Abstract
Cu2+ binding to Alzheimer’s β (Aβ) peptides in amyloid fibrils has attracted broad attention, as it was shown that Cu ion concentration elevates in Alzheimer’s senile plaque and such association of Aβ with Cu2+ triggers the production of neurotoxic reactive oxygen species (ROS) such as H2O2. However, detailed binding sites and binding structures of Cu2+ to Aβ are still largely unknown for Aβ fibrils or other aggregates of Aβ. In this work, we examined molecular details of Cu2+ binding to amyloid fibrils by detecting paramagnetic signal quenching in 1D and 2D high-resolution 13C SSNMR for full-length 40-residue Aβ(1–40). Selective quenching observed in 13C SSNMR of Cu2+-bound Aβ(1–40) suggested that primary Cu2+ binding sites in Aβ(1–40) fibrils include Nε in His-13 and His-14, and carboxyl groups in Val-40 as well as in Glu side chains (Glu-3, Glu-11, and/or Glu-22). 13C chemical shift analysis demonstrated no major structural changes upon Cu2+ binding in the hydrophobic core regions (residues 18–25 and 30–36). Although the ROS production via oxidization of Met-35 in the presence of Cu2+ has been long suspected, our SSNMR analysis of 13CεH3-S- in M35 showed little changes after Cu2+ binding, excluding the possibility of Met-35 oxidization by Cu2+ alone. Preliminary molecular dynamics (MD) simulations on Cu2+-Aβ complex in amyloid fibrils confirmed binding sites suggested by the SSNMR results and the stabilities of such bindings. The MD simulations also indicate the coexistence of a variety of Cu2+-binding modes unique in Aβ fibril, which are realized by both intra- and inter-molecular contacts and highly concentrated coordination sites due to the in-register parallel β-sheet arrangements.
Introduction
Alzheimer’s disease (AD) is characterized by the deposition of senile plaques and neural degeneration. Alzheimer’s amyloid β (Aβ) peptides are a primary component of the senile plaques.1,2 Among Aβ peptides ranging from 39 to 43 residues, 40-residue Aβ(1–40) and 42-residue Aβ(1–42) are the two major species found in plaque.2 In AD the metal ion homeostasis appears to be severely damaged, resulting in increased concentrations of Cu and Zn ions in senile plaque, which reach 400 µM and 1 mM, respectively3 or ~10 fold compared with the region outside the plaque.4,5 More interestingly, it was shown that in association with Cu2+, Aβ produces reactive oxygen species (ROS) such as H2O2 in vitro,6 reportedly through the reduction of Cu2+ to Cu+ in association with M357 and/or biochemical reductants such as ascorbate.8 The toxicity of Aβ can be greatly attenuated by H2O2 scavengers9 and metal chelator in vitro.6,10 Indeed, Aβ fibrils strongly binds Cu2+ ions.11–13 Although the mechanisms of neural cell deaths and oxidative stress in AD are still debated, these events may be explained by the ROS generated on the redox-active Cu2+ ions bound on Aβ fibrils, which was observed in vitro. Small molecules which target metal-Aβ interactions have been tested as potential therapeutic agents,14–16 including one in a clinical trial.14 Thus, intensive efforts have been made to understand the molecular details of Cu2+-binding to Aβ.8,17–33 On the other hand, most structural studies on Cu2+-Aβ binding were performed on soluble model peptides19–24,29 or monomeric Aβ25–27,33; however, some of these reports are controversial.
Solution NMR studies by Hou and Zagorski on monomeric Aβ(1–40) indicated specific binding of Cu2+ to side-chains of H6, H13, and H14 by their upfield 1H shifts, while suggesting the lack of association of D1 and Y10 with Cu2+ through little perturbation on their 1H side-chain shifts.27 The Cu2+ binding to the N-terminal region is consistent with a recent NMR study indicating paramagnetic relaxation enhancement on the residue 3–16 upon the addition of Cu2+ to monomeric Aβ(1–40).25 EPR spectra of Cu2+-bound Aβ(1–16), Aβ(1–28), and Aβ(1–42) monomers show quite similar spectral features for Cu2+19–21; it has been proposed that these EPR spectra suggests the presence of two types of Cu2+ binding sites, both of which are likely to have 4-coordination geometries such as 2N/2O and 3N/1O coordination to Cu2+.19–21 This is consistent with the solution NMR results27 since two nitrogens of His imidazole rings typically offer coordination to Cu2+.34 More detailed isotope-edited EPR studies on Aβ(1–16) suggested Cu2+ coordination to D1, H6, H13 (or H14) for one spectral component (Component I; A// =162 ± 3 G and g// = 2.272) and the second Cu2+ coordination to A2, H6, H13, H14 for the other minor spectral component at a neutral pH (Component II; A// = 148 ± 3 G and g// = 2.227).19,20 In contrast, a recent EPR study on monomeric Aβ(1–28) suggested octahedral coordination by D1, D7, H6, H13, H14 for a EPR spectrum having similar yet distinguishable EPR parameters (Component I: A// = 170 G and g// = 2.27; Component II: A// = 156 G and g// = 2.22).21 Although these Aβ monomer and fragments may have different metal binding modes, a conclusive structural model of Cu2+-Aβ complex has not been established even for the soluble model systems (see Table 1 in ref. 8 for example). More importantly, monomeric Aβ is non-toxic; a question relevant to the mechanism of AD is, therefore, Cu2+ binding structure in Aβ aggregates. Recent EPR studies on Cu2+-bound amyloid fibril of Aβ(1–40) and Aβ(1–42) report the presence of, at least, two types of binding sites that also have 4-coordination geometries.21,28,31,35 However, beyond the similarity of the EPR spectra of the Aβ fibrils with those of more well studied soluble Aβ fragments, scarce site specific information on metal binding modes has been obtained for Aβ fibrils. A recent EPR study on Aβ(1–28) showed no notable changes in the EPR by H6A or H14A mutation.21 Thus, similarity in conventional continuous wave (cw) EPR spectra among Cu(II)-bound Aβ with varied sequences may not be sufficient to deduce Cu2+ coordination structure uniquely. Rather, coordination to Cu2+ involving different His residues and other ligands can yield very similar 1D EPR spectra as long as the local environments around Cu2+ are similar. On Cu2+ binding to oligomeric Aβ, there were some interesting studies,25,35 yet the nature of these oligomeric species has not been well defined. A recent study by solid-state NMR (SSNMR), a powerful method for structural analysis of protein aggregates36–46 and other proteins,47–62 indicated Cu2+ binding to Aβ in membrane environments.63 Despite the broad attention and the intense efforts presented in the previous studies,8,17–25,27–29 the exact Cu2+ binding sites of Aβ fibrils have not been identified with any site-specificity. Lack of site-specific structural information has also severely limited our molecular-level understanding on structural changes of Aβ fibrils upon Cu2+ binding. Such detailed structural information would be highly valuable for revealing still unknown mechanisms of the toxicity for Aβ aggregates and designing metal binding inhibitors for amyloid aggregates.14–16
Table 1.
The conformational energy of simulated Cu2+-Aβ(1–40) models.
Here, we present the first systematic study to address molecular details of Cu2+ binding on amyloid fibrils of full-length Aβ(1–40) by SSNMR. With recent advance in SSNMR for paramagnetic proteins,45,47,64–67 we examine the possibility of identifying site-specific Cu2+ binding to Aβ(1–40) fibrils and molecular-level structural changes upon the binding. Our SSNMR data show a lack of perturbation to 13C chemical shifts by Cu2+ binding, and suggest that the parallel β-sheet structures in the hydrophobic core regions of Aβ(1–40) fibrils are not reorganized into different structures by Cu2+ binding for the first time. Molecular dynamics (MD) simulations that take account of the amyloid fibril structure suggest that Cu2+ binding to Aβ in fibril is likely to involve the novel binding modes that are not possible in soluble fragments or monomers of Aβ.
Results and Discussion
First, we examined the morphological changes of amyloid fibril after Cu2+ binding. Although it has been proposed that Cu2+ or Zn2+ binding to Aβ monomers may modulate misfolding kinetics,27,30,35,68,69 in this work, we focus on structural changes due to Cu2+ binding after the fibril formation. Figures 1a and 1b show transmission electron microscopy (TEM) images of Aβ fibrils without and with 0.4 mol eq CuCl2, respectively. The fibrils without Cu2+ have a diameter of ~9 nm and a length of > 1 µm, which are consistent with previous reports.70 The morphology of the Aβ fibrils is retained after Cu2+ was bound to fibrils. Although it is difficult to deduce any molecular-level structural changes from the data, it is likely that Cu2+ binding does not substantially destabilize the amyloid fibril structure. Binding of Cu2+ to Aβ fibrils was determined by photometric assay using N,N,N’,N’-tetraethylthiuram disulfide as a Cu2+ indicator.71 We confirmed that more than 95% of Cu2+ was bound to Aβ(1–40) for both CuCl2 and CuGly when the ratio of Cu2+ to Aβ (fCu/Aβ) was 0.5 or less (see SI). However, at fCu/Aβ = 1.0, 5.3 % and 15 % of Cu2+ ions were not bound to Aβ fibrils for CuCl2 and CuGly, respectively (see SI). Based on these results, we selected fCu/Aβ = 0.4 throughout the following SSNMR study as a condition where nearly all Cu2+ ions are strongly bound to Aβ fibrils.
Figure 1.
