56 ± 11 |
| 12 | A | 30 ± 10 | 57 ± 16 | 203 ± 43 | 58 ± 31 |
| 13 | A | 30 ± 10 | 73 ± 22 | 64 ± 15 | 46 ± 12 |
| 14 | A | 30 ± 10 | 46 ± 8 | 42 ± 9 | 33 ± 10 |
| 15 | A | 30 ± 10 | 30 ± 6 | 88 ± 12 | 81 ± 8 |
| 16 | G | 22 ± 9 | 16 ± 6 | 30 ± 10 | 20 ± 7 |
| 17 | C | 84 ± 13 | 30 ± 8 | 35 ± 11 | 30 ± 10 |
| 18 | C | 46 ± 10 | 32 ± 6 | 70 ± 17 | 30 ± 10 |
Figure 7.
Sequence recognition of ParB. (A) Binding constants of R3–18 and its point mutants plotted against the position of the replaced nucleotide. For the purpose of illustration, mutant G10C that shows weak ParB binding is set to have a Kd value of 300 nM. (B) The predicated and measured change in the binding free energy relative to R3–18 for 18-bp subsites. Repeats are shown as arrows and ParB binding centers as vertical lines. Each nucleotide is also labeled by its position in a repeat and distance to the central ParB binding center. N.B.: No binding; N.F. Not fit.
Interestingly, ParB also recognizes distal sequences from the preceding and succeeding repeat. Specifically, steps 1–3 favor strongly the ‘GGA’ sequence and mutations at these positions reduced the binding affinity by up to 4–7-fold. At the downstream region, steps 13,15, 17 and 18 show marginal selection of sequence with corresponding mutants causing <3-fold increase of Kd value.
To understand the affinity variation among different subsites R0–18 to R11–18 (Figure 6), we attempted to estimate the change of the ParB binding free energy (ΔΔG) of these subsites relative to R3–18 on the basis of their sequence. To the first-order of approximation, ΔΔG of a subsite sequence can be calculated as a sum of ΔΔG due to single substitution. The predicated ΔΔG values generally agree with the experimentally measured values (Figure 7B). However, subsites R9–18 and R10–18 show some discrepancy between the predicated and measured ΔΔG, likely due to coupling interaction among multiple substitutions.
Verification of the structural organization of the centromere
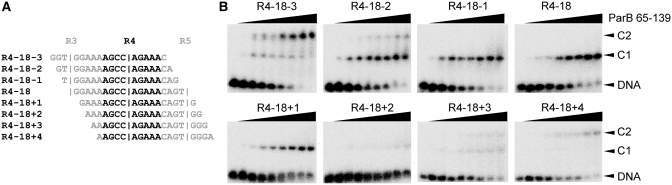
We attempted to verify by a second approach the positions of the ParB binding centers on the centromere that are predicted from the footprinting experiment. We examined the binding of the ParB RHH domain with an array of eight 18-bp probes that are consecutively shifted by one base pair to cover repeat R4 (Figure 8A). The R4–18 probe that has its midpoint at the predicted binding center of repeat R4 should display the highest affinity. If the midpoint of a probe is off the binding center, we would expect to see reduction of the binding, as one side of the binding center may become too short for optimal contact. The EMSA results showed that probes R4–18−1 and R4–18 + 1, with a midpoint 1-bp off the binding center of R4, display a binding affinity similar to that of R4–18, whereas probe R4–18 + 2, which has a midpoint 2-bp downstream of the binding center of R4, shows significantly reduced binding. These observations are consistent with the predicted ParB binding center on repeat R4.
Figure 8.
Binding of ParB to frame-shifted R4 probes. (A) Eight frame-shifted R4 probes. The R4 repeat is shown in black and its flanking sequences from repeats R3 and R5 are shown in gray. Vertical bars indicate the positions of the binding centers of R3, R4 and R5. (B) EMSA of the ParB RHH domain (65–139) and R4 probes. Lane 1 in each panel has no ParB, and the subsequent lanes have increasing amounts of protein in concentrations of 2, 8, 16, 32, 64, 128, 512 and 5120 nM. C1 and C2 denote complexes formed by one and two ParB dimers, respectively.
