Abstract
Arrestins specifically bind activated and phosphorylated G protein-coupled receptors, and orchestrate both receptor trafficking, and channel signaling to G protein-independent pathways via direct interactions with numerous non-receptor partners. Here we report the first successful use of solution NMR to map the binding sites in arrestin-1 (visual arrestin) for two polyanionic compounds that mimic phosphorylated light-activated rhodopsin: inositol hexaphosphate (IP6) and heparin. This yielded a more complete identification of residues involved in the binding with these ligands than has previously been feasible. IP6 and heparin appear to bind to the same site on arrestin-1, centered on a positively charged region in the N-domain. We present the first direct evidence that both IP6 and heparin induced a complete release of the arrestin C-tail. These observations provide novel insight into the nature of arrestin transition from basal to active state and demonstrate the potential of NMR-based methods in the study of protein-protein interactions involving members of the arrestin family.
Arrestins are soluble proteins that specifically bind to the phosphorylated forms of activated G protein-coupled receptors (GPCRs) (1), precluding further G protein activation and targeting the complex to clathrin-coated pits via direct interactions with clathrin (2) and its adaptor AP2 (3). This leads to endocytosis and receptor down-regulation. In addition to modulating signaling and trafficking of hundreds of GPCRs, arrestins also bind numerous non-receptor partners and regulate a variety of receptor-dependent and –independent signaling pathways(4). Vertebrates have four arrestin subtypes, only two of which, arrestin-2 and -32, are ubiquitously expressed(5). Arrestin-1, the first member of the family to be discovered (6), is present at very high levels in both rod (7–8) and cone (9) photoreceptors where it shuts off the signaling by light-activated opsins. Arrestin-1 was originally described as a protein that binds light-activated phosphorylated rhodopsin (6). Subsequent studies have also revealed that it has additional partners. Arrestin-1 self-associates to form dimers and tetramers (10–12) and also binds microtubules (13–14), calmodulin (15), protein kinase JNK3 and ubiquitin ligase Mdm2 (16), N-ethylmaleimide sensitive factor (17), and many other proteins. Association with active phosphorhodopsin is known to induce a global conformational change in arrestin-1 (18–19), which apparently includes the release of the C-tail. Negatively charged molecules heparin and inositol hexaphosphate (IP6) also bind arrestin-1, mimicking the phosphorylated C-terminus of rhodopsin and also increase the accessibility of the arrestin C-tail (20–21). The exposure of the C-tail upon receptor binding is highly conserved among arrestin family members, with this process facilitating subsequent GPCR internalization by making clathrin and AP2 binding sites more accessible (22).
The basal conformation of free arrestin-1 has been elucidated by X-ray crystallography (23–24) with additional structural insight also being provided by site-directed spin labeling/electronic paramagnetic resonance (EPR) spectroscopy in solution (12, 25–26). NMR and X-ray crystallography have been the main tools for determining atomic resolution structures of proteins, nucleic acids and carbohydrates. NMR has the advantage of providing insight into protein dynamics and allows studies of protein-ligand interactions under near-physiological conditions. While the application of NMR to structure determination of larger proteins such as the ~45 kDa arrestins remains challenging, technological developments such as deuteration (27–29) and transverse relaxation optimization spectroscopy (TROSY) (30) have made possible backbone chemical shift assignment and structural studies of proteins or protein complexes up to 100 kDa or even larger (31–32). Here solution NMR was used to assign ~40% of arrestin-1 backbone resonances. These assignments allowed us to map heparin and IP6 binding sites on arrestin-1 and to explore binding-induced conformational changes, including rearrangements of the important C-terminus. The dynamic nature of the C-terminus and several other segments render them “invisible” in all arrestin crystal structures (23–24, 33–35). The use of NMR spectroscopy yielded the first structural information on these elements and allowed characterization of their large-scale motions.
Arrestin interactions with dozens of binding partners regulate key signaling pathways in the cell (4–5). In most cases arrestin elements involved in the interactions with other proteins remain unknown. Their comprehensive identification reveals the molecular mechanisms of arrestin function and paves the way to targeted redesign of arrestins for therapeutic purposes (36). Here it is shown that each assigned backbone NMR resonance serves as a “reporter”, and their large number greatly facilitates comprehensive mapping of protein-protein interactions and detection of induced conformational changes at near-physiological conditions.
Materials and Methods
Preparation of Arrestin-1 NMR Sample
Arrestin-1 was expressed in E. coli and purified as previously described (37) with some modifications. Briefly, a pTrc-based plasmid encoding oligomerization-resistant arrestin-1-(F85A, F197A) (25) was transformed into BL21 (DE3) cells and plated on LB agar/ampicillin. 10 ml of overnight culture grown from a single colony in LB with 100 mg/L ampicillin (LB/A) at 30°C was inoculated into 1L of LB/A. Cells were grown at 30°C to OD600=0.8, and then induced for 18 hr using 25 μM IPTG. For 15N isotopic labeling, M9 minimum medium supplemented with 1 g/L 15NH4Cl was used. For 15N, 13C, 2H-labeled protein expression, M9 minimum medium based on 99% 2H2O with 1 g/L 15NH4Cl and 2.5 g/L 13C-glucose was used. For 15N, 2H-labeled protein, M9 minimum medium in 2H2O supplemented with 1g/L 15NH4Cl was used. The induction time for 15N, 13C, 2H-labeled or 15N, 2H-labeled protein was 24 hrs and the average cell growth time was 4 days.
For amino acid-selective isotopic labeling, a transaminase-deficient strain CT19 (generous gift of Dr. Waugh, NIH) with genetic lesions of the aspC, avtA, ilvE, trpB and tyrB genes was used to prevent 15N-labeled amino acid scrambling. This auxotroph is well-suited for selective 15N labeling with Leu, Val, Phe, Tyr and Ile (38). It was particularly important to maximize the number of assignments for lysine because of its importance in binding IP6, heparin and phosphorhodopsin. Therefore, a second transaminase-deficient cell strain S1288 (39)(CGSC #:6432, E. coli Genetic Stock Center, Yale University) was used for selective 15N-labeling with lysine, following a modified protocol (40) to optimize cell growth and protein yield. Briefly, cells were grown in 4L of LB/A supplemented with 100 mg/L kanamycin and 20 mg/L tetracycline (30°C, 225 rpm), harvested at OD600=0.6 by centrifugation at 3,000 rpm for 15 min, and resuspended in 1L of M9 minimal medium with the same antibiotics and 0.2 g/L 15N-labeled valine, leucine, isoleucine, tryptophan, phenylalanine, tyrosine, and aspartic acid individually. The cells were grown at 30°C for at least 2 h, and further incubated with 25 μM IPTG for 18 h. For S1288, the same protocol was used as for CT19 was used except that 100 mg/L ampicillin and 20 mg/L tetracycline were used as antibiotics. The M9 medium for S1288 needs to be supplemented with lysine, histidine, methonine, leucine, tryptophan and isoleucine (0.2 grams each) in 1L culture.
Protein was purified by heparin-Sepharose and Q-Sepharose chromatography, as described (37), with resulting protein purity seen to be >95% by SDS-PAGE and coomassie staining. In some cases arrestin-1 was further purified to >98% by S-Sepharose chromatography. Typical yields of pure protein were 8–15mg from a liter of culture.
Q-Sepharose fractions containing pure arrestin-1 were pooled and the NaCl concentration was immediately adjusted to 150 mM with 5M NaCl. Arrestin-1 was then concentrated to 0.5–1 mg/ml by ultrafiltration using an Amicon 30 kDa cut-off membrane, and frozen at −80°C. For each NMR experiment the buffer was exchanged from Tris (25 mM Tris-HCl, pH=7.5, 150 mM NaCl, 5 mM DTT, 1 mM EDTA) to Bis-Tris buffer (25 mM Bis-Tris, pH=6.5, 150 mM NaCl, 5 mM mercaptoethanol) using an ultrafiltration concentrator (Millipore, molecular weight cut-off=30 kDa) immediately before use. The final arrestin-1 concentrations were 0.15–0.2mM for 2D NMR and 0.5 mM for 3D NMR experiments.
Backbone NMR Resonance Assignments
Most NMR experiments were performed at 308 K on Bruker Avance 800MHz and 600MHz spectrometers equipped with cryogenic triple resonance probes with z-axis pulse field gradients. 2D HSQC spectra were obtained using a sensitivity-enhanced, phased-sensitive TROSY pulse sequence with water suppression using watergate (41– 42). Backbone assignments were made based on TROSY-based versions of HNCA, HN(CO)CA, HNCACB and HN(CO)CACB with 2H decoupling using a U-2H, 13C,15N-labeled sample (43– 44). HNCA and HNCACB were recorded on a Varian Unity Inova 900 MHz spectrometer at the Complex Carbohydrate Research Center, University of Georgia, Athens, GA. 114*80 complex points in t1 and t2 were obtained for HNCA and 90*78 for HNCACB. HN(CO)CA and HN(CO)CACB were acquired on a Bruker Avance 600MHz to minimize line broadening due to the field-dependent chemical shift anisotropy-based relaxation pathway. 116*40 and 128*40 complex points were obtained for HN(CO)CA and HN(CO)CACB respectively. All NMR data were processed with nmrPipe (45) and analyzed using Sparky (46) and NMRView (47).