(a, b) Transmission electron microscopy images of Aβ(1–40) fibrils (a) without and (b) with Cu2+, which is 0.4 molar equivalence (eq.) to Aβ. Cu2+ ions were added after the fibril formation was complete throughout this work. (c) Comparison of 1D 13C CPMAS spectra of Aβ(1–40) fibrils without (black) and with ( ) 0.4 molar eq. of Cu2+, together with signal assignments. The samples were labeled with uniformly 13C-, 15N-labeled amino acids at V12, F20, A21, I31, G33 and with 13CεH3 selectively labeled at M35. (d) Comparison of the 2D 13C/13C correlation spectra for the same fibrils samples without (black) and with (
) 0.4 molar eq. of Cu2+, together with signal assignments. The samples were labeled with uniformly 13C-, 15N-labeled amino acids at V12, F20, A21, I31, G33 and with 13CεH3 selectively labeled at M35. (d) Comparison of the 2D 13C/13C correlation spectra for the same fibrils samples without (black) and with ( ) Cu2+. The spectra were obtained at magic angle spinning (MAS) at 20 kHz. In (d), a fpRFDR sequence of 1.6 ms87 was used for 13C-13C exchange.
) Cu2+. The spectra were obtained at magic angle spinning (MAS) at 20 kHz. In (d), a fpRFDR sequence of 1.6 ms87 was used for 13C-13C exchange.
Next, we examined whether any site-specific binding of Cu2+ can be detected by SSNMR for Aβ(1–40) fibrils. Figure 1c show a comparison of the aliphatic region of 1D 13C CPMAS SSNMR spectra of Aβ(1–40) amyloid fibrils (black) without and (red) with 0.4 molar eq. of Cu2+ to Aβ. The Aβ(1–40) samples were isotope labeled with uniformly 13C- and 15N-labeled amino acids at V12, F20, A21, I31, G33, and 13Cε-selectively labeled M35 (see SI). Interestingly, most sites display no significant changes in the signal intensities. This includes 13Cε of M35 (green arrow), which has been long suspected as the site that is responsible for the production of H2O2 through interactions with Cu2+.72,73 If M35 side chain (-S-CεH3) is oxidized into -SO-CH3 as hypothesized previously,7 13Cε shift at 15.6 ppm should be altered to ~40 ppm.74 However, no major changes in the signal intensity or chemical shift were observed for 13Cε M35 after a month. In contrast, signal intensities drop by 36% and 27% selectively for 13Cγ and 13Cβ of V12 (orange arrows), respectively upon Cu2+ binding. The site is nearby to H13/H14, which were reported to interact with Cu2+ for Aβ(1–40) monomer.27 These results suggest that the effect of Cu2+ binding to fibrils is site specific, quenching the signals for sites in the vicinity of Cu2+ (V12 in this case). In a recent SSNMR study of Cu, Zn SOD, signal quenching of 13C within a ~5Å radius of Cu2+ was reported.47 Here, we utilize such paramagnetic quenching to identify Cu2+ binding sites in the aggregated form of the Aβ proteins.
Figure 1d shows a comparison of 2D 13C/13C spectra of the same Aβ fibril samples (black) without and (red) with Cu2+ bound to Aβ in a superposition. Most of the signals are well resolved without signal overlap. Remarkably, the Cu2+-bound fibril have nearly identical chemical shift positions (within ±0.2 ppm) and intensities with those for the Cu2+-free fibril, except for the V12 signals (orange arrows). Since 13C chemical shifts are a probe very sensitive to protein conformations, this presents interesting evidence that the site-specific Cu2+ binding does not alter the conformations of Aβ in the β-sheet cores, including F20, A21, I31. Additional results below support this finding. Thus, we concluded that Cu2+-binding does not introduce major conformation changes except for the N- and C-terminal regions, where binding is likely to take place, as will be discussed.
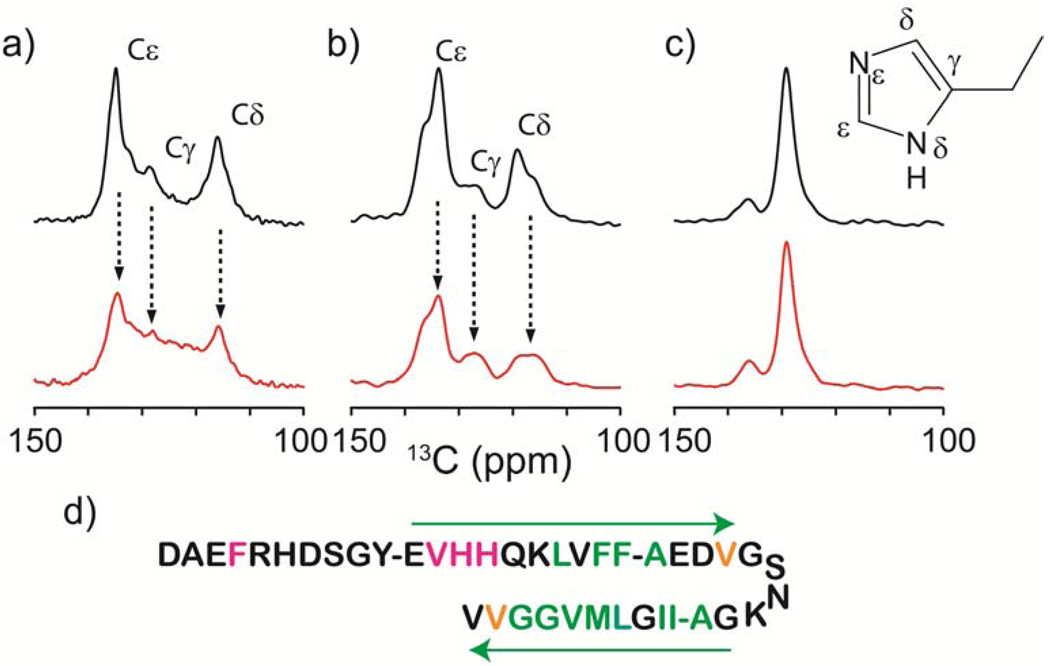
Based on the above results, we hypothesized that Cu2+ ions are interacting with H13 and H14 side chains in the Aβ fibril, and with other neighboring residues. To test this, we performed 13C SSNMR of Aβ(1–40) fibril samples in which one of these residues was uniformly 13C-labeled. Figures 2a,b show 1D 13C CPMAS spectra of (a) H13 and (b) H14 side chains for Aβ fibrils (black) without and (red) with Cu2+ bound to Aβ, together with signal assignments. Clearly, the signals for both H13 and H14 are quenched considerably by Cu2+. Spectral analysis of the aromatic side chains shows that signals for 13Cδ and 13Cε in H13/H14 are quenched by 30–60%, while those for Cγ are quenched less (by ~15%). This suggests that Cu2+ ion favors coordination to Nε in H13 and H14, which is one bond away from 13Cδ and 13Cε, over that to Nδ. As a control, we examined paramagnetic quenching for F20 (Figure 2c), which shows nearly no quenching. Previous studies indicated the involvement of Nδ/Nε of histidine in Aβ(1–40) in Cu2+ binding.26,27,75 Raman spectroscopic studies26 shows that Aβ(1–40) precipitates obtained by the addition of Cu2+ to a Aβ solution display Cu2+ coordination to Nε, in His while Aβ(1–40) monomers in a solution show Cu2+ coordination to Nδ at pH 7.4. Barnham et al.75 reported that Aβ(1–40) having N-methylated His for Nε at position His 6,13 and 14 has weaker metal-ligand interaction than WT Aβ or Aβ(1–40) having N-methylated His for Nδ. The results are consistent with our finding. To the best of our knowledge, this is the first experimental data that directly demonstrate the association of Cu2+ with H13 and H14 of Aβ(1–40) in amyloid fibrils with site resolution.
Figure 2.
Aromatic regions of 13C CPMAS spectra for (a) H13, (b) H14, and (c) F20 side chains for Aβ(1–40) fibrils in the hydrated state (black) without and ( ) with Cu2+. (d) Amino acid sequence of Aβ(1–40) and the extent of signal quenching by Cu2+ binding (green: ≤15%; orange: 15%–25%; red: ≥25%). The quenching is the average of that for all the observed 13C sites for each residue. The arrow denotes β-sheet regions reported in the previous studies by Tycko et al.77
) with Cu2+. (d) Amino acid sequence of Aβ(1–40) and the extent of signal quenching by Cu2+ binding (green: ≤15%; orange: 15%–25%; red: ≥25%). The quenching is the average of that for all the observed 13C sites for each residue. The arrow denotes β-sheet regions reported in the previous studies by Tycko et al.77
Figure 2d shows the summary of the residues quenched by Cu2+ binding for 6 Aβ samples selectively isotope labeled at different sites (see SI). Only a limited number of aliphatic side chains were quenched by Cu2+ bound to Aβ (see Figure S2 in SI). Apart from the significant changes in the N-terminal region, V24, and V39, other sites in the residues 17–38 show very limited perturbation from Cu2+ on the signal intensities. Similar signal quenching profile was obtained for Aβ(1–40) fibrils prepared at Dr. Tycko’s lab at the NIH without agitation70,76 (see SI and Figure S3). Importantly, we also found that 13C chemical shifts are not altered upon Cu2+ binding by more than 0.2 ppm for the hydrophobic core regions (residues 18–25 and 30–36) except for the moderate perturbation (~0.35 ppm) on some sites of F19 and G33 (Table S1). The results provide the first crucial experimental evidence that Cu2+ binding does not reorganize or alter the parallel β-sheet structures of the hydrophobic core regions for Aβ(1–40) fibril with site resolution.