Interestingly, when the probe midpoint was moved farther upstream (R4–18 − 2, R4–18 − 3) or downstream (R4–18 + 3, R4–18 + 4), we observed a second slow-migrating DNA–protein complex C2, which apparently corresponds to two-ParB bound complex. In these cases, the probes contain part of the R3 or R5 repeat. Despite being incomplete, the adjacent R3 and R5 sites show significant binding to a second ParB molecule. In contrast, efficient binding to a single-site probe requires a length of 9-bp at either side of a binding center. These results suggest that the ParB molecule bound at repeat R4 strongly attracts a second ParB molecule to adjacent weak sites. Comparison of probe R4–18 − 2 versus R4–18 + 2 and R4–18 − 3 versus R4–18 + 3 shows that even in their incomplete repeat forms, repeat R3 is better than R5 in cooperating R4 binding of ParB, which is consistent with the former having an ∼25-fold higher intrinsic binding ability (Figure 6). These results are fully consistent with the proposed structural model of the centromere and also indicate strong cooperativity for ParB in binding tandem repeats.
Binding cooperativity
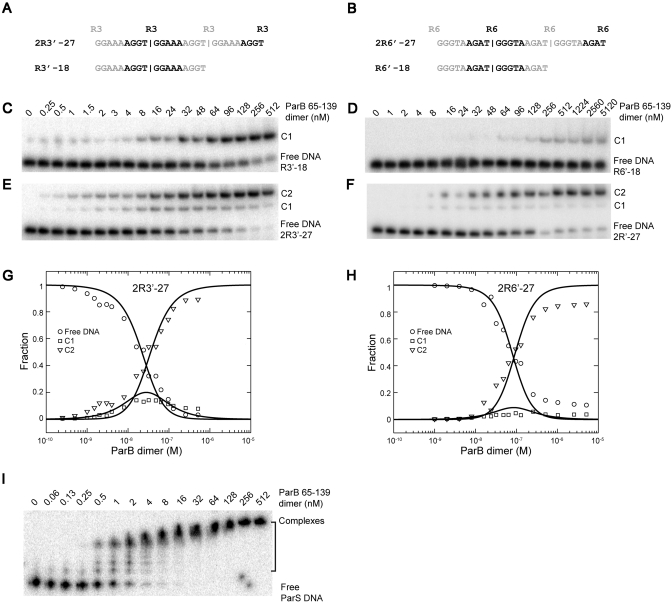
To quantify the binding cooperativity of ParB, we analyzed ParB binding to probes containing two subsites. Different affinities associated with the natural centromere binding sites would complicate measurement of cooperativity. To simplify the analysis, we engineered probes that contained two identical subsites over an 18-bp region surrounding each binding center. To construct such probes, we duplicated the sequence of a 9-bp repeat four times and selected a 27-bp segment between the two outermost binding centers. Two such probes—2R3′–27 and 2R6′–27—were derived from repeats R3 and R6, which represent a strong and a weak binding site, respectively (Figure 9A and B). The corresponding one-site 18-bp probes for R3 and R6 (R3′–18 and R6′–18) were also created.
Figure 9.
The cooperativity of ParB binding to tandem identical repeats. (A and B) Sequence of two- and one-site probes designed for repeats R3 (A) and R6 (B). (C–F) EMSA of ParB 65–139 with one-site probe R3′–18 (C) and R6′–18 (D), and two-site probes 2R3′–27 (E) and 2R6′–27 (F). C1 and C2 denote complexes formed by one and two ParB dimers, respectively. (G and H) Global analysis of the binding isotherms for 2R3′–27 (G) and 2R6′–27 (H). The lines are the best fit to Equations (2–5), with intrinsic dissociation constant Kd,int = 112 nM and cooperativity ω = 16 for 2R3′–27; Kd,int = 877 nM and ω = 110 for 2R6′–27. (I) EMSA of ParB 65–139 with a 32P-labeled 170-bp DNA covering the whole centromere region.
