Abstract
G protein-coupled receptors (GPCRs) are eukaryotic integral membrane proteins that modulate biological function by initiating cellular signaling in response to chemically diverse agonists. Despite recent progress in the structural biology of GPCRs1, the molecular basis for agonist binding and allosteric modulation of these proteins is poorly understood. Structural knowledge of agonist-bound states is essential for deciphering the mechanism of receptor activation, and for structure-guided design and optimization of ligands. However, the crystallization of agonist-bound GPCRs has been hampered by modest affinities and rapid off-rates of available agonists. Using the inactive structure of the human β2 adrenergic receptor (β2AR) as a guide, we designed a β2AR agonist that can be covalently tethered to a specific site on the receptor through a disulfide bond. The covalent β2AR-agonist complex forms efficiently, and is capable of activating a heterotrimeric G protein. We crystallized a covalent agonist-bound β2AR-T4L fusion protein in lipid bilayers through the use of the lipidic mesophase method2, and determined its structure at 3.5 Å resolution. A comparison to the inactive structure and an antibody-stabilized active structure (companion paper3) shows how binding events at both the extracellular and intracellular surfaces are required to stabilize an active conformation of the receptor. The structures are in agreement with long-timescale (up to 30 μs) molecular dynamics simulations showing that an agonist-bound active conformation spontaneously relaxes to an inactive-like conformation in the absence of a G protein or stabilizing antibody.
The relationship between agonist binding to GPCRs and the conformational changes that facilitate G protein binding and activation remains largely unknown. Characterization of GPCR activation at a molecular level has been driven by a combination of X-ray crystal structure analysis and spectroscopic approaches. Rhodopsin has served as a prototype GPCR for biophysical studies, given its ready availability and superior stability. Crystal structures have been obtained for the inactive dark state with inverse agonist 11-cis-retinal covalently bound4,5, as well as the active state mimetic low-pH opsin lacking the retinal chromophore6,7. Opsin differs from rhodopsin by outward rigid-body movements of transmembrane helices (TMs) 5 and 6 at the cytoplasmic G protein binding surface. For other GPCRs, which respond to diffusible ligands, structural information has proven more difficult to obtain. Fluorescence spectroscopy studies show that activation of the β2AR by diffusible ligands can follow multiple pathways, with a complex energy landscape of receptor conformations8. This multitude of accessible conformations has probably contributed to the difficulty in obtaining crystal structures of non-rhodopsin GPCRs.
Inactive state crystal structures of the β2AR, β1AR, and A2A adenosine receptor have been solved over the past several years with the aid of protein engineering techniques1, but agonist-bound structures for these proteins have yet to be reported. Like many GPCRs, the β2AR exhibits two agonist affinity states: a low affinity state in the absence of cognate G protein, and a high affinity state in the presence of G protein. This observation suggests that agonists can bind to two distinct receptor conformations. A more complete understanding of the processes of agonist binding and activation requires structures of both high and low affinity states. The high affinity state is challenging because a receptor-G protein complex is unstable in detergent solutions required for purification of both GPCRs and heterotrimeric G proteins. In a companion paper3, we describe the use of a conformationally selective camelid antibody (nanobody, Nb80) with G protein-like properties to obtain a structure of a high-affinity agonist-bound conformation. Obtaining a structure of the low affinity state is also challenging because of the relatively rapid association and dissociation rates of commercial β2AR agonists. Inspired by the covalent retinal-rhodopsin system, we hypothesized that the ability to crystallize an agonist-bound GPCR would be enhanced by chemically crosslinking ligand and receptor, preferably in a manner that would not inhibit conformational freedom and the capacity to activate a G protein.
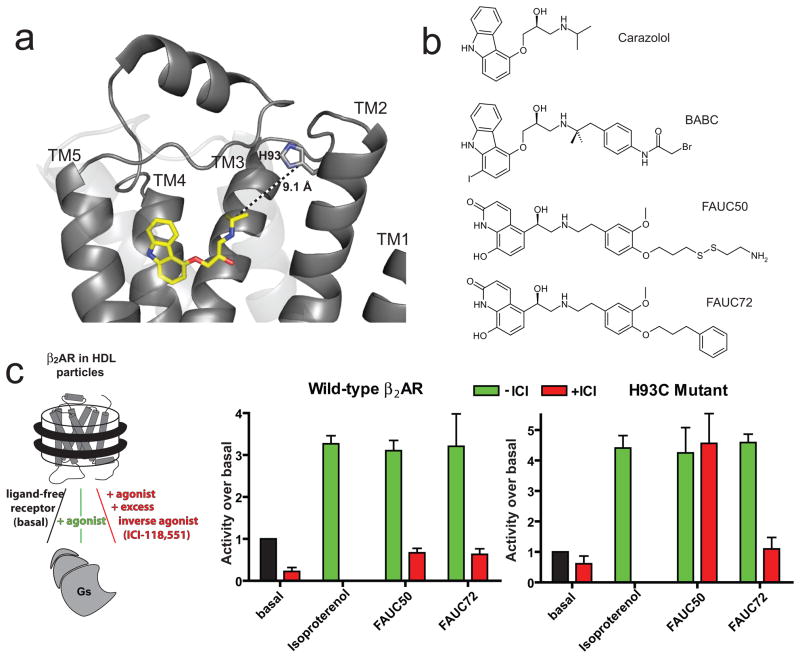
Our design strategy for a covalent β2AR agonist was to combine a β-adrenergic agonist core (procaterol) and a reactive chemical group that could be targeted to a specific residue on the receptor. Using the structure of the carazolol-bound β2AR as a template (Figure 1A), a flexible linker was added to bridge these two components such that the covalent attachment would not inhibit binding of the agonist core or conformational flexibility of the transmembrane helices. Biochemical precedent for this strategy came from the covalent labeling reagent BABC (Figure 1B), in which an electrophilic group appended to the carazolol ligand core was determined to react with His932.64 at the extracellular end of TM29 (Ballesteros-Weinstein numbering10 used in superscript). For crosslinking, we chose the reaction between a free cysteine on the receptor (introduced at position 93) and a ligand disulfide moiety, based on the mild and proximity-dependent “tethering” approach that has proven broadly applicable to different protein targets including GPCRs11. The designed covalent β agonist FAUC50 (Figure 1B) was synthesized in enantiomerically pure form12,13, along with the noncovalent analog FAUC72 (Figure 1B, Figure S1, and Supplementary Information) for use in control experiments.