Arrestin-1 backbone chemical shifts were predicted using SPARTA (48) based on the available crystal structure of the visual arrestin homotetramer (PDB ID: 1CF1) (24). It was assumed that the F85A/F197A mutant form of the protein that was used in this study has the same 3-D structure as wild type arrestin-1 because of their similarity in 2D TROSY spectra and identical biological functional properties. The predicted Cα and Cβ chemical shifts from Sparta were then used to match the experimental chemical shifts for a segment of two or more residues, which was useful for resolving ambiguous backbone resonance assignments.
PRE Measurement and Analysis
Single cysteine residues were introduced using Qiagen Quickchange into a cysteine-less arrestin-1 construct (C63A, C128S, C143S, F85A, F197A). Protein was prepared as described above. Mercaptoethanol was removed by buffer exchange using a molecular-weight-cut-off at 30 kDa Millipore centrifugal filter. Excess of MTSL (5-fold molar excess relative to arrestin-1) was mixed with protein sample and then incubated at room temperature for 10 min. Excess MTSL was immediately removed through extensive buffer exchange. Following 2-D 1H,15N-TROSY NMR measurements in the presence of the spin label, the nitroxide free radical was reduced to a diamagnetic state by adding ascorbic acid (6-fold molar excess relative to arrestin-1) and incubating at room temperature for at least two hours before again collecting an NMR spectrum. Paramagnetic relaxation rate enhancements (PREs) for each peak were determined as the intensity ratio of arrestin-1 peaks in the oxidized versus the reduced state. Distances can be derived from each PRE measurement by quantitating the increased T2 relaxation rate (R2*) and using equation 1(49–50).
| (eq. 1) |
Where K is 1.23×10−32 cm6 s−2, τc is the correlation time for electron-nuclear interaction and can be estimated from peak line-width at half height, ωh is proton Larmor frequency, r is the distance between the electron and nuclear spins. For those residues with reliable assignments, the PRE- derived distances were first used to find the position of the unpaired electron via triangulation based on the fixed coordinates of the protein. The predicted distances between the now-fixed nitroxide and amide sites for which assignments were ambiguous were then compared to the experimental distances extracted from the PREs associated with resonances with ambiguous assignments to resolve uncertainties in assignments.
Chemical Shift Perturbation by IP6 and Heparin
NMR titration experiments were performed to monitor the chemical shift perturbation of arrestin-1 by two polyanionic ligands- inositol hexaphosphate (IP6) and heparin, which had been previously shown to bind to all arrestins. IP6, also named phytic acid, was purchased from Sigma (P8810). Low-molecular-weight heparin with an average molecular weight of 4 kDa, corresponding to an average of 20 sugar units, was obtained from Fisher Scientific (BP2524100). The concentration of heparin was approximated by assuming a molecular weight of 4 kDa. IP6 and heparin were dissolved in 25mM Bis-Tris, 150mM NaCl and the pH of the stock solution was adjusted to pH=6.5 before mixing with protein samples. Different ratios of protein vs. ligand were tested until chemical shift changes reached a plateau. For IP6 and heparin, TROSY spectra at different protein-ligand ratios (1:0, 1:0.5, 1:1, 1:2, 1:5 and 1:10) and (1:0, 1:0.5, 1:1, 1:2, 1:5) were acquired at a fixed protein concentration of 0.15mM respectively. The binding site was close to saturation at protein:ligand ratios of 1:10 for IP6 and 1:5 for heparin. The chemical shift changes induced by the ligands were calculated using equation 2:
| (eq. 2) |
where ΔH and ΔN are the ligand-induced changes in the proton and nitrogen-15 chemical shifts, respectively. Chemical shift changes were plotted as a function of ligand concentrations and curves were fit to a 1:1 binding model to determine the dissociation constant using the titration analysis tool in NMRView (47).
15N NMR T2 Relaxation Time Measurements
15N transverse relaxation times T2 were measured using a Carr–Purcell–Meiboom–Gill (CPMG) sequence (51–52), which applies a high 15N pulse repetition rate in the CPMG pulse train to remove the effects of scalar coupling and a 1H 180° pulse to suppress cross-correlated relaxation effects during the relaxation period. For each cycle, 16 Hahn echoes were applied with each Hahn echo consisting of a 180 μs 180° pulse and a delay of 450 μs. Thus, one cycle takes 17.28 ms. For T2 measurements, spectra with different cycle numbers (1, 3, 4, 5, 7, 16, 32, and 64) were acquired and data were processed using the Sparky Relaxation Fitting module (46), which fits the peak heights to a decaying exponential function as in equation 3:
| (eq. 3) |
Backbone 15N T2 were measured for both ligand-free and ligand-bound arrestin-1. For the IP6-saturated arrestin-1 sample, 0.15 mM 2H,15N-arrestin-1 was mixed with 1.5 mM IP6 in 25 mM Bis-Tris buffer, 150 mM NaCl and 5 mM mercaptoethanol pH at 6.5 and spectra were acquired at 308 K using a Bruker Avance 600MHz spectrometer. For heparin-bound visual arrestin, 0.2 mM 15N, 2H-arrestin-1 was mixed with 1mM heparin in the same buffer as for IP6 and spectra were acquired at 308 K at a Bruker Avance 800MHz spectrometer.
Results
Optimization of Conditions for NMR Spectroscopy of Arrestin-1
It has long been known that wild-type arrestin-1 self-associates. Indeed the protein crystallizes in tetrameric form (24). Wild-type bovine arrestin-1 populates monomer-dimer and dimer-tetramer equilibria with dissociation constants at 37 μM and 7.5 μM respectively (25). At physiological concentrations (50–100 μM), visual arrestin is distributed between rapidly exchanging monomer, dimer, and tetramer populations. NMR studies of wild-type arrestin-1 are hindered by the severe line-broadening contributions from the large molecular weights of the dimers and tetramers, with possible additional contributions from intermediate time scale conformational exchange processes. For this reason, these studies employed a double-mutant (F85A, F197A) form of arrestin-1 that does not tetramerize and that exhibits a reduced propensity to dimerize (Kd = 0.5 mM)(25). A pull-down assay confirmed wild type-like phosphorhodopsin-binding activity for this double mutant. Acquisition of a 2-D 1H,15N-TROSY NMR spectrum that was seen to be similar to the corresponding spectrum from wild-type arrestin-1 confirmed that the double-mutation caused little or no structural perturbation (Data not shown).
Bis-Tris buffers at physiological ionic strength (>100 mM salt) were found to be suitable for acquisition of high-quality NMR spectra of arrestin-1. The optimum TROSY spectrum was obtained by preparing the sample in 25 mM Bis-Tris, 150 mM NaCl and 5 mM β-mercaptoethanol, pH=6.5. Under these conditions, arrestin-1 remained stable and functional up to 308K for 1–2 weeks, as confirmed by phosphorhodopsin pull-down assays.
Backbone NMR Chemical Shift Assignments for Arrestin-1
Arrestin-1 consists of 404 amino acids, including 25 prolines. Both in crystal and in solution it is an elongated two-domain molecule with high beta-strand content (24, 26). TROSY spectra of a fully protonated 0.2 mM protein sample exhibit more than 350 peaks, but severe peak overlap is observed between 7.8–8.2 ppm in the 1H dimension (Figure 1). Moreover, peak intensities are non-uniform with very sharp resonance from flexible loops and broad signals from most of the beta-strand residues. It has previously been demonstrated that a combination of protein perdeuteration and use of TROSY-based pulse sequences enables backbone resonance assignments to be made on large proteins (27, 53–57). We therefore pursued this strategy for arrestin-1. While successful in the assignment of flexible elements of arrestin-1 such as the C-tail (Figure 2A), this strategy was only partially successful. The difficulties faced are reflected by observation that fewer than 250 peaks were present in the 2D 1H-15N projection in the 3-D TROSY-HNCA experiment and fewer than 150 peaks in the 2D 1H-15N projection plane in the 3-D TROSY-HNCACB experiment. This dearth of peaks was further complicated by the abundance of prolines, which causes frequent interruptions in correlation chains. We therefore used a novel strategy to maximize backbone assignments by combining the available 3D data, selective amino acid labeling, chemical shift prediction, and paramagnetic relaxation enhancement methods.
Figure 1.
2-D 1H-15N TROSY spectrum of 0.2mM U-2H,15N-arrestin-1 in 25 mM Bis-Tris, 150 mM NaCl, 5 mM mercaptoethanol, pH=6.5 acquired at 308 K using a Bruker Avance 800MHz spectrometer. 152 assigned residues are labeled.
Figure 2.
(A) Crystal structure of arrestin-1 (Protein Databank code: 1CF1) with key regions labeled. The red circle represents the polar core area. The black ellipse represents the 3-element interaction involving N-terminus, C-terminus and alpha-helix 1. Numbers in parenthesis represent the start residue and the end residue. Residues for which backbone resonance assignments were made are highlighted in magenta. (B) Mapping of sites for which backbone resonance assignments were completed (highlighted in red) to the amino acid sequence of arrestin-1.
