Abstract
MicroRNAs (miRNAs) are prevalent regulatory RNAs that mediate gene silencing and play key roles in diverse cellular processes. While synthetic RNA-based regulatory systems that integrate regulatory and sensing functions have been demonstrated, the lack of detail on miRNA structure–function relationships has limited the development of integrated control systems based on miRNA silencing. Using an elucidated relationship between Drosha processing and the single-stranded nature of the miRNA basal segments, we developed a strategy for designing ligand-responsive miRNAs. We demonstrate that ligand binding to an aptamer integrated into the miRNA basal segments inhibits Drosha processing, resulting in titratable control over gene silencing. The generality of this control strategy was shown for three aptamer–small molecule ligand pairs. The platform can be extended to the design of synthetic miRNAs clusters, cis-acting miRNAs and self-targeting miRNAs that act both in cis and trans, enabling fine-tuning of the regulatory strength and dynamics. The ability of our ligand-responsive miRNA platform to respond to user-defined inputs, undergo regulatory performance tuning and display scalable combinatorial control schemes will help advance applications in biological research and applied medicine.
INTRODUCTION
MicroRNAs (miRNAs) comprise a conserved class of small non-coding RNAs that direct targeted gene silencing through the RNA interference (RNAi) pathway in humans and other eukaryotes. Most miRNAs are encoded within long transcripts transcribed from Pol II promoters (1,2). The primary (pri-)miRNA is initially processed to an ∼65-nt precursor (pre-)miRNA by the Microprocessor (3–5) composed of the cleaving enzyme Drosha and the RNA-binding protein DGCR8. Following pri-miRNA cleavage and export from the nucleus, the pre-miRNA is processed by Dicer to a 22- to 25-nt miRNA duplex. One of the duplex strands termed the mature miRNA is incorporated into the RNA-induced silencing complex (RISC), which subsequently cleaves or translationally represses the target transcript depending on the degree of complementarity between the guide sequence of the miRNA and the target. miRNA-mediated gene regulation has been implicated in diverse biological processes ranging from development to angiogenesis and may be involved in the regulation of a majority of the human genome (6).
Engineered genetic systems that display ligand control of miRNA-mediated gene silencing will provide a powerful and versatile means to control transgene and endogenous gene expression. Recently, researchers have designed synthetic RNA-based regulatory systems that integrate sensing and gene-regulatory functions, where the former are encoded in RNA aptamer sequences that recognize small molecule ligands (7,8). Such integrated ligand-responsive RNA-based control systems offer several advantages over more traditional protein-based regulatory systems in avoiding potential immunogenicity of heterologous protein components and providing a more tunable, compact control system. In addition, as aptamers can be selected against a wide range of biomolecules (9), such integrated RNA systems provide platforms for gene expression control in response to potentially any ligand.
Recently, RNA-based control systems have been developed that mediate gene silencing through the RNAi pathway in response to small molecule ligands. One system utilized an allosteric ribozyme to indirectly control miRNA silencing activity (10). Integrated systems that directly couple aptamers to intermediate substrates in the RNAi processing pathway, small hairpin RNAs (shRNAs), have also been demonstrated (11–13). These integrated designs linked small-molecule RNA aptamers to the loop region of shRNA elements to modulate the extent of Dicer processing and subsequent gene silencing through ligand-binding events based on known structural requirements for efficient Dicer processing (11,12). However, the adopted mode of ligand control inherently reduced silencing (12) and ligand regulation through Dicer processing restricts molecular sensing to cytoplasmic ligands. In addition, the in vivo toxicity of shRNAs (14–16) establish significant hurdles toward broader implementation of shRNA-based control systems. Finally, the expression architectures of all of these systems require additional promoter-RNA constructs to expand the number of ligand-responsive regulatory RNAs, which represents a challenge for therapeutic applications. Many of these practical application issues may be circumvented through the implementation of integrated ligand-responsive miRNAs (17). Unfortunately, the current shRNA-based control systems cannot be readily converted into functional miRNA-based control systems as modification of the terminal loop has been shown to limit Drosha processing in vitro and in vivo (18).
Here, we demonstrate the design of integrated ligand-responsive miRNAs that do not require modification of the miRNA terminal loop. We also demonstrate the implementation of the miRNA system in regulatory circuits that tune the resulting regulatory response. We utilized a synthetic approach to elucidate more precise structural requirements for miRNA processing, specifically that the bulge size in the miRNA basal segments dictates the extent of Drosha processing and in vivo silencing. By integrating an aptamer into the miRNA basal segments, we demonstrate that aptamer–ligand-binding interactions can be used to sufficiently increase the local structure in the miRNA basal segment region, such that Drosha processing and subsequent gene silencing are inhibited with increasing ligand concentration. The sequence flexibility of the basal segments enables the introduction of different aptamer sequences in this region, resulting in a modular design framework that allows modification of the detected ligand or target gene. We further engineered circuits comprising clusters of ligand-responsive miRNAs and self-targeting ligand-responsive miRNAs. The circuits offer tunable and stringent control over gene expression and, in the case of miRNA clusters and cis-acting miRNAs, are similar to circuits found in nature. Our integrated ligand-responsive miRNA framework offers considerable versatility for genetic control and will provide a broadly useful tool supporting basic research and new applications in biotechnology and medicine.
MATERIALS AND METHODS
Plasmid construction
The coding region of DsRed-Express was initially cloned with the consensus Kozak sequence (CGCCACC) into the NheI/XhoI restriction sites of pcDNA3.1(+) to create pCS350 (Supplementary Figure S1). pcDNA3.1(+) contains the constitutive CMV promoter upstream of a multi-cloning site. Ligand-responsive and control miRNAs reported in Supplementary Table S1 were cloned into XbaI/ApaI downstream of the DsRed-Express coding region. To construct synthetic miRNA clusters, additional miRNAs were amplified from the miRNA-containing plasmid, digested with AvrII/XhoI and iteratively inserted into XhoI/XbaI within the same plasmid. For the 2-nt spacer, the second miRNA (th3) was separately amplified without a DNA template and inserted. For all other spacer lengths, the original miRNA was amplified with a common forward primer and a reverse primer that hybridizes different distances downstream of the miRNA: Sp.fwd, 5′-GTTCCTGTAGACGGCTCTC-3′; Sp1.rev, 5′-AATACCTAGGCTGATCAGCGGGTTT-3′; Sp2.rev, 5′-AATACCTAGGAGGGGCAAACAACAG-3′; Sp3.rev, 5′-AATACCTAGGAAAGGACAGTGGGAGTG-3′. To make stable cell lines harboring the ligand-responsive miRNA constructs, the coding region of DsRed-Express was replaced with EGFP and the entire genetic construct was excised with NheI/NsiI and cloned into the same sites in pcDNA5/FRT (Invitrogen). Prior to insertion, the NsiI site was introduced into this plasmid at position 1524 using site-directed mutagenesis.
For the inducible promoter studies, the tetO-CMV promoter was PCR amplified from pTRE-Tight (Clontech) and inserted into BglII/NheI of pCS350, thereby replacing the original CMV promoter. The control miRNA (wt) was inserted into XbaI/ApaI downstream of the DsRed-Express coding region to create ptetO-wt. All restriction enzymes and T4 DNA ligase were purchased from New England Biolabs. All constructs were sequence-verified (Laragen and Elim Biopharmaceuticals).
RNA preparation
Internally radiolabeled RNAs were transcribed in vitro from an annealed template containing the T7 promoter (5′-TTCTAATACGACTCACTATAGGG-3′, where G is the first transcribed nucleotide) using the Ampliscribe T7 transcription kit (Epicentre) according to the manufacturer’s instructions with [α-32P]-GTP. Following transcription and DNase I treatment, the transcription product was purified through a NucAway clean-up column (Ambion) according to the manufacturer’s instructions and gel-purified by PAGE.
Drosha cleavage assays
In vitro assays were conducted as described previously (19). Briefly, the Drosha complex was immunopurified from 293T cells transiently transfected with pCK-Drosha-FLAG and pCK-DGCR8-FLAG (9:1 mass ratio). Two days post-transfection, cells were lysed using M-PER (Pierce) according to the manufacturer’s instructions and the resulting supernatant was incubated with anti-FLAG M2 affinity beads (Sigma Aldrich) for at least 1 h at 4°C with rotation. The beads were then washed with lysis buffer (20 mM Tris–HCl pH 8.0, 100 mM KCl, 0.2 mM EDTA) five times and evenly divided for the in vitro assays (two in vitro reactions from a 10-cm transfection dish). Radiolabeled RNAs (∼105 cpm, 3 µl) were combined with 0.75 µl RNasin (Promega), 3 µl reaction buffer (64 mM MgCl2), 8.25 µl water and 15 µl immunopurified Drosha complex. After an incubation of 90 min at 37°C, the reaction was terminated with the addition of 0.5 M sodium acetate and 0.02% sodium dodecyl sulfate (SDS), phenol:chloroform extracted and ethanol precipitated. Samples were then resuspended in 15 μl RNA loading buffer (95% formamide, 0.02% SDS, 0.025% bromophenol blue, 0.025% xylene cyanol FF) and resolved on a 12.5% denaturing polyacrylamide gel. The RNA decades ladder (Ambion) was used as a size marker.
