Abstract
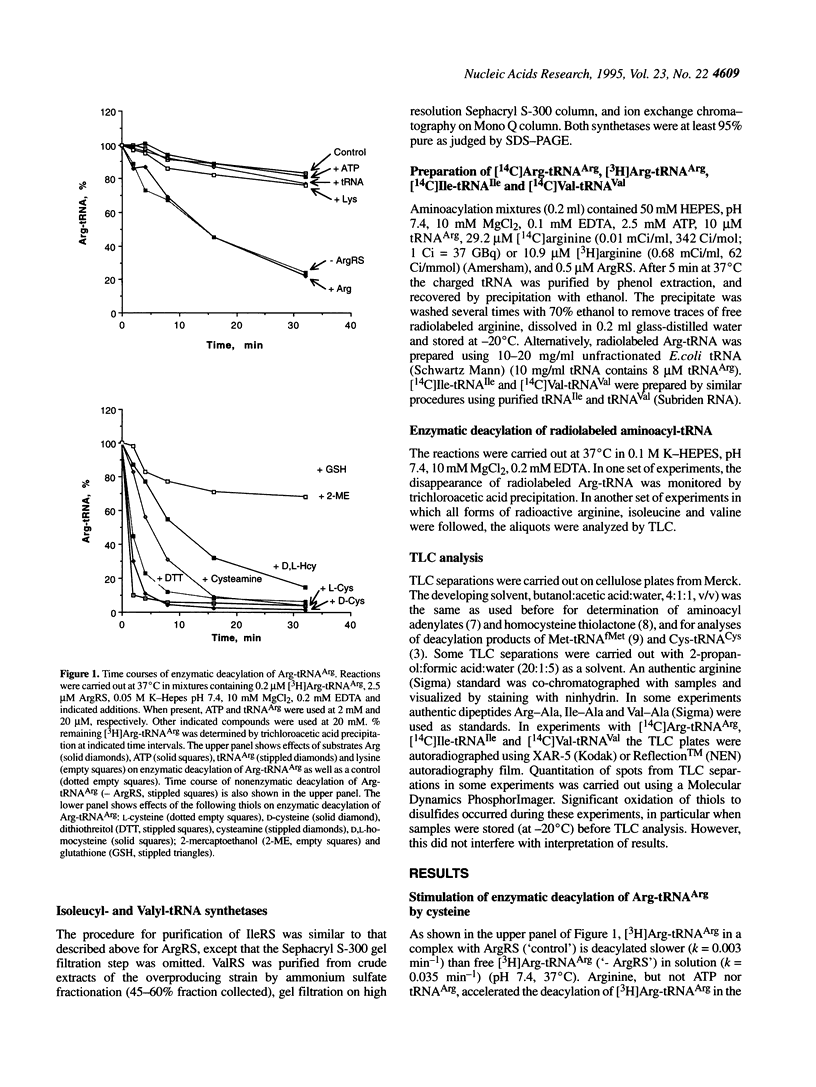
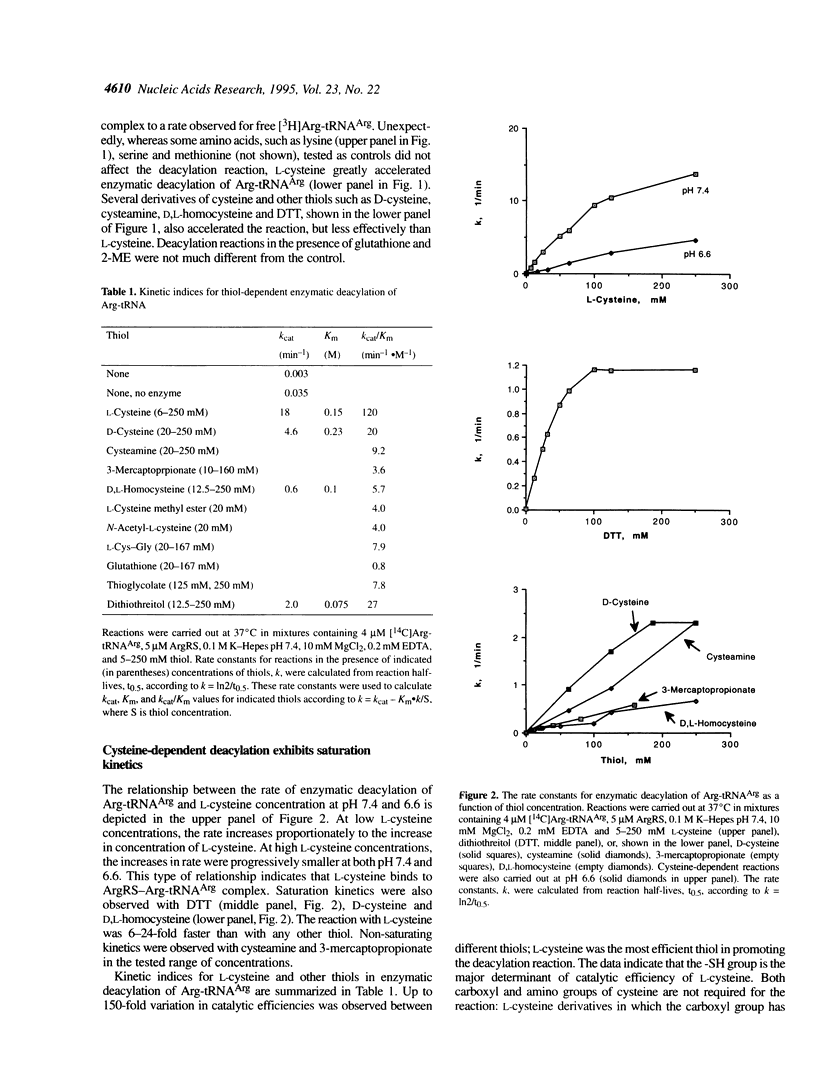
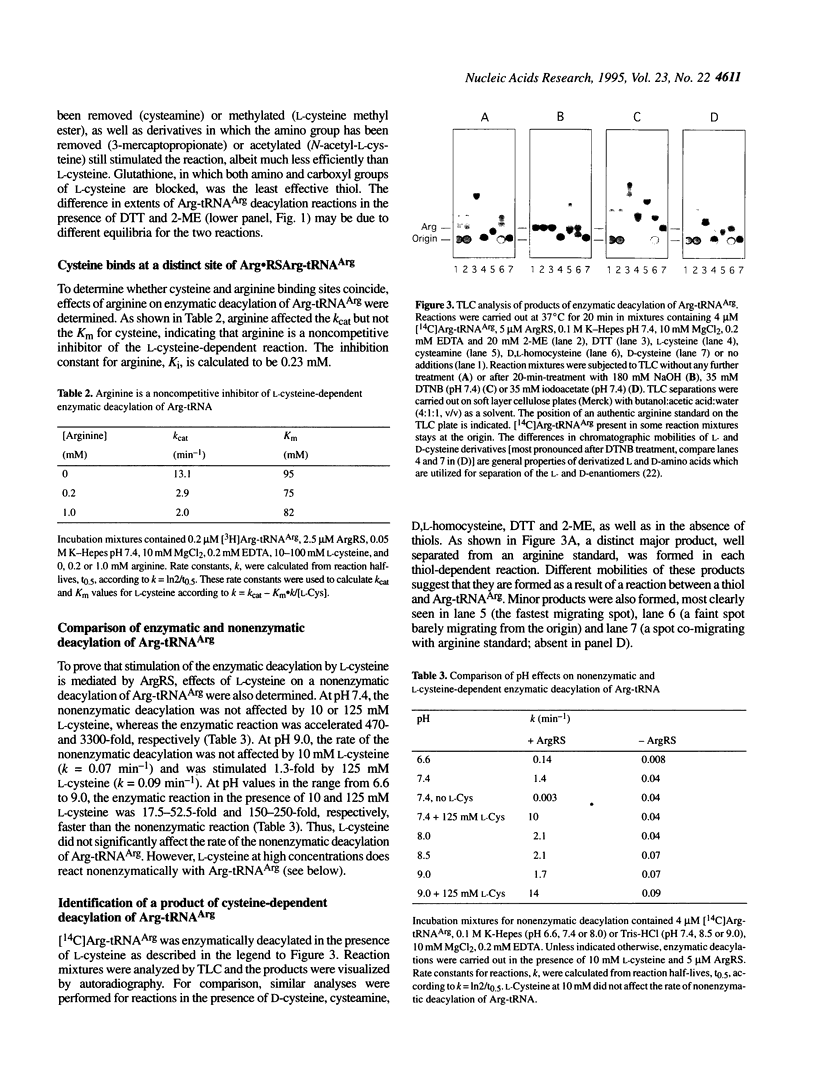
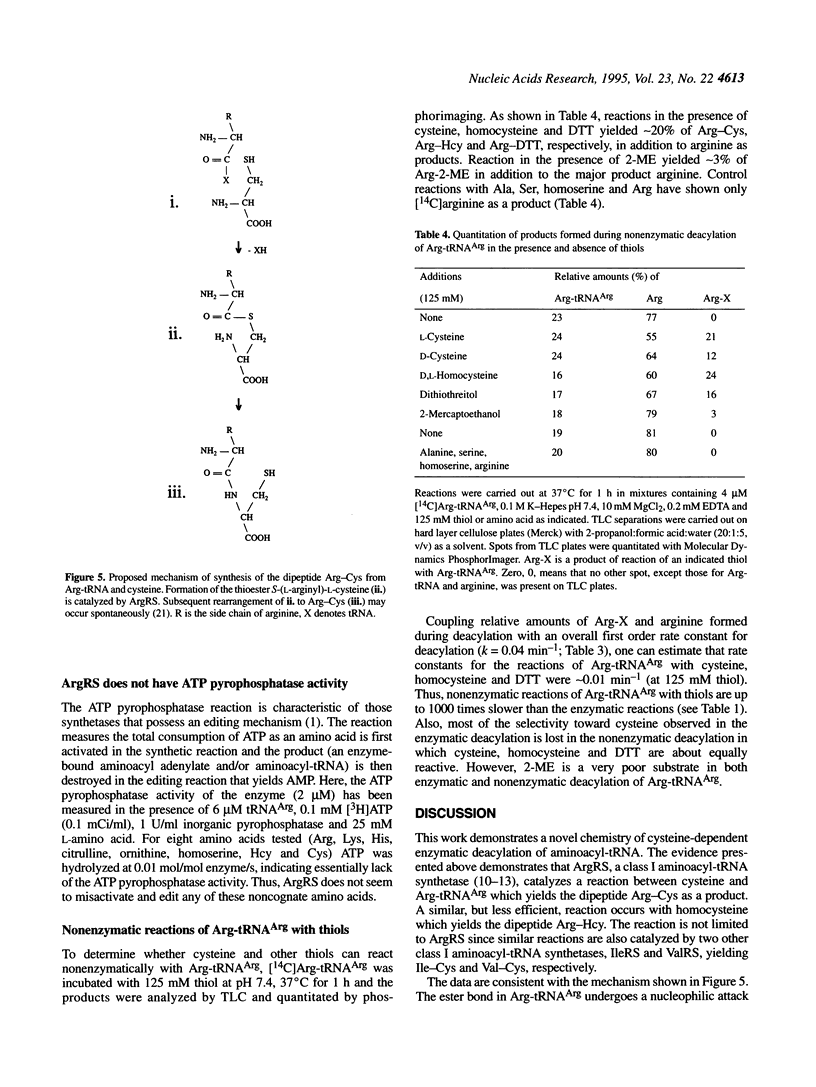
Arginyl-tRNA synthetase (ArgRS) catalyses AMP- and PPi-independent deacylation of Arg-tRNAArg in the presence of cysteine. A dipeptide, Arg-Cys, is a product of this deacylation reaction. Similar reaction with homocysteine yields Arg-Hcy. Arginine is a noncompetitive inhibitor of the cysteine-dependent deacylation which indicates that cysteine binds to the enzyme-Arg-tRNAArg complex at a site separate from the arginine binding site. In the presence of arginine, [14C]Arg-tRNAArg is deacylated at a rate similar to the rate of its spontaneous deacylation in solution and [14C]arginine is a product. Experiments with cysteine derivatives indicate that the -SH group is essential for the reaction whereas -NH2 and -COOH groups are not. Thioesters of arginine are formed with 3-mercaptopropionic acid, N-acetyl-L-cysteine and dithiothreitol. These data suggest that formation of the dipeptide Arg-Cys involves a thioester intermediate, S-(L-arginyl)-L-cysteine, which is not observed because of the rapid rearrangement to form a stable peptide bond. Facile intramolecular reaction results from the favorable geometric arrangement of the alpha-amino group of cysteine with respect to the thioester formed in the initial reaction. Similar reactions, yielding Ile-Cys and Val-Cys, are catalyzed by isoleucyl- and valyl-tRNA synthetases, respectively.