Figure 1. Design and function of a covalent agonist.
a, Structure of the carazolol-bound β2AR, receptor in gray cartoon and ligand in yellow sticks, showing distance between isopropyl group and His932.64 imidazole. b, Structures of carazolol and the related BABC ligand, covalent ligand FAUC50 and noncovalent analog FAUC72. c, G protein activation assay demonstrating that covalently bound FAUC50 activates the β2AR.
Incubation of compound FAUC50 with mutant H93C receptor led to efficient and irreversible blocking of radioligand binding (Figure S2, Supplementary Information). We sought to determine whether the tethered FAUC50-β2ARH93C complex is capable of activating a G protein. Wild-type and mutant receptor were reconstituted into High Density Lipoprotein (HDL) particles14, and then incubated with ligand alone or ligand followed by an excess of the high-affinity inverse agonist ICI-118,551. Heterotrimeric Gs protein was then added to the particles, and activation was observed by measuring GTPγ35S binding to the Gsα subunit. Figure 1C shows that the inverse agonist treatment prevented agonist-induced G protein activation by the wild type receptor. However, excess ICI-118,551 is unable to reverse FAUC50-induced coupling in the case of β2ARH93C. The noncovalent analog FAUC72 is displaced by the inverse agonist for both wild type and H93C mutant receptors. From these experiments we conclude that FAUC50 not only reacts efficiently with Cys93 to form a covalent complex, but this complex is also capable of activating a G protein.
We were motivated to develop a covalent agonist by our inability to produce diffraction-quality crystals of the previously described β2AR-T4 lysozyme chimera (β2AR-T4L) in complex with available noncovalent agonists. In contrast, the purified FAUC50-β2ARH93CT4L complex readily yielded diffraction-quality crystals in lipidic mesophases15. We overexpressed the mutant receptor in Sf9 insect cells, purified the protein to homogeneity using immunoaffinity and ligand-affinity chromatography, and introduced the FAUC50 ligand during a subsequent chromatography step. Using a cholesterol-doped monoolein cubic phase and robotic in meso technology16, we obtained 50×15×5 μm blade-shaped crystals (Figure S3) that diffracted to 3.5 Å. Combining diffraction data from 19 microcrystals (Table S1), we solved the agonist-bound structure by molecular replacement using the coordinates of carazolol-bound β2AR-T4L17 as a search model. The packing of protomers in the orthorhombic crystals is distinct from previous crystals of β2AR-T4L fusion proteins. However, the orientation of T4L relative to the receptor closely resembles the original carazolol-bound structure (PDB ID 2RH1). An omit map at the ligand binding site reveals clear electron density for the agonist (Figure S4), and the refined structure shows prominent features expected from adrenergic receptor pharmacology18: the amine and β-hydroxyl of FAUC50 contact Asp1133.32 and Asn3127.39 in a manner similar to inverse agonists17, while the aromatic moiety of FAUC50 that replaces the catechol ring forms hydrogen bonds with Ser2035.42 and Ser2075.46 (Figure 2A, left).
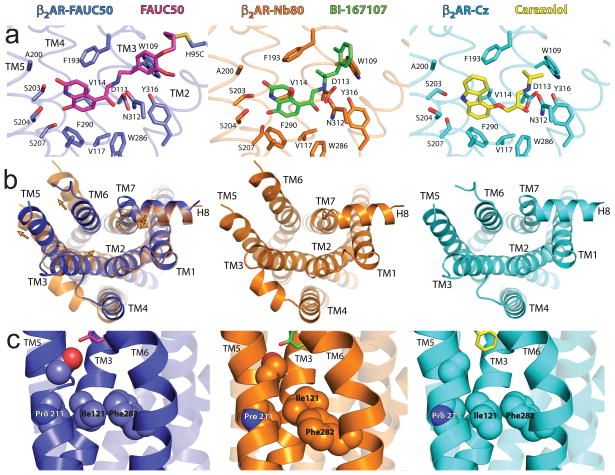
Figure 2. Comparison of agonist bound β2AR structures.
Comparison of the covalent FAUC50-bound β2ARH93CT4L (left panels, blue cartoon, ligand carbons in purple), BI-167107-bound β2AR-T4L/nanobody (β2AR-Nb80) complex (middle panels, orange cartoon, ligand carbons in green), and carazolol-bound β2AR-T4L (right panels, cyan cartoon, ligand carbons in yellow). a, Hormone binding site with interactions between ligands and receptors. TMs 6 and 7 with residues Phe289, Asn293, and Try308 are omitted for clarity. b, Comparison of the cytoplasmic surfaces showing differences in TMs 5 and 6. Superimposed β2AR-Nb80 complex is also shown in left panel as a transparent cartoon, and arrows indicate rigid body movements. c, Conformational switch region with residues Ile1213.40, Pro2115.50, and Phe2826.44 from TMs 3, 5, and 6 (other TMs transparent). Sidechains are shown in van der Waals sphere representation. A dashed line is shown at an equivalent position in each receptor.
The crystal structure of the covalent agonist-bound receptor in a lipidic mesophase does not show the conformational changes near the cytoplasmic surface associated with G protein binding, as observed in the nanobody-stabilized agonist complex (Figure 2B). This was surprising given that the covalent agonist-bound receptor activates Gs (Figure 1C) and the T4L fusion does not interfere with agonist-induced conformational changes detected with a fluorescent probe at the end of TM617. This result implies that even an agonist with zero dissociation inefficiently stabilizes an active conformation of the β2AR in a lipid bilayer environment, consistent with a higher stability of the inactive carazolol-bound conformation. Nevertheless, allosteric communication between the ligand binding pocket and the cytoplasmic G protein binding surface is a fundamental feature of GPCRs, exemplified by agonist-induced G protein stimulation and G protein-induced high-affinity agonist binding 14. For the β2AR, we can now compare an agonist-bound structure to that of a nanobody-stabilized active state mimetic (see companion paper3), to better understand conformational changes associated with activation. The superposition in Figure 2B shows that agonist binding alone is insufficient to stabilize an active conformation at the cytoplasmic surface, with the requisite outward movement of TMs 5 and 6. The largest differences proximal to the ligand binding pocket in the nanobody-stabilized active conformation involve movement of Ile1213.40 away from Pro2115.50 and into space occupied by Phe2826.44 in the inactive state (Figure 2C, middle). The concomitant outward movement of Phe2826.44 is accompanied by subtle backbone torsion changes distributed through TM6 that cause the cytoplasmic half of TM6 to be redirected outward, similar to its orientation in activated opsin6,7. In the binding pocket, the most significant difference between the two agonist-bound structures and the carazolol-bound β2AR are in the hydrogen bonding contacts with Ser2035.42 and Ser2075.46 on TM5 (Figure 2A). However, only in the nanobody-stabilized structure are these differences coordinated with further changes toward the cytoplasmic surface of the molecule (Figure 2B and 2C, middle).