Cell culture and transfection
HEK 293 cells stably expressing d2EGFP and Flp-In-293 cells were maintained in DMEM supplemented with 10% FBS at 37°C in a 5% CO2-humidified incubator. Transient transfections were conducted with FuGENE 6 (Roche) according to the manufacturer’s instructions 1 day after seeding. Immediately prior to transfection, the media was supplemented with the appropriate ligand at the specified concentration. Ligand concentrations were selected to maximize the regulatory response without severely compromising cell viability over the course of the transient assays. For characterization of ligand-regulated miRNA systems, single-plasmid transfections were performed using 250 ng of DNA plasmid in each 500-µl transfection sample. For inducible promoter studies, three-plasmid transfections were performed on 293 cells stably expressing d2EGFP using 10 ng of pCS350, 72ng of ptetO-wt and 168 ng of pTet-Off (Clontech) encoding the tetracycline transactivator (tTA). The amounts of ptetO-wt and pTet-OFF were selected to maximize expression from the ptetO promoter while maintaining a total mass of transfected plasmid of 250 ng (Supplementary Figure S2). The media was replaced 2 days post-transfection.
Cells were trypsinized and subjected to flow cytometry analysis on a Cell Lab Quanta SC MPL (Beckman Coulter) 3 days post-transfection, and the resulting data were analyzed using the FlowJo software (Tree Star). Cells were initially gated for viability by electronic volume and side scatter. GFP and DsRed fluorescence of viable cells were measured through 525- and 670-nm band-pass filters, respectively, after excitation with a 488-nm laser. Detectable DsRed levels served as a transfection control to gate between transfected and untransfected cells, where DsRed was detectable even for the constructs containing four miRNAs (Supplementary Figure S3). GFP levels were calculated as the median fluorescence of the transfected population divided by that of the untransfected population (12). This normalization approach corrected for the bias from pleiotropic effects associated with each small molecule. All ratios were normalized to the GFP value for the construct lacking a miRNA under the same conditions. Reported DsRed measurements equal the mean fluorescence value of the transfected population divided by the mean fluorescence value for the construct lacking a miRNA. Here, the mean value was selected because of the high variability associated with transient plasmid-based expression of fluorescent proteins. Reported data are averages of two or more replicates of independently transfected wells in the same cell-culture plate. Results from the cell-culture assays were similar across repeated independent experiments (Supplementary Figure S4).
Stable transfection of 293-Flp-In cell lines was performed using the Flp-In recombinase system (Invitrogen) according to the manufacturer’s instructions to generate isogenic stable cell lines. Integrants were selected using 200 μg/ml hygromycin B (Invitrogen), whereas stable cell lines were maintained in 50 μg/ml hygromycin B. The procedure described above for the fluorescence expression analysis of transiently transfected cell populations was used to analyze the stable cell lines with notable exceptions. Normalization of flow cytometry data to untransfected cells was not performed as all cells express the integrated construct, and all data were normalized to cells lacking a miRNA and grown in the absence of theophylline. Normalization to cells grown under the same conditions was not performed since theophylline differentially affected the two negative controls: no miRNA and a self-targeting miRNA lacking the theophylline aptamer. The different effects on GFP levels in the no miRNA and wt samples from theophylline addition may be attributed to differences in perturbations induced by theophylline stress on unregulated genes and genes regulated by a self-targeting miRNA.
Other methods
See Supplementary Data for remaining methods, including qRT–PCR and the design of mature miRNA sequences.
RESULTS
Extent of structure in the basal segments dictates Drosha processing and gene silencing
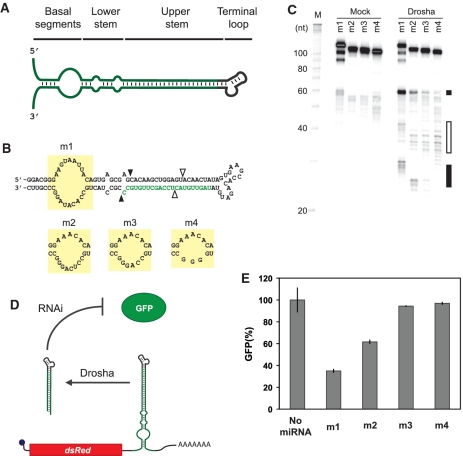
miRNAs can be partitioned into four domains: the miRNA loop, the upper stem, the lower stem and the basal segments (Figure 1A). Each domain exhibits unique structural requirements for efficient Drosha processing and RISC activation (20–23). The miRNA loop is unstructured, the upper stem encodes the mature miRNA and exhibits extensive base pairing interactions, the lower stem serves as a ruler to designate the Drosha cleavage site and exhibits fewer base pairing interactions than the upper stem, and the basal segments are generally single-stranded.
Figure 1.
Extent of structure in the basal segments dictates miRNA processing and target gene silencing. (A) General domains of a pri-miRNA. (B) Sequence and secondary structure of minimal pri-miRNAs with different bulge sizes in the basal segments. Mature miRNA sequence targeting GFP is shown in green. The bulge sequences are demarcated by yellow boxes. Black arrows indicate the predicted Drosha cleavage site for productive processing 11-bp downstream of the basal segments (23). White arrows indicate the predicted Drosha cleavage site for abortive processing 11-bp upstream of the terminal loop (23). (C) PAGE analysis of the minimal pri-miRNAs subjected to the Drosha cleavage assay. Black and white boxes mark the general locations of productive and abortive cleavage products, respectively. The three bands representing full-length m1 presumably arose from spontaneous cleavage of the unstructured bulge during or after gel extraction. (D) Schematic of the miRNA regulatory system in the cell-culture assay. GFP-targeting miRNAs were placed within the 3′ UTR of the transcript encoding DsRed-Express. 293 GFP cells transiently transfected with each construct were subjected to flow cytometry analysis. (E) Relative GFP levels obtained from the cell-culture assay for constructs harboring miRNAs with varying bulge sizes. See ‘Materials and Methods’ section for a description of data normalization. Error bars reflect the range of values from two independent transfections conducted in the same cell-culture plate. See Supplementary Figure S4A for results from an independent transfection experiment for (E).
In vitro studies probing the structural contribution of the basal segments to efficient Drosha processing have shown that mutating the sequence of this domain to form extensive base pairing interactions abolishes Drosha processing (21,23). These results suggest a mechanism for regulating Drosha processing by regulating the structure of the basal segments. To utilize this potential mechanism as a design strategy, we needed to develop a more thorough understanding of how structure—and, in particular, the extents of structure—in the basal segments affect Drosha processing and RNAi-mediated gene silencing in vivo. Therefore, we examined the relationship between bulge size in the miRNA basal segments, in vitro Drosha processing and in vivo gene silencing.
We performed in vitro Drosha cleavage assays that mimic the first step in miRNA biogenesis to examine the relationship between bulge size and Drosha processing. RNAs with varying bulge sizes in the basal segments (Figure 1B) were transcribed in vitro, incubated with immunopurified Drosha and resolved by PAGE (Figure 1C). In vitro Drosha cleavage assays using similar miRNAs were previously shown to produce a pre-miRNA of ∼61 nt (3). We found that decreasing the size of the loop from 18 to 8 nt greatly reduced the appearance of the ∼61-nt pre-miRNA (Figure 1C, black boxes). Furthermore, decreasing the size of the loop resulted in a corresponding increase in abortive processing (Figure 1C, white boxes). Previous in vitro Drosha cleavage assays also yielded abortive processing, which was attributed to DGCR8 recognition of the terminal loop instead of the miRNA basal segments (23). The in vitro assay results suggest that Drosha cleavage is sensitive to intermediate extents of structure in the basal segments.
To examine the relationship between bulge size in the basal segments and gene silencing, we developed a general cell-culture assay for miRNA activity (Figure 1D). A miRNA designed to target a transcript encoding the green fluorescent protein (GFP) was inserted into the 3′ UTR of a gene encoding the fluorescent protein DsRed-Express. Plasmid DNA encoding the DsRed-Express construct was transfected into HEK 293 cells stably expressing GFP. The level of miRNA-mediated gene silencing was determined by flow cytometry analysis. Using the cell-culture assay, miRNAs with the same bulges as in the in vitro experiments were tested for silencing efficiency. Flow cytometry results supported the data from the in vitro Drosha cleavage assays, as smaller bulge sizes showed reduced GFP silencing (Figure 1E). Negligible silencing of GFP was observed for the 11- and 8-nt bulges. These results suggest that proper Drosha processing and gene silencing correlate with the size of the bulge in the miRNA basal segments and >11 nt are required in the bulge to induce gene silencing.