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Brevet A., Chen J., Lévêque F., Plateau P., Blanquet S. In vivo synthesis of adenylylated bis(5'-nucleosidyl) tetraphosphates (Ap4N) by Escherichia coli aminoacyl-tRNA synthetases. Proc Natl Acad Sci U S A. 1989 Nov;86(21):8275–8279. doi: 10.1073/pnas.86.21.8275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buck R. H., Krummen K. High-performance liquid chromatographic determination of enantiomeric amino acids and amino alcohols after derivatization with o-phthaldialdehyde and various chiral mercaptans. Application to peptide hydrolysates. J Chromatogr. 1987 Jan 30;387:255–265. doi: 10.1016/s0021-9673(01)94529-7. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Schimmel P. Structural relationships and the classification of aminoacyl-tRNA synthetases. J Biol Chem. 1991 Sep 15;266(26):16965–16968. [PubMed] [Google Scholar]
- Dawson P. E., Muir T. W., Clark-Lewis I., Kent S. B. Synthesis of proteins by native chemical ligation. Science. 1994 Nov 4;266(5186):776–779. doi: 10.1126/science.7973629. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Dirheimer G., Gangloff J. Isolation and characterization of the gene coding for Escherichia coli arginyl-tRNA synthetase. Nucleic Acids Res. 1989 Jul 25;17(14):5725–5736. doi: 10.1093/nar/17.14.5725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fersht A. R., Dingwall C. Cysteinyl-tRNA synthetase from Escherichia coli does not need an editing mechanism to reject serine and alanine. High binding energy of small groups in specific molecular interactions. Biochemistry. 1979 Apr 3;18(7):1245–1249. doi: 10.1021/bi00574a020. [DOI] [PubMed] [Google Scholar]
- Jakubowski H. Z., Pastuzyn A., Loftfield R. B. The determination of aminoacyl adenylate by thin-layer chromatography. Anal Biochem. 1977 Sep;82(1):29–37. doi: 10.1016/0003-2697(77)90130-0. [DOI] [PubMed] [Google Scholar]
