Abstract
Objective
Proliferation of smooth muscle cells (SMC) in response to vascular injury is central to neointimal vascular remodeling. There is accumulating evidence that histone acetylation constitutes a major epigenetic modification for the transcriptional control of proliferative gene expression; however, the physiological role of histone acetylation for proliferative vascular disease remains elusive.
Methods and Results
In the present study, we investigated the role of histone deacetylase (HDAC) inhibition in SMC proliferation and neointimal remodeling. We demonstrate that mitogens induce transcription of HDAC 1, 2 and 3 in SMC. siRNA-mediated knock-down of either HDAC 1, 2 or 3 and pharmacologic inhibition of HDAC prevented mitogen-induced SMC proliferation. The mechanisms underlying this reduction of SMC proliferation by HDAC inhibition involve a growth arrest in the G1-phase of the cell cycle due to an inhibition of retinoblastoma protein phosphorylation. HDAC inhibition resulted in a transcriptional and posttranscriptional regulation of the cyclin-dependent kinase inhibitors p21Cip1 and p27Kip. Furthermore, HDAC inhibition repressed mitogen-induced cyclin D1 mRNA expression and cyclin D1 promoter activity. As a result of this differential cell cycle-regulatory gene expression by HDAC inhibition, the retinoblastoma protein retains a transcriptional repression of its downstream target genes required for S phase entry. Finally, we provide evidence that these observations are applicable in vivo by demonstrating that HDAC inhibition decreased neointima formation and expression of cyclin D1 in a murine model of vascular injury.
Conclusion
These findings identify HDAC as a critical component of a transcriptional cascade regulating SMC proliferation and suggest that HDAC might play a pivotal role in the development of proliferative vascular diseases, including atherosclerosis and in-stent restenosis.
Keywords: Vascular smooth muscle cells, HDAC, cell cycle, proliferation, cyclin D1
The majority of cardiovascular diseases are the result of vascular remodeling underlying the development of atherosclerosis and its occlusive complications.1 In addition to endothelial dysfunction and macrophage-derived inflammation, proliferation of SMC constitutes an essential component for atherosclerosis formation and neointimal remodeling.2 Vascular SMC resident in the normal arterial wall are quiescent but migrate, proliferate, and secrete extracellular matrix proteins in response to cues released at sites of vascular injury.1 Once luminal obstruction in the course of atherosclerotic remodeling occurs, revascularization procedures represent primary treatment strategies.3 Unfortunately, the clinical success of these procedures is limited by postangioplasty restenosis, transplant vasculopathy, and coronary artery bypass graft failure; and SMC proliferation has been attributed a critical role in the pathophysiology of these treatment failures.1–2 Therefore, understanding the molecular mechanisms governing SMC proliferation may yield valuable insights into the pathogenesis of all major vascular diseases that display detrimental SMC phenotype behavior.
Expression profiling of SMC has revealed a multitude of genes that are transcriptionally regulated in response to mitogenic stimulation, which suggests that there is an upper level of regulation in SMC controlling entire gene expression programs rather than a single gene.4 Transcriptional gene activation involves dynamic chromatin remodeling and histone modifications mandatory either for restricting or permitting binding of transcription factors and assembly of functional pre-initiation complexes.5 This hierarchic upper-level of chromatin remodeling provides an additional layer of transcriptional control beyond those associated with variation in the sequence of RNA or DNA itself and is referred to as epigenetic mechanism of gene regulation.6
Modification of histones by acetylation, regulated by the balance of histone acetyltransferases (HAT) and histone deacetylases (HDAC), results in the loosening of histone-DNA contacts making DNA more accessible for transcription and gene activation.7 Pharmacological HDAC inhibitors have been developed and shown to induce growth arrest and apoptosis in cancer cells, indicating that the net effect of histone acetylation is an inhibition of cell proliferation.8 Although surprisingly little is known about the epigenetic regulation of vascular gene expression programs, recent studies have indicated that histone acetylation regulates SMC differentiation,9 endothelial cell proliferation,10 and inflammatory pathways,11 processes that are critical for cardiovascular disease development. In the current study, we report that HDAC expression is increased in response to mitogenic stimulation of SMC and required for proliferation by regulating G1→S phase progression of the cell cycle. Furthermore, our in vivo experiments characterize histone acetylation as a previously unrecognized critical event for neointima formation in response to vascular injury.
