Abstract
Protein lysine methylation signaling is implicated in diverse biological and disease processes. Yet the catalytic activity and substrate specificity are unknown for many human protein lysine methyltransferases (PKMTs). We screened over forty candidate PKMTs and identified SETD6 as a methyltransferase that monomethylates chromatin-associated NF-κB RelA at lysine 310 (RelAK310me1). SETD6-mediated methylation rendered RelA inert and attenuated RelA-driven transcriptional programs, including inflammatory responses in primary immune cells. RelAK310me1 was recognized by the ankryin repeat of GLP, which under basal conditions, promoted a repressed chromatin state at RelA target genes through GLP-mediated H3K9 methylation. NF-κB activation-linked phosphorylation of RelA by PKCζ at serine 311 blocked GLP binding to RelAK310me1 and relieved target gene repression. Our findings establish a new mechanism by which chromatin signaling regulates inflammation programs.
INTRODUCTION
Chromatin dynamics regulate key cellular functions that influence survival, growth and proliferation, and disruption of chromatin homeostasis is implicated in diverse pathologic processes1. Histone lysine methylation, which is catalyzed by protein lysine methyltransferases (PKMTs), is a principal chromatin-regulatory mechanism involved in directing fundamental DNA-templated processes such as transcription and DNA repair1. Histone methylation plays a central part in orchestrating proper programming of the genome in response to various stimuli, and aberrant lysine methylation signaling is implicated in the initiation and progression of many human diseases2. Many non-histone proteins are also regulated by lysine methylation, indicating that this modification is likely a common mechanism for modulating protein-protein interactions and signaling pathways3.
NF-κB is a transcription factor and key inducer of inflammatory responses4,5. One of the principal subunits of NF-κB is RelA (p65 [http://www.signaling-gateway.org/molecule/query?afcsid=A001645]), which forms either a homodimer or a heterodimer with the structurally related p50 protein [http://www.signaling-gateway.org/molecule/query?afcsid=A002937]. Under basal conditions, the majority of NF-κB RelA is sequestered in the cytoplasm due to association with members of the IκB protein family4,5. Stimulation of cells with NF-κB-activating ligands like the cytokine tumor necrosis factor (TNF) results in degradation of IκB proteins and translocation of the released NF-κB to the nucleus where it directs various transcriptional programs5,6. In addition to this canonical pathway, there are several additional mechanisms that regulate and fine-tune NF-κB signaling and target gene activation7. For example, different post-translational modifications of RelA influence RelA target gene specificity, transcriptional activity and activation kinetics. Further, even at resting conditions, a population of RelA is present in the nucleus, bound at chromatin; however, the functional relevance of this constitutively nuclear population is not known.
Deregulation of NF-κB signaling is linked to many human diseases including cancer and autoimmune disorders8. Thus, understanding the full range of molecular mechanisms that modulate this factor in response to diverse conditions has important biological and clinical implications. Here we screened over forty known and candidate human PKMTs for in vitro methylation activity on RelA. We identify SETD6 (SET domain containing 6) as a PKMT that monomethylates RelA at K310 (RelAK310me1). The ankryin repeat of the PKMT GLP (also called G9A-like protein) functions as a recognition module for RelAK310me1, linking this mark to localized histone H3 lysine 9 (H3K9) methylation and repressed chromatin at RelAK310me1-occupied genes9,10. The SETD6-initiated lysine methylation signaling cascade acts to restrain activation of NF-κB-mediated inflammatory responses in diverse cell types. This repressive pathway is terminated by RelA phosphorylation at S311 by the atypical protein kinase C PKCζ11[http://www.signaling-gateway.org/molecule/query?afcsid=A002937], which blocks GLP recognition of RelAK310me1 to promote RelA target gene expression. Together, our findings identify SETD6 as a novel regulator of the NF-κB network, identify the ankryin repeat domain of GLP as the first known effector of methylated RelA, describe the first metazoan example of a methyl-phospho switch on a non-histone proteins, and demonstrate a new paradigm for how integrated crosstalk between modifications on transcription factors and histones modulates key physiologic and pathologic programs.
RESULTS
SETD6 monomethylates RelA at lysine 310
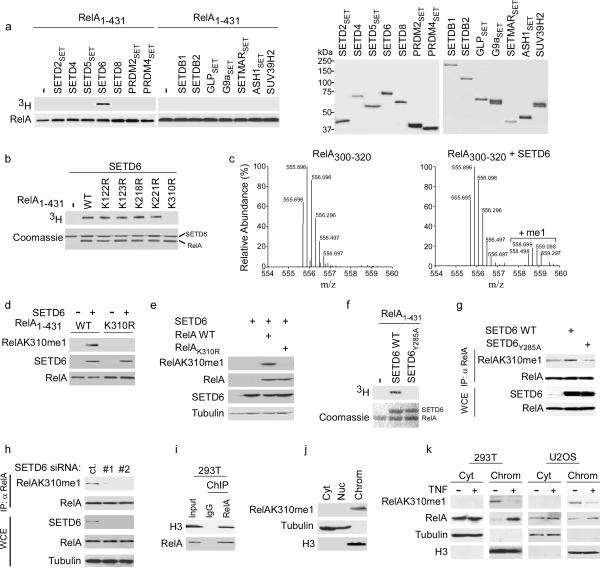
To identify novel activities for predicted PKMT enzymes and new lysine methylation events, we screened the majority of SET domain containing proteins present in the human proteome for in vitro catalytic activity on various histone and non-histone candidate substrates (Supplementary Table 1; data not shown). SETD6, a previously uncharacterized PKMT, methylated an N-terminal RelA polypeptide encompassing amino acids 1–431 (RelA1–431), but not a C-terminal polypeptide (residues 430–531) (Fig. 1a; Supplementary Fig. 1a,b). Substitution of individual lysine residues to arginines within RelA1–431 identified K310 as the target site of SETD6 (Fig. 1b). In contrast, SET7/9, which methylates RelA at several lysines12,13, was active on the RelAK310R mutant (Supplementary Fig. 1c). Mass spectrometry analysis of SETD6-catalyzed methylation assays on RelA peptides spanning K310 (RelA300–320) demonstrated that SETD6 only adds a single methyl moiety to RelA at K310 (Fig. 1c; Supplementary Fig. 2).
Figure 1. SETD6 monomethylates RelA at K310.
(a) Identification of SETD6 as a RelA lysine methyltransferase. Left, autoradiogram of methylation reactions utilizing recombinant RelA1–431 as substrate and recombinant enzymes (full-length or SET domain as indicated, see Methods). Right: Coomassie stain of recombinant enzymes used in the reactions. Representative data of three independent experiments is shown. (b) SETD6 methylates RelA at K310. Top: Autoradiogram of SETD6-catalyzed methylation assays with wild-type RelA1–431 or the indicated mutants. Bottom: Coomassie stain of proteins used in the reactions. Representative data of three independent experiments is shown. (c) SETD6 monomethylates RelA. Mass spectrometry analysis of methylation assays on RelA300–320 peptide ± SETD6 as indicated. Representative data of two independent experiments is shown. (d) In vitro monomethylation of RelA1–431 by SETD6. Immunoblot analysis with the indicated antibodies of methylation reactions with SETD6 on RelA1–431 or RelA1–431 harboring a K310R substitution. Representative data of two independent experiments is shown. (e) Exogenous SETD6 monomethylated overexpressed RelA at K310 in cells. Immunoblot analysis of whole-cell extracts (WCE) from 293T cells transfected with FLAG-SETD6 and either RelA or RelAK310R, probed with the indicated antibodies. WCE loaded represented 5% of total. Representative data of three independent experiments is shown. (f) Identification of Y285A as catalytically inactive SETD6 mutant. Top: Autoradiogram of in vitro methylation reaction with SETD6 or SETD6Y285A on RelA1–431. Bottom: Coomassie stain of recombinant proteins used in the reactions. Representative data of two independent experiments is shown. (g) Monomethylation of endogenous RelAK310 by SETD6 required an intact catalytic SET domain. Immunoblot analysis of RelA immunoprecipitations (IPs) or WCE (5% of total) from 293T cells transfected with the indicated SETD6 plasmids. Representative data of three independent experiments is shown. (h) Depletion of endogenous SETD6 decreased endogenous RelAK310me1. Immunoblot analysis as in (g) of 293T cells treated with control or two independent SETD6-targeted siRNAs. Representative data of three independent experiments is shown. (i) Association of RelA with H3 at chromatin in the absence of NF-κB stimulation. Immunoblot analysis of RelA or control IgG protein-protein ChIPs probed with the indicated antibodies. Input represented 5% of total. Representative data of two independent experiments is shown. (j) RelAK310me1 is a chromatin-associated RelA species. Immunoblot analysis of 293T cells biochemically separated into the following fractions: Cyt: cytoplasmic; Nuc: nucleoplasmic; Chrom: chromatin-enriched. Tubulin and H3 signal control for the integrity of fractionation. Representative data of two independent experiments is shown. (k) TNF treatment decreased detection of RelAK310me1 at chromatin. Immunoblot analysis of the indicated fractions as in (j) from 293T and U2OS cells ± treatment with TNF (10 ng/ml for 1 h). Representative data of two independent experiments is shown.