Aptamer integration renders miRNA processing sensitive to a small-molecule ligand
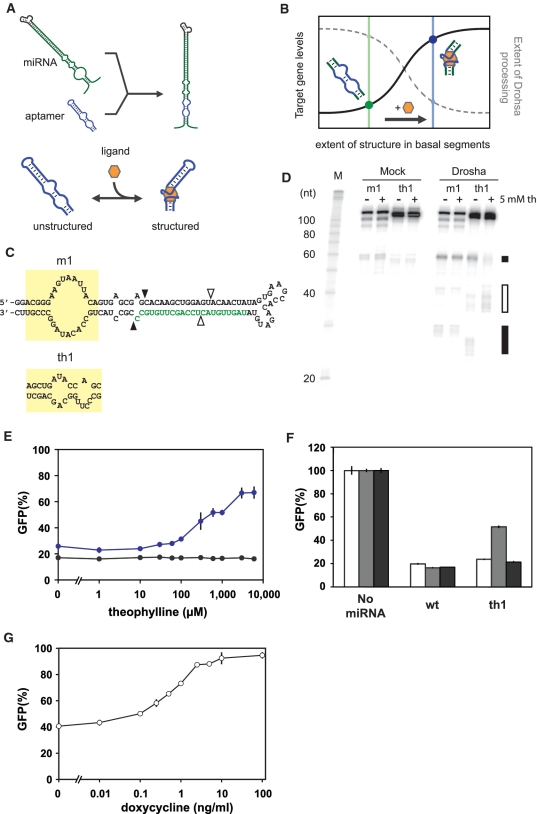
Aptamers often undergo transitions from relatively unstructured to structured conformations upon ligand binding, a phenomenon termed adaptive recognition (24). We developed a design strategy based on this phenomenon and the elucidated dependence of Drosha processing on the structure of the basal segments to introduce ligand control of miRNA-mediated gene silencing (Figure 2A). Through integration of an aptamer into the basal segments of a miRNA, we hypothesized that the aptamer–ligand-binding interactions would be able to sufficiently decrease the unstructured nature of that region, thereby inhibiting proper processing and subsequent gene silencing (Figure 2B).
Figure 2.
Ligand-responsive miRNAs enable ligand control of Drosha processing and gene silencing. (A) Design framework for ligand-responsive miRNAs. The aptamer-binding core is integrated into the miRNA basal segments adjacent to the lower stem. Ligand binding increases the structured nature of the aptamer. (B) Proposed relationship between Drosha processing (dashed gray line), target gene expression levels (black line), miRNA basal segments structure and ligand addition in the case of a ligand-responsive miRNA. Unstructured basal segments lead to efficient processing and gene silencing. Structured basal segments resulting from ligand binding inhibit Drosha processing. (C) Sequence and secondary structures of minimal pri-miRNAs with a large bulge (m1) or the theophylline aptamer (th1) inserted into the basal segments. See Figure 1B for notation. (D) PAGE analysis of m1 and th1 subjected to the Drosha cleavage assay in the presence or absence of 5 mM theophylline. See Figure 1C for notation. (E) Relative GFP levels obtained from the cell-culture assay for constructs harboring th1 (blue) or a miRNA with basal segments similar to miR-30a (wt, black) (Supplementary Figure S1). See Figure 1D and E for description of the assay. Transient transfections were conducted in the presence of varying theophylline concentrations. (F) Relative GFP levels obtained from the cell-culture assay for constructs transiently transfected in the absence (white) or presence of 1 mM theophylline (gray) or 1 mM caffeine (black). GFP levels were normalized to the construct lacking any miRNAs (No miRNA) transfected under the same conditions. (G) Relative GFP levels obtained for the tet inducible promoter system. A plasmid encoding the tetracycline transactivator was cotransfected with a plasmid encoding wt in the 3′ UTR of DsRed-Express under doxycycline-inducible control in the presence of varying doxycycline concentrations as part of the cell-culture assay. Error bars reflect the range of values from two (E and F) or three (G) independent transfections conducted in the same cell-culture plate. See Supplementary Figure S4B–D for results from independent transfection experiments for E–G, respectively.
We first examined the ability of an aptamer to mediate ligand control of Drosha processing. The theophylline aptamer (25) was inserted into the basal segments domain directly adjacent to the lower stem of a GFP-targeting miRNA (Figure 2C). The resulting miRNA (th1) was transcribed in vitro and subjected to the Drosha cleavage assay in the presence or absence of theophylline. The primary products of Drosha processing for th1 and a control miRNA (m1) were both ∼61 nt, the expected size for the pre-miRNA (3,23) (Figure 2D). In addition, the presence of theophylline inhibited proper processing of the aptamer-containing miRNA, resulting in an alternative cleavage pattern similar to that observed from miRNAs with smaller bulges (Figure 1C). The control miRNA exhibited negligible theophylline dependence, suggesting that ligand binding to an aptamer located within the basal segments can control Drosha processing.
We next examined whether theophylline regulation of Drosha processing resulted in ligand-mediated control of gene silencing by testing the silencing efficiency of the th1 miRNA using the cell-culture assay. Extensive base pairing below the bulge encoded in the aptamer sequence was incorporated to ensure proper aptamer folding. We also included a GFP-targeting miRNA with basal segments similar to the natural miRNA miR-30a as a control (wt, Supplementary Figure S5). Transient transfections were conducted in the presence of varying concentrations of theophylline. Results show that both miRNA constructs silenced GFP with comparable strength (17 and 25% basal levels for wt and th1, respectively) in the absence of theophylline (Figure 2E). Adding theophylline to cells transfected with the miRNA construct containing the theophylline aptamer resulted in a 2.4-fold increase in GFP levels. The theophylline-dependent effect was observed at a minimal concentration of 0.1 mM theophylline, as observed in other RNA-based regulatory systems using this aptamer (11,12,26). In contrast, silencing by the miRNA construct lacking the aptamer was insensitive to theophylline. Silencing by both miRNAs was insensitive to the presence of caffeine (Figure 2F), a molecule that differs from theophylline by a single methyl group and binds the theophylline aptamer with a 10 000-fold lower affinity (25). A caffeine concentration of 1 mM was used to avoid the pronounced toxicity observed at higher concentrations (11). The results demonstrate that the observed effect of theophylline on miRNA-mediated gene silencing is specific to the incorporation of the theophylline aptamer into the basal segments of the miRNA.
We also compared the quantitative response properties of our ligand-responsive miRNA system to the tet inducible promoter system, which is often used in conditional gene silencing (27). To most closely match our experimental setup, we inserted the control GFP-targeting miRNA (wt) into the 3′ UTR of the DsRed-Express gene under the control of a Tet-OFF promoter construct. This plasmid was cotransfected with the tetracycline transactivator protein expression plasmid at a plasmid ratio that had been optimized to maximize GFP silencing (Supplementary Figure S2), and GFP levels were measured in the presence of varying concentrations of the small-molecule inducer doxycycline (Figure 2G). We found that the inducible promoter system exhibits less silencing compared to the ligand-regulated miRNA system in the absence of ligand (40 versus 25% basal GFP expression levels). In addition, the dynamic range of the inducible promoter system is similar to that achieved with the ligand-regulated miRNA system at high concentrations of ligand (2.1- versus 2.4-fold), although the promoter system recovered to a higher expression level. These results demonstrate that the regulatory performance of the integrated RNA-only control system is comparable to that of an inducible promoter system under these experimental conditions. Given the practical advantages of an RNA-only system over control strategies that require the expression of a heterologous protein from a separate promoter, the ligand-regulated miRNA system represents a significant advance in conditional gene silencing technologies.
Framework modularity supports the integration of different aptamer and miRNA targeting sequences
Ligand-responsive regulatory systems that display modularity can be readily modified to change the targeted gene or recognized ligand without complete redesign. Such systems are useful in facilitating the rapid implementation of base designs in diverse applications. While most ligand-responsive RNA regulator designs can be readily modified to target different genes, only a fraction of the developed designs have been shown to support direct insertion of different aptamer sequences (12,28,29).