As expected, the one-site probes form only a single complex with ParB, whereas the two-site probes form two types of complexes corresponding to one- and two-ParB bound complexes (Figure 9C–F). For two-site probes, the two-ParB complex is more favorably formed over the one-ParB complex even at low protein concentrations. The cooperativity effect was the most dramatic for the weak repeat, R6. Global analysis of the binding isotherms yielded a cooperativity of 16 for tandem R3 subsites and 112 for tandem R6 subsites (Figure 9G and H).
The association of ParB to a 170-bp DNA covering the whole centromere region is governed by an even stronger cooperativity (Figure 9I). Majority of the DNA was transformed into a major partition complex in the presence of 8 nM ParB 65–139, while at the same concentration the two-site probes remain not fully assembled (Figure 9E–F).
DISCUSSION
The active partition system for low-copy-number plasmids is a simple and autonomous mechanism for separating duplicated DNA into daughter cells. Studies on a few model plasmids have begun to reveal the molecular mechanisms of many aspects of plasmid partitioning (5). The first step of plasmid partition is assembly of CBP and centromere repeats into the partition complex, which then recruits filament-forming ParA protein. In this study, we structurally and biochemically characterized the CBP ParB from a type Ib partition system recently reported in plasmid pCXC100 (20,21). The obtained results indicate that the pCXC100 partition system is generally similar to other type Ib systems in terms of ParB structure, centromere organization and cooperative ParB-centromere interaction, and also reveal a distinct overlapped assembly mode of ParB to the centromere repeat.
ParB structure
We show that the DNA-binding domain of ParB is located at its C-terminal half and folds into a dimeric RHH structure. The RHH motif of ParB could not be predicted based on sequence homology, as is the case with many other RHH domains. Our structure adds to the list of four known RHH structures of type Ib (omega and ParG) and type II (pSK41 ParR and pB171 ParR) CBPs (15,16,18,19), and further supports the prevalence of the RHH domain in centromere binding.
In addition to its C-terminal RHH domain, ParB contains an N-terminal domain (residues 1–65). This region is likely flexible as it is highly susceptible to trypsin digestion in both the absence and the presence of DNA (Figure 1) and is predicted to lack secondary structure (40). The presence of a flexible tail upstream the RHH domain appears to be a common feature of type Ib RHH CBPs. A shorter N-terminal flexible tail is also present in ParG (∼32 residues) and omega (∼20 residues) (15,16). The sequences of the N-terminal domains are not similar among ParB, omega and ParG, nor are they similar among ParB homologues (Figure 3). Nevertheless, the N-terminal domain may be functionally important, as the corresponding region of ParG has been shown to be involved in the assembly of the ParG-DNA higher structure for transcription repression and in the activation of polymerization and ATP-hydrolysis activity of the ParG partner ParF (41,42). The N-terminal region of omega was also critical for stimulation of ATPase activity and polymerization of its partner delta (43).
Structure of the centromere
We previously relied on sequence analysis to find a weak 9-bp periodicity within the centromere region and divided the centromere into nine 9-bp repeats (21). The phase of the repeat was arbitrarily set due to lack of strong conservation in repeat sequences. Now, we have experimentally mapped ParB binding sites on the centromere though high-resolution hydroxyl radical footprinting on both strands of the centromere (Figure 4). Based on the known RHH-DNA interaction mode, we were able to deduce the structure of the centromere and the precise location of ParB binding centers. We found that the centromere is composed of nine unspaced 9-bp direct repeats. The repeat is non-palindromic and the binding center of ParB dimer is likely located between the fourth and fifth base pairs of each repeat. The current repeats have the same 9-bp cycle as the previously defined ones, but they differ in phase and boundary. The previously defined repeats are shifted 4-bp downstream compared to the current ones and cover a region corresponding to current repeats R2-R10.