We raised antibodies against SETD6 (Supplementary Fig. 3) and the RelAK310me1 epitope (hereafter referred to as αRelAK310me1). The αRelAK310me1 antibody specifically recognized RelAK310me1 peptides and did not detect unmodified RelA, RelAK310me2/3, or numerous methylated histone peptides (Supplementary Fig. 4). Further, αRelAK310me1 detected RelA1–431 that had been methylated in vitro by SETD6, but failed to detect unmethylated RelA1–431 or RelA with a K310R substitution (Fig. 1d).
In co-transfection experiments, wild-type RelA, but not RelAK310R, was monomethylated by overexpressed SETD6 (Fig. 1e). Structure-based homology modeling predicted that SETD6 is most similar to the plant enzyme Rubisco large subunit methyltransferase (LSMT)14 (Supplementary Fig. 5). Based on this homology, Y285A of SETD6 (SETD6Y285A) was identified as a catalytic mutant in vitro (Fig. 1f; Supplementary Figs. 5,6), and overexpression of SETD6, but not SETD6Y285A, led to an increase in monomethylation of endogenous RelA at K310 (Fig. 1g). Finally, depletion of endogenous SETD6 protein in 293T cells by RNA interference (RNAi) with two independent siRNAs caused a decrease in the amount of endogenous RelAK310me1 (Fig. 1h). Based on these data we conclude that SETD6 monomethylates RelA at K310 in vitro and is required for maintenance of physiologic RelAK310me1 concentrations in cells.
RelAK310me1 is chromatin-associated under basal conditions
In unstimulated cells, while the majority of RelA is localized to the cytoplasm, a population of RelA is present in the nucleus15. In this context, protein-protein chromatin immunoprecipitation (ChIP) assays performed in the absence of stimulation demonstrated association of RelA with histone H3 (Fig. 1i). Moreover, in unstimulated cells, RelA was detected by ChIP at the promoters of several target genes in multiple cell types (Supplementary Fig. 7). SETD6 was also present in the nucleus (Supplementary Fig. 3b). RelA and SETD6 interacted in vitro and co-immunoprecipitated (IP) from cells (Supplementary Fig. 8a–d). Thus, we reasoned that in contrast to the majority of RelA, which is localized to the cytoplasm in unstimulated cells4, the population of RelA that is monomethylated at K310 might reside and function in the nucleus. In support of this hypothesis, RelAK310me1 biochemically fractionated almost exclusively with chromatin in unstimulated 293T and U2OS cells (Figs. 1j,k). TNF treatment, which activates NF-κB4, resulted in far lower amounts of RelAK310me1 at chromatin relative to no stimulation (Fig. 1k). Based on these data, we conclude that RelAK310me1 is present at chromatin in unstimulated cells.
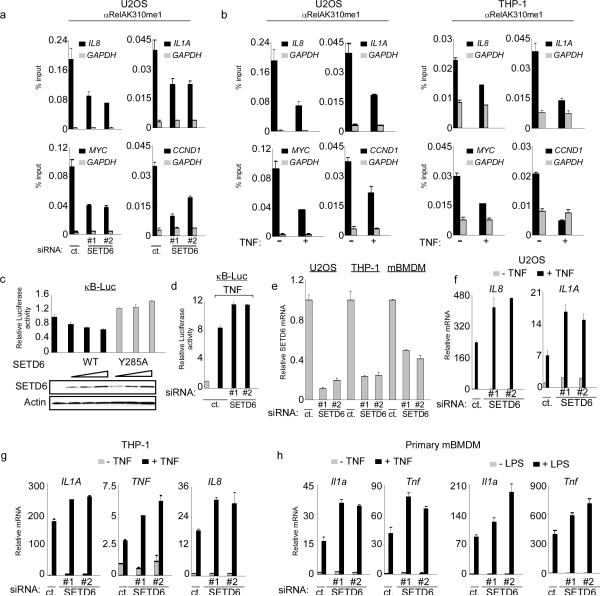
Next, ChIP assays were performed in U2OS cells under basal conditions to test if RelAK310me1 is bound to chromatin at RelA target gene promoters. RelAK310me1 occupied the promoters of several RelA-target genes (IL8, IL1A, MYC and CCND1), and the detection of this RelA species at target promoters was lost in U2OS cells treated with either of two independent siRNAs targeting SETD6 (Fig 2a; Supplementary Fig. 9a), as well as in U2OS cells stably expressing a SETD6-targeting shRNA (Supplementary Fig. 10). Consistent with the results observed in the cellular fractionation assays (see Figs. 1j,k), TNF treatment decreased RelAK310me1 occupancy at target promoters both in U2OS cells and in THP-1 cells, an acute monocytic leukemia cell line (Fig. 2b; Supplementary Fig. 9b). Thus, monomethylation of RelAK310 is a chromatin-associated modification and is inversely correlated with activation of NF-κB by TNF.
Figure 2. SETD6 monomethylation of RelA inhibits RelA transactivation activity.
(a) Enrichment of RelAK310me1 at promoters of RelA target genes required SETD6. Occupancy of RelAK310me1 in U2OS cells treated with control or SETD6 siRNAs at the promoters of IL8, IL1a, myc, ccnd1 (black bars) and gapdh (grey bars, as a control) was determined by real time (RT) PCR of ChIP samples. ChIP enrichment shown as % input (ChIP/input × 100). (b) Loss of RelAK310me1 occupancy at RelA target genes promoters in response to TNF treatment. ChIP assays as in (a) from U2OS (left) and THP-1 cells (right) ± TNF stimulation (20 ng/ml for 1hr). Negative control antibody ChIPs for (a) and (b) shown in Supplementary Fig. 9. (c) Repression of the NF-κB luciferase reporter (κB-Luc) by SETD6 was dose dependent and required an intact catalytic SET domain. κB-Luc reporter luciferase activity (normalized to Renila) and relative change compared to control transfection was determined 24 hrs after transfection of U2OS cells with increasing amounts of the indicated plasmids. Immunoblot analysis of SETD6 and SETD6Y285A expression is shown. (d) Depletion of SETD6 enhanced TNF-induced activity of an NF-κB driven reporter. Luciferase activity was determined as in (c) in 293T cells transfected with control or 2 independent SETD6 siRNAs ± TNF treatment (10 ng/ml for 1hr). (e) Efficiency of SETD6 mRNA knockdown by RNAi in the indicated cell lines was determined by RT PCR. (f–h) SETD6 depletion increased expression of RelA target genes in multiple cell types. RT PCR analysis of the indicated mRNAs from U2OS (f), THP-1 (g) and primary mouse BMDM (mBMDM) cells (h) transfected with control (ct.) or two independent SETD6 siRNAs ± TNF (20 ng/ml for 1 hr) or ± LPS (100 ng/ml for 1 hr) as indicated. Error bars in (a–h) indicate ± s.e.m. from at least three experiments.
SETD6 attenuates RelA target gene transcription
To investigate the relationship between SETD6 and the transcriptional activity of RelA, U2OS cells were co-transfected with an NF-κB-driven reporter16 and either SETD6 or SETD6Y285A. The activity of this reporter was repressed by SETD6 in a dose-dependent manner and required that the catalytic activity of SETD6 be intact (Fig. 2c). In addition, depletion of SETD6 increased reporter activity in unstimulated cells (Supplementary Fig. 11) and upon exposure to TNF (Fig. 2d). These data suggest that SETD6 represses physiologic transactivation by RelA. To test this hypothesis, SETD6 was depleted in U2OS, THP-1 and primary mouse bone marrow-derived macrophage (mBMDM) cells (Fig. 2e), and the mRNA abundance of canonical NF-κB targets was measured in the absence or presence of NF-κB stimulation (Fig. 2f–h)4. In response to TNF, SETD6 knockdown led to an increase of RelA target gene expression relative to control cells in all three cell types (Fig. 2f–2h). Similar results were observed when mBMDMs were stimulated with lipopolysaccharide (LPS) (Fig. 2h, right). In addition, SETD6 depletion resulted in higher basal expression of a subset of RelA target genes (Supplementary Fig. 12). We noted that SETD6 does not methylate the RelA partner p50 (Supplementary Fig. 1d), and in contrast to known histone lysine methyltransferases like G9a and GLP9,17, SETD6 did not methylate nucleosomes (Supplementary Fig. 1e). Finally, genome-wide gene expression analyses comparing Rela−/− MEFs18 reconstituted with mouse RelAwt or RelAK310R revealed that in the absence of stimulation, cells complemented with RelAK310R express a greater number of RelA-regulated genes and expression of these genes was higher than cells complemented with RelAwt (Supplementary Fig. 13), implicating K310 in regulating RelA target gene expression. Taken together, these results argue that SETD6-mediated methylation of RelAK310 has an inhibitory effect on expression of many RelA regulated genes.
