Abstract
Background
Administration of recombinant human (rh) interleukin (IL)-7 leads to CD4 and CD8 T cell expansions in HIV-infected individuals, demonstrating promising capacity for immune reconstitution. However, a proportion of patients treated with rhIL-7 experience transient increases in plasma HIV-RNA (“blips”), possibly reflecting “purging” of a quiescent reservoir that provides a barrier to viral eradication.
Objective
To identify the sources of HIV detected during transient viremic episodes following IL-7 administration, viral quasispecies were analyzed in a total of 281 primary sequences derived from 7 patients who experienced the episodic blips following IL-7 therapy.
Method
The C2-V3 region of the HIV-1 env gene were sequenced from HIV-1 RNA in plasma and HIV DNA from peripheral blood mononuclear cells (PBMCs) obtained at baseline (day 0 of rhIL-7 therapy), during the episode of viral blips (day 4), and at a time when levels of plasma HIV-RNA had returned to less than 50 copies/ml (day 28).
Results
The HIV sequences detected during transient viremia following IL-7 administration were closely related to those of the plasma viruses present before and after cytokine administration. All virus quasispecies detected during blips were also present in proviral sequences in PBMCs.
Conclusion
The low level viremia induced by IL-7 likely reflects predominantly transient induction or release of virus from a pre-existing pool rather than activation of silent quasispecies.
Keywords: HIV-1, quasispecies, IL-7, transient HIV viremia, virus reservoir
Introduction
IL-7 is a member of the family of common gamma-chain receptor cytokines that plays an important role in T cell homeostasis and survival [1–3]. IL-7 has long been considered a potential therapeutic agent for HIV infection, as chronic HIV infection results in progressive CD4 T cell deficiency and IL-7 can enhance the proliferation and survival of T lymphocytes [4–6]. In 2009, two separate clinical trial studies independently reported that administration of recombinant human (rh) IL-7 was well tolerated and led to CD4 and CD8 T cell expansions in HIV-infected individuals, demonstrating promising capacity for immune reconstitution in that setting [7,8]. In both studies, a proportion of patients treated with rhIL-7 experienced transient increases in plasma HIV-RNA (“blips”), possibly reflecting “purging” of a quiescent reservoir that provides a barrier to viral eradication. To better understand the short-term effect of rhIL-7 on HIV species in vivo, the current study was undertaken to examine the temporal changes in HIV quasispecies that were found after administration of rhIL-7.
Materials and methods
Study participants
The study participants were HIV-1-infected adults who had plasma HIV RNA levels less than 50 copies/ml and CD4 T cell counts ≥100 cells/µL at screening and had received highly active antiretroviral therapy (HAART) for a minimum of 12 months. The study was approved by the institutional review boards of all participating sites and written informed consent was obtained from all participants. Subjects received a single dose of subcutaneous rhIL-7 on day 0. Details of the clinical trial (AIDS Clinical Trials Group protocol 5241, NCT00099671) have been previously published [8].
Polymerase chain reaction (PCR) and sequencing
Plasma (1.5–4 ml) and PBMCs (5×106) samples were obtained at baseline (day 0 of rhIL-7 therapy), during the episode of viral blips (day 4), and at a time when levels of plasma HIV-RNA had returned to less than 50 copies/ml (day 28). Single molecules of a 425 bp fragment, encompassing the C2-V3 region of the HIV-1 envelope gene, obtained through limiting dilution, were PCR-amplified and cloned into the pCR2.1-TOPO vector (Invitrogen) for sequence analysis of individual clones, as previously described [9]. On average, sequences from 8 independent PCR-amplified clones (range: 2–19, Table 1) were obtained for each sample.
Table 1.