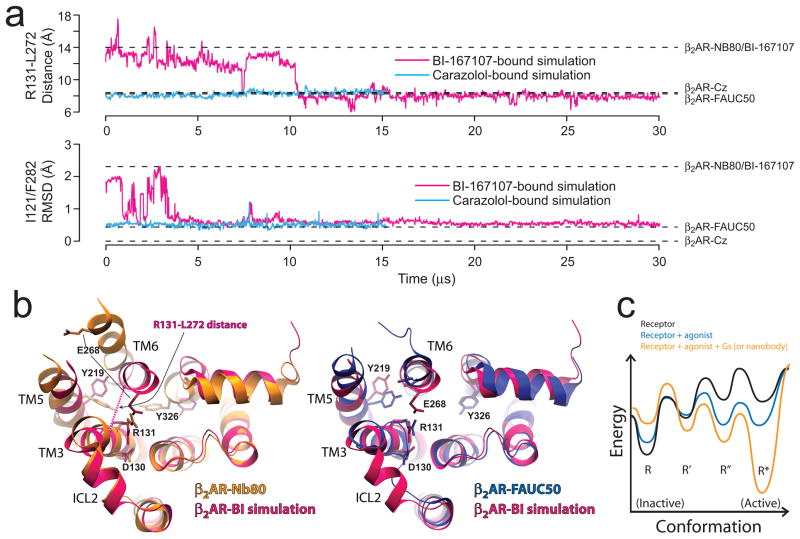
To determine whether an agonist-bound receptor would sustain a stable active conformation in the absence of a cytoplasmic binding partner, we initiated an unbiased molecular dynamics (MD) simulation of the receptor from the nanobody complex structure, but with the nanobody removed (Figure 3A). In the first several microseconds of simulated time, the intracellular ends of TM5 and TM6 exhibited high mobility (Figure S5), drifting by as much as 5 Å from their crystallographic positions. After approximately 11 μs, the agonist-bound receptor spontaneously transitioned to a more rigid conformation resembling the inactive, carazolol-bound structure and the covalent agonist-bound structure (superposition in Figure 3B), which remained stable for the remainder of the 30-μs simulation (Figure 3A, top, and Figure S6). In particular, TMs 5 and 6 reverted to the positions they adopt in the inactive structure, as did a number of side chains, including Ile1213.40 and Phe2826.44 (Figure 3A, bottom). Following the transition, the TM3–TM6 ionic lock was usually intact, as observed previously in simulations initiated from the inactive structure19. Although this 30-μs simulation is an order of magnitude longer than any previously published atomistic simulation of a membrane protein, the transition to an inactive-like conformation took place more quickly than the millisecond timescales observed experimentally for adrenergic receptor activation20. An additional simulation with different protonation states for Asp792.50 and Asp1303.49 —two conserved residues whose protonation states have been suggested to change upon receptor activation 21,22 —exhibited similar behavior (Figure S7).
Figure 3. Molecular dynamics simulations.
a, An unbiased simulation initiated from the nanobody complex (β2AR-Nb80) structure, with the agonist BI-167107 bound but the nanobody removed (magenta), and a carazolol-bound simulation initiated from the inactive structure (β2AR-Cz) (blue). (top) Distance between Cα atoms of Arg1313.50 and Leu2726.34, and (bottom) RMSD of non-symmetric non-hydrogen atoms in residues Ile1213.40 and Phe2826.44. Dashed lines indicate corresponding quantities from crystal structures. b, Cytoplasmic view of the simulated agonist-bound receptor after 30 μs, compared to the β2AR-Nb80 (left) and β2AR-FAUC50 (right) structures. The conformations of intracellular loop 2, Tyr2195.58, and Glu2686.30 shown for the agonist-bound simulation differ from β2AR-FAUC50, but have been observed in inactiveβ2AR simulations19 and in other inactive-state GPCR structures1. c, Proposed energy landscape model, in which both an agonist and a cytoplasmic binding partner are required to stabilize the fully active receptor conformation [R*] over intermediate [R′ and R″] and inactive [R] states.
In the dynamic conformational equilibrium of a GPCR that links diffusible agonist binding and G protein association, the energies of different states reflect both the ligand binding energies and the conformations of the receptor and its binding partners. This can be depicted in a hypothetical energy landscape of the β2AR as shown in Fig. 3C, where R, R′, R″ and R* represent members of the ensemble of receptor conformations along an activation pathway. The constitutive activity displayed by the β2AR implies that the energy differences and barriers between inactive (R) and active (R*) conformations are low enough to allow a significant population of active state receptors even in the absence of agonist. Agonist binding decreases the energy difference and thus increases the population of receptors in an active conformation; however, the inactive state is still the most stable conformation. Our crystallographic studies and MD simulations are compatible with fluorescence lifetime experiments on the purified receptor, demonstrating that saturating concentrations of a full agonist are incapable of pushing the β2AR conformational equilibrium toward a homogenous active state23. These results suggest that binding energy from a G protein or nanobody interaction is required to stabilize conformational changes such as those observed in the vicinity of Ile1213.40 and Phe2826.44 in the active state (Figure 2C). Alternative biophysical approaches such as NMR24,25 and further molecular dynamics simulations will be crucial to fully understand these transitions and identify potential pathways and intermediates that are compatible with these structures.