SETD6 attenuates RelA-driven inflammatory responses
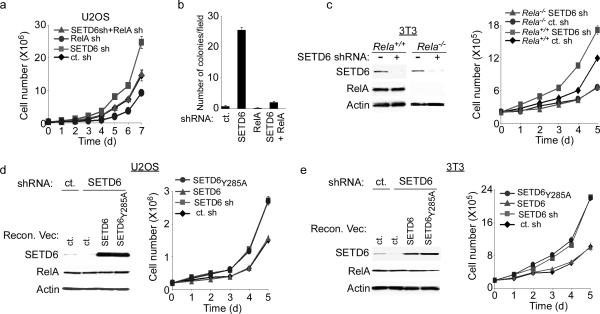
Hyperactive NF-κB has been linked to the development and progression of many types of cancer8. To investigate potential roles for SETD6 and SETD6-RelA interplay in cellular transformation, U2OS cells were established that stably expressed shRNAs targeting SETD6 alone, RelA alone, SETD6 and RelA, or a control shRNA, and tested for cell transformation-associated properties (Fig. 3a,b; Supplementary Fig. 14). Relative to the control, knockdown of SETD6 accelerated proliferation rates of cells in a RelA-dependent fashion (Fig. 3a). In addition, SETD6 depletion conferred a 25-fold increase in the ability of cells to form colonies in soft agar relative to control and RelA-depleted cells, and co-depletion of RelA with SETD6 reversed the anchorage-independent growth advantage provided by SETD6 knockdown alone (Fig. 3b). Depletion of SETD6 also led to an increase in cell proliferation rates in mouse embryonic fibroblast 3T3 cells, but not in Rela−/− 3T3 cells (Fig. 3c). Next, endogenous SETD6 was depleted in U2OS and mouse 3T3 cells employing shRNAs targeting the 3′ UTR of human or mouse SETD6, respectively (Fig. 3d,e). The SETD6-depleted cells were reconstituted with exogenous SETD6 or SETD6Y285A that lacked the 3′ UTR and was therefore shRNA-resistant. Complementation with wild-type SETD6 reestablished a normal proliferative rate, while SETD6Y285A complementation failed to do so (Fig. 3d,e). These data suggest that the enzymatic activity of SETD6 regulates a RelA-dependent effect on cell proliferation.
Figure 3. SETD6 attenuates RelA-driven cell proliferation.
(a) Increased proliferation rates of SETD6-depleted U2OS cells required RelA. Cell number in the indicated lines was determined daily for seven days. (b) Enhanced anchorage-independent growth of SETD6-depleted cells required RelA. Quantitation of colonies per field in soft agar assays with the indicated cell lines. (c) Increased proliferation rate of SETD6-depleted MEFs required RelA protein. Rela+/+ and Rela−/− MEFs stably transduced with SETD6 shRNA or control shRNA and assayed as in (a). Left panel: immunoblot analysis of WCE from the indicated cell lines. (d–e) Catalytic activity of SETD6 was required to restore a normal growth rate to SETD6-depleted cells. Growth curves and WCE immunoblot analysis of U2OS (d) and 3T3 fibroblast (e) cells reconstituted with SETD6 or SETD6Y285A as indicated. Error bars in (a–e) indicate ± s.e.m. from at least three independent experiments.
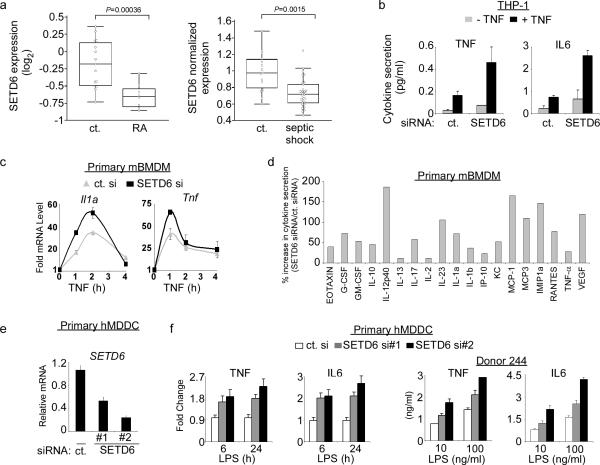
In addition to cancer, NF-κB – as a key regulator of inflammation – is also implicated in the etiology of inflammatory and autoimmune diseases5,8. Analysis of published gene expression data sets (see methods) comparing peripheral blood mononuclear cells from patients suffering from rheumatoid arthritis (RA), septic shock or juvenile idiopathic arthritis to control samples revealed downregulation of SETD6 mRNA in the disease state (Fig. 4a; Supplementary Fig. 15). In addition, SETD6 expression is lower in RA patients who respond to TNF inhibitors (suggesting a role for NF-κB in their RA disease) versus patients who are not responsive to this treatment (Supplementary Fig. 15b).
Figure 4. SETD6 attenuates RelA-driven inflammatory responses.
(a) Negative correlation between SETD6 mRNA expression and NF-kB-linked inflammatory diseases. Left: SETD6 expression level was lower (p = 0.00036, two-tailed t test) in rheumatoid arthritis (RA) patients (n = 8) compared to healthy controls (n = 15). Expression level is shown as the normalized log2 ratio of the sample compared to a common reference. Right: SETD6 expression was lower (p = 0.0015, two-tailed t test) in septic shock patients (n=30) compared to healthy controls (n = 15). See Methods for data sources and detailed filtering criteria. (b) SETD6 depletion enhanced TNF-induced cytokine secretion in THP-1 monocytic cells. THP-1 cells were transfected with control (ct.) or SETD6 siRNA ± TNF (20 ng/ml). Secretion of indicated cytokines into the culture supernatant was measured by ELISA. (c) Kinetics of SETD6-mediated inhibition of RelA target gene expression in response to TNF in mBMDM cells. Levels of IL1a and TNF mRNA measured by RT PCR at the indicated time points after TNF treatment of mBMDM cells transfected with control or SETD6 siRNA. (d) SETD6 depletion in mBMDM cells enhanced TNF-induced secretion of a large number of RelA-regulated cytokines. Primary mBMDM cells were treated as in (c) and culture supernatants were assayed for the indicated cytokines by multiplex ELISA at the 2-hr time point. y-axis: (SETD6 siRNA cytokine level/control siRNA − 1) × 100. Representative data of two independent experiments is shown. (e) siRNA-mediated depletion of SETD6 in primary human monocyte-derived dendritic cells (hMDDC). hMDDC from three individual donors were transfected with a control (ct.) siRNA or two independent SETD6 siRNAs, and the amount of SETD6 mRNA were measured by RT PCR analysis. (f) SETD6 depletion enhanced LPS-induced TNF and IL6 cytokine secretion in primary hMDDC. (Left) Average fold change of the indicated cytokines secreted from hMDDCs as in (e) and measured at 6 hrs and 24 hrs. (Right) Cytokine secretion (ng/ml) of hMDDCs isolated from an individual human donor 24 h after treatment with 10 or 100 ng/ml of LPS and transfected with the indicated siRNAs. Error bars in (b, c, e, f) indicate ± s.e.m. from at least three independent experiments.
The inverse correlation between SETD6 expression and NF-κB-linked inflammatory diseases and the observation that SETD6 attenuates RelA-dependent transactivation of cytokines such as interleukin 1α (IL-1α) and TNF suggest that SETD6 might play a role in mitigating NF-κB-driven inflammatory responses. Consistent with a negative role for SETD6 in NF-κB signaling, in monocytic THP-1 cells exposed to TNF, SETD6 knockdown increased the production of the secreted cytokines TNF and IL-6 relative to control siRNA (Fig. 4b). The relationship between SETD6 depletion and cytokine production was next tested in mouse BMDM cells. First, kinetic analysis revealed that in response to TNF and LPS, Il1a and Tnf mRNA abundance was higher in SETD6-depleted cells relative to control cells across a range of time points (Fig. 4c; Supplementary Fig. 16a; see Fig. 2e for SETD6 knockdown efficiency). Furthermore, multiplex cytokine analysis of supernatants from these cells demonstrated up-regulation of nearly twenty secreted NF-κB-regulated cytokines in SETD6-depleted cells versus control cells in response to TNF (Fig. 4d) and LPS (Supplementary Fig. 16b). Finally, depletion of SETD6 using two independent siRNAs in primary human monocyte-derived dendritic cells (hMDDCs) isolated from individual human donors (Fig. 4e) conferred a time- and dose-dependent increase in secretion of the cytokines TNF and IL-6 in response to LPS stimulation (Fig. 4f). Together, these experiments indicate that SETD6 inhibits the production of a broad array of NF-κB-regulated cytokines in diverse cell types, including antigen-presenting cells, suggesting that SETD6 is a critical repressor of RelA-mediated inflammatory responses.
GLP ankyrin repeat is a RelAK310me1 effector domain
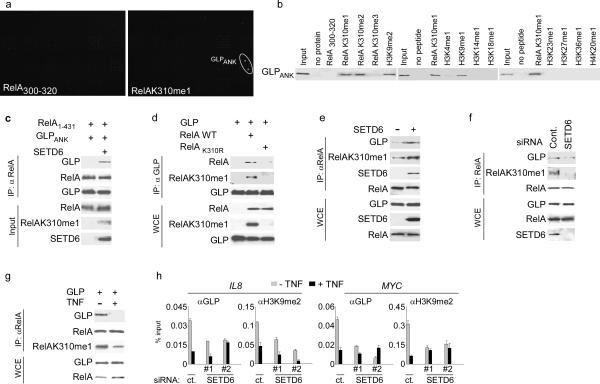
To understand the molecular basis of the repressive function associated with RelAK310me1, we screened CADOR microarrays19 for protein motifs that could potentially act as transducers of this mark (see Supplementary Fig. 17 for CADOR schematic). Of the 268 proteins on the array, the RelAK310me1 peptide bound specifically to only one: the ankyrin repeats (ANK) domain of GLP (GLPANK) (Fig. 5a). Peptide pull-down assays and measurement of dissociation constants (KD) independently confirmed and characterized the interaction between GLPANK and RelAK310me1 (Fig. 5b; Table 1). These data also demonstrate that except for the positive control H3K9me1 and H3K9me2 peptides20, other methyl histone peptides did not bind to GLPANK (Fig. 5b), and that GLPANK had similar binding affinities for RelAK310me1 and H3K9me1 (Table 1; Supplementary Fig. 18)20. In addition, GLPANK did not bind unmethylated or trimethylated RelAK310 peptides, indicating that the recognition of RelAK310 requires mono- or di-methylation (Figs. 5b; Table 1). Finally recombinant GLPANK bound recombinant RelA1–431 in co-IP experiments, but only after RelA1–431 was methylated in vitro by SETD6 (Fig. 5c). From these data, we conclude that GLPANK binds specifically to RelAK310me1 in vitro.