Backbone chemical shift assignment usually requires the sequential correlation of Cα and Cβ chemical shifts in concert with matching to the protein primary amino acid sequence. However, it can be very difficult to obtain a complete 3D correlation map for large proteins. We therefore used the method described by Langer et al (58) in combination with selective amino acid labeling to improve the number of backbone chemical shift assignments for those residues whose Cα and Cβ chemical shifts are available. A best fit algorithm was applied to find the best- segmental matches between experimental chemical shifts and predicted chemical shifts from Sparta, with the best-matched segment being the one with the minimal RMSD (58). However, the overlapped distribution of Cα and Cβ chemical shifts of several amino acids sometimes caused great degeneracy of assignments such that unique assignment cannot always be obtained.
Because ambiguity of possible assignments can be greatly minimized if the type of starting residue is know. Identifications of residue type were then obtained by the incorporation of the amino acid selective-labeling, which was used to permit the resonances to be classified according amino acid type. Selective amino acid labeling using BL21(DE3) requires the addition of 15N labeled amino acids immediately before protein induction. However, because of the very long (18 hours) post-induction time in which arrestin-1 is expressed, selective amino acid labeling of arrestin-1 using BL21(DE3) cell strains was not successful due to the scrambling of 15N from the labeled amino acid to other amino acids catalyzed by transaminases. To alleviate this problem, amino acid auxotrophic strains of E. coli harboring lesions for different transaminases were employed to avoid amino acid scrambling. Two different auxotrophs were used to selectively label Leu, Val, Tyr, Phe, Ile and Lys, with the exact number of expected peaks being observed in all cases. As an example, Figure 3 shows a spectrum of selectively-15N-labeled isoleucine arrestin-1 that exhibits the 20 Ile peaks expected based on protein sequence.
Figure 3.
2D 1H-15N TROSY spectrum of 0.2mM selectively 15N-isoleucine-labeled arrestin-1 in 25 mM Bis-Tris, 150mM NaCl and 5mM mercaptoethanol, pH=6.5 at 308 K using a Bruker Avance 800MHz spectrometer. 11 out of 20 isoleucine residue peaks were assigned, as labeled in the spectrum.
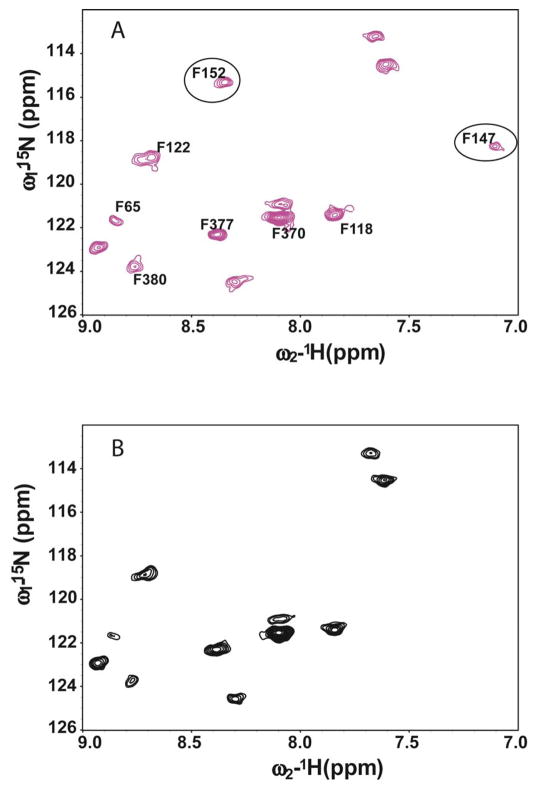
Site-directed paramagnetic relaxation enhancement (PRE) measurements were then used to validate the backbone assignments. PRE measurements have long been used to provide long-range distance restraints (15–25 Å) to complement the use of NOE distance restraints. While the application of PREs to protein backbone assignment/validation is not unprecedented (58–59), this approach has usually involved either a protein with a paramagnetic metal ion binding center or binding to the target protein of a ligand with a paramagnetic tag. For this work paramagnetic centers were introduced into fully functional cysteine-less mutant arrestin-1(26, 60) by incorporating single cysteine residues following by a thiol modification with MTSL(S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate). Single cysteines were strategically introduced at more than 1 site so that multiple sets of PRE data can be used to cross-validate the assignment. PREs were measured using selectively amino acid labeled samples, so as to simplify the spectra and to provide unambiguous identification of amino acid type. As an example, Figure 4 shows spectra of 15N-phenylalanine labeled arrestin-1 mutant (T157C, C63A, C128S, C143S, F85A, F197A) both before and after reduction of the spin label located at residue 157. It can be seen that two resonances were broadened beyond detection in the presence of the spin label, while other peaks were broadened. Based on distances estimated from the crystal structure of arrestin-1 it was straightforward to confirm that the two missing resonances correspond to F152 and F147 while the broadened peaks represent F65 and F380.
Figure 4.
Application of paramagnetic relaxation enhancement for validation of backbone NMR resonance assignments. Top: 1H,15N-TROSY spectrum of 0.15 mM selectively 15N-phenylalanine-labeled arrestin-1 (T157C, C63A, C128S, C143S, F85A, F197A mutant form with the MTSL spin label attached at C157) with reduced (diamagnetic) spin-label. Bottom: 1H,15N TROSY spectra of 0.15 mM selectively-15N-phenylalanine -labeled arrestin-1 (T157C, C63A, C128S, C143S, F85A, F197A mutant form with the MTSL spin label attached at C157) with oxidized (paramagnetic) spin-label. Peaks that disappeared (F152, F147) in the paramagnetic case are circled. Both spectra were acquired in a buffer containing 25 mM Bis-Tris, 150 mM NaCl, 5 mM mercaptoethanol, pH=6.5 at 298 K using a Bruker Avance 800MHz spectrometer.
The combination of these approaches allowed the assignment of 152 residues, i.e., ~40% of the 379 non-proline residues (Figure 1). Assigned residues are highlighted in the arrestin-1 sequence (Figure 2B) and mapped on the crystal structure (Figure 2A), where it can be seen that most structural elements of the protein are represented by at least a few assigned peaks. Assignments have been deposited to BMRB (access number17177). Most of the N-terminal residues and all of the distal C-terminus (residues 389–404) were assigned, which is notable since these elements are not observed in the available crystal structures (23–24). Residues 361–370 between the C-domain and the anchored end of the C-tail yielded very sharp peaks that were assigned using TROSY-based 3D spectra. We obtained better coverage of the N-domain (77 out of 180 residues) than of the C-domain (44 out of 180).
Binding of IP6 and Heparin to Arrestin-1
IP6 and heparin inhibit arrestin-1 binding to its physiological target, phosphorylated light-activated rhodopsin (P-Rh*) (20–21, 61), likely acting as mimics of the multi-phosphorylated rhodopsin C-terminus that occupy the site on arrestin-1 where this part of the rhodopsin normally docks. This model received strong support from the finding that at low concentrations heparin promotes arrestin-1 binding to unphosphorylated light-activated rhodopsin (20–21, 61), in a manner similar to the effect of phosphorylated C-terminal rhodopsin peptide (62). This similarity of action strongly suggests that heparin and phosphorylated rhodopsin C-terminus “activate” arestin-1 via the same mechanism, allowing its binding to unphosphorylated light-activated rhodopsin. Both heparin and IP6 appear to interact with multiple phosphate-binding residues in arrestin proteins (63–64) and induce the release of the arrestin C-tail (21, 65–66), which is known to be part of the receptor binding-induced conformational rearrangement (26, 67). IP6 also inhibits the oligomerization of arrestin-1 (68). Two independent IP6 binding sites had been identified in arrestin-2, one on the N-domain and another on the C-domain (69). Based on structure similarity (24, 33, 35) and sequence homology (5) it can be inferred that the IP6 binding site contained within the N-domain is conserved in arrestin-1. This was tested by IP6-induced NMR chemical shift changes in arrestin-1. The binding of IP6 would be expected to change the chemical shifts from residues in direct contact with the ligand and those involved in binding-induced conformational changes. Figure 5A shows a section of a TROSY spectrum of arrestin-1 upon IP6 titration, with residues showing the largest chemical shift changes labeled. Residue-specific changes in peak chemical shifts due to IP6 binding are presented in Figure 5B.
Figure 5.
Titration of arrestin-1 with IP6, as monitored by NMR. (A) Overlapping of 2D TROSY spectra of 0.15 mM 15N-labeled arrestin-1 with different concentrations of IP6. Protein:ligand molar ratios are: Black 1:0, Blue 1:0.5, Red 1:1, Purple 1:2, Magenta 1:5, Green 1:10. Samples were prepared at 25 mM Bis-Tris, 150 mM NaCl and 5 mM mercaptoethanol pH 6.5 and acquired at 308 K using a Bruker Avance 800MHz spectrometer. (B) Plot of chemical shift changes along the sequence of arrestin-1 (assigned residues) when the IP6 binding site of arrestin-1 is near saturation. Triangles represent the chemical shift changes for 0.15 mM 15N labeled arrestin-1 induced by the presence of 1.5mM IP6. The chemical shift changes are calculated as described in Materials and Methods. The magenta line indicates the 0.01 ppm experimental uncertainty associated with the chemical shift measurements. (C) Residues for which amide peaks exhibited more than 0.01 ppm chemical shift change in the present of 10-fold molar excess of IP6 were mapped onto the crystal structure of arrestin-1 (highlighted in magenta).