Cooperativity in ParB–centromere interaction
The cooperativity of ParB in binding multiple repeats is similar to that of other RHH domains. Indeed, cooperativity is a crucial feature characterizing RHH–DNA interaction (32). The majority of RHH domains have extremely weak or no binding toward a single subsite and bind efficiently only to tandem subsites as a dimer-of-dimers. In this regard, it is notable that ParB is capable of binding 18-bp single subsites with affinities up to 11 nM. Arc repressor of bacteriophage P22 is one RHH protein sharing this property. Although Arc binds as a dimer-of-dimers to a 21-bp operator that contains two subsites, it can also bind as a dimer to DNA fragments containing single subsite with nanomolar affinities (44). Arc dimer has a strong cooperativity (ω = 5900) towards full operator that literally prevents the detection of one-dimer bound intermediate complex (44). In contrast, ParB exhibits a moderate cooperativity (ω = 16–110) and one-dimer bound complex remains observable for two-site probes (Figures 5B, 8 and 9).
The binding cooperativity would allow the association of ParB with intrinsically weak sites within the centromere and spread out of the centromere core. The latter phenomenon is reflected by the detection of binding of repeat R10 in the context of the whole centromere in the footprinting experiment (Figure 4) and the observation of binding of repeat R0 in the presence of repeat R1 by EMSA (data not shown). This spreading effect was previously observed for the interaction of omega with its cognate DNA (45).
Sequence specificity of ParB interaction
A ParB dimer requires a region of 18-bp covering one central repeat as well as two surrounding half repeats for binding single subsites (Figure 5A). This is unprecedented given that an RHH domain normally contacts only ∼8–10 bp of DNA. Such an interaction mode implicates that ParB dimers bound adjacently to the centromere would contact an overlapped region. Consequently, the sequence in a repeat could be jointly specified by adjacent ParB dimers. The mutagenesis data support that ParB dimer recognizes the sequences of flanking repeats, particularly the fifth to seventh nucleotide of the preceding repeat (Table 2 and Figure 7). For instance, the sixth nucleotide of repeat, conserved as guanine, appears to be specified by the succeeding ParB rather than the immediately bound molecule. Determination of the ParB–DNA complex structure would elucidate the molecular details about their unusual interaction mode.
PDB ACCESSION CODE
The atomic coordinates and structure factors have been deposited in the Protein Data Bank under accession code 3NO7.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Chinese Ministry of Science and Technology 863 Project (grant 2008AA022310 to K.Y.) and Beijing Municipal Government support (to K.Y.); National Natural Science Foundation (grant 30770422 to Y.Z.); the National Basic Research Program 973 of China (grant 2005CB724604 to Y.Z.). Funding for open access charge: Chinese Ministry of Science and Technology.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors gratefully acknowledge support from the staff at Shanghai Synchrotron Radiation Facility beamline BL17U1. They thank She Chen and Xiaojun Ding for mass spectroscopy analysis and Anbi Xu, Shanshan Wang and Xiaobo Wan for assistance in protein purification. They are indebted to Institute of Pathogen Biology for kindly allowing them to use its ITC machine when theirs was down.