The targeted gene is specified by the mature miRNA sequence in a miRNA regulatory element. Previous work has shown that modifying the mature miRNA sequence in natural miRNAs is sufficient to target different genes (30). We found that altering the th1 miRNA to target a different location in the GFP transcript (th2) preserved silencing and the theophylline response in the cell-culture assay, while scrambling the mature miRNA sequence (th1′) abolished silencing (Supplementary Figure S6). Therefore, the mature miRNA sequence can be modified in our ligand-responsive miRNA framework without compromising the ligand-control function encoded within the aptamer sequence.
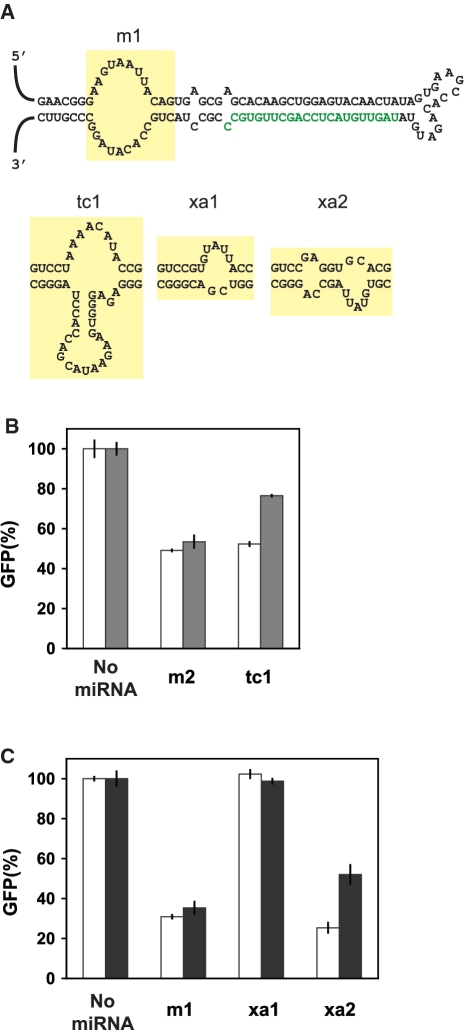
To examine the modularity of the aptamer sequence, we tested two additional aptamers that display different lengths and secondary structures: the tetracycline aptamer (31) and the xanthine aptamer (32). The binding core of each aptamer was initially integrated adjacent to the lower stem in place of the bulge (Figure 3A), and the resulting miRNAs were tested using the cell-culture assay in the presence or absence of the cognate ligand. Hypoxanthine was used as a soluble alternative to xanthine that binds the aptamer with comparable affinity (32). Control miRNAs that produced similar GFP basal levels to the ligand-responsive miRNAs were also tested to determine any non-specific impact of ligand addition on GFP levels. The miRNA harboring the tetracycline aptamer (tc1) reduced GFP basal levels to 52% and mediated a 1.5-fold increase in GFP levels in the presence of tetracycline (Figure 3B). The control miRNA (m2) delivered a similar extent of silencing with negligible tetracycline dependence, indicating that insertion of the tetracycline aptamer rendered gene silencing sensitive to tetracycline. However, compared to the theophylline aptamer, insertion of the tetracycline aptamer imparted reduced silencing and ligand sensitivity. The altered silencing in the absence of ligand may be attributed to the nature of the unbound aptamer structure, where the tetracycline aptamer folds into a preformed pocket (33). The altered regulatory response may be attributed to aptamer affinity, the relative membrane permeability of each small molecule and the extent to which each aptamer adopts a more structured conformation upon ligand binding.
Figure 3.
Ligand-responsive miRNAs can accommodate different aptamers to tailor the input-responsiveness of the regulatory system. (A) Sequence and secondary structure of GFP-targeting miRNAs with the tetracycline (tc1) or hypoxanthine (xa1, xa2) aptamers inserted into the basal segments. tc1 and xa1 contain the aptamer-binding core, xa2 contains the aptamer-binding core and an adjacent internal loop present in the original aptamer. See Figure 1B for notation. (B and C) Relative GFP levels obtained from the cell-culture assay for constructs transiently transfected in the absence (white) or presence of either 100 μM tetracycline (gray) or 5 mM hypoxanthine (black). See Figure 1D and E for description of the assay. m1 and m2 were used as negative controls as they result in similar levels of GFP silencing as xa2 and tc1, respectively, in the absence of ligand. Error bars reflect the range of values from two independent transfections conducted in the same cell-culture plate. See Supplementary Figure S4E and F for results from independent transfection experiments for (B) and (C), respectively.
Insertion of the binding core of the xanthine aptamer (xa1) completely abolished silencing (Figure 3C). The size of the bulge in the basal segments of xa1 was similar to that in m4, the miRNA with the smallest bulge tested (Figure 1B). Therefore, the small size of the xanthine-binding core may similarly prevent proper Drosha processing. We hypothesized that an aptamer with a small binding core bulge can be made more unstructured by including additional bulges, thereby restoring proper processing. Most aptamers selected in vitro contain loops that are separate from the binding core. To decrease the structure of the basal segments without compromising binding activity, we included the loop of the original xanthine aptamer and inverted the aptamer sequence to ensure hypoxanthine binding was proximal to the lower stem (xa2). Results from the cell-culture assay show that inserting the aptamer loop restored GFP silencing (25% basal levels) and mediated a 2.1-fold increase in GFP levels in the presence of hypoxanthine (Figure 3C), whereas hypoxanthine had no effect on a control miRNA with a similar silencing efficiency (m1). These studies support the generality of our design strategy based on the demonstrated ability of aptamer–ligand-binding interactions to modulate miRNA processing and subsequent targeted gene silencing.
Ligand-responsive miRNA clusters can be engineered to achieve tunable genetic control
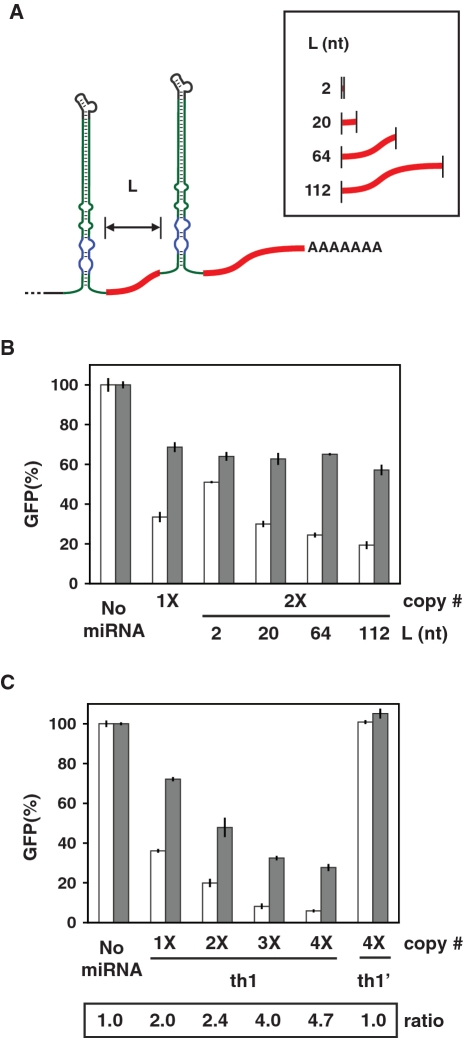
Multiple miRNAs have been found in natural clusters within a single transcript (34), allowing cells to efficiently regulate multiple miRNAs from a single promoter. By integrating multiple miRNAs into the same transcript, researchers have exploited this architecture to target multiple genes or tune gene silencing (35–37). Integrating ligand-responsive miRNAs into clusters would allow tuning of the regulatory response, simultaneous regulation of different targets and multi-input control of gene expression.
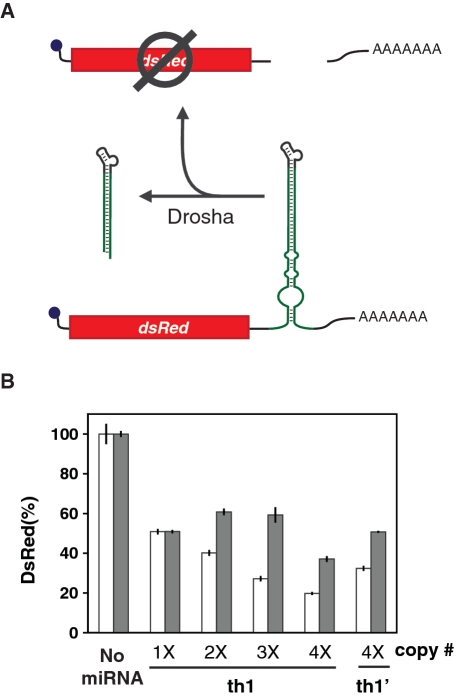
Evidence from natural miRNA clusters suggests that the spacer length between miRNAs can affect miRNA levels (38). To examine the effects of spacer length on gene silencing and ligand control in ligand-responsive miRNA clusters, we inserted a second copy of the th1 miRNA upstream from the first copy in the 3′ UTR of the transcript encoding DsRed-Express. Different spacer sequences were inserted between the two miRNA copies that ranged from 2 to 112 nt and were selected from the region downstream of the first miRNA copy (Figure 4A, Supplementary Figure S1). We then performed the cell-culture assay with each construct in the presence or absence of theophylline. Adjacent placement of the miRNAs compromised both silencing and the response to theophylline, potentially due to miRNA misfolding or steric hindrance of Drosha processing (Figure 4B). Increasing the spacer length to 20 nt restored GFP silencing, and extending the spacer length to 112 nt further improved silencing (19 versus 34% basal levels) and the theophylline response (3.0- versus 2.1-fold increase) as compared to the single-copy construct. These results suggest that separating identical miRNAs by at least 20 nt improves processing and gene silencing.