- Jakubowski H. Editing function of Escherichia coli cysteinyl-tRNA synthetase: cyclization of cysteine to cysteine thiolactone. Nucleic Acids Res. 1994 Apr 11;22(7):1155–1160. doi: 10.1093/nar/22.7.1155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H., Fersht A. R. Alternative pathways for editing non-cognate amino acids by aminoacyl-tRNA synthetases. Nucleic Acids Res. 1981 Jul 10;9(13):3105–3117. doi: 10.1093/nar/9.13.3105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H., Goldman E. Editing of errors in selection of amino acids for protein synthesis. Microbiol Rev. 1992 Sep;56(3):412–429. doi: 10.1128/mr.56.3.412-429.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H., Goldman E. Synthesis of homocysteine thiolactone by methionyl-tRNA synthetase in cultured mammalian cells. FEBS Lett. 1993 Feb 15;317(3):237–240. doi: 10.1016/0014-5793(93)81283-6. [DOI] [PubMed] [Google Scholar]
- Jakubowski H. Proofreading and the evolution of a methyl donor function. Cyclization of methionine to S-methyl homocysteine thiolactone by Escherichia coli methionyl-tRNA synthetase. J Biol Chem. 1993 Mar 25;268(9):6549–6553. [PubMed] [Google Scholar]
- Jakubowski H. Proofreading in vivo. Editing of homocysteine by aminoacyl-tRNA synthetases in Escherichia coli. J Biol Chem. 1995 Jul 28;270(30):17672–17673. [PubMed] [Google Scholar]
- Jakubowski H. Proofreading in vivo: editing of homocysteine by methionyl-tRNA synthetase in Escherichia coli. Proc Natl Acad Sci U S A. 1990 Jun;87(12):4504–4508. doi: 10.1073/pnas.87.12.4504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H. Proofreading in vivo: editing of homocysteine by methionyl-tRNA synthetase in the yeast Saccharomyces cerevisiae. EMBO J. 1991 Mar;10(3):593–598. doi: 10.1002/j.1460-2075.1991.tb07986.