Characteristics of study participants
| Subject ID | Dose of rhIL-7 |
Antiretroviral regimen |
Days | Total CD4 (cell/µl) |
HIV RNA in Plasma (copies/ml) |
HIV DNA in PBMC (copies/106 cells) |
Sequences detected by nested PCR |
Genetic diversity of plasma virus (θ) |
95% confidence interval |
|
|---|---|---|---|---|---|---|---|---|---|---|
| HIV DNA in PBMC |
HIV RNA in plasma |
|||||||||
| 250223 | 10 µg/kg | RTV, TDF, ATV, ddI | 0 | 362 | <50 | 26.1 | + (12) | + (4) | 0.012 | 0.003–0.058 |
| 4 | 427 | 125 | 16.8 | + (11) | + (16) | 0.043 | 0.022–0.086 | |||
| 28 | 504 | 96 | 17.2 | + (12) | + (12) | 0.028 | 0.013–0.064 | |||
| 251104 | 10 µg/kg | TDF, ddI, LPV/RTV | 0 | 1174 | <50 | 6.8 | + (11) | + (6) | 0.091 | 0.039–0.266 |
| 4 | 1317 | 56 | 3.8 | + (9) | + (11) | 0.096 | 0.050–0.202 | |||
| 28 | 1476 | <50 | 15.1 | + (5) | − | |||||
| 251536 | 10 µg/kg | ABC, FTC, ATV | 0 | 694 | <50 | 28.5 | + (13) | + (6) | 0.015 | 0.005–0.050 |
| 4 | 745 | 73 | 26.4 | + (6) | + (10) | 0.031 | 0.014–0.075 | |||
| 28 | 796 | <50 | 35.3 | + (9) | + (10) | 0.003 | 0.001–0.010 | |||
| 272661 | 30 µg/kg | TDF, FTC, EFV | 0 | 972 | <50 | 2.2 | + (15) | − | ||
| 4 | n.a. | 154 | 18.2 | + (6) | + (9) | 0.018 | 0.007–0.051 | |||
| 28 | 1843 | 61 | 8.7 | + (15) | − | |||||
| 381546 | 30 µg/kg | NVP, FTC, TDF | 0 | 240 | <50 | N.D. | N.D. | − | ||
| 4 | n.a. | 79 | N.D. | N.D. | + (6) | 0.026 | 0.010–0.083 | |||
| 28 | 453 | <50 | N.D. | N.D. | + (4) | 0.015 | 0.004–0.070 | |||
| 381666 | 30 µg/kg | EFV, 3TC, TDF | 0 | 212 | <50 | N.D. | N.D. | + (2) | 0.096 | 0.020–1.714 |
| 4 | 298 | 83 | N.D. | N.D. | + (19) | 0.042 | 0.022–0.080 | |||
| 28 | 446 | <50 | N.D. | N.D. | + (6) | 0.004 | 0.000–0.017 | |||
| 252400 | 60 µg/kg | FTC/TDF, EFV | 0 | 660 | <50 | 7.3 | + (2) | + (2) | 0.000 | 0.000–0.000 |
| 4 | 629 | 66 | 5.8 | + (10) | + (9) | 0.004 | 0.001–0.017 | |||
| 28 | n.a. | <50 | 5.9 | + (5) | + (6) | 0.016 | 0.005–0.054 | |||
Abbreviations: ABC, abacavir; ATV, atazanavir; ddI, didanosine; EFV, efavirenz; FTC, emtricitabine; FTC/TDF, truvada; LPV/RTV, kaletra; NVP, nevirapine; RTV, ritonavir; TDF, tenofovir disoproxil fumerate; 3TC, lamivudine.
N.D., not done; n.a., not available
Numbers in paretheses represent number of sequences obtained for the envelope genes.
+, Sequence detectable; −, sequence undetectable
Genotypic analyses
Phylogenetic relationships among the HIV envelope sequences were estimated, by use of the neighbor-joining method, with the PAUP* program (Swofford, DL. 2003. PAUP* Phylogenetic analysis using parsimony and other methods. Version 4.0b10. Sinauer Associates). The sequence data used in this study have been deposited in GenBank and are available under the accession numbers HM118852-HM119132. To elucidate the genealogical relationships among the HIV envelope sequences, a haplotype network was constructed for subjects 251536 and 250223 using TCS v1.21 [10]. Genetic diversity was estimated with the use of a phylogenetic approach to estimate nucleotide diversity implemented in LAMARC v.2.1.3 [11].
Measurement of HIV-1 DNA copy numbers
Levels of HIV DNA in the PBMC were measured by quantitative real-time PCR as described elsewhere [12].
Results
Transient “blips” in plasma viremia were detected during the days of observed peak T cell proliferation (day 4) and increased T cell counts (day 14) in 7 of 15 rhIL-7 recipients with HIV-RNA <50 copies/ml at study entry (Table 1) [8]. The magnitude of the blips was low (median: 79 copies/ml, range: 56–154 copies/ml) and plasma HIV levels returned to less than 50 copies/ml by day 28 in all subjects except subjects 272661 and 250223 in whom suppression to <50 copies/mL was achieved subsequently as reported before [8].