Figure 5. The GLP ankyrin repeat domain specifically binds RelAK310me1.
(a) Identification of the GLP ankyrin repeat domain as a RelAK310me1 binding partner. CADOR microarrays containing 268 unique domains (Supplementary Fig. 17) were probed with the indicated peptides. Representative data of three independent experiments is shown. (b) Validation of CADOR array results by peptide-binding assays. Biotinylated peptide pull-down assay with GST-GLPANK. Representative data of three independent experiments is shown. (c) GLPANK binding to RelA in vitro requires monomethylation of RelA by SETD6. Immunoblot for αRelA IPs with mock or SETD6-methylated RelA1-431 and GLPANK and probed with the indicated antibodies. Input: 10% of starting material used for the IPs. Representative data of two independent experiments is shown. (d) Wild-type RelA, but not RelAK310R, coimmunoprecipitates with GLP. Immunoblot analysis probed with the indicated antibodies of αFlag-GLP immunoprecipitates and WCE from 293T cells transfected with the indicated plasmid. WCE: 10% of total. Representative data of two independent experiments is shown. (e) SETD6 overexpression drove the endogenous interaction between GLP and RelA. Immunoblot analysis of αRelA IPs and WCE of 293T cells ± SETD6 transfection. WCE: 10% of total. Representative data of two independent experiments is shown. (f) Endogenous SETD6 was required for the physiologic RelA-GLP interaction. Immunoblot analysis as in (e)) ± SETD6 siRNA. Representative data of two independent experiments is shown. (g) TNF treatment inhibited the interaction between RelA and GLP. Immunoblot analysis as in (e) of U2OS cells transfected with GLP ± TNF treatment (10 ng/ml). Representative data of two independent experiments is shown. (h) GLP and H3K9me2 occupancy at promoters of RelA target genes required SETD6 and was inhibited by TNF treatment. Occupancy of GLP and H3K9me2 at the IL8 and MYC promoters in U2OS cells treated with control (ct.) or SETD6 siRNAs ± TNF treatment (20 ng/ml) was determined as in Fig. 2a. Negative control antibody ChIPs are shown in Supplementary Fig. 19. Error bars indicate ± s.e.m. from at least three experiments.
Table 1.
Summary of ITC binding assays of GLPANK with the indicated peptides. KD: dissociation constants; NB: no binding. Representative data of three independent experiments is shown.
| Peptide | Ligand | KD (μm) |
|---|---|---|
| RelA300–320 | GLP | NB |
| RelAK310me1 | GLP | 4.8 -/+ 0.4 |
| RelAK310me2 | GLP | 5.4 -/+ 0.5 |
| RelAK310me3 | GLP | NB |
| RelAK310me1S311ph | GLP | NB |
| H3K9me1 | GLP | 5.0 -/+ 0.3 |
Next, the ability of RelAK310me1 to be recognized by GLP in cells was investigated. Exogenous GLP coimmunoprecipitated overexpressed wild-type RelA, but not RelAK310R (Fig.5d; Supplementary Fig. 8e). In addition, SETD6 expression enhanced the interaction between endogenous GLP and RelA (Fig. 5e). Decreasing RelAK310me1 abundance via RNAi-mediated depletion of SETD6 or TNF treatment inhibited GLP association with RelA (Fig. 5f,g). These data suggest that in the absence of NF-κB activation, RelA and GLP directly interact, and this interaction requires SETD6-dependent monomethylation of RelA at K310.
GLP and its heterodimeric partner G9a generate mono- and di-methylated H3K9 at euchromatin to repress transcription9,21, and H3K9 methylation suppresses expression of inducible inflammatory genes10. Our observations that SETD6 methylation of RelA inhibits expression of NF-κB target genes and that GLP binds to RelAK310me1, suggest a model in which H3K9me2 content is increased at RelAK310me1-occupied NF-κB target genes in unstimulated cells due to stabilization of GLP via its interaction with RelAK310me1. Two predictions of this model are: (i) under basal conditions and in a SETD6-dependent manner, chromatin of distinct RelA-regulated gene promoters should be enriched for GLP and H3K9me2, and (ii) GLP should be required for SETD6 to inhibit expression of these RelA-regulated genes. In support of the first prediction, ChIP assays demonstrated a reduction of GLP and H3K9me2 occupancy at the promoters of the IL8 and MYC genes in response to TNF stimulation (Fig. 5h; Supplementary Fig. 19), and knockdown of SETD6 with two independent siRNAs largely eliminated the baseline enrichment of GLP and H3K9me2 at these promoters (Fig. 5h; Supplementary Fig. 19). Similar results were observed when SETD6 was stably depleted using an shRNA approach (Supplementary Fig. 20). In addition, induction of RelAK310me1 via SETD6 overexpression increased GLP and H3K9me2 occupancy at two RelA target promoters (Supplementary Fig. 21a). Thus, these data demonstrate a role for SETD6 and RelAK310me1 in stabilizing GLP activity at specific RelA target genes.
To investigate the functional interplay between GLP and SETD6-mediated attenuation of RelA transcriptional activity, GLP-depleted cells were challenged with TNF and found to have more IL8 and MYC mRNA relative to control cells (Supplementary Fig. 22a). Moreover, the ability of SETD6 overexpression to suppress baseline IL8 and MYC mRNA expression was largely abrogated in GLP-depleted cells (Supplementary Fig. 22b). These data argue that SETD6 inhibition of NF-κB signaling occurs at chromatin and is mediated by a lysine methylation network that connects SETD6 activity on RelA to GLP activity on H3K9.
RelA phosphorylation blocks GLP-RelAK310me1 interaction
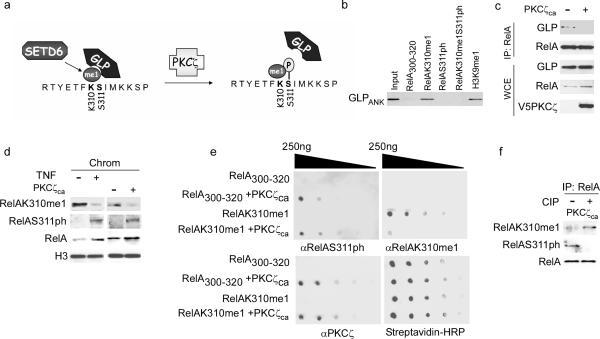
TNF stimulation initiates several activating phosphorylation events on RelA7 including phosphorylation at serine 311 (RelAS311ph) by the atypical protein kinase C PKCζ11. The molecular mechanism by which S311ph activates RelA is not known11. Because K310 methylation and S311 phosphorylation are coupled to opposing biological outcomes and the two modifications are in close physical proximity, we postulated that S311ph functionally inhibits GLP recognition of RelAK310me1 (Fig. 6a). In this regard, the ability of GLPANK to bind RelAK310me1 peptides was abolished when S311 is phosphorylated (RelAK310me1S311ph), as determined by isothermal titration calorimetry (Table 1) and in peptide pull-down assays (Fig. 6b). Moreover, overexpression of constitutively active PKCζ (PKCζca)22 disrupted the endogenous interaction between RelA and GLP (Fig. 6c). Consistent with a physiologic role for RelAS311ph in regulating GLP binding to RelA at chromatin, TNF treatment (and PKCζca overexpression) increased RelAS311ph signal at chromatin, whereas RelAK310me1 signal decreased (Fig. 6d). Thus, we propose that phosphorylation of S311 masks RelAK310me1 to prevent its recognition by GLPANK (Fig. 6a).
Figure 6. Phosphorylation of RelA at S311 by PKCζ blocks GLP recognition of RelAK310me1.
(a) Schematic of methyl-phospho switch at RelA K310 and S311 regulating GLP recognition of RelAK310me1. (b) RelAS311 phosphorylation blocked GLP binding to RelAK310me1. Biotinylated peptide pull-down assays as in Fig. 5b with GST-GLPANK. Representative data of two independent experiments is shown. (c) PKCζca overexpression disrupted the endogenous interaction between GLP and RelA. Immunoblot analysis of αRelA IPs and WCE from 293T cells ± transfection with PKCζca. 5% of total WCE loaded. Representative data of two independent experiments is shown. (d) TNF treatment and overexpression of PKCζca increased RelAS311ph amounts at chromatin and decreased amounts of RelAK310me1 detected at chromatin. Immunoblot analysis of the chromatin-enriched fraction isolated from 293T ± TNF treatment or ± PKCζca transfection and probed with the indicated antibodies. Representative data of three independent experiments is shown. (e) In-vitro kinase reactions with recombinant PKCζca and the indicated peptides were subjected to dot blot analysis and probed with αRelAS311ph, αRelAK310me1 and αPKCζ antibodies. Peptides were spotted at 0.25 ug/ul followed by 5X serial dilutions. HRP-conjugated streptavidin is shown as a loading control. Representative data of two independent experiments is shown. (f) Evidence that RelAK310me1 and RelAS311ph were present on the same molecule. RelA immunoprecipitated from 293T cells transfected with constitutively active PKCζ (PKCζca) were treated ± CIP for 1 hour and then probed with the indicated antibodies. Representative data of two independent experiments is shown.