To identify other binding sites, we further performed 2D 13C/13C correlation SSNMR (Figure 3a) of uniformly 13C- and 15N-labeled Aβ(1–40) fibrils (black) without and (red) with Cu2+. Surprisingly, as shown in the slices (Figure 3b–d), considerable signal quenching by 45±11 % and 59±9 % due to Cu2+ (orange arrows) was observed for 13CO2− of (d) V40 in the C-terminus and (b) Glu side chains, respectively. The extent of the quenching due to Cu2+ is comparable to those for His-13 and His-14. Although Glu signals were not assigned to individual residues, the results clearly suggest Cu2+ bindings to the carboxyl terminus of V40 and side chains of Glu. Whereas binding to Glu was hypothesized in previous studies,8 such Cu2+ binding, including that to the C-terminus, has been difficult to examine for Aβ fibrils until this work. Although site directed mutations may be effective for short Aβ fragments,29 such mutations on the full-length Aβ may alter the structure of amyloid fibril. The data in Figure 3a also indicate signals for other side-chain carboxyl groups at 175–180 ppm are likely quenched by Cu2+ binding; further studies will be needed for signal assignments and detailed analysis. With these SSNMR results, we conclude that the N-terminal region including H13, H14, and Glu side chains and the CO2− in the C-termius are highly likely to play major roles in Cu2+ binding to the Aβ(1–40) fibril. The Cu2+ binding is likely to make very little or no changes on the basic structural units in the hydrophobic cores of the Aβ amyloid fibril.
Figure 3.
(a) 2D 13C/13C SSNMR spectra between aliphatic and 13CO regions for uniformly 13C- and 15N-labeled Aβ(1–40) fibrils (black) without and ( ) with Cu2+, with 1D slices at ω1 = (b) 34.6, (c) 43.9, (d) 59.8 ppm. The spectra were obtained at MAS of 40 kHz with a fpRFDR mixing of 1.6 ms.87
) with Cu2+, with 1D slices at ω1 = (b) 34.6, (c) 43.9, (d) 59.8 ppm. The spectra were obtained at MAS of 40 kHz with a fpRFDR mixing of 1.6 ms.87
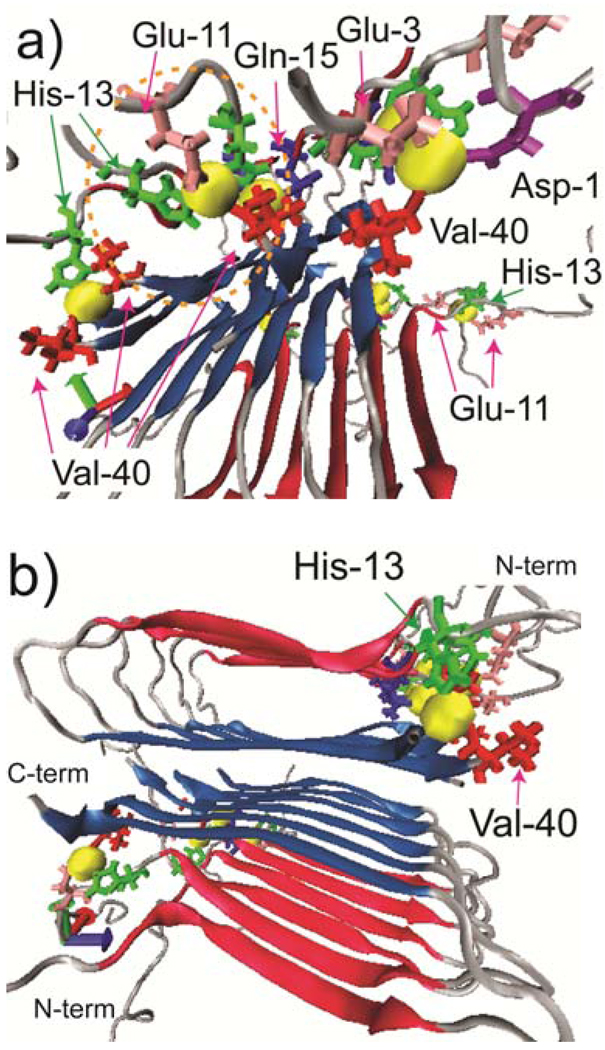
To elucidate possible Cu2+ binding structures to Aβ, we performed two sets of MD simulations of Aβ(1–40) fibrils with Cu2+. As the initial structure, we adopted a structural model from Tycko’s group77 with a fixed geometry for the residues 15–39 for oligomeric Aβ. In the first set of the MD simulation, we used cationic dummy atom (CaDa) approach for Cu2+ ions in order to identify possible Cu2+ binding ligands.78 Based on our SSNMR results, Cu2+ ions were non-covalently bound to Nε of H13 (or H14 in Figure 5) by ionic interactions in the initial structure. Indeed, the final structure with implicit solvents after three thermal annealing cycles demonstrated that CO2−/CO side chains of D1, E3, E11, Q15, and the CO2− terminal of V40 coordinate to Cu2+, despite the absence of such interactions in the initial structures (Figure 4). While previous EPR studies for monomeric Aβ fragments8,19,20,27 predicted two major coordination modes, here we suggest that a variety of coordination structures are likely to coexist in the amyloid fibril. Many of these observed binding modes are realized by the parallel β-sheet arrangements, in which Cu2+-coordination sites at the N-terminal are concentrated via aggregation. For example, some Cu2+ ions were bridged by two H13 (or H14) rings of each two peptides (orange circles in Figure 4a and Figure 5a). Such binding mode is not possible in a monomer, although the possibility of Cu2+-mediated inter-molecular His bridge was proposed for an Aβ dimer previously.69 Also, the Cu2+ ions bridge carboxyl terminal of V40 and H13 (or E11); such a binding has not been reported for monomeric Aβ. It should be noted that the above MD simulation results do not necessarily exclude the previously proposed coordination models by EPR and other studies. Our SSNMR results indeed indicate substantial relaxation on F4, which is consistent with the Cu2+ coordination to H6, which was proposed by the previous EPR and NMR studies.19,27,28 In our preliminary MD simulations in Fig. 4–5, such long-range coordination may not be completely reproduced probably because of the restricted time frame of the simulations (20 ns) and the limited ensemble of the initial structures. Our MD simulation starting from a different initial structure with some Cu2+ ions coordinated to Nε of H6, H13, H14 showed that a common Cu2+ ion can be coordinated to H6 and H14 of Aβ fibril (data not shown). It is possible that the various unique coordination modes presented here for the amyloid fibril coexist with those proposed for the monomeric/fragment Aβ at different population and perhaps in a dynamically exchangeable manner.27
Figure 5.
(a, b) The final MD structure of Cu2+ bound Aβ(1–40) fibril viewed from (a) one of the N-terminal sides and (b) the fibril axis after three thermal annealing cycles. In the initial structures, Cu2+ were bound to Nε of His-14 via non-covalent interactions. Blue and red arrows indicate β-sheet regions in residue 16–24 (red) and residue 30–39 (blue) of Aβ. The MD structure show that Cu2+ (yellow) are bound to His-14 (green), Glu-3, 11 (pink), Asp-1 (purple), Gln-15(blue), and Val-40 (red) after the thermal cycles. The radius of the yellow sphere is ~2 Å from the Cu2+ ion.
Figure 4.
(a, b) The final MD structure of Cu2+ bound Aβ(1–40) fibril viewed from (a) one of the N-terminal sides and (b) the fibril axis after three thermal annealing cycles (see SI). In the initial structures, Cu2+ were bound to Nε of H13 via non-covalent interactions. Blue and red arrows indicate β-sheet regions in residue 16–24 (red) and residue 30–39 (blue) of Aβ. The MD structure shows that Cu2+ (yellow) are bound to H13 (green), E3, E11 (pink), D1 (purple), Q15 (blue), and V40 (red) after the thermal cycles. The radius of the Cu2+ ions’ sphere is ~2 Å. The distances between Cu2+ and some of the ligands are shown in Fig. S4 in SI.
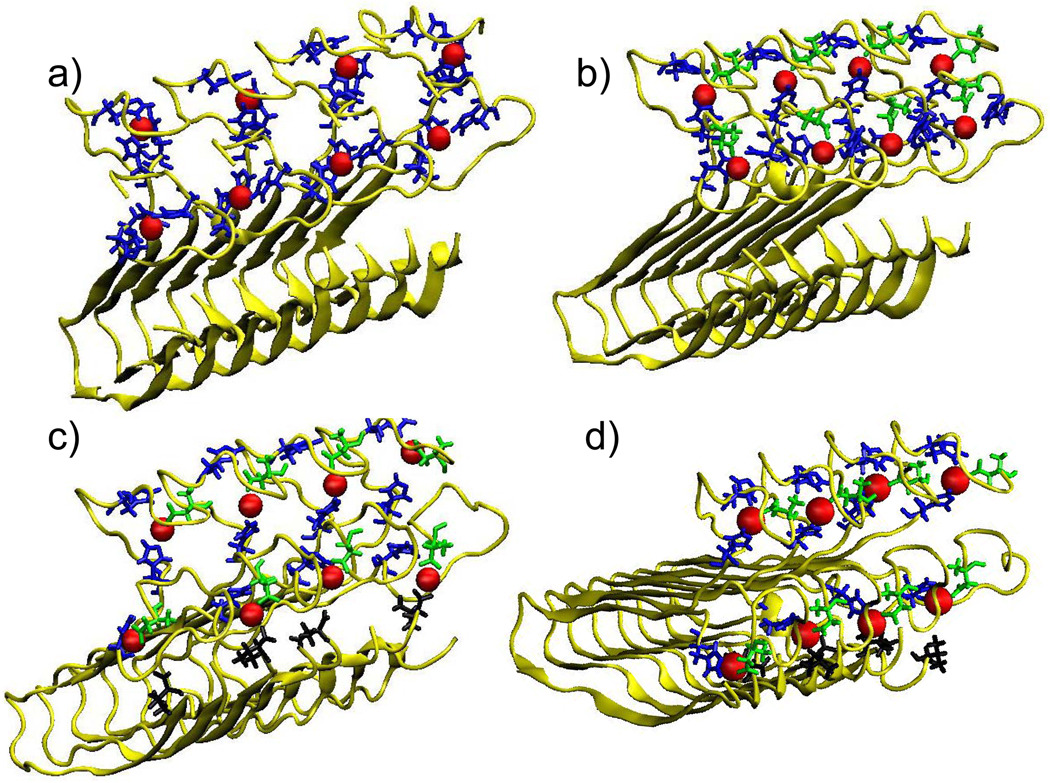
To confirm the presence of various coordination modes, we examined the stability and the conformational energies for Aβ(1–40) fibril models of three different Cu2+-binding modes (Table 1, Figure 6) with a more elaborate MD simulations using explicit solvents based on a recent study on Zn2+-Aβ oligomer association.68 The simulations show that such Cu2+-coordination modes are all stable over 20 ns. In Conformer 1, Cu2+ binds to His-6, His-13 and His-14 (Fig. 6a) and in Confomer 2, Cu2+ binds to Asp-1, His-6 and His-14 (Fig. 6b). As shown in Table 1, Conformer 1 (Fig. 6a) is more stable than Conformer 2 (Fig. 6b); this indicates that the interaction of Cu2+ with His is likely to stabilize the model more than Asp, although the energy difference is within standard deviation due to thermal fluctuation. In Conformer 3, 4 Cu2+ ions interact with D1, H6 and H14 and 4 Cu2+ ions bind to D1, H14 and V40 (Fig. 6c); in Conformer 4, the latter set of ligands are D1, H13 and V40 (Fig. 6d). Conformer 4, which has Cu2+ coordination to −CO2− of V40 and side chains of D1 and H13, shows higher stability than Conformer 2. The coordination of Cu2+ to D1, H14, V40 in Conformer 3, also has a slightly higher stability than that of D1, H6 and H14 in Conformer 2. These results suggest that Cu2+ may indeed interact with the C-termini, as suggested by our SSNMR analysis.