Proviral DNA was present in all PBMC samples analyzed. Levels of proviral DNA remained stable during the 28-day sampling period: day 0, median 7.3 copies/106 cells (range 2.2–28.5); day 4, median 16.8 copies/106 cells (range 3.8–26.4); day 28, median 15.1 copies/106 cells (range 5.9–35.3) (d0 vs d4, p=0.50; d4 vs d28, p=0.35; d0 vs d28, p=0.69, Wilcoxon signed-rank test). Baseline level of provirus did not appear to predict the likelihood of blipping during IL-7 therapy, as the highest level of blips was seen in a patient with the lowest HIV proviral DNA (subject 272661). By use of a nested PCR technique, plasma virus could be detected in the majority of plasma samples including samples with viral RNA levels <50 copies/ml. Overall, The genetic diversity of the plasma virus population appeared to be fairly stable in each subject and did not show a trend towards an increase or decrease during the sampling period: day 0, median θ=0.014 (range 0.000–0.096); day 4, median θ=0.037 (range 0.004–0.043); day 28, median θ=0.010 (range 0.003–0.028)(d0 vs d4, p=0.72; d4 vs d28, p=0.14; d0 vs d28, p=1.00). The genetic diversity was independent of number of sequences obtained (r=−0.33, p=0.24) and of the magnitude of blips (r=0.10, p=0.34). Indeed, the highest genetic diversity was found in subject 251104 who had the lowest level of HIV RNA in the blips. Additionally, we found no association between increases in cell cycling and viral evolution (the minimum change, 0.76% divergence from day 0, was observed in subject 252400 who had the highest increase in cell cycling) [8].
To examine the temporal changes in HIV quasispecies that were found after administration of IL-7, phylogenetic relationships between viral quasispecies derived from plasma and PBMCs were examined. Two representative patterns are shown in Fig. 1. Overall, the plasma virus detected on day 4 was indistinguishable from the viral quasispecies present at day 0 and 28 within an individual patient (Fig. 1b and 1d).
Fig. 1.
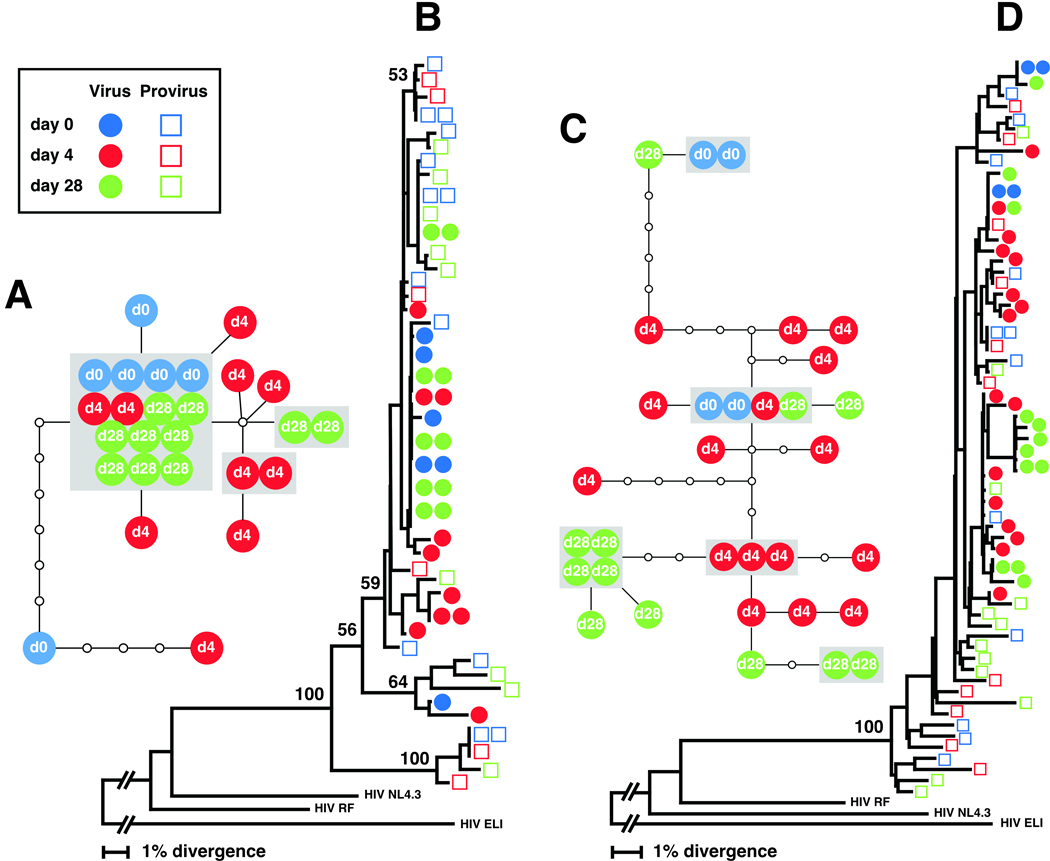
Phylogenetic analysis of the HIV-1 envelope sequences obtained from plasma virion RNA and proviral DNA in PBMC at baseline (day 0 of rhIL-7 therapy, when plasma HIV-RNA levels <50 copies/ml), during the time point of viral relapse (day 4), and a time point when levels of plasma HIV-RNA levels returned to below 50 copies/ml (day 28) from subjects 251536 (a, b) and 250223 (c, d). Phylogenetic relationships among the C2-V3 region of the env gene were estimated using the neighbor-joining method (b: subject 251536, d: subject 250223). Bootstrap percentile values from 1000 replications are shown at nodes defining major grouping of sequences. Statistical support of 50% or greater is shown. Colors identify times when samples were obtained. An additional phylogenetic tree analysis, which involved the envelope sequences obtained from all 7 patients, revealed that isolates from each patient were well confined within distinct patient-specific clusters that were divergent from each other and from known laboratory strains of HIV-1, confirming the absence of cross-patient contamination (data not shown). HIV-1 ELI was used as an outgroup. Genotype network analysis was also conducted for the HIV-1 envelope sequences obtained from plasma virus at days 0, 4, and 28 (a: subject 251536, c: subject 250223). Each colored circle represents an actual cloned sequence. The colored circles inside the grey box represent an identical sequence. A small open circle represents putative quasispecies in the evolutionary pathway. Each solid line represents one mutational step and connects two quasispecies with at least a 95% degree of confidence.