We note that an antibody against the RelAS311ph epitope recognized this mark irrespective of methylation at K310 (Supplementary Fig. 23). In contrast, recognition of RelAK310me1 by αRelAK310me1 was disrupted by S311 phosphorylation as observed in dot blot assays with a dual-modified peptide containing K310me1 and S311ph (Supplementary Fig. 23b) or on a RelAK310me1 peptide phosphorylated at S311 in vitro by recombinant PKCζ (Fig. 6e). These results also demonstrate that PKCζ phosphorylated S311 regardless of K310 monomethyl status (Fig. 6e). Furthermore, detection of endogenous RelAK310me1 on RelA immunoprecipitated from cells overexpressing PKCζca strongly increased after in vitro dephosphorylation of the immunoprecipitated protein, indicating that the RelAK310me1 epitope becomes exposed after removal of RelAS311ph, and therefore demonstrating that the two marks likely co-occupy the same molecule (Fig. 6f; Supplementary Fig. 23c). Together, these findings suggest that TNF-induced phosphorylation of chromatin-bound RelAK310me1 at S311 disrupts the association of GLP with RelA, thereby promoting activation of the population of NF-κB target genes occupied by RelAK310me1.
RelA methyl-phospho switch regulates NF-κB signaling
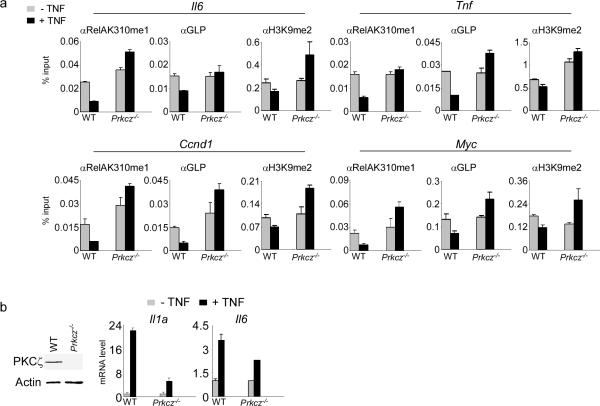
Consistent with this hypothesis, co-expression of PKCζca with SETD6 abrogated the increased occupancy of RelAK310me1, GLP, and H3K9me2 at RelA target genes induced by SETD6 expression alone (Supplementary Fig. 21a). In addition, SETD6 failed to inhibit IL8 and MYC expression when co-expressed with PKCζca (Supplementary Fig. 22b). Indeed, PKCζca expression induced increased IL8 and MYC expression above baseline, whereas the capacity of PKCζca to antagonize SETD6 and stimulate RelA target gene activation was abrogated in GLP-depleted cells (Supplementary Fig. 22b). Finally, the occupancy of RelAK310me1, GLP and H3K9me2 at the promoters of four different RelA target genes involved in cell proliferation and inflammation decreased in response to TNF stimulation in PKCζ-sufficient MEFs, but these changes were not observed in Prkcz−/− MEFs23 (Fig. 7a; Supplementary Fig. 24). In agreement with the ChIP results and as previously reported23, TNF induction of Il1a and Il6 expression was largely attenuated in Prkcz−/− cells (Fig 7b). These results support a model in which the chromatin environment of NF-κB target genes can be regulated by competing modifications of RelA by SETD6 and PKCζ, with the inert state linked to SETD6-mediated GLP binding to RelAK310me1, and the active state linked to PKCζ-mediated disassociation of GLP from RelAK310me1S311ph (Supplementary Fig. 25).
Figure 7. A RelA methyl-phospho switch at chromatin regulates NF-κB signaling.
(a) Requirement for PKCζ to decrease occupancy of RelAK310me1, GLP, and H3K9me2 at chromatin of RelA target gene promoters in response to NF-κB stimulation. ChIP assays of RelAK310me1, GLP, and H3K9me2 at promoters in Prkcz+/+ and Prkcz−/− MEFs23 ± TNF (20 ng/ml). y-axis: % input (see Fig. 2a). Negative control antibody ChIPs are shown in Supplementary Fig. 24. (b) Requirement for PKCζ in TNF-dependent induction of RelA target gene expression. Left: immunoblot analysis showing the absence of PKCζ protein in Prkcz−/− MEFs23. Right: amounts of Il1a and Il6 mRNA ± TNF (20 ng/ml) in the indicated cell lines were measured by RT PCR analysis. Error bars indicate ± s.e.m. from at least three experiments.
DISCUSSION
In summary, we report the discovery of a new lysine methylation event occurring on the transcription factor RelA, which is catalyzed by the Rubisco LSMT-like enzyme SETD6 and regulates the clinically important NF-κB pathway. SETD6 monomethylation of nuclear RelA at K310 attenuates NF-κB signaling by docking GLP (via its ankyrin repeats) at target genes to generate a silent chromatin state, effectively rendering chromatin-bound RelA inert. As deregulation of NF-κB is linked to pathologic inflammatory processes and cancer8 and SETD6 inhibits NF-κB signaling in diverse cell types, including primary human cells, SETD6 may provide a new link by which protein lysine methylation and chromatin regulation influence tumor suppression and anti-inflammatory responses2,5.
SET7/9 is a well-characterized PKMT with numerous protein substrates3, including TNF-dependent methylation of RelA at three different lysines12,13. In contrast, SETD6 methylates RelA at a single residue (K310), which occurs in the absence of stimulation and is functionally suppressed by TNF-induced RelAS311 phosphorylation. Thus, RelAK310me1 represents a specialized population of RelA that, under basal conditions, is not sequestered in the cytoplasm but rather bound and quiescent at target promoters. We postulate that transcription factor methylation can aid in the rapid and dynamic modulation of specific RelA gene expression programs by establishing transcriptional memory at marked genes24. In addition, the SETD6-RelA-GLP-H3K9me2 network delineated here constitutes the first description of a lysine methylation signaling cascade. We have also demonstrated that TNF-induced phosphorylation of RelAS311 terminates SETD6 action by blocking GLP recognition of RelAK310me1, which in turn leads to chromatin relaxation and expression of RelA target genes. These findings provide the first metazoan example of a regulatory methyl-phospho switch on a non-histone protein25–27. Together, our results highlight how the convergence and integration of multiple signals at chromatin can modify important biologic and disease pathways.
METHODS
Plasmids and reagents
For over-expression in mammalian cells, the plasmids used were: pCAG Flag-SETD6 wt, pCAG Flag-SETD6Y285A, pCAG Flag-Smyd1, pCAG Flag-Smyd2, pCAG Flag-NSD2, pcDNA-RelA, pcDNA-RelAK310R, and pcDNA Flag-GLP. pcDNA3.1/GSV5-Catalytic PKCζca was a gift from J. Smith (University of Alabama Birmingham). For in vitro assays, RelA1–431 and RelA430–551 were sub-cloned into pGEX-6P1. Single or double mutations of RelA1–431 were generated using the QuikChange site-directed mutagenesis kit (Stratagene), and sequences were confirmed by DNA sequencing. The pGEX-derived plasmids generated by mutagenesis were: pGEX-RelAK122R, pGEX-RelAK123R, pGEX-RelAK218R, pGEX-RelAK221R, pGEX-RelAK310R, pGEX-RelAK314/315R and pGEX-RelA430–551. p50 was sub-cloned into pGEX-6P1 using standard methods. A list of all the enzymes present in the PKMT library is shown in Supplementary Table 1 and a summary of the enzymes used in Figures 1a and S1 is shown in Supplementary Table 2. NSD1SET was a gift from D. Reinberg (NYU).
For insect cell expression, the cDNAs of full length SETD6 and various additional PKMTs were first cloned into pENTR3C and recombined into Gateway pDEST20 using the Gateway LR Clonase II system (Invitrogen). Recombinant baculovirus were generated according to the manufacturer's protocol (Invitrogen). Briefly, DH10Bac E. coli were transfected with pDEST20 to generate recombinant bacmid DNA. Sf9 cells were then transfected with 2 μg of bacmid DNA using Cellfection II reagent (Invitrogen), and baculovirus were amplified 3 times to obtain the optimal viral titer. For protein expression, baculovirus stocks were added to Sf9 cells grown in suspension at 10^6 cells/ml, and transduced Sf9 cells were collected 2 days after transduction. Sf9 cells were maintained in Sf-900 II SFM media supplemented with 0.5% penicillin/streptomycin.
Murine RelA (mRelA) cDNA (Open Biosystems) was first cloned into pENTR3C vector (Invitrogen) and then recombined into pBABE-FLAG-HA vector, using the Gateway system as described above. RelAK310R was than generated using the QuikChange site-directed mutagenesis kit (Stratagene).