As can be seen in Figure 5C the residues affected by IP6 are located only in the N-domain and in the C-tail. One region that shows many relatively large changes in chemical shift in response to IP6 binding is in the receptor-binding cavity of the N-domain (70–71), which contains multiple positive charges (Figure 2). It includes the loop between beta-strands IX and X (residues 161 through 168, three of which are lysines) and beta-strand I (residues 11 through 20, which contains three lysines and one arginine). Most of the positive charges in these two areas have been previously implicated in the interactions with receptor-attached phosphates (35, 63–64, 72). A second region is the “finger loop” (residues 68–78), also previously implicated in receptor binding (26), and a third is the C-tail (residues 390–404) released upon binding to the phosphorylated receptor (21, 26, 73). In addition, several perturbations were observed in beta-strands IV and IX (residue 55, 64, 65, 172), as well as in the “polar core” (residues 174 and 299), which is thought to be critical to phosphate recognition by all arrestins (35, 74–78). C-domain residues that participate in IP6 binding in arrestin-2 (69) are not conserved in arrestin-1. Indeed, we did not detect IP6 binding site in this element. Similar titration experiments were performed for heparin, a polyanionic sulfated glycosaminoglycan. A series of TROSY spectra were acquired at different concentrations of heparin as illustrated in Figure 6A. The heparin-induced chemical shift changes are presented on a residue-specific basis in Figure 6B. The localization of residues showing significant chemical shifts in response to heparin (Figure 6C) is generally similar to those that respond to IP6 binding, although there are significant differences. Several regions that were not affected by IP6 binding responded to heparin. Among these was a short helix (residues 283 to 287) in the C-domain and several residues in the beta-strand II in the N-domain. Moreover, heparin binding but not IP6 induced significant changes in the peaks from K109 and K110 in the helix I participating in the “three-element interaction” that anchors the C-tail to the body of the N-domain (24) and that serves as a secondary phosphate sensor (79) (Figures 5C &6C). Both IP6 and heparin induce widespread chemical shift changes, which suggest that in addition to contributions to shift changes due to direct residue-ligand interactions that there are also likely some arrestin conformational changes upon ligand binding that are localized primarily to the N-domain and C-tail. Indeed, this conformational change involves the release of the C-tail as evidenced by NMR T2 relaxation results described later in this paper. This release further perturbs the finger loop and the polar core through allosteric propagation.
Figure 6.
Titration of arrestin-1 with heparin, as monitored by NMR. (A) Overlapping of 2D TROSY spectra of 0.1mM 2H,15N-labeled arrestin-1 with different concentrations of heparin. Protein:ligand molar ratios are: Green 1:0, Red 1:0.5, Yellow 1:1, Blue 1:2 and Magenta 1:5. Samples were prepared and acquired in the same conditions as in Figure 6A. (B) Plot of chemical shift changes for assigned resonances along the sequence of arrestin-1 when the heparin binding site of arrestin-1 is near saturation. Black circles illustrate the chemical shift changes for 0.1 mM 15N labeled arrestin-1 induced by the presence of 0.5mM heparin. The chemical shift changes are calculated as described in Materials and Methods. The magenta line indicates experimental uncertainty associated with the chemical shift measurements. (C) Residues for which amide peaks exhibited more than 0.01 ppm chemical shift changes in the present of 5-fold molar excess of IP6 were mapped onto the crystal structure of arrestin-1 (highlighted in magenta).
The observed changes in chemical shifts in response to increasing concentrations of IP6 or heparin are monotonous (Figure 7), indicating that free and arrestin-bound IP6 and heparin are in fast exchange on the NMR timescale. The chemical shift changes were seen to plateau upon reaching 5- and 10-fold molar excess of heparin or IP6, respectively, relative to the 0.1–0.2 mM arrestin-1. Saturability of the binding indicates specificity of recognition. Dissociation constants were determined by fitting the concentration-dependent chemical shift changes as a function of ligand concentration (Figures 7A &7B). The Kd for IP6 was 160±60 μM based on averaging residue-specific individual values. While the heparin used for the titration represented by Figures 7 and 7B was not completely homogenous but rather was a mixture of heparins of different lengths (average, ca. 20 sugar units), it is possible to calculate an apparent dissociation constant based on assuming an average molecular weight for this ligand. The apparent dissociation constant Kd,app was found to be roughly 10 μM. Higher affinity of arrestin-1 for heparin than IP6 likely reflects the larger number of residues and surface area involved in its binding site relative to IP6, as indicated by Figures 5C and 6C. It is important to note that (1,3,5)-inositol triphosphate and a heparin disaccharide with only three sulfate groups do not appear to bind arrestin-1 as judged by NMR titrations (data not shown). These findings are consistent with previous reports (61, 69) and recent studies showing that at least three receptor-attached phosphates are necessary for arrestin-1 activation (80).
Figure 7.
Determination of ligand-arrestin-1 dissociation constants from the NMR titration data. (A) Chemical shift changes of residues (A64, I16, L172, Y67) are plotted as a function of IP6 concentration, to which was fit a 1:1 binding model using NMRView titration analysis module to obtain dissociation constants. (B) Chemical shift changes of residues (A64, I16, L172, Y67) are plotted as a function of heparin concentration, with the same analysis of the data and the determination of apparent dissociation constants Kd,app being performed as in (A).
Evaluation of ligand-induced increase in the C-tail mobility by T2 measurement
While 15N NMR T1 relaxation times provide information of motions only on the pico- to nanosecond time scale, T2 relaxation times are sensitive not only to high frequency motions but also to protein conformational changes that occur on the micro- to millisecond time scale(81). Conformational changes in proteins usually involve motions on the microsecond to millisecond time scale (82). T2 is also sensitive to chemical/conformational exchange, if present. Here we used a CPMG experiment to measure the T2 for the backbone amide 15N of arrestin-1 in the absence and presence of IP6 and heparin. To enhance the ease and accuracy of T2 measurements, uniformly-2H,15N-labeled protein samples were employed to remove the relaxation contribution from dipole-dipole interactions involving remote protons.
Figure 8A shows the measured arrestin-1 backbone 15N T2 in the absence and presence of saturating heparin. For the free protein, most of the residues from the core exhibit T2 around 25–35 ms, indicative of low mobility, while both the N-terminus (K5) and the C-terminus (residues 392–404) are seen to be very flexible, with T2 in the range of 100–500 ms. Residues 362–370 are also very flexible. These observations are consistent with the crystal structure of arrestin-1, where these highly flexible elements are invisible in electron density maps (23–24). The presence of heparin does not affect the transverse relaxation of the N-terminus (K5) or 362–370 loop (Figure 8A), which remains in the range of 100–150 ms. However, heparin substantially increased transverse relaxation times in the C-terminus, with T2 for residues at the end of the protein jumping to >1 second in the presence of heparin. The increase of T2 gradually increases starting with residues K386 and A387 and extending to the extreme C-terminus. These data suggest that in the presence of heparin the C-terminal tail becomes completely disordered. A similar pattern was observed upon IP6 binding (Figure 8B). CD spectroscopy previously indicated that binding of IP6 or heparin does not change the secondary structure of arrestin-1 (83). Our T2 relaxation data are in agreement with this conclusion, but highlight a dramatic increase in C-terminus flexibility upon binding of either IP6 or heparin. This finding further supports the hypothesis (20) that the C-terminus is likely to interact with positively charged elements in the cavity of the N-domain when arrestin-1 resides in its basal state.
Figure 8.
Release of the arrestin-1 C-tail upon association of arrestin-1 heparin or IP6, as revealed by T2 relaxation rate changes. (a) Transverse relaxation times (T2) for arrestin-1 backbone amide 15N in the absence (blue bar) and in the presence of 5-fold molar excess of heparin (red bar), as measured at 308 K and 800 MHz. (b) Transverse relaxation times (T2) for arrestin-1 backbone amide 15N in the absence (blue bar) and in the presence of 10-fold molar excess of IP6 (red bar) at 308 K and 800 MHz.