The phylogenetic tree for subject 251536 is segregated into three separate clusterings, supported by 59, 64, and 100 bootstrap values, which are statistical estimates of the reliability of a given cluster in a tree. Approximately 92% of plasma virus sequences (24 out of 26) of subject 251536 can be found in the main trunk of the tree with the bootstrap value of 59, in which the plasma virus quasispecies are found in an intermingled manner, irrespective of sampling time points (Fig. 1b). Short branch lengths seen for the plasma virus sequences indicate that the virus population is genetically homogeneous. Similarly, all plasma virus sequences derived from subject 250223 are intermixed and found distributed throughout the main trunk of the phylogenetic tree (Fig. 1d). Additionally, all plasma viruses detected in the 7 subjects were predicted to be CCR5-tropic.
Overall, provirus quasispecies are more genetically heterogeneous than the plasma virus population, as evidenced by the presence of multiple branches within the monophyletic group and the longer branch lengths in both subject cases. All proviruses are found distributed throughout the phylogenetic tree. The episodic blips did not have any substantial impact on the distribution pattern. Instead, the diverse provirus population present at baseline persisted throughout the sampling period. In both subjects, the day 4 virus sequences from the blipping time point clustered with proviral DNA sequences. This indicates that the viruses emerging during the episode of blips were not only similar to the virus quasispecies detected before and after the rhIL-7 therapy but also were genetically indistinguishable from provirus quasispecies detected during the sampling time point.
To further elucidate the genealogical relationships among the plasma virus sequences, a haplotype network was constructed (Fig. 1a and 1c). The genotype network diagram inferred from plasma virus sequences derived from subject 251536 clearly demonstrates that plasma virus population is genetically homogenous (Fig. 1a). Approximately half of the plasma virus sequences (54%) are found to be identical at the nucleotide level, reflected as a hub formation in the center of the diagram. The rest of plasma virus sequences cluster around the main hub, being separated only by 1–2 nucleotide changes. The plasma virus population in subject 250223 is less homogeneous, as reflected by the presence of increased number of steps connecting two virus quasispecies (Fig. 1c). However, all day 4 virus quasispecies from subject 250223 are still found within a maximum of 7-nucleotide substitution away from the day 0 quasispecies. In addition, no shift in the genealogy of plasma virus population was observed during the sampling period.