Figure 4.
Synthetic ligand-responsive miRNA clusters allow tuning of the regulatory response. (A) Schematic of a synthetic miRNA cluster in which multiple ligand-responsive miRNAs are placed into the 3′ UTR of a transgene-encoded transcript. The spacer sequence downstream of each miRNA (indicated in red) was kept consistent, and the spacer length (L) was varied between 2 and 112 nt. Multiple copies (from 1 to 4) of a single miRNA were sequentially inserted. (B) The impact of spacer length between two theophylline-responsive GFP-targeting miRNAs (2×, th1) on gene silencing in the presence (gray) or absence (white) of 5 mM theophylline. The GFP-targeting miRNAs were cloned into the plasmid constructs and characterized through the cell-culture assays described in Figure 1D. GFP levels are reported as described in Figure 2F. The GFP silencing from a single-copy theophylline-responsive miRNA construct (1×, th1) is shown for comparison. (C) The impact of ligand-responsive miRNA copy number on gene silencing and dynamic range. Multiple copies (#×) of the GFP-targeting (th1) or non-targeting (th1′, Figure S6) theophylline-responsive miRNAs were cloned into the plasmid constructs described in Figure 1D using the largest spacer length tested (112 nt). GFP levels were characterized and reported as described in Figure 1D, and the dynamic range is reported as the ratio of GFP levels in the presence and absence of theophylline. Error bars reflect the range of values from two independent transfections conducted in the same cell-culture plate.
Gene silencing and the dynamic range improved when two copies of a ligand-responsive miRNA were separated by the longest spacer tested (112 nt). As miRNAs in natural clusters are assumed to be individually processed, we expected that inserting additional copies separated by appropriate spacer lengths would further improve silencing and the theophylline response. Constructs harboring up to four copies of the theophylline-responsive, GFP-targeting miRNA (th1) or four copies of the non-targeting variant (th1′) separated by the longest spacer sequence were subjected to the cell-culture assay. Each additional copy of th1 improved GFP silencing (36% basal levels for one copy versus 6% basal levels for four copies) and the theophylline response (2.0-fold increase for one copy versus 4.7-fold increase for four copies), whereas four copies of th1′ had no effect on GFP levels (Figure 4C). The addition of each miRNA copy increased silencing and the dynamic range (measured as the ratio of GFP levels in the presence or absence of theophylline) by providing more miRNAs for Drosha recognition and more opportunities to inhibit processing. Despite the improved dynamic range, the addition of each miRNA copy decreased GFP levels in the presence of theophylline. This decrease may be attributed to the inability to access higher theophylline concentrations due to cytotoxicity (39) or incomplete inhibition of Drosha processing when theophylline is bound to the aptamer. Overall, changing the copy number of ligand-responsive miRNAs provides one approach to coordinately tune gene silencing and the dynamic range.
Ligand-responsive miRNA clusters can regulate endogenous genes
Most of the synthetic ligand-responsive RNA-based regulatory systems are encoded in the target transcript, providing regulation in cis (7,8). However, the regulation of endogenous genes through cis-regulatory strategies is currently limited by the lack of directed recombination technologies. To test whether ligand-responsive miRNAs provide a trans-regulatory strategy for the effective regulation of endogenous genes, we incorporated a mature miRNA sequence against the endogenous La gene into the control miRNA wt (La1) and the theophylline aptamer-containing miRNA th1 (La2) (Supplementary Figure S7). Four copies of each miRNA separated by the longest spacer sequence were inserted into the 3′-UTR of the transcript encoding DsRed-Express, and the resulting constructs were stably integrated into a single site in 293-Flp-In cells. Stable cell lines were grown in the presence or absence of 1.5 mM theophylline for over 1 week and assayed for relative La transcript levels by qRT–PCR. The reduced theophylline concentration from 5 to 1.5 mM was used for all stable expression studies to maintain 293 growth for extended periods of time (data not shown). Four copies of either La1 or La2 resulted in relatively strong silencing of the La target (19 and 22% basal levels, respectively), and only La2 mediated a theophylline-dependent increase of 2.2-fold in La transcript levels (Supplementary Figure S7). The observed fold-change represents a fraction of the full dynamic range since higher theophylline concentrations could not be used. These results demonstrate that ligand-responsive miRNAs can control endogenous genetic targets, providing a control strategy that does not physically disrupt the locus of the target gene.
Ligand-responsive miRNAs can control gene expression in cis
Drosha cleavage is anticipated to separate the coding region from the poly(A) tail when the miRNA is located in the 3′ UTR, resulting in the prevention of translation and facilitation of mRNA degradation (Figure 5A). Drosha was recently shown to cleave a naturally occurring pseudo-miRNA in the transcript of DGCR8 to regulate the activity of the Microprocessor (40). In addition, introducing a 3′ UTR-encoded miRNA was shown to downregulate expression from the transcript harboring the miRNA (41). From these insights we hypothesized that ligand-responsive miRNAs could be utilized to regulate gene expression in cis.
Figure 5.
Ligand-responsive miRNAs can control transgene expression in cis. (A) Schematic of gene regulation in cis through miRNA cleavage. Drosha processing of the miRNA located in the 3′ UTR of a transcript encoding DsRed Express separates the coding region from the poly(A) tail, thereby inactivating the transcript. (B) The impact of ligand-responsive miRNA copy number on expression of the transgene through regulation in cis in the presence (gray) or absence (white) of 5 mM theophylline. DsRed-Express levels of the constructs tested in Figure 4C were characterized through identical cell-culture assays. The population mean of DsRed-Express fluorescence from transiently transfected HEK 293 cells stably expressing GFP was normalized to that from a construct lacking any miRNAs (no miRNA) transfected under the same conditions. Error bars reflect the range of values from two independent transfections conducted in the same cell-culture plate.
We first examined the effects of ligand-responsive GFP-targeting miRNAs encoded in the 3′ UTR of the DsRed-Express gene on DsRed expression. DsRed-Express levels were quantified by flow cytometry under similar conditions as in the trans-targeted gene silencing experiments. The presence of one copy of th1 reduced DsRed levels to 51% and introducing additional copies with a spacer length of 112 nt further reduced DsRed levels (21% for four copies). Furthermore, two copies were sufficient to introduce ligand regulation, with a maximum observed fold-induction of 2.2 for three miRNA copies (Figure 5B). Similar effects were observed with four copies of the non-targeting miRNA (th1′), indicating that DsRed transcript silencing and ligand control are independent of mature miRNA activity and GFP targeting. qRT–PCR analysis confirmed that mRNA destabilization is the predominant regulatory mechanism of ligand-responsive miRNAs acting in cis (Supplementary Figure S8). A similar analysis of the La-targeting miRNAs revealed that four copies of either La1 or La2 reduced DsRed-Express levels ∼25-fold in the absence of theophylline, again supporting the notion that this cis-effect is not affected by RNAi-related downstream events (Supplementary Figure S7C). In addition, the miRNA containing the theophylline aptamer (La2) conferred a 3-fold increase in DsRed levels in the presence of theophylline that was not observed for the construct lacking the theophylline aptamer (La1) (Supplementary Figure S7C). These results suggest that ligand-responsive miRNAs can regulate transcripts in cis by modulating Drosha cleavage.