Discussion
Use of NMR Spectroscopy to Characterize Arrestin Structure, Dynamics, and Interactions
There are two major challenges in the elucidation of the structural basis of arrestin function: precise identification of the binding sites for numerous arrestin-binding proteins and elucidation of non-basal arrestin conformations, particularly the structures of the important receptor- and microtubule-bound forms. Theoretically, crystal structures of protein-protein or protein-ligand complexes can yield this information. Unfortunately, only two structures of arrestin complexes have been determined so far: arrestin-2 in complex with a clathrin domain (84) and with IP6 (69). Moreover, crystallography is an imperfect tool for exploring the dynamics of the interactions, and may also be subject to structural artifacts resulting from crystal contacts/packing. For example, it was recently shown that the shape of the biologically relevant solution tetramer of arrestin-1 is very different from that found in crystal (12, 25). A second approach previously used to characterize arrestin structures and interactions is EPR spectroscopy, which can be used to map binding sites and to follow the dynamics of the interactions and conformational changes in proteins at physiological conditions in solution (85- 86). However, because each measurement requires introduction of 1–2 site-directed spin labels into engineered cysteine sites, numerous mutants must be generated and purified to comprehensively map a binding site (13, 15, 26, 73) or to elucidate the interacting surfaces of multi-protein complexes (12, 25).
Solution NMR has not previously been applied to arrestins. Here we were able to assign chemical shifts for ~40% of arrestin-1 residues localized in functionally important elements of the protein. Importantly, we obtained particularly good coverage in flexible regions that were not resolved in the crystal structure, including the C-tail, parts of the three-element interaction, hinge region, finger loop and several residues in the polar core (Figure 1B). We showed that this coverage is sufficient to map the interaction sites of two negatively charged arrestin-1 ligands, heparin and IP6. Both ligands bind to the same site, although, heparin binding involves a higher surface area and number of residues than IP6, including the involvement of a few residues from the C-domain and the hinge region between the N- and C-domains. The binding site for both ligands is clearly centered at the concave side of the N-domain, which includes a number of conserved positively charged residues. For IP6 this is expected based on the crystal structure for the complex between IP6 and the homologous arrestin-2, since all three positive charges implicated in IP6 binding to the N-domain of arrestin-2 (K157, R160, K161) are also present in arrestin-1 (K163, K166, K167, respectively). IP6 was also seen to bind via five positively charged residues (K232, R236, K250, K324, K326) in the arrestin-2 C-domain (69). We did not detect any interaction of IP6 with the C-domain of arrestin-1, likely because arrestin-1 has no IP6 binding site in this region: one residue in a homologous position is neutral hydrophobic (I256 corresponds to K250), and the other is negatively charged (E242 corresponds to K236). NMR also readily detected the conformational changes induced by these phosphoreceptor mimics, demonstrating that both displace the arrestin C-tail.
Novel Insight into Arrestin Interactions with IP6, Heparin, and Phosphorylated Rhodopsin
Arrestin-1 was originally discovered as a protein that binds active phosphorhodopsin (6). After the non-visual arrestin-2 (termed beta-arrestin at the time) was cloned and shown to bind the beta2-adrenergic receptor (87–88), GPCRs were believed to be the only targets of arrestins. However, subsequent studies found that arrestins bind dozens of other proteins, placing them at a cross-roads for multiple signaling pathways, as reviewed in (4–5). Arrestins assume at least three distinct conformations: a basal state in solution (25–26, 73) that is believed to be reflected by the crystal structure (23–24, 33–35), a receptor-bound “active” (26, 73) state, and a microtubule-bound state (13). Thus, the understanding of the molecular mechanisms of arrestin function requires identification of the binding sites of its partners and of the conformational preferences of its interaction partners (22). Other than the crystal structure of IP6-complexed arrestin-2, only the binding sites for clathrin (2, 84, 89), AP2 (3, 89), microtubules (13), Ca2+-liganded calmodulin (15), and PDE4D (90) have been mapped on arrestins with any degree of certainty. Since arrestins are not large enough to accommodate more than 5–6 partners simultaneously, certain binding sites likely overlap and the proteins that recognize them must compete (22). Indeed, the competition for arrestin binding by GPCRs with microtubules and calmodulin (15) due to overlapping binding sites has been demonstrated experimentally. Similarly, the solution tetramer of arrestin-1 in which receptor-binding elements are shielded by adjacent subunits (25) cannot bind rhodopsin (12). However, for most proteins that bind to arrestins virtually nothing is known about the locations of binding sites. The sites of protein-protein interactions can be mapped by detection of decreased residue mobility within binding interface. Site-directed spin labeling/EPR accomplishes by making one measurement at a time (25–26, 60, 67, 85–86), which requires expression and purification of dozens of single-cysteine mutants for spin labeling to define an interaction site. In NMR each residue with an assigned peak can serve as a reporter, so that extensive multi-residue binding sites can be mapped in a single experiment.
Arrestin elements directly engaged by the active phosphoreceptor (26, 64, 70–71, 91–93) invariably map to the concave sides of the two arrestin domains (Figures 5C &6C). Phosphate-binding positively charged residues, which constitute part of the receptor-binding site, are localized on the N-domain, whereas other parts of the activated receptor are believed to be engaged by the C-domain (35, 63–65, 72, 79, 94). It was also shown early on that phosphoreceptor binding induces a global conformational change in arrestin (18). The crystal structure of arrestin-1 identified key intramolecular interactions that stabilize the basal state (24). Receptor-attached phosphates disrupt two of these: the main phosphate sensor termed the polar core (an arrangement of five solvent-excluded charged residues containing Arg-Asp salt bridge that the phosphates disrupt) (74–76), and the three-element interaction between beta-strand I, alpha-helix I, and the C-tail (35, 79, 95). The disruption of these interactions allows arrestins to transition into the receptor-binding state, which appears to involve the movement of the two domains towards the receptor (1) requiring an extended inter-domain hinge (13, 65). Judging by the effects of hinge deletions, microtubule binding likely involves domain movement in the opposite direction. The disruption of the interaction anchoring the C-tail to the body of the N-domain induces its release, which was the first receptor-induced conformational rearrangement deduced on the basis of its increased susceptibility to proteolysis (21) and later directly observed using site-directed spin labeling and distance measurements (26, 73). However, despite of a wealth of deductions and a few observations, the actual conformation of receptor- or microtubule-bound arrestin remains to be elucidated.
The C-tail is displaced upon binding to phosphorhodopsin (26), increasing the mobility of its residues. Our data directly demonstrate for the first time that phosphoreceptor mimics heparin and IP6 also fully displace the C-tail, significantly increasing the mobility of residues 387–404, with a clear gradient of the change, which increases from proximal to distal residues (Figure 4). In addition, the observed mobility gradient strongly suggests that in the active state of arrestin the C-tail does not have a fixed alternative conformation but is fully released and disordered. The flexible finger loop (residues 68–78) has distinct conformations in different monomers within crystal tetramer (24) and was implicated in the binding of rhodopsin, microtubules, and calmodulin (13, 15, 26), making the residues assigned in this region useful as reporters for the interactions with these partners. Our data demonstrate the finger loop’s involvement in the binding of negatively-charged phosphoreceptor mimics, heparin and IP6 (Figures 5C and 6C). The polar core and the three element interaction are believed to undergo significant conformational rearrangements in the process of the arrestin-1 transition into the active state. The availability of partial backbone assignment in these areas paves the way for further NMR studies to elucidate the detailed conformational and dynamic properties of these regions within the active form of arrestin-1. This is particularly important considering the absence of the crystal structure of any arrestin-receptor complex.
Even with partial resonance assignments solution NMR yields a wealth of information on the localization of ligand-binding sites on arrestin-1 and allows simultaneous detection of binding-induced conformational changes in different areas of the molecule. This approach is particularly attractive for multi-functional arrestin proteins that bind a variety of partners and exist in multiple conformations. Systematic use of solution NMR holds promise for mapping of arrestin binding sites for its partners and elucidation of the effects of these interactions on arrestin conformation.
Abbreviations
- CPMG
Carr-Purcell-Meiboom-Gill
- DTT
dithiothreitol
- EDTA
ethylenediaminetetraacetic acid
- EPR
electron paramagnetic resonance
- GPCR
G protein-coupled receptor
- IP6
inositol hexaphosphate
- IPTG
isopropylthiogalactoside
- LB
Luria broth
- MTSL
S-(2,2,5,5-tetramethyl-2,5-dihydro-1H-pyrrol-3-yl)methyl methanesulfonothioate
- NMR
nuclear magnetic resonance
- P-Rh*
phosphorylated light-activated rhodopsin
- PRE
paramagnetic relaxation enhancement
- T2
transverse NMR relaxation time
- TROSY
transverse relaxation-optimized NMR spectroscopy
Footnotes
This study was supported by NIH grants GM081756 (ARRA supplement), EY011500, and GM077561 to VVG, and by PO1 GM08513 and Roadmap RO1 GM081816 to CRS.
We use systematic names of arrestin proteins: arrestin-1 (historic names S-antigen, 48 kDa protein, visual or rod arrestin), arrestin-2 (β-arrestin or β-arrestin1), arrestin-3 (β-arrestin2), and arrestin-4 (cone or X-arrestin; for unclear reasons its gene is called “arrestin 3” in HUGO database).