Discussion
In the present study, we have clearly demonstrated that plasma viruses detected during episodic HIV viremia following administration of IL-7 resemble the viruses present in the plasma immediately before and after rhIL-7 therapy. By examining the temporal relationship among plasma virus quasispecies derived from baseline (when plasma HIV-RNA levels were <50 copies/ml), during the episode of blips, and the time when levels of plasma HIV-RNA returned to less than 50 copies/ml, we were able to demonstrate that the viruses detected at the times of episodic HIV viremia were derived from the same source(s) giving rise to viruses in plasma before and after rhIL-7 therapy.
Plasma viruses detected during HIV viral blips were not only similar to the viral quasispecies present before and after the initiation of rhIL-7 therapy but also genetically indistinguishable from provirus quasispecies detected during the sampling period. Levels of genetic diversity for the plasma virus quasispecies population remained fairly stable within a given patient. In addition, no shift in the genealogical relationships among the plasma virus quasispecies was associated with the initiation of rhIL-7 therapy. Given these observations, it is likely that rhIL-7 is inducing a transient viral burst primarily by amplifying virus present before IL-7 therapy, rather than inducing production from previously silent reservoir(s) [13,14].
Had silent reservoir(s) contributed to these transient viral bursts, an increased genetic diversity would be anticipated following the initiation of rhIL-7 therapy. However, the genetic diversity of the plasma virus quasispecies population remained fairly stable within a given patient with no particular trend towards an increase or decrease during the sampling period. In addition, unusual formation of sequence cluster that consists only of the blipping viruses was not observed in the phylogenetic trees presented in this study, further supporting the notion that activation of previously silent quasispecies played a minimal, if any, role in the HIV blips induced by rhIL-7. However, our observations are limited, since the sampling was only from peripheral blood and CD4 subsets were not studied due to limited sample availability.
Transient HIV viremia raises concerns that these episodes could reflect activation and further seeding of cellular reservoirs. In the present study, plasma virus returned to the baseline level by day 28 in 5 out of 7 patients and within no more than two months in the other two who had experienced blips after rhIL-7 administration. In addition, there was no significant increase in levels of proviral HIV DNA in PBMC after rhIL-7 in any studied subjects. It is therefore unlikely that the rhIL-7-induced HIV blips led to substantial reseeding or spread of viral reservoirs particularly in so far as these patients were receiving suppressive anti-retroviral therapy.
Taken together, these results indicate that transient low level increases in plasma HIV-RNA in response to a single dose of rhIL-7 do not result in substantial changes in viral quasispecies. Additional long-term follow up study on patients receiving multiple doses of rhIL-7 may be necessary to evaluate the long-term impact of rhIL-7 administration on HIV quasispecies.
Supplementary Material
Acknowledgements
This project has been funded in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract HHSN261200800001E, the National institute of Allergy and infectious Disease (NIAID), National Institutes of Health, under contract HHSN261200800001E and by the Intramural Research Program of NIAID. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
This study was supported in part by the AIDS Clinical Trials Group funded by the National Institute of Allergy and Infectious Diseases and the individual AIDS Clinical Trials Units at Case Western Reserve University (AI 25879), Rush University (AI 68636), Northwestern University (AI 25915), University of California, Davis Medical Center (AI 38858), and University of Miami (AI 27675). This work was also supported in part by the Intramural Research Program of the National Institutes of Health, National Institute of Allergy and Infectious Diseases. The authors thank all study participants and the ACTG5214 study team.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
M.M.L. has received research support from Cytheris. All other authors have declared no conflict of interest.
None of this material has been published or is under consideration for publication elsewhere.
Author’s contributions.
H.I. performed the experiments, analyzed the data, and wrote the paper. G.D. performed experiments. D.V.A., M.A.F., and A.L.L. contributed in study planning, data interpretation, and paper preparation. M.M.L. and I.S. contributed in study planning, data interpretation, and wrote parts of the paper. All the authors have read and approved the text as submitted to AIDS.