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakubowski H. Valyl-tRNA synthetase form yellow lupin seeds: hydrolysis of the enzyme-bound noncognate aminoacyl adenylate as a possible mechanism of increasing specificity of the aminoacyl-tRNA synthetase. Biochemistry. 1980 Oct 28;19(22):5071–5078. doi: 10.1021/bi00563a021. [DOI] [PubMed] [Google Scholar]
- Kleinkauf H., von Döhren H. Biosynthesis of peptide antibiotics. Annu Rev Microbiol. 1987;41:259–289. doi: 10.1146/annurev.mi.41.100187.001355. [DOI] [PubMed] [Google Scholar]
- Landès C., Perona J. J., Brunie S., Rould M. A., Zelwer C., Steitz T. A., Risler J. L. A structure-based multiple sequence alignment of all class I aminoacyl-tRNA synthetases. Biochimie. 1995;77(3):194–203. doi: 10.1016/0300-9084(96)88125-9. [DOI] [PubMed] [Google Scholar]
- Nagel G. M., Doolittle R. F. Evolution and relatedness in two aminoacyl-tRNA synthetase families. Proc Natl Acad Sci U S A. 1991 Sep 15;88(18):8121–8125. doi: 10.1073/pnas.88.18.8121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stein T., Vater J., Kruft V., Wittmann-Liebold B., Franke P., Panico M., Mc Dowell R., Morris H. R. Detection of 4'-phosphopantetheine at the thioester binding site for L-valine of gramicidinS synthetase 2. FEBS Lett. 1994 Feb 28;340(1-2):39–44. doi: 10.1016/0014-5793(94)80169-x. [DOI] [PubMed] [Google Scholar]
- Webster T., Tsai H., Kula M., Mackie G. A., Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984 Dec 14;226(4680):1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]