Materials and Methods
Cell culture
Rat aortic SMC were isolated and cultured as described previously.12 Cells were serum-deprived in 0.01% fetal bovine serum (FBS, Gibco) for 48 h, pretreated with 6 μM of the broad-spectrum HDAC-Inhibitor Scriptaid13 (Enzo Life Sciences) or DMSO for 30 min followed by stimulation with 10% FBS or 50 ng/ml PDGF as indicated. For all data shown, cells were used between passages 3 and 7, and individual experiments were repeated at least 3 times with different cell preparations.
Cell proliferation assays
SMC proliferation was analyzed by cell counting using a hemocytometer. In addition, proliferation was analyzed by means of cell division using 5(6)-carboxyfluorescein diacetate N-succinimidyl ester (CFSE, Sigma). Cells were labeled with CFSE at 37° C for 10 min and washed. One plate was immediately fixed in 10% formalin and stored in 80 % ethanol for baseline analysis. The remaining cells were split at a density of 6 × 103 cells/cm2 and allowed to adhere overnight. The following day cells were starved for 24 h, pretreated with Scriptaid or DMSO for 30 min and stimulated with 10 % FBS. After an additional 48 h, cells were analyzed by FACS to determine fluorescence of the daughter cells.
Apoptosis-Assay
Serum-deprived SMC were pretreated with Scriptaid or DMSO and stimulated with 10 % FBS. Apoptosis was analyzed after 24h using the Alexa Fluor 488 annexin V/Dead Cell Apoptosis Kit (Invitrogen).
Cell cycle analysis
Serum-deprived SMC were pretreated with Scriptaid or DMSO and stimulated with 10 % FBS or 50 ng/ml PDGF. At baseline and 24 h after stimulation cells were fixed in ice cold 70% ethanol. Cells were incubated in PBS containing 40 μg/ml RNase for 30 min at 37°C and resuspended in PBS containing 50 μg/ml propidium iodide. Cell cycle distribution was analyzed using a FACSCalibursorting system (Becton Dickinson).
RNA isolation and quantitative real-time RT-PCR
RNA was isolated and reverse transcribed as described.12 Quantitative real-time PCR analysis of target gene expression was performed using an iCycler and SYBR Green I system (Bio-Rad) as described.12 Each samplewas analyzed in triplicate and normalized to mRNA expression of the house-keeping gene rpl13a. Primer sequences are available on request.
Western blotting
Western Blotting for protein expression was performed as described previously.12 SMC were harvested in cell lysis buffer, and whole cell proteins were subjected to immunoblotting using the following antibodies: HDAC 1 (5356, Cell Signaling), HDAC 2 (5113, Cell Signaling), HDAC 3 (3949, Cell Signaling), p21 (ab7960, Abcam), p27 (ab7961, Abcam), Cyclin D1 (2922, Cell Signaling), GAPDH (sc-25778, Santa Cruz), Phospho-Rb (Ser807/811) (9308S, Cell Signaling) and Total Rb (554136, BD Pharmingen). Quantification of the Western blots was performedby densitometry and normalization to GAPDH.
HDAC siRNA transfection
SMC were transfected twice during 24 h with 100 nM siRNA against HDAC 1, HDAC 2, HDAC 3 or HDAC 6 (Dharmacon) using Lipofectamine 2000 (Invitrogen). Following serum-deprivation for 24 h, synchronized cells were stimulated with 10 % FBS and used for analysis of cell proliferation or mRNA expression.