Figure 6.
(a) Conformer 1 for which His-6, His-13, His-14 residues are coordinated to copper ions. (b) Conformer 2 for which Asp-1 (green), His-6 (blue), His-14 (blue) residues are coordinated to a copper ion. (c) Conformer 3 for which Asp-1 (green), His-6 (blue), His-14 (blue) residues are coordinated to a copper ion and Asp-1 (green), His-14 (blue), Val-40 (black) are coordinated to another copper ion. (d) Conformer 4 for which Asp-1 (green), His-6 (blue), His-14 (blue) residues are coordinated to a copper ion and Asp-1 (green), His-13 (blue), Val-40 (black) are coordinated to another copper ion. The magnified coordination geometries are shown in Fig. S6 in SI.
The simulations presented here are intended to provide insights into the possible ligands coordinated to Cu2+ in the Aβ amyloid fibril. At this point, universal force fields are not available for MD simulations involving Cu2+ ions.18,79,80 Development of more optimized force fields is likely needed to reproduce more accurate coordination geometries for detailed comparison with EPR spectra. Despite the limitation, the MD simulation results allowed for the evaluation of Cu2+ coordination modes for more realistic Cu2+-Aβ fibril model based on our SSNMR results, which suggested that the structural changes by Cu2+ binding are restricted to the N-term and C-term regions. It is encouraging that the two separate sets of MD simulations both suggested the previously unexpected unique coordination modes that are consistent with our SSNMR relaxation data.
In conclusion, this study has presented molecular details of Cu2+-bound Aβ(1–40) amyloid fibril system by SSNMR. Three chemically and biologically novel aspects are presented in our work. First, this study has demonstrated the first systematic approach to examine binding of Cu2+ to insoluble amyloid aggregates at site specific level by SSNMR, based on the recent progress in SSNMR for small paramagnetic compounds66,81–84 and for paramagnetic proteins45,47,49,67 by our groups and other. We have shown that uses of paramagnetic quenching and chemical-shift perturbation in SSNMR can be effectively employed in order to examine possible Cu2+ binding sites and structural changes upon Cu2+ binding for insoluble amyloid system. Second, this study has, for the first time, provided detailed site-specific structural information on Cu2+ bound full-length Aβ(1–40) in amyloid fibrils. Despite the linkage of the Cu2+-Aβ fibril with AD and its significance as a potential pharmaceutical target,14–16 Cu2+-Aβ association has been examined mostly for smaller and early-stage model systems such as soluble Aβ fragments or Aβ monomers. Our study has offered a solution to this situation by overcoming the difficulties in the sample preparation and the lack of suitable structural analysis methods for Cu2+-Aβ in amyloid fibrils, for which structural information has been very scarce. Third, our SSNMR data experimentally demonstrated various novel features of Cu2+ coordination to the Aβ fibril. Besides offering experimental evidence of the Cu2+ coordination to Nε of H13 and H14, this study has revealed previously unexpected ligands such as the carboxyl group of V40 at the C-terminus and Glu side chains. Our chemical-shift perturbation data suggested very little structural changes for the hydrophobic core regions (residues 18–25 and 30–36) of the Aβ fibrils upon Cu2+ binding. The unique coordination modes suggested by SSNMR were confirmed by the two sets of MD simulations on sophisticated Aβ oligomer models reflecting realistic amyloid fibril structures. Unlike previous MD studies on Cu2+-Aβ binding,18 both MD simulations incorporated intermolecular parallel β-sheet structure that is characteristic of the Aβ(1–40) amyloid fibril as well as the non-convalent bonding models for Cu2+-ligand interactions. Although further studies are needed to fully understand Cu2+ binding structure to Aβ, including its relation to the previous EPR studies and a ROS production mechanism, the combination of SSNMR analyses and MD simulations have provided insights into previously unknown detailed structural features of Cu2+-bound Aβ(1–40) in amyloid fibrils.
The new SSNMR approach presented here may be also applicable to examine binding of small paramagnetic ligands, such as drugs, to amyloid proteins. It has been proposed that metal ions such as Cu2+ and Zn2+ may trigger the formation of Aβ oligomer or fibril, modulating misfolding kinetics and aggregation states of Aβ.25,30,85 In this initial work, we focused on establishing SSNMR structural analysis of Aβ(1–40) fibril upon Cu2+ binding after fibril formation. A similar approach is likely useful for SSNMR characterization of the structures of Aβ oligomer and Aβ fibril that are formed in the presence of Cu2+ or other metal ions.
Materials and Methods
Synthesis and purification of Aβ(1–40) peptide
Aβ(1–40) peptide (NH2-DAEFRHDSGY-EVHHQKLVFF-AEDVGSNKGA-IIGLMVGGVV-COOH) was synthesized and purified as reported previously. Briefly, Aβ(1–40) was synthesized using solid-phase peptide synthesis with standard Fmoc synthesis and cleavage protocols.40,76 The crude peptide was purified by HPLC using acetonitrile and water gradient with 0.1% trifluoroaceticacid. 13C- and 15N-labeling was introduced as described previously by incorporating Fmoc-protected uniformly 13C- and 15N labeled amino acids at selected residues. The Fmoc protection of the uniformly 13C- and 15N- labeled amino acids (Isotec/Sigma-Aldrich, Miamisburg, OH) was performed at the Research Resource Center (RRC) at University of Illinois at Chicago (UIC) using the protocol of Fields et al.86 The purity of the peptide samples were determined by MALDI-TOF mass spectra performed at UIC RRC and the purity were approximately 95% after the HPLC purification. The purified samples were stored at −20°C before when it was used. The labeling schemes for the 6 samples used are as follows. 1) His-14, Ile-32, Val-36, Gly-37; 2) His-13, Ala-30, Gly-38, Val-39; 3) Val-12, Phe-20, Ala-21, Ile-31,Gly-33, Met-35(13CεH3); 4) Phe-4, Gly-9, Val-12, Leu-17, Ala-21; 5) Phe-19, Val-24, Gly-25, Ala-30, Leu-34; 6) Val-18, Phe-19, Ala-21, Ile-31, Gly-33.
Uniformly 13C- and 15N-labeled Aβ(1–40) was expressed as a fusion protein with Glutathione S-Transferase (GST) tag connected by a Factor Xa recognition site (IEGR▼) in E. Coli. The GST tag was removed by Factor Xa enzymatic cleavage, and then, the peptide was purified by HPLC as described above. The yield of uniformly 15N- and/or 13C-protein Aβ(1–40) was ~1.5 mg from 1 L of the cell culture. The details of the protocol will be discussed elsewhere.
Preparation of Aβ(1–40) fibrils for Cu2+-binding assay and SSNMR experiments
For Cu2+-binding assay with Aβ(1–40) fibrils using UV-Vis spectroscopy, a solution of 5 mM Aβ(1–40) was prepared by first dissolving Aβ(1–40) peptide in 50 mM NaOH.45 The peptide mixture was then briefly vortexed, and diluted to a final peptide concentration of 500 µM with deionized water containing 0.02 % NaN3. The pH of the solution was adjusted to 7.4 by using 100 mM HCl. The Aβ(1–40) solution was sonicated for 5 minutes in an ice bath, and this solution was then centrifuged at 16.1 × 103 g for 5 minutes to remove any preformed aggregates. The peptide solution was divided into several 1.5-mL microvials for Cu2+-binding assay. The concentration of Aβ(1–40) was measured using UV-VIS spectroscopy.40 The Aβ(1–40) solution was then incubated for 14 days with constant agitation in the sample tube at room temperature. Aβ(1–40) fibril formation was confirmed by thioflavin T (ThT) fluorescence assay.