References
- 1.Gurevich VV, Gurevich EV. The molecular acrobatics of arrestin activation. Trends Pharmacol Sci. 2004;25:59–112. doi: 10.1016/j.tips.2003.12.008. [DOI] [PubMed] [Google Scholar]
- 2.Goodman OB, Krupnick JG, Santini F, Gurevich VV, Penn RB, Gagnon AW, Keen JH, Benovic JL. Beta-arrestin acts as a clathrin adaptor in endocytosis of the beta2-adrenergic receptor. Nature. 1996;383:447–450. doi: 10.1038/383447a0. [DOI] [PubMed] [Google Scholar]
- 3.Laporte SA, Oakley RH, Holt JA, Barak LS, Caron MG. The interaction of beta-arrestin with the AP-2 adaptor is required for the clustering of beta 2-adrenergic receptor into clathrin-coated pits. J Biol Chem. 2000;275:23120–23126. doi: 10.1074/jbc.M002581200. [DOI] [PubMed] [Google Scholar]
- 4.DeWire SM, Ahn S, Lefkowitz RJ, Shenoy SK. beta-arrestins and cell signaling. Annu Rev Physiol. 2007;69:483–510. doi: 10.1146/annurev.physiol.69.022405.154749. [DOI] [PubMed] [Google Scholar]
- 5.Gurevich EV, Gurevich VV. Arrestins: ubiquitous regulators of cellular signaling pathways. Genome Biol. 2006;7 doi: 10.1186/gb-2006-7-9-236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kuhn H, Hall SW, Wilden U. Light-induced binding of 48-kDa protein to photoreceptor membranes is highly enhanced by phosphorylation of rhodopsin. FEBS Lett. 1984;176:473–478. doi: 10.1016/0014-5793(84)81221-1. [DOI] [PubMed] [Google Scholar]
- 7.Strissel KJ, Sokolov M, Trieu LH, Arshavsky VY. Arrestin translocation is induced at a critical threshold of visual signaling and is superstoichiometric to bleached rhodopsin. J Neurosci. 2006;26:1146–1153. doi: 10.1523/JNEUROSCI.4289-05.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song X, Gurevich VV. Each rhodopsin molecule binds its own arrestin. Proc Nat Acad Sci USA. 2007 doi: 10.1073/pnas.0610886104. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nikonov SS, Brown BM, Davis JA, Zuniga FI, Bragin A, Pugh ENJ, Craft CM. Mouse cones require an arrestin for normal inactivation of phototransduction. Neuron. 2008;59:462–474. doi: 10.1016/j.neuron.2008.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schubert C, Hirsch JA, Gurevich VV, Engelman DM, Sigler PB, Fleming KG. Visual arrestin activity may be regulated by self-association. J Biol Chem. 1999;274:21186–21190. doi: 10.1074/jbc.274.30.21186. [DOI] [PubMed] [Google Scholar]
- 11.Imamoto Y, Tamura C, Kamikubo H, Kataoka M. Concentration-dependent tetramerization of bovine visual arrestin. Biophys J. 2003;85:1186–1195. doi: 10.1016/S0006-3495(03)74554-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hanson SM, Gurevich EV, Vishnivetskiy SA, Ahmed MR, Song XF, Gurevich VV. Each rhodopsin molecule binds its own arrestin. P Natl Acad Sci USA. 2007;104:3125–3128. doi: 10.1073/pnas.0610886104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hanson SM, Cleghorn WM, Francis DJ, Vishnivetskiy SA, Raman D, Song S, Nair KS, Slepak VZ, Klug CS, Gurevich VV. Arrestin mobilizes signaling proteins to the cytoskeleton and redirects their activity. J Mol Biol. 2007;368:375–387. doi: 10.1016/j.jmb.2007.02.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nair KS, Hanson SM, Kennedy MJ, Hurley JB, Gurevich VV, Slepak VZ. Direct binding of visual arrestin to microtubules determines the differential subcellular localization of its splice variants in rod photoreceptors. J Biol Chem. 2004;279:41240–41248. doi: 10.1074/jbc.M406768200. [DOI] [PubMed] [Google Scholar]
- 15.Wu N, Hanson SM, Francis DJ, Vishnivetskiy SA, Thibonnier M, Klug CS, Shoham M, Gurevich VV. Arrestin binding to calmodulin: a direct interaction between two ubiquitous signaling proteins. J Mol Biol. 2006;364:955–963. doi: 10.1016/j.jmb.2006.09.075. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Song X, Raman D, Gurevich EV, Vishnivetskiy SA, Gurevich VV. Visual and both non-visual arrestins in their “inactive” conformation bind JNK3 and Mdm2 and relocalize them from the nucleus to the cytoplasm. J Biol Chem. 2006;281:21491–21499. doi: 10.1074/jbc.M603659200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang SP, Brown BM, Craft CM. Visual Arrestin 1 acts as a modulator for N-ethylmaleimide-sensitive factor in the photoreceptor synapse. J Neurosci. 2010;30:9381–9391. doi: 10.1523/JNEUROSCI.1207-10.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Schleicher A, Kuhn H, Hofmann KP. Kinetics, binding constant, and activation energy of the 48-kDa protein-rhodopsin complex by extra-metarhodopsin II. Biochemistry. 1989;28:1770–1775. doi: 10.1021/bi00430a052. [DOI] [PubMed] [Google Scholar]
- 19.Gurevich VV, Benovic JL. Visual arrestin interaction with rhodopsin: Sequential multisite binding ensures strict selectivity towards light-activated phosphorylated rhodopsin. J Biol Chem. 1993;268:11628–11638. [PubMed] [Google Scholar]
- 20.Gurevich VV, Chen CY, Kim CM, Benovic JL. Visual arrestin binding to rhodopsin: Intramolecular interaction between the basic N-terminus and acidic C-terminus of arrestin may regulate binding selectivity. J Biol Chem. 1994;269:8721–8727. [PubMed] [Google Scholar]
- 21.Palczewski K, Pulvermuller A, Buczylko J, Hofmann KP. Phosphorylated rhodopsin and heparin induce similar conformational changes in arrestin. J Biol Chem. 1991;266:18649–18654. [PubMed] [Google Scholar]
- 22.Gurevich VV, Gurevich EV. The structural basis of arrestin-mediated regulation of G-protein-coupled receptors. Pharmacol Therapeut. 2006;110:465–502. doi: 10.1016/j.pharmthera.2005.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Granzin J, Wilden U, Choe HW, Labahn J, Krafft B, Buldt G. X-ray crystal structure of arrestin from bovine rod outer segments. Nature. 1998;391:918–921. doi: 10.1038/36147. [DOI] [PubMed] [Google Scholar]
- 24.Hirsch JA, Schubert C, Gurevich VV, Sigler PB. The 2.8 angstrom crystal structure of visual arrestin: A model for arrestin’s regulation. Cell. 1999;97:257–269. doi: 10.1016/s0092-8674(00)80735-7. [DOI] [PubMed] [Google Scholar]
- 25.Hanson SM, Dawson ES, Francis DJ, Van Eps N, Klug CS, Hubbell WL, Meiler J, Gurevich VV. A model for the solution structure of the rod arrestin tetramer. Structure. 2008;16:924–934. doi: 10.1016/j.str.2008.03.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hanson SM, Francis DJ, Vishnivetskiy SA, Kolobova EA, Hubbell WL, Klug CS, Gurevich VV. Differential interaction of spin-labeled arrestin with inactive and active phosphorhodopsin. P Natl Acad Sci USA. 2006;103:4900–4905. doi: 10.1073/pnas.0600733103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lemaster DM, Richards FM. NMR Sequential Assignment of Escherichia-Coli Thioredoxin Utilizing Random Fractional Deuteriation. Biochemistry. 1988;27:142–150. doi: 10.1021/bi00401a022. [DOI] [PubMed] [Google Scholar]
- 28.Gardner KH, Kay LE. The use of H-2, C-13, N-15 multidimensional NMR to study the structure and dynamics of proteins. Annu Rev Bioph Biom. 1998;27:357–406. doi: 10.1146/annurev.biophys.27.1.357. [DOI] [PubMed] [Google Scholar]
- 29.Tugarinov V, Hwang PM, Kay LE. Nuclear magnetic resonance spectroscopy of high-molecular-weight proteins. Annu Rev Biochem. 2004;73:107–146. doi: 10.1146/annurev.biochem.73.011303.074004. [DOI] [PubMed] [Google Scholar]
- 30.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. P Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Fiaux J, Bertelsen EB, Horwich AL, Wuthrich K. NMR analysis of a 900K GroEL-GroES complex. Nature. 2002;418:207–211. doi: 10.1038/nature00860. [DOI] [PubMed] [Google Scholar]
- 32.Religa TL, Sprangers R, Kay LE. Dynamic Regulation of Archaeal Proteasome Gate Opening As Studied by TROSY NMR. Science. 2010;328:98–102. doi: 10.1126/science.1184991. [DOI] [PubMed] [Google Scholar]
- 33.Han M, Gurevich VV, Vishnivetskiy SA, Sigler PB, Schubert C. Crystal structure of beta-arrestin at 1.9 A: possible mechanism of receptor binding and membrane translocation. Structure. 2001;9:869–880. doi: 10.1016/s0969-2126(01)00644-x. [DOI] [PubMed] [Google Scholar]