METHODS SUMMARY
Synthetic chemistry methodology to generate FAUC50 and FAUC72 are described in Supplementary Information. Protocols for radioligand binding and Gs activation assays to characterize ligands, are described in Methods.
Crystallization
FAUC50-β2ARH93CT4L was expressed, purified and crystallized as described in Methods. The receptor was crystallized in a cholesterol-doped (10%) monoolein cubic phase overlaid with precipitant in glass sandwich plates. Optimized precipitant consisted of 24–27 %(v/v) PEG400, 200 mM Li2SO4, 4 %(v/v) DMSO, 3.5 %(v/v) 1,4-butanediol, 100 mM MES pH 6.7. After 3–5 days of growth, crystals were harvested after adding an excess of precipitant solution for cryoprotection, and flash frozen in liquid nitrogen.
Data collection, structure solution and refinement
Diffraction data were collected at beamline 23-ID (GM/CA-CAT) of the Advanced Photon Source, using a 10 μm diameter collimated microbeam. Oscillation data were measured in 1.0° frames with 10 sec or 25 sec exposures using 2x or 5x attenuated beam, respectively. The complete data set consisted of 106° of data from 19 crystals. The structure of FAUC50-β2ARH93CT4L was solved by molecular replacement, and refined by group B factor and TLS refinement. Full details are provided in Methods.
Molecular dynamics
All-atom classical molecular dynamics simulations with explicitly represented lipids and water were performed using the CHARMM force field26 on Anton27, a special-purpose computer that accelerates such simulations by orders of magnitude; details are provided in the Methods.
METHODS
G protein activation assay with ICI-118,551 reversal
Wild-type β2AR or β2ARH93C were purified in unliganded form28. Samples of these receptors were reconstituted into rHDL particles as described14. For GTPγS binding assay, receptor-rHDL particles were pre-incubated with 5 μM FAUC50 or 5 μM FAUC72 for 4 hrs at 4 °C. Samples were diluted 20-fold into binding buffer and split. Half of the samples were used in GTPγS binding assays without ICI-118,551 competition, while the other half was incubated with 6 μM ICI-118,551 at room temperature for 1 hr with shaking. Control samples of receptor-rHDL particles with no ligand, 10 μM Isoproterenol, or 10 μM ICI-118,551 were also prepared. Purified Gs heterotrimer29 was added to each sample and incubated for 10 minutes at 23 °C. The final concentrations of reconstituted receptor and Gs were 100 nM and 600 nM, respectively. GTPγS binding reactions were initiated by the addition of 0.4 nM [35S]GTPγS. Free [35S]GTPγS was removed by rapid filtration of the particles using glass fiber filters. Filter bound radioactivity was determined by liquid scintillation counting using a Beckman LS6000 scintillation counter. Data shown in Figure 1C are from three independent experiments each performed in triplicate.
Purification of β2ARH93CT4L
The construct used for crystallography consisted of theβ2AR-T4L fusion protein17 with the following modifications: a TEV site was inserted after residue 23 of the human β2AR sequence; Histidine 93 was mutated to cysteine; the construct was terminated by a 6xHis tag after residue 348 of human β2AR. This construct was cloned into the baculovirus transfer vector pVL1392, and the resulting vector was used to make a high-titer baculovirus stock using the Bestbac system (Expression Systems). The β2ARH93CT4L receptor was expressed in Sf9 insect cell cultures infected with this baculovirus, and Sf9 membranes were solubilized as described17. Purification was achieved using M1 FLAG affinity chromatography (Sigma), functional alprenolol-sepharose chromatography30, and a second M1 chromatography concentrating step. While the receptor was still bound to the second M1 FLAG column, bound alprenolol was washed out and replaced with the covalent ligand using 3 hrs of washing with 15 column volumes of buffer containing 30 μM FAUC50 (10-fold total molar excess over receptor). Likewise, the dodecylmaltoside detergent used in all previous steps was exchanged for 0.1% (w/v) MNG-3 amphiphile 31. Covalent agonist-bound and detergent-exchanged β2ARH93CT4L was eluted in [20 mM HEPES pH 7.5, 100 mM NaCl, 0.1% (w/v) MNG-3, 30 μM FAUC50]. In contrast to previously published protocols, receptor was not alkylated prior to alprenolol-sepharose chromatography. Instead, receptor was alkylated after agonist exchange by treatment with 2 mM iodoacetamide for 30 minutes at 4 °C. Incubation of the eluate with AcTEV protease (Invitrogen) succeeded in removing the N-terminus of the receptor (23 amino acids plus TEV site, leaving a glycine scar preceding residue 24), as verified by SDS-PAGE analysis. The sample at this stage was further purified and concentrated using Ni chelating chromatography, taking advantage of the C-terminal 6xHis tag. Receptor was bound to a 0.7 ml column Ni-sepharose column, and eluted in [20 mM HEPES pH 7.5, 100 mM NaCl, 0.1% (w/v) MNG-3, and 30 μM FAUC50, 200 mM Imidazole]. Finally, the receptor was concentrated to 50 mg/ml with a 100 kDa cutoff Vivaspin concentrator (Vivascience).
Crystallization
Purified FAUC50-β2ARH93CT4L was crystallized using the in meso method2. Cubic phase reconstitution consisted of 2 parts 50 mg/ml receptor and 3 parts molten lipid mixture (10:1 monoolein:cholesterol by mass, lipids purchased from Sigma). Note that cholesterol is required for successful crystallization, and one cholesterol molecule is included in the refined model. Aqueous and lipid components were combined at room temperature using a syringe mixing apparatus32. Crystallization experiments in glass sandwich plates, set up by either by hand or using an in meso robot16, consisted of 30–50 nL cubic phase overlaid with 800 nL precipitant. Precipitant conditions producing diffraction quality crystals were identified by screening around previous conditions17, and testing additives and alternative buffers. Final optimized conditions consisted of 24–27 %(v/v) PEG400, 200 mM Li2SO4, 4 %(v/v) DMSO, 3.5 %(v/v) 1,4-butanediol, 100 mM MES pH 6.7. Crystals grew at 20 °C to a maximum size of 50 × 15 × 5 μm3 within 3 to 5 days (see Figure S3, Supplementary Materials). For harvesting and cryocooling, exposed crystallization drops were overlaid with an excess of precipitant solution, and cryoloops (MiTeGen) containing single crystals were flash frozen in liquid nitrogen.