For SSNMR studies, amyloid fibril samples were prepared as described above, but with selectively isotope labeled or uniformly 13C- and 15N-labeled Aβ(1–40) peptides. For the samples used for Fig. 1, 2, after incubation of an Aβ solution for fibril formation, the Aβ(1–40) fibril with Cu2+ was prepared by adding CuCl2 solution (pH7.4) to the Aβ solution in the mole ratio of 0.4:1.0 (Cu2+:Aβ(1–40)), and incubated at 4°C for 24h with initial vortexing. The control sample without Cu2+ was prepared in the same manner except for the addition of CuCl2. The samples were then centrifuged at 16.1 × 103 g for about 1 h (20min at a time), and the supernatant was removed. The gel like pellet at the bottom was then lyophilized. The lyophilized Aβ(1–40) fibril samples with and without Cu2+ were packed in to 2.5mm MAS rotors with rubber O-rings on the spacer and cap. The lyophilized samples were rehydrated with deionized water (3µl water/mg Aβ(1–40)) in the rotor with 3µL of water added at one time, and centrifuged at 2.0 × 103 g for 2 min. The hydrated samples were incubated overnight at 4°C before running the SSNMR experiments.
For the uniformly 13C- and 15N-labeled Aβ(1–40) fibril sample used for Fig. 3, after incubation for fibril formation, we centrifuged the sample at 16.1 × 103 g for ~1.0 h (20 min at a time), and removed the supernatant. The gel-like pellet at the bottom of the centrifugal vial was transferred into the 1.8mm MAS rotor of 10µL volume by centrifugation. The fibrils were then packed into a 1.8 mm rotor by fitting a 200 µL-pipette tip to the rotor with the fibrils and centrifuging the rotor-pipette tip mounted in a micro centrifuge tube with minimum amount of the supernatent for 2 – 4 min at 6 × 103 g. Then, the rotor cap was glued with Krazy Glue (Krazy Glue, Columbus OH) with a small piece of Teflon tape between the cap and the sample to avoid interaction of the glue with the peptide. This cap can be removed easily by immersing it in liquid nitrogen. The Aβ(1–40) fibrils with Cu2+ was prepared by adding a 26.5 mM CuCl2 solution in the mole ratio of 0.4:1.0 (Cu2+ : Aβ(1–40)) into the rotor. Firstly, the hydrated Aβ(1–40) sample in a rotor was lyophilized, and then a CuCl2 solution was introduced directly into the 1.8mm rotor by centrifugation and the cap was glued as described above. The rotor was then incubated at 4°C for 24h before the SSNMR experiments.
SSNMR experiments
All the SSNMR experiments were performed with an Infinityplus SSNMR spectrometer from Varian (Fort Collins, CO) with a homebuilt 2.5-mm MAS triple-resonance magic-angle-spinning (MAS) probe or a 1.8-mm MAS triple-resonance MAS probe developed at Dr. Samoson’s lab (National Institutes of Chemical Physics and Biophysics, Tallinn, Estonia) at 9.4 T (1H frequency of 400.2 MHz) in the double-configuration. For the data in Fig. 1 and 2, the spinning speed was set to 20,000 ± 3 Hz throughout the experiments with cooling air at −10°C supplied through a Varian VT stack at a flow rate of ~140 standard-cubic-feet per hour (scfh), which kept the sample temperature at ~11 °C. Approximately, 2.0–3.5 mg of labeled Aβ(1–40) fibril samples were used. In the 1D 13C CPMAS experiments for Fig. 1 and 2, adiabatic CP transfer was used. During the CP period, the 13C RF field amplitude was linearly swept from 45 kHz to 65 kHz during a contact time of 1.0 ms while the 1H RF amplitude was kept constant at 75 kHz. During the detection period, 1H TPPM decoupling of 90 kHz was employed. The recycle delays for the 1D experiments were 1.8 s. The 1D spectra in Fig.1c and Fig.2c were collected with 1024 scans, and were processed with Gaussian broadening of 20Hz and 150 Hz, respectively. The 1D spectra in Fig. 2a, b were collected with 4096 scans and processed with Gaussian broadening of 150Hz.
In the 2D experiments for Fig. 1d, a 2D 13C/13C correlation pulse sequence with the fpRFDR mixing was used.76,87 After the adiabatic CP, signals were recorded during the t1 period, and then a real or imaginary component of the magnetization was stored along the z axis. Then, during a mixing period, a fpRFDR 13C-13C dipolar-recoupling sequence with a mixing time of 1.6 ms and a 13C π-pulse width of 15 µs was used. 1H TPPM decoupling of 90 kHz was employed during the t1 and t2 periods, while cw decoupling of the same amplitude was used during the mixing period. For each t1 point, 192 scans of signals were accumulated with an acquisition period of 10 ms. A total of 180 complex t1 points were recorded with a t1 increment of 33.4 µs. The obtained NMR data were processed by NMRPipe software.88 Gaussian window functions of 110 Hz and 90 Hz were applied along the t1 and t2 time domains. An overall experimental time was 35 hours. The experiments were performed for the 6 Aβ samples listed above in which different residues are uniformly 13C- and 15N-labeled. 15N-labeling was introduced for experiments to be performed in the future.
The spectra in Fig. 3 were acquired at a spinning speed of 40,000 ± 10 Hz with cooling air at −20°C supplied through a Varian VT stack at a flow rate of ~140 scfh with cooled bearing air (1°C), which kept sample temperature at ~12°C. The 2D 13C/13C correlation spectra were obtained using fpRFDR mixing of 1.6 ms with 13C π- pulse widths of 13 µs. Approximate amount of the Aβ fibril sample (excluding water) was ~0.6 mg. The π/2-pulse for proton excitation was 2.5 µs. During the 1 ms CP period, the 13C rf field was swept from 48 kHz to 76 kHz, while the 1H rf field was kept at 102 kHz. The signal was collected during an acquisition period of 10 ms with low-power TPPM (lpTPPM) 1H decoupling with the rf field intensity (ω1/2π) at 7 kHz was applied with phase alternation between ±17°. A total of 126 complex t1 points were recorded with a t1 increment 48 µs. For each t1 point, 728 scans of signals were accumulated. For the uniformly 13C- and 15N-labeled Aβ fibril sample without Cu2+, total experimental time was 72 hours with recycle delays of 1.4 s. For the sample with 0.4 molar eq. of Cu2+, the experiment was collected using PACC method 45 with a total experimental time of 8 hours and recycle delays of 150 ms. The details of the pulse program and lpTPPM are described elsewhere.45,89 The obtained NMR data were processed by NMR pipe software. Lorentz to Gaussian window functions with inverse exponential width of 30Hz and Gaussian broadening of 80Hz were applied for both t1 and t2 time domains.
Molecular dynamics (MD) simulations protocol
In order to identify possible Cu2+ binding structures in Aβ(1–40) fibrils and verify their structural stabilities, we have performed two sets of molecular dynamics (MD) simulations. In the first set of the MD simulations related to the structure shown in Figs. 4–5, we used the structural refinement within AMBER 890. MD simulations were performed using a modified AMBER ff99sb force-field91 with generalized Born (GB) implicit solvation92 on an Argo Beowulf PC cluster at the Academic Computing and Communications Center at UIC (http://www.uic.edu/depts/accc/hardware/argo/index.html). The AMBER refinement scheme used herein is a slightly modified version which has been used for a number of NMR structure refinements.93 Force-fields for Cu2+ were adopted from those for Zn2+ denoted as tetrahedral divalent cation model and used with a minor modification with the differences in the atomic mass (65.360) and vdW radius for copper (r* = 3.100 Å).78 Although it is possible that Cu2+ bound to Aβ takes other coordinates such as a distorted planer geometries suggested by EPR studies,19,21 we assumed this tetrahedral geometry in order to identify possible metal binding structures for one of the most common Cu2+ geometries. It is interesting to note, however, that for the undistorted tetrahedral model of Cu2+ used herein, the ligands can readily sample and coordinate in a distorted fashion (Table S2). Thus, there is indeed some flexibility with regard to how the ligands can arrange around the Cu ions in the CaDa model. In a recent study to simulate Cu2+-binding to Aβ(1–40), Baik et al showed that the optimization of Cu2+ coordination to His by ab-initio calculations resulted in the distorted tetrahedral symmetry.18 A comparison of the coordination geometries for the MD structures for Cu, Zn SOD obtained with different types of force fields with X-ray structure showed that the structural parameters obtained by the tetrahedral model show reasonable agreement with those for the X-ray structure (see Table S2 in SI). A recent X-ray study suggested that Cu, Zn-superoxide dismutase (SOD), a type II Cu2+ protein, which has a distorted square-planar Cu2+ coordination geometry, show very limited changes in the coordination structure by replacement of Cu2+ with Zn2+ (i.e. Zn, Zn SOD).94 This is consistent with our MD simulation results with the Cu-CaDa tetrahedral model in SI. The initial structure of the Aβ(1–40) fibril was adopted from Tycko’s model77 and the missing 8 N-terminal amino acids were built back into the molecule using Molmol.95 Based on our NMR data, two sets of initial Cu2+ locations (i.e at His-13 or His-14) were assumed; Cu2+ was non-covalently attached to Nε of His-13 for Fig. 4 (or His-14 for Fig. 5) by replacement of the Hε and held in position by the incorporation of an NOE between the Cu2+ atom and Nε of His with a square well potential varying from 1.6–3.0 Å and a force constant of 10 kcal/mol. Protons at the Nδ positions of Cu2+-bound His-13/14 were removed, while those for other histidines were retained. All hydrogen atom bond lengths were constrained using the SHAKE algorithm.96 A cutoff of 12 Å was used for nonbonded interactions. Steepest descentminimization was performed for 2000 steps prior to and after MD refinement. The final MD structures were obtained after three thermal annealing cycles (0 K → 1000 K → 0 K) for a total of 60 ps run time. During the MD simulations the coordinates of residues 16–39 were fixed using a harmonic potential of 20 kcal/mol. Based on our NMR results, the structure of these regions is likely to be unaffected by Cu2+-binding. As shown in Fig. 4a and Fig. 5a, respectively, many Cu2+ ions remained attached to His-13 and His-14 via ionic interactions even after three thermal cycles, while some ions dissociated from the positions. Although previous EPR studies predicted only two major Cu2+ binding structures, coexistence of a variety of binding modes was observed in our MD simulations. The simulations provided insights into the possible metal binding structures.