Cell lines, transfection, and retro- and lentiviral transduction
Human embryonic kidney 293T cells, human osteosarcoma U2OS cells, mouse 3T3 cells (wt and RelA−/−, gift from M. Covert (Stanford University)), and wt and PKCζ−/− mouse embryonic fibroblasts (MEFs, gift from J. Moscat (University of Cincinnati College of Medicine) were grown in Dulbecco's modified Eagle's medium (DMEM; GIBCO) supplemented with 10% fetal calf serum (FCS, GIBCO), 100 units/ml penicillin and L-glutamine. The human monocytic THP-1 cell line was obtained from the American Type Culture Collection (ATCC) and cultured under the same conditions in Roswell Park Memorial Institute medium (RPMI-1640, GIBCO) supplemented with 0.05 mM β-mercaptoethanol and 1X streptomycin. All cells were cultured at 37°C in a humidified incubator with 5% CO2. Cells were transfected with TransIT transfection reagent (Mirus) for plasmids and with DharmaFECT reagent (Dharmacon) for siRNAs, according to the manufacturer's protocols. Human SETD6 siRNAs sequences were as follows: ACCTATGCCACAGACTTATT 3' for SETD6si#1 and GACCACCACACTAAAGGTATT for SETD6 si#2.
Isolation of mouse primary cells was performed under protocol 07064 at Rockefeller University and IACUC 9982 at Stanford University. The human biological samples were sourced ethically and their research use was in accord with the terms of the informed consent received from each donor per Hertfordshire Ethics Committee Code: 07/H0311/ 103. Mouse primary bone marrow-derived macrophages (BMDMs) were generated as described previously28. Briefly, C57BL/6 bone marrow cells from femur and tibia were cultured for 7 to 8 days at 37 °C and 5% CO2 in presence of 5ng/mL recombinant M-CSF and IL-3 (Peprotech). For knockdown experiments, siRNAs directed against mSetD6 or a control siRNA were transfected using HiPerFect Transfection Reagent (Qiagen) following the manufacturer's protocol, and stimulation experiments were performed 48 hours after transfection. The sequences for the SETD6 siRNAs used in BMDMs were: GAACAAAGGATGAAACTGA for siRNA #1 and GTGAGGAGGTGCTGACTGA for siRNA #2. For the human primary monocyte-derived dendritic cells (mDdc), CD14+ cells were separated using Miltenyi (MACS) CD14 beads (positive selection), following the manufacturer's protocols. After separation, CD14+ cells were resuspended at 1×106 cells/ml RPMI 1640 medium (plus L-Glutamine and 10 % heat inactivated FCS) containing hrGMCSF (30 ng/ml) and hrIL4 (20 ng/ml). Cells were differentiated for five days before transfection. Nucleofection of siRNA was carried out according to the manufacturer's protocol (Amaxa) with minor alterations. Briefly, siRNAs were pre-plated into a 96 well U'bottom plate such that a final concentration of 2uM would be achieved. Monocyte-derived dendritic cells (DC) were resuspended in Amaxa nucleofecter buffer such that each 20 ul contained 100,000 cells, and 20ul of cells were added to the siRNA. The plate was placed into the Amaxa and the monocyte program EA-100 was applied to all wells. After removal of the Amaxa plate, 100ul of pre-warmed RPMI medium (plus 10% heat-inactivated FCS, penicillin and L-glutamine) was added to each well, and the cells were immediately removed from the Amaxa plate and added to a second flat bottom plate containing an additional 100ul pre-warmed media. The sequences for the SETD6 siRNAs used in mDdc were: TAATGCTGCCTCACGAACTGT for siRNA #1 and TAGGAAATCCCAGCGCTCGTA for siRNA #2. Murine dendritic cells (DC) were isolated as described in29. The FL-B16 cells used to make the conditioned media were a gift from E. Engleman (Stanford University).
Retro- and lentiviral transductions were performed as previously described30. Lentivirus for control, SETD6 and GLP shRNAs were purchased from Santa Cruz Biotechnology. The human RelA shRNA target sequence is 5′-GATTGAGGAGAAACGTAAA-3′. To generate the reconstituted cell lines, shRNAs directed against the 3'UTR of SETD6 were cloned into the shRNA vector pLentiLox3.7, and wt and Y285A SETD6 were cloned into pWZL-3FLAG-hygro as AscI-PacI cassettes. U2OS and 3T3 cells were first transduced with pLentiLox-SETD6 shRNAs and selected with 2μg/mL puromycin. The puromycin-resistant cells were than transduced with pWZL-3Flag-SETD6 (wt or Y285A) or with empty pWZL-hygro, and cells were selected with hygromycin-B (250 μg/ml, Invitrogen) for 4 days. SETD6 shRNA target sequences are: 5' CCTGTTCCCTGAAGGAACAGCAATA 3' (human) and 5' TGCTATTTGGCAGTTAGAATCAAAG 3' (mouse). Where indicated, cells were stimulated with 10–20ng/mL mTNF-α (R&D Systems) or 10–100ng/mL LPS (Sigma).
ELISA and Luminex bead-based cytokine assays
ELISA assays were performed as previously described31. The antibodies used were anti-IL6 (MP5-20F3; eBioscience) and TNF (1F3F3D4; eBioscience). Plates were scanned using a SpectraMax 190 (Molecular Devices). Luminex standards were analyzed by multiplex bead-based arrays using the Mouse 26-Plex Multi-Cytokine Detection System (Panomics/Affymetrix) and analyzed with the Luminex 200 instrument, according to the manufacturer's protocol. Cytokine arrays were performed at Human Immune Monitoring Center (HIMC- Stanford).
Gene expression profiling
Gene expression arrays were performed at the Stanford Functional Genomics Facility (SFGF) using the MOUSE-REF-8 all-genome array (Illumina) according to the manufacturer's protocol. The expression array experiments were done in two independent biological replicates. Genes were selected for each group if the mean fold change was higher than 1.5. (Δ>1.5). p-values for the Venn diagram and the pie diagrams were calculated by Fisher's Exact Test (http://www.langsrud.com/fisher.htm) and the Chi-Square Test (http://people.ku.edu/~preacher/chisq/chisq.htm), respectively. The list of all genes analyzed is provided in the supplementary dataset.
SETD6 mRNA expression analysis in Rheumatoid Arthritis (RA), Septic Shock, and Juvenile Idiopathic Arthritis (JIA) diseases
To determine if SETD6 mRNA expression was significantly different in human patients suffering from RA, septic shock, and JIA, we conducted a review of available literature involving microarray studies for these diseases. Four studies that met the criteria described below were identified32–37. For RA, data were retrieved from SMD using filtering criteria implemented in32 and from GEO (accession GDS362833). Data were retrieved from GEO for septic shock35,37 (accession GSE8121) and JIA34 (accessions GSE13501 and GSE13849). Studies were only considered for microarray platforms that contained SETD6 probes36. In van der Pouw Kraan TC et al.32, which employed custom cDNA microarrays deposited in SMD, initial analysis revealed that 2 of the 3 SETD6 probes on the array (IMAGE 213233 and IMAGE 66711) were of poor quality due to low signal to background ratio in both channels (mean = 3.7 and 5.1, respectively, which resulted in many data points being removed by the 2.5 ratio filtering threshold employed by the publication), and were thus removed from the analysis. The remaining probe (IMAGE 1699773) had a considerably higher mean ratio (13.5), well above this threshold, and was thus used in the analysis. Final normalized data were retrieved from either the Gene Expression Omnibus database (GEO) or the Stanford Microarray Database (SMD) using filtering criteria described in their respective publications36. Any datasets containing multiple high quality probes representing SETD6 were averaged prior to statistical analysis. To determine significance, a 2-tailed independent t test with unequal variance was applied to each study containing a comparison between disease group vs health control group, or in one case, RA patients that responded to infliximab versus patients that did not respond to infliximab treatment.
In vitro lysine methylation assay
Assays were performed as previously described38. Briefly, recombinant RelA derivatives and peptides, recombinant nucleosomes (a gift from R. Kingston and M. Simon, Harvard Medical School), or purified HeLa nucleosomes39 were incubated with recombinant protein lysine methyltransferases and 0.1 mM S-adenosyl-methionine (AdoMet, Sigma) or 2 mCi 3H-AdoMet (Amersham) in methylation buffer containing 50 mM Tris-HCl (pH 8.0), 10 % glycerol, 20 mM KCl, 5 mM MgCl2, and 1 mM PMSF at 30°C for over-night. The reaction mixture was resolved by SDS-PAGE, followed by either autoradiography, immunoblot analysis or Coomassie stain (Pierce). The reactions with peptides were subjected to dot-blot or mass spectrometry analysis.
Mass spectrometry (MS)
Prior to MS analysis, peptides were diluted in 0.1% acetic acid for desalting using homemade C18 STAGE tips as previously described40. Samples were then loaded by an Eksigent AS2 autosampler into 75 μm ID fused silica capillary columns packed with 15 cm of C18 reversed phase resin (Magic C18, 5 μm particles, 200Å pore size, Michrom BioResources). Capillary columns were constructed with an electrospray tip for nanoflow reversed-phase high performance liquid chromatography tandem mass spectrometry on a hybrid linear quadrupole ion trap-Orbitrap mass spectrometer (Thermo Electron). Peptides were resolved with a gradient from 5 to 35% Buffer B in 110 minute gradient (Buffer A = 0.1 M acetic acid; Buffer B = 70% acetonitrile in 0.1 M acetic acid) with a flow rate of approximately 200 nl/min on an Agilent 1200 binary HPLC system.