Data collection and processing
Diffraction data were collected at beamline 23-ID of the General Medicine and Cancer Institutes Collaborative Access Team (GM/CA-CAT) of the Advanced Photon Source, Argonne IL. All data was acquired using a 10 μm diameter collimated microbeam. Attenuated 1.0° rotation images were used to locate and center crystals within the opaque mesophase in each cryoloop. Oscillation data were measured in 1.0° frames with 10 sec or 25 sec exposures using 2x or 5x attenuated beam, respectively. Significant radiation damage caused decay in the signal that prevented merging more than the first 5–10° of oscillation data from each crystal. A total of 106° of data from 19 crystals were integrated with Mosflm33 and merged with Scala34.
Structure solution and refinement
Molecular replacement to obtain initial phases was performed with the program Phaser35. The separated structures of the receptor and T4L components of the high-resolution carazolol-bound β2AR structure (PDB ID 2RH1 with all non protein atoms removed), were used as search models. The model was refined in Phenix36 and Buster37, using group B factor refinement (one B factor per residue) followed by TLS refinement (using two TLS groups, one for the receptor and one for T4L). Refinement statistics are given in Table S1 of Supplementary Information. The crystallographic data was strongly anisotropic, as seen in the anisotropic B factor corrections, however the electron density was clear enough for the placement of side chains. While there was clear electron density present for the agonist (see omit map in Figure S4, Supplementary Information), we did observe a small discontinuity at the end of the linker connecting to Cys93. This discontinuity could arise from flexibility of the polymethylene component of the linker, as well as potential radiation damage38 localized to the disulfide bond. As a control, we carried out refinement in which the ligand was modeled as a non-covalent species without a disulfide connection to the receptor. No significant differences in final model R/Rfree or 2Fo-Fc and Fo-Fc electron density maps were observed.
Methods for molecular dynamics simulations
In all simulations, β2AR was embedded in a hydrated lipid bilayer, with all atoms, including those in the lipids and water, represented explicitly. The BI-167107-bound β2AR simulations were initiated with the receptor and ligand in the conformation of the β2AR-NB80 crystal structure (companion paper), with the nanobody (NB80) removed. The carazolol-bound β2AR simulations were initiated with the receptor and ligand in the conformation of the high-resolution carazolol-bound crystal structure17. Both crystal structures were determined using a β2AR-T4L fusion protein, in which intracellular loop 3 (ICL3) of the receptor was replaced by T4 lysozyme (although the T4L was not resolved in the BI-167107-bound nanobody complex structure). We omitted the T4L in all simulations. Experimentally, removal of the bulk of ICL3 by partial tryptic digest does not appear to affect receptor function37.
Production simulations were performed on Anton27, a special-purpose computer designed to accelerate standard molecular dynamics simulations by orders of magnitude relative to the previous state of the art. Prior to production simulation, systems were equilibrated using Desmond39 on a commodity cluster, according to the protocol described below.
System setup and simulation protocol
Hydrogens were added to the crystal structures of carazolol-bound β2AR (β2AR-Cz; PDB entry 2RH1) and β2AR-NB80/BI-167107 (companion paper) using Maestro (Schrödinger LLC, New York NY) as described previously19. T4L and NB80 were deleted, and chain termini were capped with neutral groups (acetyl and methylamino).
All titratable residues other than Glu1223.41, Asp792.50, and Asp1303.49 were left in the dominant protonation state at pH 7.0. Glu1223.41 was protonated in all simulations. It faces the lipid bilayer and is thus likely protonated40; in addition, a similarly positioned residue in rhodopsin (Glu1223.37) has been found to be protonated during the entire photocycle22.
We performed both BI-167107-bound and carazolol-bound simulations using two different sets of protonation states for Asp792.50 and Asp1303.49. Previous studies have suggested that Asp1303.49 is protonated upon activation22, and FTIR data has shown that this is the case for the corresponding residue of rhodopsin, Glu1343.49-41. Asp792.50 is homologous to Asp832..50 of rhodopsin, which has been found by FTIR spectroscopy to remain protonated during the entire photocycle40. On the other hand, neutralization of Asp792.50 in β2AR by mutation to asparagine uncouples agonist binding from G protein activation 42, and a recent study suggested that Asp79 may be deprotonated upon activation21. Thus, we repeated each simulation with two sets of protonation states: first with Asp79 protonated and Asp130 deprotonated (“Ash79/Asp130,” representing potential protonation states in the inactive receptor), and then with Asp79 deprotonated and Asp130 protonated (“Asp79/Ash130,” representing possible protonation states in the active receptor). Results shown in Figures 3, S5, and S6 are for the Ash79/Asp130 simulations, while results in Figure S7 are for the Asp79/Ash130 simulations.
β2AR residues that were truncated or not resolved in the crystal structures were omitted from the simulations. In particular, N-terminal residues 1–28, C-terminal residues 343–413, and ICL3 residues 231–262 were omitted from the carazolol-bound system, while N-terminal residues 1–22, C-terminal residues 345–413, and ICL3 residues 228–265 were omitted from the BI-167107-bound system. Both simulations included a glutamate at position 187, reflecting an Asn187Glu mutation made in both crystallization constructs to eliminate a glycosylation site.
We prepared the carazolol-bound Ash79/Asp130 β2AR system following our previously described protocol19, and the other systems according to an updated protocol (details below). We do not expect the differences in the simulation setup protocols to affect the quantities of interest in this study. As a control, however, we performed an additional 5 μs simulation of a carazolol-bound Ash79/Asp130 β2AR system prepared according to the new protocol and obtained results (not shown) similar to those of the 15 μs carazolol-bound Ash79/Asp130 β2AR simulation prepared according to the old protocol
Prepared protein structures were inserted into an equilibrated phospholipid bilayer solvated with 0.15 M NaCl as described previously19. Carazolol-bound Ash79/Asp130 β2AR was simulated in a POPE bilayer system initially measuring 85 × 73 × 88 Å3, containing 170 lipid molecules, 8,798 water molecules, 19 sodium ions, and 24 chloride ions, for a total of 52,396 atoms. Carazolol-bound Asp79/Ash130 β2AR was simulated in a POPC bilayer system initially measuring 83 × 83 × 87 Å3, containing 160 lipid molecules, 11,313 water molecules, 30 sodium ions, and 35 chloride ions, for a total of 60,153 atoms. (The 5 μs carazolol-bound Ash79/Asp130 control simulation prepared according to the new protocol also used a POPC bilayer.) BI-167107-bound β2AR (both Ash79/Asp130 and Asp79/Ash130) was simulated in a POPC bilayer system initially measuring 79 × 79 × 87 Å3, containing 138 lipid molecules, 10,116 water molecules, 26 sodium ions, and 31 chloride ions, for a total of 53,603 atoms.