The second set of MD simulations in Fig. 6 was used to rigorously compare the stability of the Cu2+ binding structures for three possible binding modes. For the conformation energy calculations in Table 1, MD simulations of solvated 8Cu2+-Aβ(1–40) oligomers were also performed in NPT ensemble using the NAMD program97 and the CHARMM package98 with the all-atom CHARMM27 force field. The oligomers were explicitly solvated with TIP3P water molecules. Constant temperature (300 K) and constant pressure (1 atm) were controlled by Langevin thermostat with a damping coefficient of 10 ps−1, using the NAMD program. For simulations using the NAMD program97, the Langevin piston method97,99,100 with a decay period of 100 fs, and a damping time of 50 fs was used to maintain a constant pressure of 1 atm. The short-range van der Waals (VDW) interactions were calculated using the switching function, with a twin range cutoff of 10.0 and 12.0 Å. Long-range electrostatic interactions were calculated using the particle mesh Ewald method with a cut-off of 12.0 Å for all simulations. The equations of motion were integrated using the leapfrog integrator with a step of 1 fs. All initial 8Cu2+-Aβ(1–40) oligomer was minimized and then solvated in a TIP3P water box with a minimum distance of 15 Å from any edge of the box to any Aβ atom. Any water molecule within 2.5 Å of the Aβ was removed. Even with Cu2+ binding, the overall charge for Cu2+-Aβ(1–40) is still -1. Therefore, counter ions (Na+) were added into solution at random locations to neutralize the charge of Cu2+-Aβ(1–40). The solvated systems were minimized for 2000 conjugated gradient steps, with the distance between the β-sheets fixed in the range 2.2 – 2.5 Å. The VDW parameters of Cu2+ ion and all Cu2+-ligands distances were constrained to the estimated distances,101 both along the minimization process and during all dynamics simulation timescales. Counter ions, peptides and water molecules were all allowed to move. The hydrogen atoms were constrained to the equilibrium bond using the SHAKE algorithm.96 The minimized solvated systems were quickly heated to 250 K, where all atoms were allowed to move. Then, the systems were heated from 250 K to 300 K for 300 ps and equilibrated at 300 K for 300 ps. Simulations ran for 20 ns and structures were saved every 10 ps for analysis.
Copper-binding octamer was constructed from Tycko’s amyloid fibril model.77 We used one monomer from the fibril conformation of Aβ(9–40). Residues G9-K16 were removed from each monomer and the N-terminal fragment peptide (D1–K16) was linked binding to Cu2+. The initial conformation of Cu2+-Aβ(1–16) was combined with the Aβ(17–40) as follows. The Zn2+ ions in the solution NMR structure of Zn2+−Aβ(1–16) complex from Zirah et al.102 were replaced with the Cu2+ ion. Then, all Cu2+-ligands distances were constrained to the estimated distances with the residues that coordinate with the ions. Based on previous EPR and NMR studies,19–21,27 two conformers were tested where Cu2+ binds to residues in the N-terminal: in the first model, Cu2+ binds to H6, H13 and H14 (Conformer 1; Figure 6a) and in the second model, Cu2+ interacts with D1, H6 and H14 (Conformer 2; Figure 6b). To test interactions of the copper ions with the C-termini of Aβ suggested in the SSNMR data and CaDa MD simulation, we constructed a model with 4 Cu2+ ions coordinating to D1, H14 and V40; and 4 Cu2+ ions to D1, H6 and H14 (Conformer 3; Figure 6c) and another model with 4 Cu2+ ions coordinating to D1, H14 and V40; and 4 Cu2+ ions to D1, H6 and H13 (Conformer 4; Figure 6d).
Analysis details for MD simulations
To obtain the relative structural stability of the 8Cu2+-Aβ(1–40) oligomers listed in Table 1, the Aβ trajectories of the last 5 ns were first extracted from the explicit MD simulation excluding water molecules. The solvation energies of all systems were calculated using the Generalized Born Method with Molecular Volume (GBMV).103,104 In the GBMV calculations, the dielectric constant of water was set to 80.0 and no distance cutoff was used. The hydrophobic solvent-accessible surface area (SASA) term factor was set to 0.00592 kcal/mol·Å2. Each conformer is minimized for 1000 cycles and the conformation energy is evaluated by grid-based GBMV. The minimization does not change the conformations of each conformer, but only relaxed the local geometries due to thermal fluctuation which occurred during the MD simulations.
The relative conformational stabilities of the oligomers were measured by root mean-squared deviation (RMSD) of the C-terminal region (residues Leu17-Val40), the N-terminal region (residues Ala2-Gln15) and for the U-turn region (residues Glu22-Gly29) with respect to the initial minimized structure throughout the simulations. We also examined the stability of the oligomers by following changes in the number of hydrogen bonds between the β strands with the hydrogen bond cutoff set to 2.5 Å (Fig. S7a) and by monitoring the change in the inter-sheet distance (Cα backbone-backbone distance) in the core domains of all oligomers (Ala-21-Ile-31 distance; Fig. S7b). The latter distance is the averaged distance between each two β-sheet regions near to the salt bridge in each monomer for each conformer.
Supplementary Material
ACKNOWLEDGMENT
This study was supported primarily by NIH RO1 program (AG028490), and the Alzheimer’s Association grant (IIRG 08-91256). The SSNMR methodology development in this work was also supported by the NSF (CHE 449952, CHE 957793), and the Dreyfus Foundation Teacher-Scholar Award program. This project has been funded in part with Federal funds from the NCI, NIH under contract number HHSN261200800001E. Simulations were also performed on the Biowulf cluster at the NIH (http://biowulf.nih.gov). We thank Dr. Robert Tycko at the NIH for providing a pdb file of the structural model for Aβ(1–40) fibril and the Aβ fibrils samples used for Fig. S3 in SI. We are also grateful to Prof. Michael Zagorski at the Case Western Reserve University for kind clarifications on his works.
Footnotes
Supporting Information Available: Details of sample preparation, Cu2+ binding assays, and other SSNMR and MD data are available free of charge via the Internet at http://pubs.acs.org/.
References
- 1.Mattson MP. Nature. 2004;430:631–639. doi: 10.1038/nature02621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Selkoe DJ. Nat. Cell. Biol. 2004;6:1054–1061. doi: 10.1038/ncb1104-1054. [DOI] [PubMed] [Google Scholar]
- 3.Lovell MA, Robertson JD, Teesdale WJ, Campbell JL, Markesbery WR. Journal of the Neurological Sciences. 1998;158:47–52. doi: 10.1016/s0022-510x(98)00092-6. [DOI] [PubMed] [Google Scholar]
- 4.Leskovjan AC, Lanzirotti A, Miller LM. Neuroimage. 2009;47:1215–1220. doi: 10.1016/j.neuroimage.2009.05.063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Miller LM, Wang Q, Telivala TP, Smith RJ, Lanzirotti A, Miklossy J. J. Struct. Biol. 2006;155:30–37. doi: 10.1016/j.jsb.2005.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Huang XD, Atwood CS, Hartshorn MA, Multhaup G, Goldstein LE, Scarpa RC, Cuajungco MP, Gray DN, Lim J, Moir RD, Tanzi RE, Bush AI. Biochemistry. 1999;38:7609–7616. doi: 10.1021/bi990438f. [DOI] [PubMed] [Google Scholar]
- 7.Ali FE, Separovic F, Barrow CJ, Cherny RA, Fraser F, Bush AI, Masters CL, Barnham KJ. J. Pept. Sci. 2005;11:353–360. doi: 10.1002/psc.626. [DOI] [PubMed] [Google Scholar]