The Orbitrap was operated with a resolution of 30,000 for a full MS spectrum, followed by 7 subsequent data-dependent MS/MS spectra collected in the ion trap after electron transfer dissociation of peptides. Peptides selected for MS/MS interrogation were placed on an exclusion list for 60 seconds to avoid duplicate MS/MS spectra. All MS and MS/MS spectra were analyzed with Qual Browser (version 2.0.7, Thermo Scientific), and the identities of all peptides were confirmed by manual inspection of MS/MS spectra.
Interaction studies, immunoblot analysis, CIP assays, and antibodies
Cell extracts were prepared by lysis of PBS-washed cells in RIPA lysis buffer (50 mM Tris-HCl, pH 8, 150 mM NaCl, 1% Nonidet P-40 (v/v), 0.5% deoxycholate (v/v), 0.1% SDS (v/v), 1 mM dithiothreitol (DTT) and Sigma protease inhibitor cocktail (P8340 diluted 1:100)). Protein concentration was determined using Bradford reagent (Bio-Rad). Equal amounts of protein were mixed with Laemmli sample buffer (4% SDS, 20% glycerol, 10% 2-mercaptoethanol and 0.125 M Tris·HCl pH 6.8), heated at 95°C for 5 min, and loaded onto a polyacrylamide-SDS gel.
To test if K310me1 and S311ph are present on the same molecule, Immunoprecipitated endogenous RelA from 293T cells transfected with constitutively active PKCζ (PKCζca) was washed 3 times with RIPA buffer. The immunoprecipitated RelA bound on protein A/G beads was divided in half and treated ± Calf Intestinal Alkaline Phosphatase (CIP, New England Biolabs) for 1 hour at 37°C prior to immunoblot analysis. Biochemical fractionation, GST pull-down assays, immunofluorescence staining, and protein–protein ChIP experiments were performed as previously described38,41,42.
The antibodies used were as follows: GLP (sc-81260; Santa Cruz Biotechnology); p65 (sc-8008 AC, Santa Cruz Biotechnology); Flag (F3165, Sigma-Aldrich); GLP (09-078, Millipore); GLP (A301-642A-1, Bethyl); GST-HRP (ab3416, Abcam); H3 (ab1791, Abcam); V5 HRP (ab1325, Abcam); p65 (ab16502, Abcam); p65-phospho S311 (ab51059, Abcam); H3K9me2 (ab1220, Abcam); PKCζ (ab4137, Abcam); p65 phospho S311 (ab-311, GenScript); β-tubulin (05-661, Millipore); β-Actin (A1978, Sigma-Aldrich). RelAK310me1 rabbit polyclonal antibody was raised against the corresponding peptide: TYETFKme1SIMKKSPC (Century biochemical). SETD6 rabbit polyclonal antibody was raised against GST-SETD6 protein (Covance).
Chromatin immunoprecipitation (ChIP)
ChIP was performed according to the protocol of Ainbinder et al.43. Briefly, formaldehyde cross-linked protein-DNA complexes were precipitated by incubation overnight with the indicated antibodies or with rabbit IgG as a negative control. Precipitated DNA fragments were isolated using Chelex 100 resin (BioRad) as described44 and amplified by Quantitative real-time PCR (ABI PRISM 7700 Sequence Detection System). Primers used in the study are shown in Supplementary Table 3.
Luciferase assay
Cells were transiently transfected with the indicated combinations of plasmids. The total amount of transfected DNA in each dish was kept constant by the addition of empty vector wherever necessary. Cell extracts were prepared 24 hours after transfection, and luciferase acitivty was measured using the Dual-Glo Luciferase Assay System (Promega). Luciferase activity was measured and normalized to Renilla. The κB-luc reporter plasmid was kindly provided by W.C. Greene (UCSF).
RNA extraction and reverse transcription
RNA was prepared using the RNeasy plus kit (Qiagen) and reverse-transcribed using the SuperScript III first strand synthesis system (Invitrogen). Quantitative real-time RT-PCR was performed in triplicate on the ABI PRISM 7700 Sequence Detection System using Taqman GeneExpression Assay primer/probe sets (Applied Biosystems). Data were normalized to gapdh abundance.
Isothermal titration calorimetry (ITC)
The GLPANK protein was concentrated to 24 μM in 150 mM NaCl and 20 mM Tris PH 8.0. ITC measurements were carried out with 370–475 μM peptide on a MicroCal VP-ITC instrument at 25 °C. Binding constants were calculated by fitting the data using the ITC data analysis module of Origin 7.0 (OriginLab Corporation).
Homology modeling of SETD6
Homology modeling for SETD6 was done using the PHYRE server23 (http://www.sbg.bio.ic.ac.uk/phyre)45. Among the models with the highest confidence scores (E-value: 1.73e-28), a homology model of SETD6 readily gave a solution using the structure of the Rubisco large subunit methyltransferase (LSMT) bound to AdoMet and Lysine (PDB 2H2E)46.
Peptide pull-down and peptide/protein array
Biotinylated histone peptides, peptide microarray experiments, and peptide pull-down assays were performed as previously described47. CADOR protein microarrays were generated as described48. RelA peptides were synthesized at the Yale W.M. Keck facility. The sequence of the peptides (RelA amino acids 300–320) is: Biotin-E-K-R-K-R-T-Y-E-T-F K#-S*-I-M-K-K-S-P-F-S G. K310 (denoted with a #) was either unmodified, monomethylated, dimethylated, or trimethylated as indicated, and S311 (denoted with an *) was either unmodified or phosphorylated as indicated.
Growth curve and soft agar assay
For growth curves, 5–20×104 cells were plated in triplicate, and cell number was determined every day for a 5–7 days using a hemocytometer. For soft agar assays, 2×104 cells were plated in triplcate in 0.8% agarose on 3% agarose base agar medium. Colony formation was quantified by counting the numbers of colonies per field after 21 days.
Supplementary Material
Acknowledgements
We thank R. Kingston and M. Simon for recombinant nucleosomes, J. Smith for the PKCζca plasmid, W.C. Greene for RelA1-431 cDNA and the κB-Luc reporter plasmid, D. Reinberg for the NSD1SET plasmid, M. Covert and T.D Gilmore for the mouse 3T3 WT and Rela−/− cells, and J. Moscat for the WT and Prkcz−/− MEFs. We thank E. Green for critical reading of the manuscript and A. Alizadeh for helpful comments. This work was supported in part by grants from the NIH to OG and MTB (DA025800), XC (GM068680), an American Society for Mass Spectrometry Research Award BAG. PJU was funded by a gift from the Floren Family Trust, CLL by NIH F32AI080086, AJK a Genentech Foundation Predoctoral Fellowship, DL by EMBO LTF, Human Frontier Science Program, and the Machiah Fellowship. XC is a Georgia Research Alliance Eminent Scholar. OG is a recipient of an Ellison Senior Scholar Award.
Footnotes
METHODS Methods and any associated references are available in the online version of the paper at http://www.nature.com/natureimmunology/.