Chromatin immunoprecipitation assay (ChIP)
ChIP-assays were performed using the MAGnify ChIP-Kit (Invitrogen) according to the manufacturer’s instructions. Briefly, serum-deprived SMC were pretreated with Scriptaid or DMSO and stimulated with 10 % FBS for two hours. Cells were harvested and soluble chromatin was prepared. Chromatin was immunoprecipitated using an antibody directed against histone H3 acetylated at lysine 9 (9649S, Cell signaling) or control IgG (Invitrogen). Final DNA extractions were PCR-amplified using the following primer pairs that cover the E-box or EGR1 binding sites in the rat cyclin D1 promoter: E-box forward: 5′-CTT CGC CTG TTA CAC AGT TCC TGA AT -3′ and reverse: 5′-ACA CCC TAT ACT TAA GCG GAG AGA ATG -3′; EGR1 forward: 5′-CGG CGA TTT GCA TAT CTA CGA AGG-3′ and reverse: 5′-AAG CCG GGC AGA GAA AAA GGA G-3′.
Endovascular femoral artery guide wire injury
Guide wire endothelial denudation injuries were performed in C57BL/6J mice (The Jackson Laboratory) at 8 weeks of age as described.12 Mice were injected with Scriptaid (3.5 μg/g body weight i.p.) or DMSO the day before injury, on the day of the injury, and every other day thereafter for 28 days. The dose of Scriptaid used in this study was based on the doses used in previous murine studies.14–15 Vessels for analysis of mRNA expression were collected in TRIzol 48 h after injury. The intimal and medialareas were measured after 28 days by computerized morphometry using the Image-Pro Plus 5.0 software (MediaCybernetics) as described.12