- 8.Faller P. Chembiochem. 2009;10:2837–2845. doi: 10.1002/cbic.200900321. [DOI] [PubMed] [Google Scholar]
- 9.Opazo C, Huang XD, Cherny RA, Moir RD, Roher AE, White AR, Cappai R, Masters CL, Tanzi RE, Inestrosa NC, Bush AI. J. Biol. Chem. 2002;277:40302–40308. doi: 10.1074/jbc.M206428200. [DOI] [PubMed] [Google Scholar]
- 10.Scott LE, Orvig C. Chem. Rev. 2009;109:4885–4910. doi: 10.1021/cr9000176. [DOI] [PubMed] [Google Scholar]
- 11.Atwood CS, Scarpa RC, Huang XD, Moir RD, Jones WD, Fairlie DP, Tanzi RE, Bush AI. J. Neurochem. 2000;75:1219–1233. doi: 10.1046/j.1471-4159.2000.0751219.x. [DOI] [PubMed] [Google Scholar]
- 12.Atwood CS, Moir RD, Huang XD, Scarpa RC, Bacarra NME, Romano DM, Hartshorn MK, Tanzi RE, Bush AI. J. Biol. Chem. 1998;273:12817–12826. doi: 10.1074/jbc.273.21.12817. [DOI] [PubMed] [Google Scholar]
- 13.Bush AI, Tanzi RE. Proc. Natl. Acad. Sci. U. S. A. 2002;99:7317–7319. doi: 10.1073/pnas.122249699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bush AI. J. Alz. Dis. 2008;15:223–240. doi: 10.3233/jad-2008-15208. [DOI] [PubMed] [Google Scholar]
- 15.Barnham KJ, Kenche VB, Ciccotosto GD, Smith DP, Tew DJ, Liu X, Perez K, Cranston GA, Johanssen TJ, Volitakis I, Bush AI, Masters CL, White AR, Smith JP, Cherny RA, Cappai R. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6813–6818. doi: 10.1073/pnas.0800712105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi J-S, Braymer JJ, Nanga RPR, Ramamoorthy A, Lim MH. Proc. Natl. Acad. Sci. U. S. A. 2010;107:21990–21995. doi: 10.1073/pnas.1006091107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jun S, Gillespie JR, Shin B-k, Saxena S. Biochemistry. 2009;48:10724–10732. doi: 10.1021/bi9012935. [DOI] [PubMed] [Google Scholar]
- 18.Mantri Y, Fioroni M, Baik MH. J. Biol. Inorg. Chem. 2008;13:1197–1204. doi: 10.1007/s00775-008-0403-6. [DOI] [PubMed] [Google Scholar]
- 19.Drew SC, Noble CJ, Masters CL, Hanson GR, Barnham KJ. J. Am. Chem. Soc. 2009;131:1195–1207. doi: 10.1021/ja808073b. [DOI] [PubMed] [Google Scholar]
- 20.Drew SC, Masters CL, Barnham KJ. J. Am. Chem. Soc. 2009;131:8760–8761. doi: 10.1021/ja903669a. [DOI] [PubMed] [Google Scholar]
- 21.Sarell CJ, Syme CD, Rigby SEJ, Viles JH. Biochemistry. 2009;48:4388–4402. doi: 10.1021/bi900254n. [DOI] [PubMed] [Google Scholar]
- 22.Minicozzi V, Stellato F, Comai M, Serra MD, Potrich C, Meyer-Klaucke W, Morante S. J. Biol. Chem. 2008;283:10784–10792. doi: 10.1074/jbc.M707109200. [DOI] [PubMed] [Google Scholar]
- 23.Ali FE, Separovic F, Barrow CJ, Yao SG, Barnham KJ. Int. J. Pept. Res. Ther. 2006;12:153–164. [Google Scholar]
- 24.Gaggelli E, Grzonka Z, Kozlowski H, Migliorini C, Molteni E, Valensin D, Valensin G. Chem. Commun. 2008:341–343. doi: 10.1039/b713453c. [DOI] [PubMed] [Google Scholar]
- 25.Lim KH, Kim YK, Chang YT. Biochemistry. 2007;46:13523–13532. doi: 10.1021/bi701112z. [DOI] [PubMed] [Google Scholar]
- 26.Miura T, Suzuki K, Kohata N, Takeuchi H. Biochemistry. 2000;39:7024–7031. doi: 10.1021/bi0002479. [DOI] [PubMed] [Google Scholar]
- 27.Hou LM, Zagorski MG. J. Am. Chem. Soc. 2006;128:9260–9261. doi: 10.1021/ja046032u. [DOI] [PubMed] [Google Scholar]
- 28.Karr JW, Kaupp LJ, Szalai VA. J. Am. Chem. Soc. 2004;126:13534–13538. doi: 10.1021/ja0488028. [DOI] [PubMed] [Google Scholar]
- 29.Karr JW, Szalai VA. J. Am. Chem. Soc. 2007;129 doi: 10.1021/ja068952d. 3796-+ [DOI] [PubMed] [Google Scholar]
- 30.Liu ST, Howlett G, Barrow CJ. Biochemistry. 1999;38:9373–9378. doi: 10.1021/bi990205o. [DOI] [PubMed] [Google Scholar]
- 31.Antzutkin ON. Magn. Reson. Chem. 2004;42:231–246. doi: 10.1002/mrc.1341. [DOI] [PubMed] [Google Scholar]
- 32.Hindo SS, Mancino AM, Braymer JJ, Liu YH, Vivekanandan S, Ramamoorthy A, Lim MH. J. Am. Chem. Soc. 2009;131 doi: 10.1021/ja907045h. 16663-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Olofsson A, Lindhagen-Persson M, Vestling M, Sauer-Eriksson AE, Ohman A. Febs Journal. 2009;276:4051–4060. doi: 10.1111/j.1742-4658.2009.07113.x. [DOI] [PubMed] [Google Scholar]
- 34.Zoroddu MA, Medici S, Peana M. J. Inorg. Biochem. 2009;103:1214–1220. doi: 10.1016/j.jinorgbio.2009.06.008. [DOI] [PubMed] [Google Scholar]
- 35.Karr JW, Szalai VA. Biochemistry. 2008;47:5006–5016. doi: 10.1021/bi702423h. [DOI] [PubMed] [Google Scholar]
- 36.Petkova A, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Heise H, Hoyer W, Becker S, Andronesi OC, Riedel D, Baldus M. Proc. Natl. Acad. Sci. U. S. A. 2005;102:15871–15876. doi: 10.1073/pnas.0506109102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chimon S, Ishii Y. J. Am. Chem. Soc. 2005;127:13472–13473. doi: 10.1021/ja054039l. [DOI] [PubMed] [Google Scholar]
- 39.Sciarretta KL, Gordon DJ, Petkova AT, Tycko R, Meredith SC. Biochemistry. 2005;44:6003–6014. doi: 10.1021/bi0474867. [DOI] [PubMed] [Google Scholar]
- 40.Chimon S, Shaibat MA, Jones CR, Calero DC, Aizezi B, Ishii Y. Nat. Struct. Mol. Biol. 2007;14:1157–1164. doi: 10.1038/nsmb1345. [DOI] [PubMed] [Google Scholar]
- 41.Wasmer C, Soragni A, Sabate R, Lange A, Riek R, Meier BH. Angew. Chem. Int. Edit. 2008;47:5839–5841. doi: 10.1002/anie.200704896. [DOI] [PubMed] [Google Scholar]
- 42.Helmus JJ, Surewicz K, Nadaud PS, Surewicz WK, Jaroniec CP. Proc. Natl. Acad. Sci. U. S. A. 2008;105:6284–6289. doi: 10.1073/pnas.0711716105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Curtis-Fisk J, Spencer RM, Weliky DP. J. Am. Chem. Soc. 2008;130 doi: 10.1021/ja8039426. 12568-+ [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaroniec CP, MacPhee CE, Astrof NS, Dobson CM, Griffin RG. Proc. Natl. Acad. Sci. U. S. A. 2002;99:16748–16753. doi: 10.1073/pnas.252625999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wickramasinghe NP, Parthasarathy S, Jones CR, Bhardwaj C, Long F, Kotecha M, Mehboob S, Fung LWM, Past J, Samoson A, Ishii Y. Nat. Methods. 2009;6:215–218. doi: 10.1038/nmeth.1300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Masuda Y, Uemura S, Ohashi R, Nakanishi A, Takegoshi K, Shimizu T, Shirasawa T, Irie K. Chembiochem. 2009;10:287–295. doi: 10.1002/cbic.200800411. [DOI] [PubMed] [Google Scholar]
- 47.Pintacuda G, Giraud N, Picrattelli R, Bockmann A, Bertini I, Emsley L. Angew. Chem. Int. Edit. 2007;46:1079–1082. doi: 10.1002/anie.200603093. [DOI] [PubMed] [Google Scholar]
- 48.Balayssac S, Bertini I, Lelli M, Luchinat C, Maletta M. J. Am. Chem. Soc. 2007;129:2218–2219. doi: 10.1021/ja068105a. [DOI] [PubMed] [Google Scholar]
- 49.Balayssac S, Bertini I, Bhaumik A, Lelli M, Luchinat C. Proc. Natl. Acad. Sci. U. S. A. 2008;105:17284–17289. doi: 10.1073/pnas.0708460105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pooransingh-Margolis N, Renirie R, Hasan Z, Wever R, Vega AJ, Polenova T. J. Am. Chem. Soc. 2006;128:5190–5208. doi: 10.1021/ja060480f. [DOI] [PubMed] [Google Scholar]
- 51.Castellani F, van Rossum B, Diehl A, Schubert M, Rehbein K, Oschkinat H. Nature. 2002;420:98–102. doi: 10.1038/nature01070. [DOI] [PubMed] [Google Scholar]
- 52.Morcombe CR, Gaponenko V, Byrd RA, Zilm KW. J. Am. Chem. Soc. 2005;127:397–404. doi: 10.1021/ja045581x. [DOI] [PubMed] [Google Scholar]
- 53.Lorieau JL, Day LA, McDermott AE. Proc. Natl. Acad. Sci. U. S. A. 2008;105:10366–10371. doi: 10.1073/pnas.0800405105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Qiang W, Bodner ML, Weliky DP. J. Am. Chem. Soc. 2008;130:5459–5471. doi: 10.1021/ja077302m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Lange A, Giller K, Hornig S, Martin-Eauclaire MF, Pongs O, Becker S, Baldus M. Nature. 2006;440:959–962. doi: 10.1038/nature04649. [DOI] [PubMed] [Google Scholar]