REFERENCES
- 1.Kouzarides T. Chromatin modifications and their function. Cell. 2007;128:693–705. doi: 10.1016/j.cell.2007.02.005. [DOI] [PubMed] [Google Scholar]
- 2.Albert M, Helin K. Histone methyltransferases in cancer. Semin Cell Dev Biol. 2009:S1084–9521. doi: 10.1016/j.semcdb.2009.10.007. [DOI] [PubMed] [Google Scholar]
- 3.Huang J, Berger SL. The emerging field of dynamic lysine methylation of nonhistone proteins. Curr Opin Genet Dev. 2008;18:152–158. doi: 10.1016/j.gde.2008.01.012. [DOI] [PubMed] [Google Scholar]
- 4.Hoffmann A, Natoli G, Ghosh G. Transcriptional regulation via the NF-kappaB signaling module. Oncogene. 2006;25:6706–6716. doi: 10.1038/sj.onc.1209933. [DOI] [PubMed] [Google Scholar]
- 5.Natoli G. Control of NF-kappaB-dependent transcriptional responses by chromatin organization. Cold Spring Harb Perspect Biol. 2009;1:a000224. doi: 10.1101/cshperspect.a000224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ghosh S, Hayden MS. New regulators of NF-kappaB in inflammation. Nat Rev Immunol. 2008;8:837–848. doi: 10.1038/nri2423. [DOI] [PubMed] [Google Scholar]
- 7.Perkins ND. Post-translational modifications regulating the activity and function of the nuclear factor kappa B pathway. Oncogene. 2006;25:6717–6730. doi: 10.1038/sj.onc.1209937. [DOI] [PubMed] [Google Scholar]
- 8.Grivennikov SI, Greten FR, Karin M. Immunity, inflammation, and cancer. Cell. 2010;140:883–899. doi: 10.1016/j.cell.2010.01.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tachibana M, et al. Histone methyltransferases G9a and GLP form heteromeric complexes and are both crucial for methylation of euchromatin at H3-K9. Genes Dev. 2005;19:815–826. doi: 10.1101/gad.1284005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Saccani S, Natoli G. Dynamic changes in histone H3 Lys 9 methylation occurring at tightly regulated inducible inflammatory genes. Genes Dev. 2002;16:2219–2224. doi: 10.1101/gad.232502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Duran A, Diaz-Meco MT, Moscat J. Essential role of RelA Ser311 phosphorylation by zetaPKC in NF-kappaB transcriptional activation. Embo J. 2003;22:3910–3918. doi: 10.1093/emboj/cdg370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ea CK, Baltimore D. Regulation of NF-{kappa}B activity through lysine monomethylation of p65. Proc Natl Acad Sci U S A. 2009;106:18972–18977. doi: 10.1073/pnas.0910439106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang XD, et al. Negative regulation of NF-kappaB action by Set9-mediated lysine methylation of the RelA subunit. Embo J. 2009;28:1055–1066. doi: 10.1038/emboj.2009.55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Trievel RC, Flynn EM, Houtz RL, Hurley JH. Mechanism of multiple lysine methylation by the SET domain enzyme Rubisco LSMT. Nat Struct Biol. 2003;10:545–552. doi: 10.1038/nsb946. [DOI] [PubMed] [Google Scholar]
- 15.Dong J, Jimi E, Zeiss C, Hayden MS, Ghosh S. Constitutively active NF-kappaB triggers systemic TNFalpha-dependent inflammation and localized TNFalpha-independent inflammatory disease. Genes Dev. 2010;24:1709–1717. doi: 10.1101/gad.1958410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Fischle W, Verdin E, Greene WC. Duration of nuclear NF-kappaB action regulated by reversible acetylation. Science. 2001;293:1653–1657. doi: 10.1126/science.1062374. [DOI] [PubMed] [Google Scholar]
- 17.Tachibana M, Sugimoto K, Fukushima T, Shinkai Y. Set domain-containing protein, G9a, is a novel lysine-preferring mammalian histone methyltransferase with hyperactivity and specific selectivity to lysines 9 and 27 of histone H3. J Biol Chem. 2001;276:25309–25317. doi: 10.1074/jbc.M101914200. [DOI] [PubMed] [Google Scholar]
- 18.Buerki C, et al. Functional relevance of novel p300-mediated lysine 314 and 315 acetylation of RelA/p65. Nucleic Acids Res. 2008;36:1665–1680. doi: 10.1093/nar/gkn003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kim J, et al. Tudor, MBT and chromo domains gauge the degree of lysine methylation. EMBO Rep. 2006;7:397–403. doi: 10.1038/sj.embor.7400625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Collins RE, et al. The ankyrin repeats of G9a and GLP histone methyltransferases are mono- and dimethyllysine binding modules. Nat Struct Mol Biol. 2008;15:245–250. doi: 10.1038/nsmb.1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tachibana M, Matsumura Y, Fukuda M, Kimura H, Shinkai Y. G9a/GLP complexes independently mediate H3K9 and DNA methylation to silence transcription. Embo J. 2008;27:2681–2690. doi: 10.1038/emboj.2008.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith L, et al. Activation of atypical protein kinase C zeta by caspase processing and degradation by the ubiquitin-proteasome system. J Biol Chem. 2000;275:40620–40627. doi: 10.1074/jbc.M908517199. [DOI] [PubMed] [Google Scholar]
- 23.Leitges M, et al. Targeted disruption of the zetaPKC gene results in the impairment of the NF-kappaB pathway. Mol Cell. 2001;8:771–780. doi: 10.1016/s1097-2765(01)00361-6. [DOI] [PubMed] [Google Scholar]
- 24.Su IH, Tarakhovsky A. Lysine methylation and `signaling memory'. Curr Opin Immunol. 2006;18:152–157. doi: 10.1016/j.coi.2006.01.012. [DOI] [PubMed] [Google Scholar]
- 25.Hirota T, Lipp JJ, Toh BH, Peters JM. Histone H3 serine 10 phosphorylation by Aurora B causes HP1 dissociation from heterochromatin. Nature. 2005;438:1176–1180. doi: 10.1038/nature04254. [DOI] [PubMed] [Google Scholar]
- 26.Fischle W, et al. Regulation of HP1-chromatin binding by histone H3 methylation and phosphorylation. Nature. 2005;438:1116–1122. doi: 10.1038/nature04219. [DOI] [PubMed] [Google Scholar]
- 27.Zhang K, et al. The Set1 methyltransferase opposes Ipl1 aurora kinase functions in chromosome segregation. Cell. 2005;122:723–734. doi: 10.1016/j.cell.2005.06.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Toney LM, et al. BCL-6 regulates chemokine gene transcription in macrophages. Nat Immunol. 2000;1:214–220. doi: 10.1038/79749. [DOI] [PubMed] [Google Scholar]
- 29.Yasuda K, et al. Murine dendritic cell type I IFN production induced by human IgGRNA immune complexes is IFN regulatory factor (IRF)5 and IRF7 dependent and is required for IL-6 production. J Immunol. 2007;178:6876–6885. doi: 10.4049/jimmunol.178.11.6876. [DOI] [PubMed] [Google Scholar]
- 30.Michishita E, et al. SIRT6 is a histone H3 lysine 9 deacetylase that modulates telomeric chromatin. Nature. 2008;452:492–496. doi: 10.1038/nature06736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kattah MG, Coller J, Cheung RK, Oshidary N, Utz PJ. HIT: a versatile proteomics platform for multianalyte phenotyping of cytokines, intracellular proteins and surface molecules. Nat Med. 2008;14:1284–1289. doi: 10.1038/nm.1755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.van der Pouw Kraan TC, et al. Rheumatoid arthritis subtypes identified by genomic profiling of peripheral blood cells: assignment of a type I interferon signature in a subpopulation of patients. Ann Rheum Dis. 2007;66:1008–1014. doi: 10.1136/ard.2006.063412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Julia A, et al. An eight-gene blood expression profile predicts the response to infliximab in rheumatoid arthritis. PLoS One. 2009;4:e7556. doi: 10.1371/journal.pone.0007556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barnes MG, et al. Subtype-specific peripheral blood gene expression profiles in recent-onset juvenile idiopathic arthritis. Arthritis Rheum. 2009;60:2102–2112. doi: 10.1002/art.24601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Wong HR, et al. Genome-level expression profiles in pediatric septic shock indicate a role for altered zinc homeostasis in poor outcome. Physiol Genomics. 2007;30:146–155. doi: 10.1152/physiolgenomics.00024.2007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Demeter J, et al. The Stanford Microarray Database: implementation of new analysis tools and open source release of software. Nucleic Acids Res. 2007;35:D766–770. doi: 10.1093/nar/gkl1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Shanley TP, et al. Genome-level longitudinal expression of signaling pathways and gene networks in pediatric septic shock. Mol Med. 2007;13:495–508. doi: 10.2119/2007-00065.Shanley. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Shi X, et al. ING2 PHD domain links histone H3 lysine 4 methylation to active gene repression. Nature. 2006;442:96–99. doi: 10.1038/nature04835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Schnitzler GR. Isolation of histones and nucleosome cores from mammalian cells. Curr Protoc Mol Biol. 2001;Chapter 21(Unit 21):25. doi: 10.1002/0471142727.mb2105s50. [DOI] [PubMed] [Google Scholar]
- 40.Rappsilber J, Ishihama Y, Mann M. Stop and go extraction tips for matrix-assisted laser desorption/ionization, nanoelectrospray, and LC/MS sample pretreatment in proteomics. Anal Chem. 2003;75:663–670. doi: 10.1021/ac026117i. [DOI] [PubMed] [Google Scholar]
- 41.Mendez J, Stillman B. Chromatin association of human origin recognition complex, cdc6, and minichromosome maintenance proteins during the cell cycle: assembly of prereplication complexes in late mitosis. Mol Cell Biol. 2000;20:8602–8612. doi: 10.1128/mcb.20.22.8602-8612.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Michishita E, Park JY, Burneskis JM, Barrett JC, Horikawa I. Evolutionarily conserved and nonconserved cellular localizations and functions of human SIRT proteins. Mol Biol Cell. 2005;16:4623–4635. doi: 10.1091/mbc.E05-01-0033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Ainbinder E, et al. Mechanism of rapid transcriptional induction of tumor necrosis factor alpha-responsive genes by NF-kappaB. Mol Cell Biol. 2002;22:6354–6362. doi: 10.1128/MCB.22.18.6354-6362.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Nelson JD, Denisenko O, Bomsztyk K. Protocol for the fast chromatin immunoprecipitation (ChIP) method. Nat Protoc. 2006;1:179–185. doi: 10.1038/nprot.2006.27. [DOI] [PubMed] [Google Scholar]
- 45.Kelley LA, Sternberg MJ. Protein structure prediction on the Web: a case study using the Phyre server. Nat Protoc. 2009;4:363–371. doi: 10.1038/nprot.2009.2. [DOI] [PubMed] [Google Scholar]
- 46.Couture JF, Hauk G, Thompson MJ, Blackburn GM, Trievel RC. Catalytic roles for carbon-oxygen hydrogen bonding in SET domain lysine methyltransferases. J Biol Chem. 2006;281:19280–19287. doi: 10.1074/jbc.M602257200. [DOI] [PubMed] [Google Scholar]
- 47.Bua DJ, et al. Epigenome microarray platform for proteome-wide dissection of chromatin-signaling networks. PLoS One. 2009;4:e6789. doi: 10.1371/journal.pone.0006789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Espejo A, Cote J, Bednarek A, Richard S, Bedford MT. A protein-domain microarray identifies novel protein-protein interactions. Biochem J. 2002;367:697–702. doi: 10.1042/BJ20020860. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.