All systems were equilibrated in the NPT ensemble at 310 K and 1 bar using the Berendsen coupling scheme with 5 kcal mol−1 Å−2 harmonic position restraints applied to all non-hydrogen atoms of the protein; these restraints were tapered off linearly over 5 ns. Unrestrained systems were then simulated for an additional 5 ns to further equilibrate the aspect ratio of the simulation box. During the equilibration process, Van der Waals and short-range electrostatic interactions were cut off at 9 Å for the carazolol-bound Ash79/Asp130 β2AR system and at 12 Å for the other systems. Long-range electrostatic interactions were computed using the Particle Mesh Ewald method43, with a 64 × 64 × 64 grid and σ = 2.26 Å for the carazolol-bound Ash79/Asp130 β2AR system, and with a 32 × 32 × 32 grid and σ = 3.23 Å for the other systems; fifth-order B-splines were used for interpolation in both cases. All bond lengths to hydrogen atoms were constrained using M-SHAKE44. A RESPA integrator45 was used with a timestep of 2 fs, and long-range electrostatics were computed every 6 fs.
Production simulations on Anton were initiated from the final snapshot of each corresponding equilibration runs on Desmond, using the same integration schemes. Van der Waals and short-range electrostatic interactions were cut off at 9 Å for the carazolol-bound Ash79/Asp130 β2AR system and at 13.5 Å for the other systems. Long-range electrostatics were computed using the k-space Gaussian Split Ewald method46, with a 64 × 64 × 64 grid, σ = 2.01 Å, and σs = 1.41 Å for the carazolol-bound Ash79/Asp130 β2AR system, and with a 32 × 32 × 32 grid, σ = 3.33 Å, and σs = 2.35 Å for the other systems.
Force field parameters
The CHARMM2726 parameter set (with CMAP terms47) and the CHARMM TIP3P48 water model were used for all protein molecules, POPE lipid molecules, water molecules, and salt ions. A modified CHARMM lipid force field49, which became available after we performed the 15-μs simulation of carazolol-bound β2AR in POPE, was used for POPC lipids. Force field parameters for carazolol and palmitoyl-cysteine were designed previously19. Force field parameters for BI-167107 were transferred from previously parameterized model compounds: parameters for the hydroxyethylamine “tail” group were transferred from the alkylamine parameters we previously designed for carazolol19, and parameters for the benzoxazine “head” group were transferred from the model compounds anisole and p-phenol acetamide from the CHARMM General Force Field50. Full parameter sets are available upon request.
Analysis protocols
Trajectory snapshots, each containing a record of all atom positions at a particular instant in time, were saved every 180 ps during production simulation. Distance and RMSD measurements were computed using the HiMach parallel analysis framework51.
The distance and RMSD measurements shown in Figures 3A, S5, and S7 are smoothed; the smoothed time series were computed from the original time series by a weighted running average, using a filter kernel of half-width 12.51 ns whose shape corresponded to that of the cosine function from −π/2 to π/2.
VMD52 was used to visualize trajectories and to produce the molecular renderings of Figure 3B.
Supplementary Material
Acknowledgments
We acknowledge support from National Institutes of Health Grants NS028471 and GM083118 (B.K.K.), GM56169 (W.I.W.), P01 GM75913 (S.H.G), GM75915 and P50GM073210 (M.C), and P60DK-20572 (R.K.S.), a grant from Boerhinger Ingelheim (B.K.K.), the Mathers Foundation (B.K.K and W.I.W), the Lundbeck Foundation (Junior Group Leader Fellowship, S.G.F.R.), Science Foundation Ireland (07/IN.1/B1836) and FP7 COST Action CM0902 (M.C.), the Bavaria California Technology Center (P.G.), and the University of Michigan Biomedical Sciences Scholars Program (R.K.S). We thank Stefan Löber, Harald Hübner, Albert Pan and Paul Maragakis for discussion and suggestions.
Footnotes
Coordinates and structure factors for β2AR-FAUC50 are deposited in the Protein Data Bank (accession code 3PDS).
Reprints and permissions information is available at www.nature.com/reprints.
Author Contributions D.M.R. designed project and agonists, did binding assays to characterize agonists, developed purification, optimized crystallization conditions, optimized construct, purified protein, grew and harvested crystals, collected data, solved structure, refined structure, wrote manuscript. C.Z. performed G protein activation assays for covalent agonist, prepared recombinant baculovirus, performed large-scale expression of recombinant β2AR in insect cells, purified protein, grew crystals. J.L. helped to optimize crystallization conditions, grew and harvested crystals, collected data. D.A. was involved in LCP optimization, harvested crystals, collected data. R.H. synthesized agonists. S.G.F.R. identified the use of MNG3 detergent for β2AR stabilization and assisted with manuscript preparation. H.J.C. assisted with data processing and refinement. R.K.S. and B.D. provided ApoA1 and Gs protein for functional characterization of the covalent ligand. P.S.C. and S.G. provided MNG3 detergent for stabilization of purified β2AR. D.H.A. and R.O.D. performed and analyzed MD simulations; R.O.D. and D.E.S. oversaw MD simulations and analysis. W.I.W. assisted with data processing and refinement and with manuscript preparation. M.C. helped to guide the LCP crystallization efforts at Stanford and in Ireland, and oversaw automated lipidic cubic phase crystallography screens. P.G. designed the strategy for the synthesis of the covalent agonist. B.K. was responsible for the overall project strategy and management, oversaw manuscript preparation, and assisted with synchrotron data collection.