REFERENCES
- 1.Leonard TA, Moller-Jensen J, Lowe J. Towards understanding the molecular basis of bacterial DNA segregation. Philos. Trans. R. Soc. Lond. B Biol. Sci. 2005;360:523–535. doi: 10.1098/rstb.2004.1608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu. Rev. Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 3.Hayes F, Barilla D. The bacterial segrosome: a dynamic nucleoprotein machine for DNA trafficking and segregation. Nat. Rev. Microbiol. 2006;4:133–143. doi: 10.1038/nrmicro1342. [DOI] [PubMed] [Google Scholar]
- 4.Ghosh SK, Hajra S, Paek A, Jayaram M. Mechanisms for chromosome and plasmid segregation. Annu. Rev. Biochem. 2006;75:211–241. doi: 10.1146/annurev.biochem.75.101304.124037. [DOI] [PubMed] [Google Scholar]
- 5.Schumacher MA. Structural biology of plasmid partition: uncovering the molecular mechanisms of DNA segregation. Biochem. J. 2008;412:1–18. doi: 10.1042/BJ20080359. [DOI] [PubMed] [Google Scholar]
- 6.Gerdes K, Moller-Jensen J, Bugge Jensen R. Plasmid and chromosome partitioning: surprises from phylogeny. Mol. Microbiol. 2000;37:455–466. doi: 10.1046/j.1365-2958.2000.01975.x. [DOI] [PubMed] [Google Scholar]
- 7.Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc. Natl Acad. Sci. USA. 2010;107:11763–11768. doi: 10.1073/pnas.1003817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Tang M, Bideshi DK, Park HW, Federici BA. Iteron-binding ORF157 and FtsZ-like ORF156 proteins encoded by pBtoxis play a role in its replication in Bacillus thuringiensis subsp. israelensis. J. Bacteriol. 2007;189:8053–8058. doi: 10.1128/JB.00908-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Larsen RA, Cusumano C, Fujioka A, Lim-Fong G, Patterson P, Pogliano J. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21:1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Tinsley E, Khan SA. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J. Bacteriol. 2006;188:2829–2835. doi: 10.1128/JB.188.8.2829-2835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Anand SP, Akhtar P, Tinsley E, Watkins SC, Khan SA. GTP-dependent polymerization of the tubulin-like RepX replication protein encoded by the pXO1 plasmid of Bacillus anthracis. Mol. Microbiol. 2008;67:881–890. doi: 10.1111/j.1365-2958.2007.06100.x. [DOI] [PubMed] [Google Scholar]
- 12.Schumacher MA, Funnell BE. Structures of ParB bound to DNA reveal mechanism of partition complex formation. Nature. 2005;438:516–519. doi: 10.1038/nature04149. [DOI] [PubMed] [Google Scholar]
- 13.Leonard TA, Butler PJ, Lowe J. Structural analysis of the chromosome segregation protein Spo0J from Thermus thermophilus. Mol. Microbiol. 2004;53:419–432. doi: 10.1111/j.1365-2958.2004.04133.x. [DOI] [PubMed] [Google Scholar]
- 14.Khare D, Ziegelin G, Lanka E, Heinemann U. Sequence-specific DNA binding determined by contacts outside the helix-turn-helix motif of the ParB homolog KorB. Nat. Struct. Mol. Biol. 2004;11:656–663. doi: 10.1038/nsmb773. [DOI] [PubMed] [Google Scholar]
- 15.Murayama K, Orth P, de la Hoz AB, Alonso JC, Saenger W. Crystal structure of omega transcriptional repressor encoded by Streptococcus pyogenes plasmid pSM19035 at 1.5 A resolution. J. Mol. Biol. 2001;314:789–796. doi: 10.1006/jmbi.2001.5157. [DOI] [PubMed] [Google Scholar]
- 16.Golovanov AP, Barilla D, Golovanova M, Hayes F, Lian LY. ParG, a protein required for active partition of bacterial plasmids, has a dimeric ribbon-helix-helix structure. Mol. Microbiol. 2003;50:1141–1153. doi: 10.1046/j.1365-2958.2003.03750.x. [DOI] [PubMed] [Google Scholar]