Histology
Paraffin sections were stained with an elastic Verhoeff-van Gieson staining kit (Sigma-Aldrich) to visualize the internal and external elastic lamina. Immunohistochemistry for PCNA was performed as described.12 Paraffin sections were incubated with a primary antibody (ab2426, Abcam) followed by incubation with a biotinylated goat anti-rabbit IgG (BA-1000, Vector Laboratories).
Statistics
One-way ANOVA or 2-way ANOVA was used to compare multiple groups as appropriate with Bonferroni’s t test for post hoc analysis. Unpaired t test was performed for statistical analysis of two groups. Data were reported as means ± SEM. P values < 0.05 were considered statistically significant.
Results
Expression of HDAC in response to mitogenic stimulation of SMC
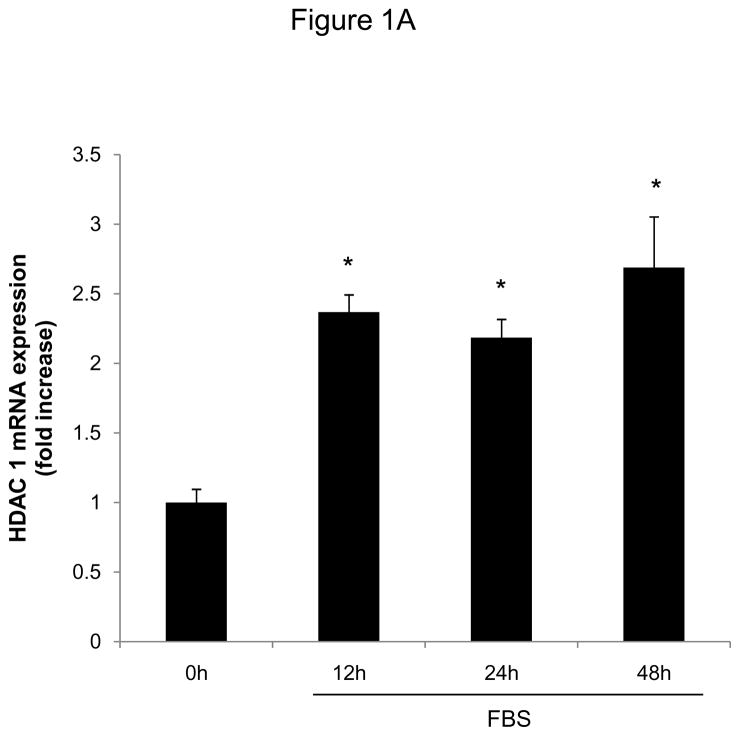
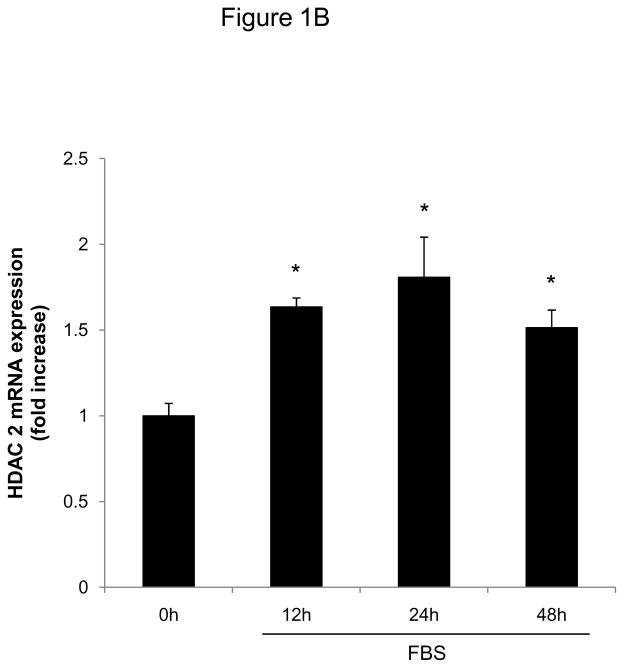
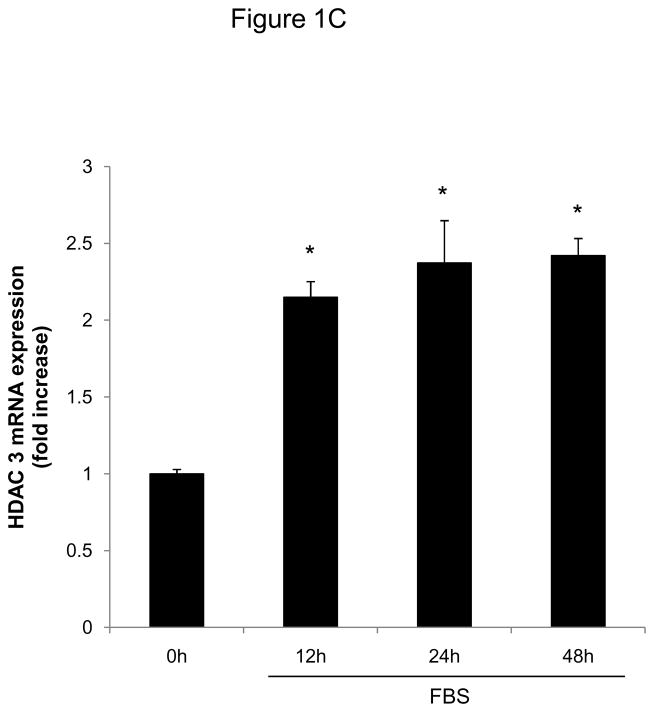
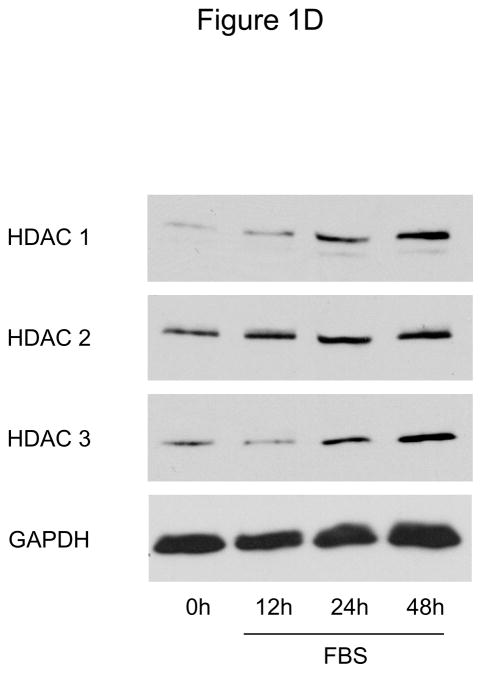
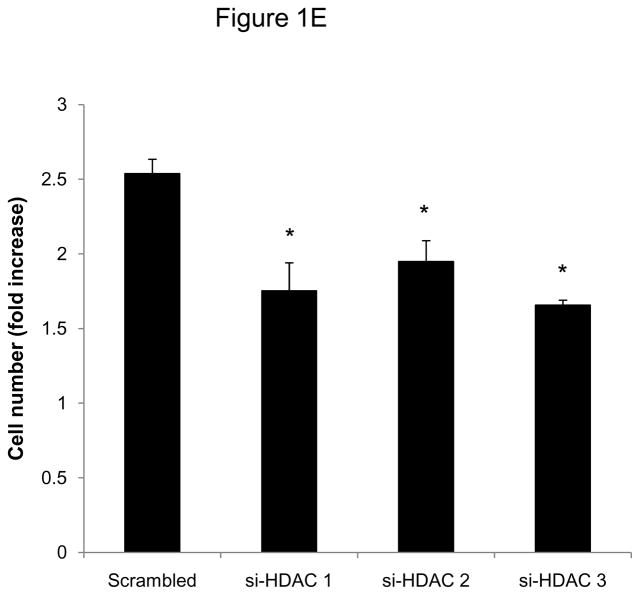
To investigate whether HDAC regulation might modulate SMC proliferation, we first assessed expression levels of HDAC members in primary rat aortic SMC. As depicted in Figure 1A-D, mitogenic stimulation of quiescent SMC resulted in increased transcript and protein levels