LITERATURE CITED
- 1.Rosenbaum DM, Rasmussen SG, Kobilka BK. The structure and function of G-protein-coupled receptors. Nature. 2009;459:356–363. doi: 10.1038/nature08144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caffrey M. Crystallizing membrane proteins for structure determination: use of lipidic mesophases. Annu Rev Biophys. 2009;38:29–51. doi: 10.1146/annurev.biophys.050708.133655. [DOI] [PubMed] [Google Scholar]
- 3.Rasmussen SG, et al. Structure of a nanobody-stabilized active state of the β2 adrenoceptor. Nature. doi: 10.1038/nature09648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li J, Edwards PC, Burghammer M, Villa C, Schertler GF. Structure of bovine rhodopsin in a trigonal crystal form. J Mol Biol. 2004;343:1409–1438. doi: 10.1016/j.jmb.2004.08.090. [DOI] [PubMed] [Google Scholar]
- 5.Palczewski K, et al. Crystal structure of rhodopsin: A G protein-coupled receptor. Science. 2000;289:739–745. doi: 10.1126/science.289.5480.739. [DOI] [PubMed] [Google Scholar]
- 6.Park JH, Scheerer P, Hofmann KP, Choe HW, Ernst OP. Crystal structure of the ligand-free G-protein-coupled receptor opsin. Nature. 2008;454:183–187. doi: 10.1038/nature07063. [DOI] [PubMed] [Google Scholar]
- 7.Scheerer P, et al. Crystal structure of opsin in its G-protein-interacting conformation. Nature. 2008;455:497–502. doi: 10.1038/nature07330. [DOI] [PubMed] [Google Scholar]
- 8.Deupi X, Kobilka BK. Energy landscapes as a tool to integrate GPCR structure, dynamics, and function. Physiology (Bethesda) 2010;25:293–303. doi: 10.1152/physiol.00002.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dohlman HG, Caron MG, Strader CD, Amlaiky N, Lefkowitz RJ. Identification and sequence of a binding site peptide of the beta 2 adrenergic receptor. Biochemistry. 1988;27:1813–1817. doi: 10.1021/bi00406a002. [DOI] [PubMed] [Google Scholar]
- 10.Ballesteros JA, Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein coupled receptors. Meth Neurosci. 1995;25:366–428. [Google Scholar]
- 11.Buck E, Wells JA. Disulfide trapping to localize small-molecule agonists and antagonists for a G protein-coupled receptor. Proc Natl Acad Sci U S A. 2005;102:2719–2724. doi: 10.1073/pnas.0500016102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kolb HC, VanNieuwenhze MS, Sharpless KB. Catalytic Asymmetric Dihydroxylation. Chemical Reviews. 1994;94:2483–2547. [Google Scholar]
- 13.Martinelli MJ, et al. Catalytic regioselective sulfonylation of alpha-chelatable alcohols: scope and mechanistic insight. J Am Chem Soc. 2002;124:3578–3585. doi: 10.1021/ja016031r. [DOI] [PubMed] [Google Scholar]
- 14.Whorton MR, et al. A monomeric G protein-coupled receptor isolated in a high-density lipoprotein particle efficiently activates its G protein. Proc Natl Acad Sci U S A. 2007;104:7682–7687. doi: 10.1073/pnas.0611448104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Caffrey M, Cherezov V. Crystallizing membrane proteins using lipidic mesophases. Nat Protoc. 2009;4:706–731. doi: 10.1038/nprot.2009.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Cherezov V, Peddi A, Muthusubramaniam L, Zheng YF, Caffrey M. A robotic system for crystallizing membrane and soluble proteins in lipidic mesophases. Acta Crystallogr D Biol Crystallogr. 2004;60:1795–1807. doi: 10.1107/S0907444904019109. [DOI] [PubMed] [Google Scholar]
- 17.Rosenbaum DM, et al. GPCR engineering yields high-resolution structural insights into beta2-adrenergic receptor function. Science. 2007;318:1266–1273. doi: 10.1126/science.1150609. [DOI] [PubMed] [Google Scholar]
- 18.Tota MR, Candelore MR, Dixon RA, Strader CD. Biophysical and genetic analysis of the ligand-binding site of the beta-adrenoceptor. Trends Pharmacol Sci. 1991;12:4–6. doi: 10.1016/0165-6147(91)90479-c. [DOI] [PubMed] [Google Scholar]
- 19.Dror RO, et al. Identification of two distinct inactive conformations of the beta2-adrenergic receptor reconciles structural and biochemical observations. Proc Natl Acad Sci U S A. 2009;106:4689–4694. doi: 10.1073/pnas.0811065106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Vilardaga JP, Bunemann M, Krasel C, Castro M, Lohse MJ. Measurement of the millisecond activation switch of G protein-coupled receptors in living cells. Nat Biotechnol. 2003;21:807–812. doi: 10.1038/nbt838. [DOI] [PubMed] [Google Scholar]
- 21.Vanni S, Neri M, Tavernelli I, Rothlisberger U. A conserved protonation-induced switch can trigger “ionic-lock” formation in adrenergic receptors. J Mol Biol. 2010;397:1339–1349. doi: 10.1016/j.jmb.2010.01.060. [DOI] [PubMed] [Google Scholar]
- 22.Ghanouni P, et al. The Effect of pH on beta(2) Adrenoceptor Function. Evidence for protonation-dependent activation. J Biol Chem. 2000;275:3121–3127. doi: 10.1074/jbc.275.5.3121. [DOI] [PubMed] [Google Scholar]
- 23.Ghanouni P, et al. Functionally different agonists induce distinct conformations in the G protein coupling domain of the beta 2 adrenergic receptor. J Biol Chem. 2001;276:24433–24436. doi: 10.1074/jbc.C100162200. [DOI] [PubMed] [Google Scholar]
- 24.Ahuja S, et al. Helix movement is coupled to displacement of the second extracellular loop in rhodopsin activation. Nat Struct Mol Biol. 2009;16:168–175. doi: 10.1038/nsmb.1549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bokoch MP, et al. Ligand-specific regulation of the extracellular surface of a G-protein-coupled receptor. Nature. 2010;463 doi: 10.1038/nature08650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.MacKerell AD, et al. All-atom empirical potential for molecular modeling and dynamics studies of proteins. J Phys Chem B. 1998;102:3586–3616. doi: 10.1021/jp973084f. [DOI] [PubMed] [Google Scholar]