- 17.Weihofen WA, Cicek A, Pratto F, Alonso JC, Saenger W. Structures of omega repressors bound to direct and inverted DNA repeats explain modulation of transcription. Nucleic Acids Res. 2006;34:1450–1458. doi: 10.1093/nar/gkl015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schumacher MA, Glover TC, Brzoska AJ, Jensen SO, Dunham TD, Skurray RA, Firth N. Segrosome structure revealed by a complex of ParR with centromere DNA. Nature. 2007;450:1268–1271. doi: 10.1038/nature06392. [DOI] [PubMed] [Google Scholar]
- 19.Moller-Jensen J, Ringgaard S, Mercogliano CP, Gerdes K, Lowe J. Structural analysis of the ParR/parC plasmid partition complex. EMBO J. 2007;26:4413–4422. doi: 10.1038/sj.emboj.7601864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li TY, Yin P, Zhou Y, Zhang Y, Zhang YY, Chen TA. Characterization of the replicon of a 51-kb native plasmid from the gram-positive bacterium Leifsonia xyli subsp. cynodontis. FEMS Microbiol. Lett. 2004;236:33–39. doi: 10.1016/j.femsle.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 21.Yin P, Li TY, Xie MH, Jiang L, Zhang Y. A Type Ib ParB protein involved in plasmid partitioning in a gram-positive bacterium. J. Bacteriol. 2006;188:8103–8108. doi: 10.1128/JB.01232-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dong A, Xu X, Edwards AM, Chang C, Chruszcz M, Cuff M, Cymborowski M, Di Leo R, Egorova O, Evdokimova E, et al. In situ proteolysis for protein crystallization and structure determination. Nat. Methods. 2007;4:1019–1021. doi: 10.1038/nmeth1118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Otwinowski Z, Minor W. Processing of X-ray diffraction data collected in oscillation mode. Methods Enzymol. 1997;276:307–326. doi: 10.1016/S0076-6879(97)76066-X. [DOI] [PubMed] [Google Scholar]
- 24.Sheldrick GM. A short history of SHELX. Acta Crystallogr. A. 2008;64:112–122. doi: 10.1107/S0108767307043930. [DOI] [PubMed] [Google Scholar]
- 25.Vonrhein C, Blanc E, Roversi P, Bricogne G. Automated structure solution with autoSHARP. Methods Mol. Biol. 2007;364:215–230. doi: 10.1385/1-59745-266-1:215. [DOI] [PubMed] [Google Scholar]
- 26.Langer G, Cohen SX, Lamzin VS, Perrakis A. Automated macromolecular model building for X-ray crystallography using ARP/wARP version 7. Nat. Protoc. 2008;3:1171–1179. doi: 10.1038/nprot.2008.91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 28.Murshudov GN, Vagin AA, Lebedev A, Wilson KS, Dodson EJ. Efficient anisotropic refinement of macromolecular structures using FFT. Acta Crystallogr. D Biol. Crystallogr. 1999;55:247–255. doi: 10.1107/S090744499801405X. [DOI] [PubMed] [Google Scholar]
- 29.Jain SS, Tullius TD. Footprinting protein-DNA complexes using the hydroxyl radical. Nat. Protoc. 2008;3:1092–1100. doi: 10.1038/nprot.2008.72. [DOI] [PubMed] [Google Scholar]
- 30.Laederach A, Das R, Vicens Q, Pearlman SM, Brenowitz M, Herschlag D, Altman RB. Semiautomated and rapid quantification of nucleic acid footprinting and structure mapping experiments. Nat. Protoc. 2008;3:1395–1401. doi: 10.1038/nprot.2008.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Senear DF, Brenowitz M. Determination of binding constants for cooperative site-specific protein-DNA interactions using the gel mobility-shift assay. J. Biol. Chem. 1991;266:13661–13671. [PubMed] [Google Scholar]
- 32.Schreiter ER, Drennan CL. Ribbon-helix-helix transcription factors: variations on a theme. Nat. Rev. Microbiol. 2007;5:710–720. doi: 10.1038/nrmicro1717. [DOI] [PubMed] [Google Scholar]
- 33.Somers WS, Phillips SE. Crystal structure of the met repressor-operator complex at 2.8 A resolution reveals DNA recognition by beta-strands. Nature. 1992;359:387–393. doi: 10.1038/359387a0. [DOI] [PubMed] [Google Scholar]