of the class I HDAC 1, 2, and 3. Considering this mitogenic induction of HDAC 1–3 expression, we hypothesized that HDAC might contribute to the coordinate regulation of SMC proliferation. To further address this question, we performed cell counting and CFSE proliferation assays following siRNA-mediated knock down of class I HDAC members (Supplemental Figure I). As depicted in Figure 1E and Supplemental Figure II, knock-down of either HDAC 1, 2, or 3 reduced proliferation of serum-stimulated SMC. In contrast knock down of HDAC 6, a class II HDAC, did not alter SMC proliferation (Supplemental Figures I and III). These data demonstrate differential functions of HDAC in SMC and establish an essential role of class I HDAC in SMC proliferation.
Figure 1.
Mitogenic stimulation of SMC induces class I HDAC expression. A–D: Serum-deprived rat aortic SMC were stimulated with 10% FBS. HDAC mRNA and protein expression was analyzed at the indicated time points. The autoradiograms shown are representative of three independently performed experiments using different cell preparations. E: Cells were transfected twice with siRNA against HDAC 1, 2, 3 and serum-deprived for 24 h. Following this starvation period, synchronized cells were stimulated with 10 % FBS for 48 h and counted using a hemocytometer. Data is presented as mean ± SEM (* p < 0.05 vs. baseline or scrambled siRNA).