Self-targeting miRNAs combine trans- and cis-regulation for a tighter control system
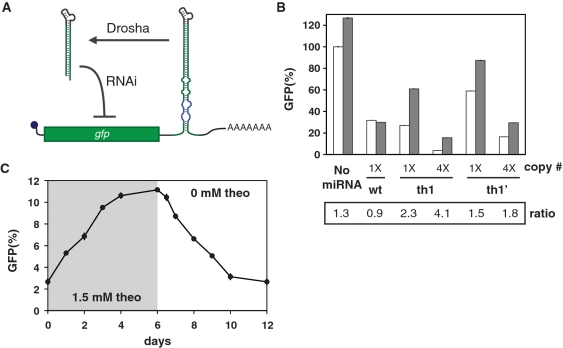
Ligand-responsive miRNAs can regulate genes in trans through RISC targeting and in cis through Drosha cleavage. A control system based on combining both modes of regulation into ‘self-targeting miRNAs’ may offer tighter regulation while still operating within the 3′ UTR of the transgene-encoding transcript. We developed a dual-acting miRNA circuit based on the insertion of ligand-responsive miRNAs or miRNA clusters into the 3′ UTR of a GFP transcript targeted by these miRNAs (Figure 6A). Here, the presence of ligand is expected to modulate direct destabilization of the cleaved transcript and subsequent RISC-mediated inactivation of other target transcripts. We tested five constructs to assess the relative impact of copy number and the cis and trans-regulatory mechanisms (Figure 6B). The resulting constructs were stably integrated into 293-Flp-In cells to ensure consistent expression. Cells were grown in the presence or absence of 1.5 mM theophylline for over a week and analyzed by flow cytometry (Figure 6B and Supplementary Figure S9).
Figure 6.
Self-targeting miRNAs provide an enhanced regulatory response. (A) Schematic of the miRNA regulatory circuit associated with self-targeting miRNAs. Self-targeting miRNAs are located in the 3′ UTR of the targeted transcript encoding GFP such that Drosha cleavage and RISC targeting downregulate expression. Both events are inhibited by ligand binding to the aptamer contained in the miRNA basal segments. (B) Relative GFP levels for cells stably expressing the self-targeting miRNA constructs grown in the presence (gray) or absence (white) of 1.5 mM theophylline for over a week. Gene silencing from one (1×) or four (4×) copies of a theophylline-responsive self-targeting miRNA (th1) and one or four copies of a theophylline-responsive non-targeting miRNA (th1′) were determined, where multiple copies were separated by the largest spacer length (112 nt). One copy of a self-targeting miRNA with basal segments similar to miR-30a (wt) was used as a negative control. The dynamic range is reported as the ratio of GFP levels in the presence and absence of theophylline. (C) Temporal response of the relative GFP levels to a change in ligand concentration for cell lines stably expressing ligand-responsive miRNAs. Representative time course data is shown for cells expressing the miRNA construct containing four copies of th1. Cells were grown in the presence of 1.5 mM theophylline for 6 days and then transferred to media without theophylline for 6 days. Error bars reflect the range of values from cells grown in three separate wells in the same cell-culture plate. See Supplementary Figure S4G for results from an independent experiment for (B).
Flow cytometry results revealed that each parameter (copy number, cis-regulation, cis and trans regulation) contributed to gene silencing and the theophylline response. The non-targeting miRNAs (th1′) had the weakest impact, with 59% GFP basal levels and a 1.5-fold induction from theophylline with single copy. The self-targeting miRNAs (th1) had a much stronger impact, with 27% basal levels and a 2.3-fold induction from theophylline at single copy. Increasing the copy number of the non-targeting and self-targeting miRNAs from one to four improved both GFP silencing and the theophylline response, with 4% basal levels and a 4.1-fold induction from theophylline for four copies of the self-targeting miRNA. These results demonstrate the diverse tuning capabilities when encoding ligand-responsive miRNAs in a transcript 3′-UTR by implementing self-targeting and non-targeting miRNAs at different copy numbers.
We performed time course studies on the dual-acting miRNA circuits to examine the dynamic properties of these regulatory systems. Cell lines were grown in the presence of theophylline for 6 days and then grown in media without theophylline for another 6 days. Cell lines harboring th1 or th1′ in single or four copies exhibited increasing GFP levels when grown in the presence of theophylline and reached steady-state levels by Day 6 (Figure 6C and Supplementary Figure S10). GFP levels decreased upon removal of theophylline and returned to original levels after 4–6 days of growth in the absence of theophylline, indicating that the genetic control exerted by ligand-responsive miRNAs is reversible. After theophylline addition, cells containing the self-targeting miRNAs required an extra day to reach steady-state in comparison to cells containing the non-targeting miRNAs, likely due to the slow turnover of activated RISC molecules (Supplementary Figure S10) (42). The particular response dynamics associated with the cis and cis/trans-regulatory mechanisms offer yet another design parameter when specifying the regulatory performance of ligand-responsive miRNAs.
DISCUSSION
The construction of synthetic ligand-responsive miRNAs provides new insights into miRNA structure–function relationships
In this work we explored the structural requirements for Drosha processing and utilized this mechanistic insight to develop a regulatory framework based on the control of Drosha cleavage. While previous work had demonstrated that both strands of the basal segments must be present and unpaired for efficient Drosha processing (3,21,23), our work evaluated the effects of bulge size in the basal segments on Drosha processing and subsequent gene silencing. In particular, we show that Drosha processing and gene silencing can be modulated by making small changes to the bulge size, revealing a graded structure–function relationship.
Our work also demonstrates that the structural changes that occur upon aptamer–small molecule ligand binding are sufficient to drive the miRNA structure into a less-processable conformation. This finding provides an important contribution to the field by demonstrating how unique properties of RNA structure and molecular binding can impart complex functional properties onto an important class of regulatory molecules. The use of three aptamer–ligand pairs supports the ability of our integrated platform to control miRNA activity with diverse molecules, such as metabolites and disease markers, against which aptamers can be selected. In addition to selected aptamers, aptamers present in natural riboswitches may also provide sequences that can be integrated into such synthetic regulatory platforms (43). While most aptamers contain a bulge and stem-loop, alternative structures (44–46) may be incompatible with the described ligand-responsive miRNA framework. To address such broader implementation challenges, future work may focus on the development of modified frameworks that accept more diverse aptamer structures, the modification of aptamer selection procedures to preferentially select bulge-loop structures or the development of in vivo screening strategies for RNA-based regulatory systems that identify sequences that function as integrated sensing components (47).
Our results also indicate that the variable spacing and secondary structure within intervening sequences of natural-occurring miRNA clusters may be a key factor in miRNA processability and an evolutionary factor to tune miRNA processing and subsequent gene silencing activity. Future studies that correlate the secondary structures of natural miRNA clusters with the extent of processing of each miRNA will provide further insight into this relationship. From this enhanced understanding, molecular engineers may be able to design synthetic cluster expression platforms that minimize cluster length, while maximizing or even tuning the processing of the encoded miRNAs (37). Scalable synthetic cluster expression platforms will be important tools in the development of gene regulatory circuits based on simultaneous implementation of multiple ligand-responsive miRNAs to achieve finely tuned, combinatorial gene expression control schemes.
Finally, the synthetic ligand-responsive miRNA systems reported here may foreshadow the discovery of a prevalent strategy used by natural systems to achieve sophisticated control over miRNA regulatory networks. Since natural riboswitches functioning as miRNA regulators have not yet been discovered, the design of synthetic systems reported here suggests relevant architectures to assist in the identification of natural counterparts. For instance, bioinformatics searches for conserved sequences and secondary structures in the miRNA basal segments may uncover molecular recognition elements that modulate Drosha processing. Researchers have previously identified mechanisms that modulate miRNA activity based on binding interactions between RNA-binding proteins and miRNA loops (48) and target sites (49), lending further support to the likelihood of an as yet undiscovered natural regulatory strategy based on small-molecule control of miRNA activity.
Ligand-responsive miRNAs provide unique capabilities as research tools and therapeutic molecules
Given the considerable interest in using miRNAs as research tools and for therapeutic interventions (50,51), the ability to implement user-defined molecular control over miRNA regulatory effects is critical to facilitating applications in basic research and applied medicine. Previous strategies have expressed miRNAs from inducible promoter systems to achieve control over miRNA-mediated gene silencing (52). However, a programmable, integrated RNA-only platform provides several unique advantages that will support new studies and applications of miRNA regulatory systems. For example, strategies based on inducible promoter systems require the expression of additional transgenic proteins encoding transcriptional regulators. The stable, long-term expression of multiple heterologous proteins represents a significant challenge in primary cells and organisms. In addition, the clinical application of such systems is limited by concerns associated with potential nonspecific immunogenicity of the heterologous protein components. In contrast, the ligand-controlled miRNA platform integrates the desired regulatory function into a compact RNA-only system that does not require separate expression constructs for the control system, providing a significant implementation advantage.