- 34.Milano SK, Pace HC, Kim YM, Brenner C, Benovic JL. Scaffolding functions of arrestin-2 revealed by crystal structure and mutagenesis. Biochemistry. 2002;41:3321–3328. doi: 10.1021/bi015905j. [DOI] [PubMed] [Google Scholar]
- 35.Sutton RB, Vishnivetskiy SA, Robert J, Hanson SM, Raman D, Knox BE, Kono M, Navarro J, Gurevich VV. Crystal Structure of Cone Arrestin at 2.3Å: Evolution of Receptor Specificity. J Mol Biol. 2005;354:1069–1080. doi: 10.1016/j.jmb.2005.10.023. [DOI] [PubMed] [Google Scholar]
- 36.Gurevich VV, Gurevich EV. Custom-designed proteins as novel therapeutic tools? The case of arrestins. Expert Rev Mol Med. 2010;12 doi: 10.1017/S1462399410001444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gurevich VV, Benovic JL. Arrestin: mutagenesis, expression, purification, and functional characterization. Methods Enzymol. 2000;315:422–437. doi: 10.1016/s0076-6879(00)15859-8. [DOI] [PubMed] [Google Scholar]
- 38.Waugh DS. Genetic tools for selective labeling of proteins with alpha-15N-amino acids. J Biomol NMR. 1996;8:184–192. doi: 10.1007/BF00211164. [DOI] [PubMed] [Google Scholar]
- 39.Delcampillo-Campbell A, Campbell A. Molybdenum Cofactor Requirement for Biotin Sulfoxide Reduction in Escherichia-Coli. J Bacteriol. 1982;149:469–478. doi: 10.1128/jb.149.2.469-478.1982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Marley J, Lu M, Bracken C. A method for efficient isotopic labeling of recombinant proteins. Journal of Biomolecular NMR. 2001;20:71–75. doi: 10.1023/a:1011254402785. [DOI] [PubMed] [Google Scholar]
- 41.Weigelt J. Single scan, sensitivity- and gradient-enhanced TROSY for multidimensional NMR experiments. Journal of the American Chemical Society. 1998;120:10778–10779. [Google Scholar]
- 42.Zhu G, Kong XM, Sze KH. Gradient and sensitivity enhancement of 2D TROSY with water flip-back, 3D NOESY-TROSY and TOCSY-TROSY experiments. Journal of Biomolecular Nmr. 1999;13:77–81. doi: 10.1023/A:1008398227519. [DOI] [PubMed] [Google Scholar]
- 43.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. TROSY in triple-resonance experiments: New perspectives for sequential NMR assignment of large proteins. P Natl Acad Sci USA. 1998;95:13585–13590. doi: 10.1073/pnas.95.23.13585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Salzmann M, Wider G, Pervushin K, Senn H, Wuthrich K. TROSY-type triple-resonance experiments for sequential NMR assignments of large proteins. Journal of the American Chemical Society. 1999;121:844–848. [Google Scholar]
- 45.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. Nmrpipe - a Multidimensional Spectral Processing System Based on Unix Pipes. Journal of Biomolecular Nmr. 1995;6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 46.Goddard TD, Kneller DG. SPARKY 3. University of California; San Francisco: [Google Scholar]
- 47.Johnson BA, Blevins RA. Nmr View - a Computer-Program for the Visualization and Analysis of Nmr Data. Journal of Biomolecular Nmr. 1994;4:603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 48.Shen Y, Bax A. Protein backbone chemical shifts predicted from searching a database for torsion angle and sequence homology. Journal of Biomolecular Nmr. 2007;38:289–302. doi: 10.1007/s10858-007-9166-6. [DOI] [PubMed] [Google Scholar]
- 49.Krugh TR. In: Spin Labeling: Theory and Applications. Berliner LJ, editor. Academic Press; New York: 1976. [Google Scholar]
- 50.Battiste JL, Wagner G. Utilization of site-directed spin labeling and high-resolution heteronuclear nuclear magnetic resonance for global fold determination of large proteins with limited nuclear overhauser effect data. Biochemistry. 2000;39:5355–5365. doi: 10.1021/bi000060h. [DOI] [PubMed] [Google Scholar]
- 51.Carr HY, Purcell EM. Effects of diffusion on free precession in Nuclear Magnetic Resonance experiments. Physical Review. 1954;94 [Google Scholar]
- 52.Meiboom S, Gill D. Compensation for pulse imperfections in Carr-Purcell NMR experiments. Review of Scientific Instruments. 1958;29 [Google Scholar]
- 53.Pervushin K, Riek R, Wider G, Wuthrich K. Attenuated T-2 relaxation by mutual cancellation of dipole-dipole coupling and chemical shift anisotropy indicates an avenue to NMR structures of very large biological macromolecules in solution. P Natl Acad Sci USA. 1997;94:12366–12371. doi: 10.1073/pnas.94.23.12366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Mulder FAA, Ayed A, Yang DW, Arrowsmith CH, Kay LE. Assignment of H-1(N), N-15, C-13(alpha), (CO)-C-13 and C-13(beta) resonances in a 67 kDa p53 dimer using 4D-TROSY NMR spectroscopy. J Biomol Nmr. 2000;18:173–176. doi: 10.1023/a:1008317825976. [DOI] [PubMed] [Google Scholar]
- 55.Salzmann M, Pervushin K, Wider G, Senn H, Wuthrich K. NMR assignment and secondary structure determination of an octameric 110 kDa protein using TROSY in triple resonance experiments. J Am Chem Soc. 2000;122:7543–7548. [Google Scholar]
- 56.Tugarinov V, Muhandiram R, Ayed A, Kay LE. Four-dimensional NMR spectroscopy of a 723-residue protein: Chemical shift assignments and secondary structure of malate synthase G. J Am Chem Soc. 2002;124:10025–10035. doi: 10.1021/ja0205636. [DOI] [PubMed] [Google Scholar]
- 57.McElroy C, Manfredo A, Wendt A, Gollnick P, Foster M. TROSY-NMR studies of the 91 kDa TRAP protein reveal allosteric control of a gene regulatory protein by ligand-altered flexibility. Journal of Molecular Biology. 2002;323:463–473. doi: 10.1016/s0022-2836(02)00940-3. [DOI] [PubMed] [Google Scholar]
- 58.Langer T, Vogtherr M, Elshorst B, Betz M, Schieborr U, Saxena K, Schwalbe H. NMR backbone assignment of a protein kinase catalytic domain by a combination of several approaches: Application to the catalytic subunit of cAMP-dependent protein kinase. Chembiochem. 2004;5:1508–1516. doi: 10.1002/cbic.200400129. [DOI] [PubMed] [Google Scholar]
- 59.Schmitz C, John M, Park AY, Dixon NE, Otting G, Pintacuda G, Huber T. Efficient chi-tensor determination and NH assignment of paramagnetic proteins. Journal of Biomolecular Nmr. 2006;35:79–87. doi: 10.1007/s10858-006-9002-4. [DOI] [PubMed] [Google Scholar]
- 60.Hanson SM, Van Eps N, Francis DJ, Altenbach C, Vishnivetskiy SA, Arshavsky VY, Klug CS, Hubbell WL, Gurevich VV. Structure and function of the visual arrestin oligomer. Embo J. 2007;26:1726–1736. doi: 10.1038/sj.emboj.7601614. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Palczewski K, Pulvermüller A, Buczylko J, Gutmann C, Hofmann KP. Binding of inositol phosphates to arrestin. FEBS Lett. 1991;295:195–199. doi: 10.1016/0014-5793(91)81416-6. [DOI] [PubMed] [Google Scholar]
- 62.Puig J, Arendt A, Tomson FL, Abdulaeva G, Miller R, Hargrave PA, Mcdowell JH. Synthetic Phosphopeptide from Rhodopsin Sequence Induces Retinal Arrestin Binding to Photoactivated Unphosphorylated Rhodopsin. Febs Lett. 1995;362:185–188. doi: 10.1016/0014-5793(95)00225-x. [DOI] [PubMed] [Google Scholar]
- 63.Gurevich VV, Benovic JL. Visual arrestin binding to rhodopsin: diverse functional roles of positively charged residues within the phosphorylation-recignition region of arrestin. J Biol Chem. 1995;270:6010–6016. doi: 10.1074/jbc.270.11.6010. [DOI] [PubMed] [Google Scholar]
- 64.Hanson SM, Gurevich VV. The differential engagement of arrestin surface charges by the various functional forms of the receptor. J Biol Chem. 2006;281:3458–3462. doi: 10.1074/jbc.M512148200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Vishnivetskiy SA, Hirsch JA, Velez MG, Gurevich YV, Gurevich VV. Transition of arrestin into the active receptor-binding state requires an extended interdomain hinge. J Biol Chem. 2002;277:43961–43967. doi: 10.1074/jbc.M206951200. [DOI] [PubMed] [Google Scholar]
- 66.Xiao K, Shenoy SK, Nobles K, Lefkowitz RJ. Activation-dependent conformational changes in {beta}-arrestin 2. J Biol Chem. 2004;279:55744–55753. doi: 10.1074/jbc.M409785200. [DOI] [PubMed] [Google Scholar]