- 56.Cady SD, Schmidt-Rohr K, Wang J, Soto CS, DeGrado WF, Hong M. Nature. 2010;463 doi: 10.1038/nature08722. 689-U127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hu FH, Luo WB, Hong M. Science. 2010;330:505–508. doi: 10.1126/science.1191714. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sharma M, Yi MG, Dong H, Qin HJ, Peterson E, Busath DD, Zhou HX, Cross TA. Science. 2010;330:509–512. doi: 10.1126/science.1191750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Griffin RG. Nat. Struct. Biol. 1998;5:508–512. doi: 10.1038/749. [DOI] [PubMed] [Google Scholar]
- 60.Opella SJ. Nat. Struct. Biol. 1997;4:845–848. [PubMed] [Google Scholar]
- 61.McDermott AE. Curr. Opin. Struct. Biol. 2004;14:554–561. doi: 10.1016/j.sbi.2004.09.007. [DOI] [PubMed] [Google Scholar]
- 62.Baldus M. Curr. Opin. Struct. Biol. 2006;16:618–623. doi: 10.1016/j.sbi.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 63.Gehman JD, O'Brien CC, Shabanpoor F, Wade JD, Separovic F. Eur. Biophys. J. Biophys. Lett. 2008;37:333–344. doi: 10.1007/s00249-007-0251-2. [DOI] [PubMed] [Google Scholar]
- 64.Bertini I, Emsley L, Lelli M, Luchinat C, Mao JF, Pintacuda G. J. Am. Chem. Soc. 2010;132 doi: 10.1021/ja100398q. 5558-+ [DOI] [PubMed] [Google Scholar]
- 65.Jovanovic T, McDermott AE. J. Am. Chem. Soc. 2005;127:13816–13821. doi: 10.1021/ja0438314. [DOI] [PubMed] [Google Scholar]
- 66.Wickramasinghe NP, Shaibat MA, Ishii Y. J. Phys. Chem. B. 2007;111:9693–9696. doi: 10.1021/jp0727289. [DOI] [PubMed] [Google Scholar]
- 67.Nadaud PS, Helmus JJ, Kall SL, Jaroniec CP. J. Am. Chem. Soc. 2009;131:8108–8120. doi: 10.1021/ja900224z. [DOI] [PubMed] [Google Scholar]
- 68.Miller Y, Ma BY, Nussinov R. Proc. Natl. Acad. Sci. U. S. A. 2010;107:9490–9495. doi: 10.1073/pnas.0913114107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Smith DP, Smith DG, Curtain CC, Boas JF, Pilbrow JR, Ciccotosto GD, Lau TL, Tew DJ, Perez K, Wade JD, Bush AI, Drew SC, Separovic F, Masters CL, Cappai R, Barnham KJ. J. Biol. Chem. 2006;281:15145–15154. doi: 10.1074/jbc.M600417200. [DOI] [PubMed] [Google Scholar]
- 70.Petkova AT, Leapman RD, Guo ZH, Yau WM, Mattson MP, Tycko R. Science. 2005;307:262–265. doi: 10.1126/science.1105850. [DOI] [PubMed] [Google Scholar]
- 71.Matsuba Y, Takahash Y. Analytical Biochemistry. 1970;36 doi: 10.1016/0003-2697(70)90346-5. 182-&. [DOI] [PubMed] [Google Scholar]
- 72.Varadarajan S, Kanski J, Aksenova M, Lauderback C, Butterfield DA. J. Am. Chem. Soc. 2001;123:5625–5631. doi: 10.1021/ja010452r. [DOI] [PubMed] [Google Scholar]
- 73.Schoneich C, Pogocki D, Hug GL, Bobrowski K. J. Am. Chem. Soc. 2003;125:13700–13713. doi: 10.1021/ja036733b. [DOI] [PubMed] [Google Scholar]
- 74.Barnham KJ, Ciccotosto GD, Tickler AK, Ali FE, Smith DG, Williamson NA, Lam YH, Carrington D, Tew D, Kocak G, Volitakis I, Separovic F, Barrow CJ, Wade JD, Masters CL, Cherny RA, Curtain CC, Bush AI, Cappai R. J. Biol. Chem. 2003;278:42959–42965. doi: 10.1074/jbc.M305494200. [DOI] [PubMed] [Google Scholar]
- 75.Tickler AK, Smith DG, Ciccotosto GD, Tew DJ, Curtain CC, Carrington D, Masters CL, Bush AI, Cherny RA, Cappai R, Wade JD, Barnham KJ. J. Biol. Chem. 2005;280:13355–13363. doi: 10.1074/jbc.M414178200. [DOI] [PubMed] [Google Scholar]
- 76.Petkova A, Ishii Y, Balbach JJ, Antzutkin ON, Leapman RD, Delaglio F, Tycko R. Proc. Natl. Acad. Sci. U.S.A. 2002;99:16742–16747. doi: 10.1073/pnas.262663499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Petkova AT, Yau WM, Tycko R. Biochemistry. 2006;45:498–512. doi: 10.1021/bi051952q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Pang YP. J. Mol. Modeling. 1999;5:196–202. [Google Scholar]
- 79.Branco RJF, Fernandes PA, Ramos MJ. J. Phys. Chem. B. 2006;110:16754–16762. doi: 10.1021/jp056855l. [DOI] [PubMed] [Google Scholar]
- 80.Peters MB, Yang Y, Wang B, Fusti-Molnar L, Weaver MN, Merz KM. J. Chem. Theory Comput. 2010;6:2935–2947. doi: 10.1021/ct1002626. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Ishii Y, Chimon S, Wickramasinghe NP. J. Am. Chem. Soc. 2003;125:3438–3439. doi: 10.1021/ja0291742. [DOI] [PubMed] [Google Scholar]
- 82.Wickramasinghe NP, Shaibat M, Ishii Y. J. Am. Chem. Soc. 2005;127:5796–5797. doi: 10.1021/ja042188i. [DOI] [PubMed] [Google Scholar]
- 83.Shaibat MA, Casabianca LB, Wickramasinghe NP, Guggenheim S, de Dios AC, Ishii Y. J. Am. Chem. Soc. 2007;129 doi: 10.1021/ja0703980. 10968-+ [DOI] [PubMed] [Google Scholar]
- 84.Kervern G, Pintacuda G, Zhang Y, Oldfield E, Roukoss C, Kuntz E, Herdtweck E, Basset JM, Cadars S, Lesage A, Coperet C, Emsley L. J. Am. Chem. Soc. 2006;128:13545–13552. doi: 10.1021/ja063510n. [DOI] [PubMed] [Google Scholar]
- 85.Miller Y, Ma B, Nussinov R. Chem. Rev. 2010;110:4820–4838. doi: 10.1021/cr900377t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Fields CA, Fields GB, Noble RL, Cross TA. Int. J. Peptide Protein Res. 1989;33:298–303. doi: 10.1111/j.1399-3011.1989.tb01285.x. [DOI] [PubMed] [Google Scholar]
- 87.Ishii Y. J. Chem. Phys. 2001;114:8473–8483. [Google Scholar]
- 88.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 89.Kotecha M, Wickramasinghe NP, Ishii Y. Magn. Reson. Chem. 2007;45:S221–S230. doi: 10.1002/mrc.2151. [DOI] [PubMed] [Google Scholar]
- 90.Case DAD, Cheatham ITE, Simmerling CL, Wang J, Duke RE, Luo R, Merz KM, Wang B, Pearlman DA, Crowley M, Brozell S, Tsui V, Gohlke H, Mongan J, Hornak V, Cui G, Beroza P, Schafmeister C, Caldwell JW, Ross WS, Kollman PA. San Fransisco: University of California; 2004. [Google Scholar]
- 91.Hornak V, Abel R, Okur A, Strockbine B, Roitberg A, Simmerling C. Proteins-Struct. Funct. Bioinf. 2006;65:712–725. doi: 10.1002/prot.21123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Tsui V, Case DA. J. Am. Chem. Soc. 2000;122:2489–2498. [Google Scholar]
- 93.Perez-Alvarado GC, Martinez-Yamout M, Allen MM, Grosschedl R, Dyson HJ, Wright PE. Biochemistry. 2003;42:7348–7357. doi: 10.1021/bi034062o. [DOI] [PubMed] [Google Scholar]
- 94.Strange RW, Antonyuk SV, Hough MA, Doucette PA, Valentine JS, Hasnain SS. J. Mol. Biol. 2006;356:1152–1162. doi: 10.1016/j.jmb.2005.11.081. [DOI] [PubMed] [Google Scholar]
- 95.Koradi R, Billeter M, Wuthrich K. J. Mol. Graph. 1996;14 doi: 10.1016/0263-7855(96)00009-4. 51-&. [DOI] [PubMed] [Google Scholar]
- 96.Ryckaert JP, Ciccotti G, Berendsen HJC. J. Comput. Phys. 1977;23:327–341. [Google Scholar]
- 97.Kale L, Skeel R, Bhandarkar M, Brunner R, Gursoy A, Krawetz N, Phillips J, Shinozaki A, Varadarajan K, Schulten K. J. Comput. Phys. 1999;151:283–312. [Google Scholar]
- 98.Brooks BR, Bruccoleri RE, Olafson BD, States DJ, Swaminathan S, Karplus M. J. Comput. Chem. 1983;4:187–217. [Google Scholar]
- 99.Martyna GJ, Tobias DJ, Klein ML. J. Chem. Phys. 1994;101:4177–4189. [Google Scholar]
- 100.Feller SE, Zhang YH, Pastor RW, Brooks BR. J. Chem. Phys. 1995;103:4613–4621. [Google Scholar]
- 101.Deng NJ, Yan L, Singh D, Cieplak P. Biophys. J. 2006;90:3865–3879. doi: 10.1529/biophysj.105.064444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Zirah S, Kozin SA, Mazur AK, Blond A, Cheminant M, Segalas-Milazzo I, Debey P, Rebuffat S. J. Biol. Chem. 2006;281:2151–2161. doi: 10.1074/jbc.M504454200. [DOI] [PubMed] [Google Scholar]
- 103.Lee MS, Salsbury FR, Brooks CL. J. Chem. Phys. 2002;116:10606–10614. [Google Scholar]
- 104.Lee MS, Feig M, Salsbury FR, Brooks CL. J. Comput. Chem. 2003;24:1348–1356. doi: 10.1002/jcc.10272. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.