The dynamic range of the ligand-responsive miRNA system (∼2- to 5-fold) was limited in part by the inability to fully inhibit Drosha processing. While it is possible that ligand binding to the miRNA basal segments does not fully inhibit processing, another likely contribution is the upper limit on the applied ligand concentration for the systems reported here. Incorporating higher-affinity aptamers or using ligands with desirable pharmacokinetic properties such as high cell permeability and low cytotoxicity may improve the dynamic range of the miRNA switches and fully inhibit processing. However, even with a limited dynamic range, other studies have shown such gene regulatory activities to be sufficient to control a number of physiological processes (29,53,54). The ability to finely tune the regulatory performance of our platform will aid in optimizing the system response in the presence and absence of selected ligands.
We demonstrated that the ligand-responsive miRNA platform is modular such that different aptamers can be directly integrated into the basal segment region to change the effector controlling gene silencing. Because new aptamers can be generated through standard selection schemes (55), the integrated RNA control system supports the direct construction of systems that regulate miRNA silencing through many different user-defined effectors (a property that is not shared with commonly used protein-based regulatory systems). For example, by integrating aptamers that bind pharmaceutical drugs, systems can be built that control miRNA silencing activities with clinically relevant drug molecules to enable safer and more effective therapeutic strategies for cell-based and gene therapies. In addition, the described platform can be used to build systems that control miRNA activities in response to endogenous metabolites, proteins and hormones to enable targeting of gene silencing to cell-specific states, including stem cells, tissue-specific cells, cancer cells, cells suffering from metabolic diseases and cells infected with pathogens. By implementing ligand-responsive miRNAs that respond to endogenous molecules, these regulators will provide a platform for reprogramming the cellular state according to the intracellular environment, supporting autonomous approaches to tissue engineering and disease treatment.
Finally, the ligand-responsive miRNA system provides a scalable platform for implementing multi-input, multi-output control schemes in mammalian cells. Specifically, the described system can be extended to the design of miRNA clusters composed of different miRNA silencing activities controlled by independent input signals in a compact platform. Such systems will allow for the simultaneous and independent perturbation of different components within a biological network, supporting the complex probing and programming of cellular function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Caltech Joseph Jacobs Institute for Molecular Engineering for Medicine (to C.D.S.); Alfred P. Sloan Foundation, fellowship (to C.D.S.); Bill and Melinda Gates Foundation (to C.D.S.); Department of Defense (W81XWH-06-1-0250 to C.D.S.); National Institutes of Health (fellowship to K.G.H, GM091298 to C.D.S.); National Science Foundation (fellowship to C.L.B.). Funding for open access charge: National Institutes of Health (GM091298 to C.D.S.).
Conflict of interest statement. We declare competing financial interests in the form of a pending patent application whose value may be affected by the publication of this article.
ACKNOWLEDGEMENTS
The authors thank A. Brown, M. Greenwood-Goodwin and J. Vowles for constructive comments on the article and V.N. Kim for providing the pCK-Drosha-FLAG and pCK-DGCR8-FLAG plasmids. C.L.B. and C.D.S. designed the experiments, Y.Y.C. performed the inducible promoter experiments, S.J.C. prepared the stable cell lines, K.G.H. helped perform the Drosha cleavage assays, C.L.B. performed all other experiments and C.L.B. and C.D.S. wrote the article.
REFERENCES
- 1.Cai X, Hagedorn CH, Cullen BR. Human microRNAs are processed from capped, polyadenylated transcripts that can also function as mRNAs. RNA. 2004;10:1957–1966. doi: 10.1261/rna.7135204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Lee Y, Jeon K, Lee JT, Kim S, Kim VN. MicroRNA maturation: stepwise processing and subcellular localization. EMBO J. 2002;21:4663–4670. doi: 10.1093/emboj/cdf476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lee Y, Ahn C, Han J, Choi H, Kim J, Yim J, Lee J, Provost P, Radmark O, Kim S, et al. The nuclear RNase III Drosha initiates microRNA processing. Nature. 2003;425:415–419. doi: 10.1038/nature01957. [DOI] [PubMed] [Google Scholar]
- 4.Gregory RI, Yan KP, Amuthan G, Chendrimada T, Doratotaj B, Cooch N, Shiekhattar R. The Microprocessor complex mediates the genesis of microRNAs. Nature. 2004;432:235–240. doi: 10.1038/nature03120. [DOI] [PubMed] [Google Scholar]
- 5.Han J, Lee Y, Yeom KH, Kim YK, Jin H, Kim VN. The Drosha-DGCR8 complex in primary microRNA processing. Genes Dev. 2004;18:3016–3027. doi: 10.1101/gad.1262504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Friedman RC, Farh KK, Burge CB, Bartel DP. Most mammalian mRNAs are conserved targets of microRNAs. Genome Res. 2009;19:92–105. doi: 10.1101/gr.082701.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Suess B, Weigand JE. Engineered riboswitches: overview, problems and trends. RNA Biol. 2008;5:24–29. doi: 10.4161/rna.5.1.5955. [DOI] [PubMed] [Google Scholar]
- 8.Win MN, Liang JC, Smolke CD. Frameworks for programming biological function through RNA parts and devices. Chem. Biol. 2009;16:298–310. doi: 10.1016/j.chembiol.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Osborne SE, Ellington AD. Nucleic acid selection and the challenge of combinatorial chemistry. Chem. Rev. 1997;97:349–370. doi: 10.1021/cr960009c. [DOI] [PubMed] [Google Scholar]
- 10.Kumar D, An CI, Yokobayashi Y. Conditional RNA interference mediated by allosteric ribozyme. J. Am. Chem. Soc. 2009;131:13906–13907. doi: 10.1021/ja905596t. [DOI] [PubMed] [Google Scholar]
- 11.An CI, Trinh VB, Yokobayashi Y. Artificial control of gene expression in mammalian cells by modulating RNA interference through aptamer-small molecule interaction. RNA. 2006;12:710–716. doi: 10.1261/rna.2299306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Beisel CL, Bayer TS, Hoff KG, Smolke CD. Model-guided design of ligand-regulated RNAi for programmable control of gene expression. Mol. Syst. Biol. 2008;4:224. doi: 10.1038/msb.2008.62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tuleuova N, An CI, Ramanculov E, Revzin A, Yokobayashi Y. Modulating endogenous gene expression of mammalian cells via RNA-small molecule interaction. Biochem. Biophys. Res. Commun. 2008;376:169–173. doi: 10.1016/j.bbrc.2008.08.112. [DOI] [PubMed] [Google Scholar]
- 14.Boudreau RL, Martins I, Davidson BL. Artificial microRNAs as siRNA shuttles: improved safety as compared to shRNAs in vitro and in vivo. Mol. Ther. 2009;17:169–175. doi: 10.1038/mt.2008.231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Grimm D, Streetz KL, Jopling CL, Storm TA, Pandey K, Davis CR, Marion P, Salazar F, Kay MA. Fatality in mice due to oversaturation of cellular microRNA/short hairpin RNA pathways. Nature. 2006;441:537–541. doi: 10.1038/nature04791. [DOI] [PubMed] [Google Scholar]
- 16.McBride JL, Boudreau RL, Harper SQ, Staber PD, Monteys AM, Martins I, Gilmore BL, Burstein H, Peluso RW, Polisky B, et al. Artificial miRNAs mitigate shRNA-mediated toxicity in the brain: implications for the therapeutic development of RNAi. Proc. Natl Acad. Sci. USA. 2008;105:5868–5873. doi: 10.1073/pnas.0801775105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bauer M, Kinkl N, Meixner A, Kremmer E, Riemenschneider M, Forstl H, Gasser T, Ueffing M. Prevention of interferon-stimulated gene expression using microRNA-designed hairpins. Gene. Ther. 2009;16:142–147. doi: 10.1038/gt.2008.123. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X, Zeng Y. The terminal loop region controls microRNA processing by Drosha and Dicer. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq645. doi:10.1093/nar/gkq1645 [Epub ahead of print, 21 July 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lee Y, Kim VN. In vitro and in vivo assays for the activity of Drosha complex. Methods Enzymol. 2007;427:89–106. doi: 10.1016/S0076-6879(07)27005-3. [DOI] [PubMed] [Google Scholar]