References
- 1.Fry TJ, Mackall CL. Interleukin-7: from bench to clinic. Blood. 2002;99:3892–3904. doi: 10.1182/blood.v99.11.3892. [DOI] [PubMed] [Google Scholar]
- 2.Mazzucchelli R, Durum SK. Interleukin-7 receptor expression: intelligent design. Nat Rev Immunol. 2007;7:144–154. doi: 10.1038/nri2023. [DOI] [PubMed] [Google Scholar]
- 3.Sasson SC, Zaunders JJ, Kelleher AD. The IL-7/IL-7 receptor axis: understanding its central role in T-cell homeostasis and the challenges facing its utilization as a novel therapy. Curr Drug Targets. 2006;7:1571–1582. doi: 10.2174/138945006779025365. [DOI] [PubMed] [Google Scholar]
- 4.Beq S, Nugeyre MT, Ho Tsong Fang R, Gautier D, Legrand R, Schmitt N, et al. IL-7 induces immunological improvement in SIV-infected rhesus macaques under antiviral therapy. J Immunol. 2006;176:914–922. doi: 10.4049/jimmunol.176.2.914. [DOI] [PubMed] [Google Scholar]
- 5.Nugeyre MT, Monceaux V, Beq S, et al. IL-7 stimulates T cell renewal without increasing viral replication in simian immunodeficiency virus-infected macaques. J Immunol. 2003;171:4447–4453. doi: 10.4049/jimmunol.171.8.4447. [DOI] [PubMed] [Google Scholar]
- 6.Nugeyre MT, Monceaux V, Beq S, Cumont MC, Ho Tsong Fang R, Chêne L, et al. IL-7 administration to humans leads to expansion of CD8+ and CD4+ cells but a relative decrease of CD4+ T-regulatory cells. J Immunother. 2006;29:313–319. doi: 10.1097/01.cji.0000210386.55951.c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Levy Y, Lacabaratz C, Weiss L, Viard JP, Goujard C, Lelièvre JD, et al. Enhanced T cell recovery in HIV-1-infected adults through IL-7 treatment. J Clin Invest. 2009;119:997–1007. doi: 10.1172/JCI38052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sereti I, Dunham RM, Spritzler J, Aga E, Proschan MA, Medvik K, et al. IL-7 administration drives T cell-cycle entry and expansion in HIV-1 infection. Blood. 2009;113:6304–6314. doi: 10.1182/blood-2008-10-186601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Imamichi H, Crandall KA, Natarajan V, Jiang MK, Dewar RL, Berg S, et al. Human Immunodeficiency virus type 1 quasi species that rebound after discontinuation of highly active antiretroviral therapy are similar to the viral quasi species present before initiation of therapy. J Infect Dis. 2001;183:36–50. doi: 10.1086/317641. [DOI] [PubMed] [Google Scholar]
- 10.Clement M, Posada D, Crandall KA. TCS: a computer program to estimate gene genealogies. Mol Evol. 2000;9:1657–1659. doi: 10.1046/j.1365-294x.2000.01020.x. [DOI] [PubMed] [Google Scholar]
- 11.Kuhner MK. LAMARC 2.0: maximum likelihood and Bayesian estimation of population parameters. Bioinformatics. 2006;22:768–770. doi: 10.1093/bioinformatics/btk051. [DOI] [PubMed] [Google Scholar]
- 12.Pasternak AO, Adema KW, Bakker M, Jurriaans S, Berkhout M, Cornelissen M, et al. Highly sensitive methods based on seminested real-time reverse transcription-PCR for quantitation of human immunodeficiency virus type 1 unspliced and multiply spliced RNA and proviral DNA. J Clin Microbiol. 2008;46:2206–2211. doi: 10.1128/JCM.00055-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wang FX, Xu Y, Sullivan J, Souder E, Argyris EG, Acheampong EA, et al. IL-7 is a potent and proviral strain-specific inducer of latent HIV-1 cellular reservoirs of infected individuals on virally suppressive HAART. J Clin Invest. 2005;115:128–137. doi: 10.1172/JCI22574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Scripture-Adams DD, Brooks DG, Korin YD, Zack JA. Interleukin-7 induces expression of latent human immunodeficiency virus type 1 with minimal effects on T-cell phenotype. J Virol. 2002;76:13077–13082. doi: 10.1128/JVI.76.24.13077-13082.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