- 27.Shaw DE, et al. Proceedings of the Conference on High Performance Computing, Networking, Storage and Analysis. ACM; Portland, Oregon: 2009. [Google Scholar]
- 28.Yao X, et al. Coupling ligand structure to specific conformational switches in the beta2-adrenoceptor. Nat Chem Biol. 2006;2:417–422. doi: 10.1038/nchembio801. [DOI] [PubMed] [Google Scholar]
- 29.Kozasa T, Gilman AG. Purification of recombinant G proteins from Sf9 cells by hexahistidine tagging of associated subunits. Characterization of alpha 12 and inhibition of adenylyl cyclase by alpha z. J Biol Chem. 1995;270:1734–1741. doi: 10.1074/jbc.270.4.1734. [DOI] [PubMed] [Google Scholar]
- 30.Kobilka BK. Amino and carboxyl terminal modifications to facilitate the production and purification of a G protein-coupled receptor. Anal Biochem. 1995;231:269–271. doi: 10.1006/abio.1995.1533. [DOI] [PubMed] [Google Scholar]
- 31.Chae PS, et al. Accessible new amphiphiles for solubilization, stabilization and crystallization of membrane proteins. Nature Methods. 2010 doi: 10.1038/nmeth.1526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng A, Hummel B, Qiu H, Caffrey M. A simple mechanical mixer for small viscous lipid-containing samples. Chem Phys Lipids. 1998;95:11–21. doi: 10.1016/s0009-3084(98)00060-7. [DOI] [PubMed] [Google Scholar]
- 33.Leslie AG. The integration of macromolecular diffraction data. Acta Crystallogr D Biol Crystallogr. 2006;62:48–57. doi: 10.1107/S0907444905039107. [DOI] [PubMed] [Google Scholar]
- 34.The CCP4 suite: programs for protein crystallography. Acta Crystallogr D Biol Crystallogr. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 35.McCoy AJ, et al. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Afonine PV, Grosse-Kunstleve RW, Adams PD. A robust bulk-solvent correction and anisotropic scaling procedure. Acta Crystallogr D Biol Crystallogr. 2005;61:850–855. doi: 10.1107/S0907444905007894. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Blanc E, et al. Refinement of severely incomplete structures with maximum likelihood in BUSTER-TNT. Acta Crystallogr D Biol Crystallogr. 2004;60:2210–2221. doi: 10.1107/S0907444904016427. [DOI] [PubMed] [Google Scholar]
- 38.Weik M, et al. Specific chemical and structural damage to proteins produced by synchrotron radiation. Proc Natl Acad Sci U S A. 2000;97:623–628. doi: 10.1073/pnas.97.2.623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bowers KJ, et al. Proceedings of the 2006 ACM/IEEE Conference on Supercomputing. ACM; Tampa, Florida: 2006. [Google Scholar]
- 40.Fahmy K, et al. Protonation states of membrane-embedded carboxylic acid groups in rhodopsin and metarhodopsin II: a Fourier-transform infrared spectroscopy study of site-directed mutants. Proc Natl Acad Sci U S A. 1993;90:10206–10210. doi: 10.1073/pnas.90.21.10206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vogel R, et al. Functional role of the “ionic lock”--an interhelical hydrogen-bond network in family A heptahelical receptors. J Mol Biol. 2008;380:648–655. doi: 10.1016/j.jmb.2008.05.022. [DOI] [PubMed] [Google Scholar]
- 42.Liapakis G, Chan WC, Papadokostaki M, Javitch JA. Synergistic contributions of the functional groups of epinephrine to its affinity and efficacy at the beta2 adrenergic receptor. Mol Pharmacol. 2004;65:1181–1190. doi: 10.1124/mol.65.5.1181. [DOI] [PubMed] [Google Scholar]
- 43.Darden T, York D, Pedersen L. Particle mesh Ewald: An N•Σlog(N) method for Ewald sums in large systems. Journal of Chemical Physics. 1993;98:10089. [Google Scholar]
- 44.Kräutler V, van Gunsteren WF, Hünenberger PH. A fast SHAKE algorithm to solve distance constraint equations for small molecules in molecular dynamics simulations. Journal of Computational Chemistry. 2001;22:501–508. [Google Scholar]
- 45.Tuckerman M, Berne BJ, Martyna GJ. Reversible multiple time scale molecular dynamics. Journal of Chemical Physics. 1992;97:1990. [Google Scholar]
- 46.Shan Y, Klepeis JL, Eastwood MP, Dror RO, Shaw DE. Gaussian split Ewald: A fast Ewald mesh method for molecular simulation. J Chem Phys. 2005;122 doi: 10.1063/1.1839571. [DOI] [PubMed] [Google Scholar]
- 47.Mackerell AD, Jr, Feig M, Brooks CL., 3rd Extending the treatment of backbone energetics in protein force fields: limitations of gas-phase quantum mechanics in reproducing protein conformational distributions in molecular dynamics simulations. J Comput Chem. 2004;25:1400–1415. doi: 10.1002/jcc.20065. [DOI] [PubMed] [Google Scholar]
- 48.Beglov D, Roux B. Finite representation of an infinite bulk system: solvent boundary potential for computer simulations. J Chem Phys. 1994;100:9050–9063. [Google Scholar]
- 49.Klauda JB, et al. Update of the CHARMM all-atom additive force field for lipids: validation on six lipid types. J Phys Chem B. 2010;114:7830–7843. doi: 10.1021/jp101759q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Vanommeslaeghe K, et al. CHARMM general force field: A force field for drug-like molecules compatible with the CHARMM all-atom additive biological force fields. Journal of Computational Chemistry. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tu T, et al. Proceedings of the 2008 ACM/IEEE Conference on Supercomputing. IEEE Press; Austin, Texas: 2008. [Google Scholar]
- 52.Humphrey W, Dalke A, Schulten K. VMD: visual molecular dynamics. J Mol Graph. 1996;14:33–38. 27–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.