- 20.Zeng Y, Cullen BR. Sequence requirements for micro RNA processing and function in human cells. RNA. 2003;9:112–123. doi: 10.1261/rna.2780503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zeng Y, Cullen BR. Efficient processing of primary microRNA hairpins by Drosha requires flanking nonstructured RNA sequences. J. Biol. Chem. 2005;280:27595–27603. doi: 10.1074/jbc.M504714200. [DOI] [PubMed] [Google Scholar]
- 22.Zeng Y, Yi R, Cullen BR. Recognition and cleavage of primary microRNA precursors by the nuclear processing enzyme Drosha. EMBO J. 2005;24:138–148. doi: 10.1038/sj.emboj.7600491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Han J, Lee Y, Yeom KH, Nam JW, Heo I, Rhee JK, Sohn SY, Cho Y, Zhang BT, Kim VN. Molecular basis for the recognition of primary microRNAs by the Drosha-DGCR8 complex. Cell. 2006;125:887–901. doi: 10.1016/j.cell.2006.03.043. [DOI] [PubMed] [Google Scholar]
- 24.Hermann T, Patel DJ. Adaptive recognition by nucleic acid aptamers. Science. 2000;287:820–825. doi: 10.1126/science.287.5454.820. [DOI] [PubMed] [Google Scholar]
- 25.Jenison RD, Gill SC, Pardi A, Polisky B. High-resolution molecular discrimination by RNA. Science. 1994;263:1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- 26.Desai SK, Gallivan JP. Genetic screens and selections for small molecules based on a synthetic riboswitch that activates protein translation. J. Am. Chem. Soc. 2004;126:13247–13254. doi: 10.1021/ja048634j. [DOI] [PubMed] [Google Scholar]
- 27.Dickins RA, McJunkin K, Hernando E, Premsrirut PK, Krizhanovsky V, Burgess DJ, Kim SY, Cordon-Cardo C, Zender L, Hannon GJ, et al. Tissue-specific and reversible RNA interference in transgenic mice. Nat. Genet. 2007;39:914–921. doi: 10.1038/ng2045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bayer TS, Smolke CD. Programmable ligand-controlled riboregulators of eukaryotic gene expression. Nat. Biotechnol. 2005;23:337–343. doi: 10.1038/nbt1069. [DOI] [PubMed] [Google Scholar]
- 29.Win MN, Smolke CD. A modular and extensible RNA-based gene-regulatory platform for engineering cellular function. Proc. Natl Acad. Sci. USA. 2007;104:14283–14288. doi: 10.1073/pnas.0703961104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zeng Y, Wagner EJ, Cullen BR. Both natural and designed micro RNAs can inhibit the expression of cognate mRNAs when expressed in human cells. Mol. Cell. 2002;9:1327–1333. doi: 10.1016/s1097-2765(02)00541-5. [DOI] [PubMed] [Google Scholar]
- 31.Berens C, Thain A, Schroeder R. A tetracycline-binding RNA aptamer. Bioorg. Med. Chem. 2001;9:2549–2556. doi: 10.1016/s0968-0896(01)00063-3. [DOI] [PubMed] [Google Scholar]
- 32.Kiga D, Futamura Y, Sakamoto K, Yokoyama S. An RNA aptamer to the xanthine/guanine base with a distinctive mode of purine recognition. Nucleic Acids Res. 1998;26:1755–1760. doi: 10.1093/nar/26.7.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Muller M, Weigand JE, Weichenrieder O, Suess B. Thermodynamic characterization of an engineered tetracycline-binding riboswitch. Nucleic Acids Res. 2006;34:2607–2617. doi: 10.1093/nar/gkl347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Zhang Y, Zhang R, Su B. Diversity and evolution of MicroRNA gene clusters. Sci. China C Life Sci. 2009;52:261–266. doi: 10.1007/s11427-009-0032-5. [DOI] [PubMed] [Google Scholar]
- 35.Sun D, Melegari M, Sridhar S, Rogler CE, Zhu L. Multi-miRNA hairpin method that improves gene knockdown efficiency and provides linked multi-gene knockdown. Biotechniques. 2006;41:59–63. doi: 10.2144/000112203. [DOI] [PubMed] [Google Scholar]
- 36.Xia XG, Zhou H, Xu Z. Multiple shRNAs expressed by an inducible pol II promoter can knock down the expression of multiple target genes. Biotechniques. 2006;41:64–68. doi: 10.2144/000112198. [DOI] [PubMed] [Google Scholar]
- 37.Aagaard LA, Zhang J, von Eije KJ, Li H, Saetrom P, Amarzguioui M, Rossi JJ. Engineering and optimization of the miR-106b cluster for ectopic expression of multiplexed anti-HIV RNAs. Gene Ther. 2008;15:1536–1549. doi: 10.1038/gt.2008.147. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ballarino M, Pagano F, Girardi E, Morlando M, Cacchiarelli D, Marchioni M, Proudfoot NJ, Bozzoni I. Coupled RNA processing and transcription of intergenic primary microRNAs. Mol. Cell Biol. 2009;29:5632–5638. doi: 10.1128/MCB.00664-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Beisel CL, Smolke CD. Design principles for riboswitch function. PLoS Comput. Biol. 2009;5:e1000363. doi: 10.1371/journal.pcbi.1000363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Han J, Pedersen JS, Kwon SC, Belair CD, Kim YK, Yeom KH, Yang WY, Haussler D, Blelloch R, Kim VN. Posttranscriptional crossregulation between Drosha and DGCR8. Cell. 2009;136:75–84. doi: 10.1016/j.cell.2008.10.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stern P, Astrof S, Erkeland SJ, Schustak J, Sharp PA, Hynes RO. A system for Cre-regulated RNA interference in vivo. Proc. Natl Acad. Sci. USA. 2008;105:13895–13900. doi: 10.1073/pnas.0806907105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bartlett DW, Davis ME. Insights into the kinetics of siRNA-mediated gene silencing from live-cell and live-animal bioluminescent imaging. Nucleic Acids Res. 2006;34:322–333. doi: 10.1093/nar/gkj439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wieland M, Benz A, Klauser B, Hartig JS. Artificial ribozyme switches containing natural riboswitch aptamer domains. Angew. Chem. Int. Ed Engl. 2009;48:2715–2718. doi: 10.1002/anie.200805311. [DOI] [PubMed] [Google Scholar]
- 44.Tuerk C, MacDougal S, Gold L. RNA pseudoknots that inhibit human immunodeficiency virus type 1 reverse transcriptase. Proc. Natl Acad. Sci. USA. 1992;89:6988–6992. doi: 10.1073/pnas.89.15.6988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wilson C, Nix J, Szostak J. Functional requirements for specific ligand recognition by a biotin-binding RNA pseudoknot. Biochemistry. 1998;37:14410–14419. doi: 10.1021/bi981371j. [DOI] [PubMed] [Google Scholar]
- 46.Koizumi M, Breaker RR. Molecular recognition of cAMP by an RNA aptamer. Biochemistry. 2000;39:8983–8992. doi: 10.1021/bi000149n. [DOI] [PubMed] [Google Scholar]
- 47.Weigand JE, Sanchez M, Gunnesch EB, Zeiher S, Schroeder R, Suess B. Screening for engineered neomycin riboswitches that control translation initiation. RNA. 2008;14:89–97. doi: 10.1261/rna.772408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Guil S, Caceres JF. The multifunctional RNA-binding protein hnRNP A1 is required for processing of miR-18a. Nat. Struct. Mol. Biol. 2007;14:591–596. doi: 10.1038/nsmb1250. [DOI] [PubMed] [Google Scholar]
- 49.Kedde M, Strasser MJ, Boldajipour B, Oude Vrielink JA, Slanchev K, le Sage C, Nagel R, Voorhoeve PM, van Duijse J, Orom UA, et al. RNA-binding protein Dnd1 inhibits microRNA access to target mRNA. Cell. 2007;131:1273–1286. doi: 10.1016/j.cell.2007.11.034. [DOI] [PubMed] [Google Scholar]
- 50.Wang M, Xie H, Mi S, Chen J. Recent patents on the identification and clinical application of microRNAs and target genes. Recent Pat. DNA Gene Seq. 2007;1:116–124. doi: 10.2174/187221507780887063. [DOI] [PubMed] [Google Scholar]
- 51.Wang V, Wu W. MicroRNA-based therapeutics for cancer. BioDrugs. 2009;23:15–23. doi: 10.2165/00063030-200923010-00002. [DOI] [PubMed] [Google Scholar]
- 52.Wiznerowicz M, Szulc J, Trono D. Tuning silence: conditional systems for RNA interference. Nat. Methods. 2006;3:682–688. doi: 10.1038/nmeth914. [DOI] [PubMed] [Google Scholar]
- 53.Chen YY, Jensen MC, Smolke CD. Genetic control of mammalian T-cell proliferation with synthetic RNA regulatory systems. Proc. Natl Acad. Sci. USA. 2010;107:8531–8536. doi: 10.1073/pnas.1001721107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Sinha J, Reyes SJ, Gallivan JP. Reprogramming bacteria to seek and destroy an herbicide. Nat. Chem. Biol. 2010;6:464–470. doi: 10.1038/nchembio.369. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 55.Stoltenburg R, Reinemann C, Strehlitz B. SELEX–a (r)evolutionary method to generate high-affinity nucleic acid ligands. Biomol. Eng. 2007;24:381–403. doi: 10.1016/j.bioeng.2007.06.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.