- 34.Raumann BE, Rould MA, Pabo CO, Sauer RT. DNA recognition by beta-sheets in the Arc repressor-operator crystal structure. Nature. 1994;367:754–757. doi: 10.1038/367754a0. [DOI] [PubMed] [Google Scholar]
- 35.Schreiter ER, Wang SC, Zamble DB, Drennan CL. NikR-operator complex structure and the mechanism of repressor activation by metal ions. Proc. Natl Acad. Sci. USA. 2006;103:13676–13681. doi: 10.1073/pnas.0606247103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mattison K, Wilbur JS, So M, Brennan RG. Structure of FitAB from Neisseria gonorrhoeae bound to DNA reveals a tetramer of toxin-antitoxin heterodimers containing pin domains and ribbon-helix-helix motifs. J. Biol. Chem. 2006;281:37942–37951. doi: 10.1074/jbc.M605198200. [DOI] [PubMed] [Google Scholar]
- 37.Zhou Y, Larson JD, Bottoms CA, Arturo EC, Henzl MT, Jenkins JL, Nix JC, Becker DF, Tanner JJ. Structural basis of the transcriptional regulation of the proline utilization regulon by multifunctional PutA. J. Mol. Biol. 2008;381:174–188. doi: 10.1016/j.jmb.2008.05.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ni L, Jensen SO, Ky Tonthat N, Berg T, Kwong SM, Guan FH, Brown MH, Skurray RA, Firth N, Schumacher MA. The Staphylococcus aureus pSK41 plasmid-encoded ArtA protein is a master regulator of plasmid transmission genes and contains a RHH motif used in alternate DNA-binding modes. Nucleic Acids Res. 2009;37:6970–6983. doi: 10.1093/nar/gkp756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gomis-Ruth FX, Sola M, Acebo P, Parraga A, Guasch A, Eritja R, Gonzalez A, Espinosa M, del Solar G, Coll M. The structure of plasmid-encoded transcriptional repressor CopG unliganded and bound to its operator. EMBO J. 1998;17:7404–7415. doi: 10.1093/emboj/17.24.7404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cole C, Barber JD, Barton GJ. The Jpred 3 secondary structure prediction server. Nucleic Acids Res. 2008;36:W197–W201. doi: 10.1093/nar/gkn238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Barilla D, Carmelo E, Hayes F. The tail of the ParG DNA segregation protein remodels ParF polymers and enhances ATP hydrolysis via an arginine finger-like motif. Proc. Natl Acad. Sci. USA. 2007;104:1811–1816. doi: 10.1073/pnas.0607216104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Carmelo E, Barilla D, Golovanov AP, Lian LY, Derome A, Hayes F. The unstructured N-terminal tail of ParG modulates assembly of a quaternary nucleoprotein complex in transcription repression. J. Biol. Chem. 2005;280:28683–28691. doi: 10.1074/jbc.M501173200. [DOI] [PubMed] [Google Scholar]
- 43.Pratto F, Cicek A, Weihofen WA, Lurz R, Saenger W, Alonso JC. Streptococcus pyogenes pSM19035 requires dynamic assembly of ATP-bound ParA and ParB on parS DNA during plasmid segregation. Nucleic Acids Res. 2008;36:3676–3689. doi: 10.1093/nar/gkn170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Brown BM, Sauer RT. Assembly of the Arc repressor-operator complex: cooperative interactions between DNA-bound dimers. Biochemistry. 1993;32:1354–1363. doi: 10.1021/bi00056a022. [DOI] [PubMed] [Google Scholar]
- 45.de la Hoz AB, Pratto F, Misselwitz R, Speck C, Weihofen W, Welfle K, Saenger W, Welfle H, Alonso JC. Recognition of DNA by omega protein from the broad-host range Streptococcus pyogenes plasmid pSM19035: analysis of binding to operator DNA with one to four heptad repeats. Nucleic Acids Res. 2004;32:3136–3147. doi: 10.1093/nar/gkh633. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.