- 67.Vishnivetskiy SA, Francis D, Van Eps N, Kim M, Hanson SM, Klug CS, Hubbell WL, Gurevich VV. The Role of Arrestin alpha-Helix I in Receptor Binding. Journal of Molecular Biology. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Hanson SM, Vishnivetskiy SA, Hubbell WL, Gurevich VV. Opposing effects of inositol hexakisphosphate on rod arrestin and arrestin2 self-association. Biochemistry. 2008;47:1070–1075. doi: 10.1021/bi7021359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Milano SK, Kim YM, Stefano FP, Benovic JL, Brenner C. Nonvisual arrestin oligomerization and cellular localization are regulated by inositol hexakisphosphate binding. Journal of Biological Chemistry. 2006;281:9812–9823. doi: 10.1074/jbc.M512703200. [DOI] [PubMed] [Google Scholar]
- 70.Gurevich VV, Dion SB, Onorato JJ, Ptasienski J, Kim CM, Sterne-Marr R, Hosey MM, Benovic JL. Arrestin interaction with G protein-coupled receptors. Direct binding studies of wild type and mutant arrestins with rhodopsin, b2-adrenergic, and m2 muscarinic cholinergic receptors. J Biol Chem. 1995;270:720–731. doi: 10.1074/jbc.270.2.720. [DOI] [PubMed] [Google Scholar]
- 71.Vishnivetskiy SA, Hosey MM, Benovic JL, Gurevich VV. Mapping the arrestin-receptor interface: structural elements responsible for receptor specificity of arrestin proteins. J Biol Chem. 2004;279:1262–1268. doi: 10.1074/jbc.M308834200. [DOI] [PubMed] [Google Scholar]
- 72.Gurevich VV, Benovic JL. Mechanism of phosphorylation-recognition by visual arrestin and the transition of arrestin into a high affinity binding state. Mol Pharmacol. 1997;51:161–169. doi: 10.1124/mol.51.1.161. [DOI] [PubMed] [Google Scholar]
- 73.Vishnivetskiy SA, Francis DJ, Van Eps N, Kim M, Hanson SM, Klug CS, Hubbell WL, Gurevich VV. The role of arrestin alpha-helix I in receptor binding. J Mol Biol. 2010;395:42–54. doi: 10.1016/j.jmb.2009.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Vishnivetskiy SA, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. How does arrestin respond to the phosphorylated state of rhodopsin? J Biol Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 75.Celver J, Vishnivetskiy SA, Chavkin C, Gurevich VV. Conservation of the phosphate-sensitive elements in the arrestin family of proteins. J Biol Chem. 2002;277:9043–9048. doi: 10.1074/jbc.M107400200. [DOI] [PubMed] [Google Scholar]
- 76.Kovoor A, Celver J, Abdryashitov RI, Chavkin C, Gurevich VV. Targeted construction of phosphorylation-independent b-arrestin mutants with constitutive activity in cells. J Biol Chem. 1999;274:6831–6834. doi: 10.1074/jbc.274.11.6831. [DOI] [PubMed] [Google Scholar]
- 77.Song X, Vishnivetskiy SA, Gross OP, Emelianoff K, Mendez A, Chen J, Gurevich EV, Burns ME, Gurevich VV. Enhanced Arrestin Facilitates Recovery and Protects Rod Photoreceptors Deficient in Rhodopsin Phosphorylation. Curr Biol. 2009;19:700–705. doi: 10.1016/j.cub.2009.02.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Gray-Keller MP, Detwiler PB, Benovic JL, Gurevich VV. Arrestin with a single amino acid sustitution quenches light-activated rhodopsin in a phosphorylation0independent fasion. Biochemistry. 1997;36:7058–7063. doi: 10.1021/bi963110k. [DOI] [PubMed] [Google Scholar]
- 79.Vishnivetskiy SA, Schubert C, Climaco GC, Gurevich YV, Velez MG, Gurevich VV. An additional phosphate-binding element in arrestin molecule. Implications for the mechanism of arrestin activation. J Biol Chem. 2000;275:41049–41057. doi: 10.1074/jbc.M007159200. [DOI] [PubMed] [Google Scholar]
- 80.Vishnivetskiy SA, Raman D, Wei J, Kennedy MJ, Hurley JB, Gurevich VV. Regulation of arrestin binding by rhodopsin phosphorylation level. J Biol Chem. 2007;282:32075–32083. doi: 10.1074/jbc.M706057200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Kay LE, Torchia DA, Bax A. Backbone Dynamics of Proteins as Studied by N-15 Inverse Detected Heteronuclear Nmr-Spectroscopy - Application to Staphylococcal Nuclease. Biochemistry. 1989;28:8972–8979. doi: 10.1021/bi00449a003. [DOI] [PubMed] [Google Scholar]
- 82.Akke M. NMR methods for characterizing microsecond to millisecond dynamics in recognition and catalysis. Curr Opin Struc Biol. 2002;12:642–647. doi: 10.1016/s0959-440x(02)00369-x. [DOI] [PubMed] [Google Scholar]
- 83.Wilson CJ, Copeland RA. Spectroscopic characterization of arrestin interactions with competitive ligands: study of heparin and phytic acid binding. J Protein Chem. 1997;16:755–763. doi: 10.1023/a:1026311832660. [DOI] [PubMed] [Google Scholar]
- 84.Kang DS, Kern RC, Puthenveedu MA, von Zastrow M, Williams JC, Benovic JL. Structure of an arrestin2/clathrin complex reveals a novel clathrin binding domain that modulates receptor trafficking. J Biol Chem. 2009;284:29860–29872. doi: 10.1074/jbc.M109.023366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altenbach C, Froncisz W, Hemker R, Mchaourab H, Hubbell WL. Accessibility of nitroxide side chains: Absolute Heisenberg exchange rates from power saturation EPR. Biophysical Journal. 2005;89:2103–2112. doi: 10.1529/biophysj.105.059063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Altenbach C, Oh KJ, Trabanino RJ, Hideg K, Hubbell WL. Estimation of inter-residue distances in spin labeled proteins at physiological temperatures: Experimental strategies and practical limitations. Biochemistry. 2001;40:15471–15482. doi: 10.1021/bi011544w. [DOI] [PubMed] [Google Scholar]
- 87.Lohse MJ, Benovic JL, Codina J, Caron MG, Lefkowitz RJ. beta-Arrestin: a protein that regulates beta-adrenergic receptor function. Science. 1990;248:1547–1550. doi: 10.1126/science.2163110. [DOI] [PubMed] [Google Scholar]
- 88.Lohse MJ, Andexinger S, Pitcher J, Trukawinski S, Codina J, Faure JP, Caron MG, Lefkowitz RJ. Receptor-specific desensitization with purified proteins. Kinase dependence and receptor specificity of beta-arrestin and arrestin in the beta 2-adrenergic receptor and rhodopsin systems. J Biol Chem. 1992;267:8558–8564. [PubMed] [Google Scholar]
- 89.Kim YM, Benovic JL. Differential roles of arrestin-2 interaction with clathrin and adaptor protein 2 in G protein-coupled receptor trafficking. J Biol Chem. 2002;277:30760–30768. doi: 10.1074/jbc.M204528200. [DOI] [PubMed] [Google Scholar]
- 90.Baillie GS, Adams DR, Bhari N, Houslay TM, Vadrevu S, Meng D, Li X, Dunlop A, Milligan G, Bolger GB, Klussmann E, Houslay MD. Mapping binding sites for the PDE4D5 cAMP-specific phosphodiesterase to the N- and C-domains of beta-arrestin using spot-immobilized peptide arrays. Biochem J. 2007;404:71–80. doi: 10.1042/BJ20070005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Ohguro H, Palczewski K, Walsh KA, Johnson RS. Topographic study of arrestin using differential chemical modifications and hydrogen/deuterium exchange. Protein Sci. 1994:2428–2434. doi: 10.1002/pro.5560031226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Pulvermuller A, Schroder K, Fischer T, Hofmann KP. Interactions of metarhodopsin II. Arrestin peptides compete with arrestin and transducin. J Biol Chem. 2000;275:37679–37685. doi: 10.1074/jbc.M006776200. [DOI] [PubMed] [Google Scholar]
- 93.Dinculescu A, McDowell JH, Amici SA, Dugger DR, Richards N, Hargrave PA, Smith WC. Insertional mutagenesis and immunochemical analysis of visual arrestin interaction with rhodopsin. J Biol Chem. 2002;277:11703–11708. doi: 10.1074/jbc.M111833200. [DOI] [PubMed] [Google Scholar]
- 94.Vishnivetskiy SK, Paz CL, Schubert C, Hirsch JA, Sigler PB, Gurevich VV. How does arrestin respond to the phosphorylated state of rhodopsin? J Biol Chem. 1999;274:11451–11454. doi: 10.1074/jbc.274.17.11451. [DOI] [PubMed] [Google Scholar]
- 95.Gurevich VV. The selectivity of visual arrestin for light-activated phosphorhodopsin is controlled by multiple nonredundant mechanisms. J Biol Chem. 1998;273:15501–15506. doi: 10.1074/jbc.273.25.15501. [DOI] [PubMed] [Google Scholar]