Pharmacological HDAC inhibition prevents mitogen-induced SMC proliferation
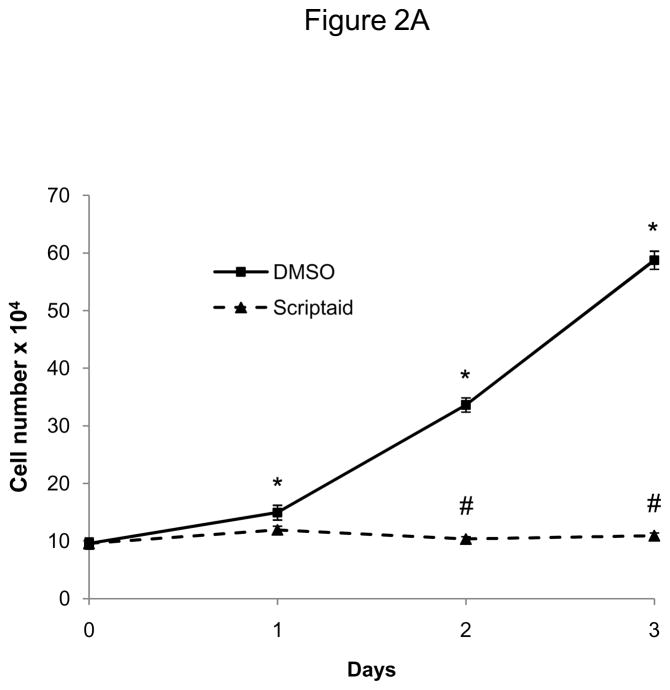
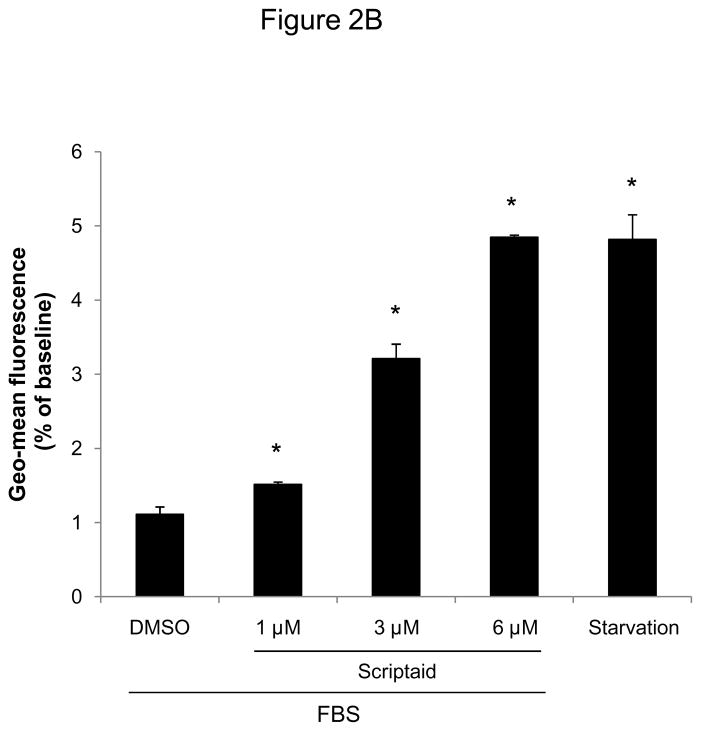
To induce a broad inhibition of HDAC activity, we next employed a pharmacological approach using the HDAC inhibitor Scriptaid.13 Scriptaid, structurally similar to the widely studied HDAC inhibitor trichostatin A (TSA), confers a higher efficiency in inhibiting endogenous histone deacetylation while exhibiting less cytotoxic activity compared to TSA.13 As depicted in Figure 2A, pretreatment of quiescent SMC with Scriptaid completely prevented mitogen-induced cell proliferation. This inhibition of SMC proliferation by Scriptaid was dose-dependent and revealed a maximal inhibition of proliferation at 6 μM, as reflected by the increase in CFSE geo-mean fluorescence (Figure 2B). As illustrated in Supplemental Figure IV, this concentration did not induce apoptosis in SMC and corresponds to the reported maximal HDAC inhibition of more than 100-fold at a dose of 6 μM Scriptaid.13
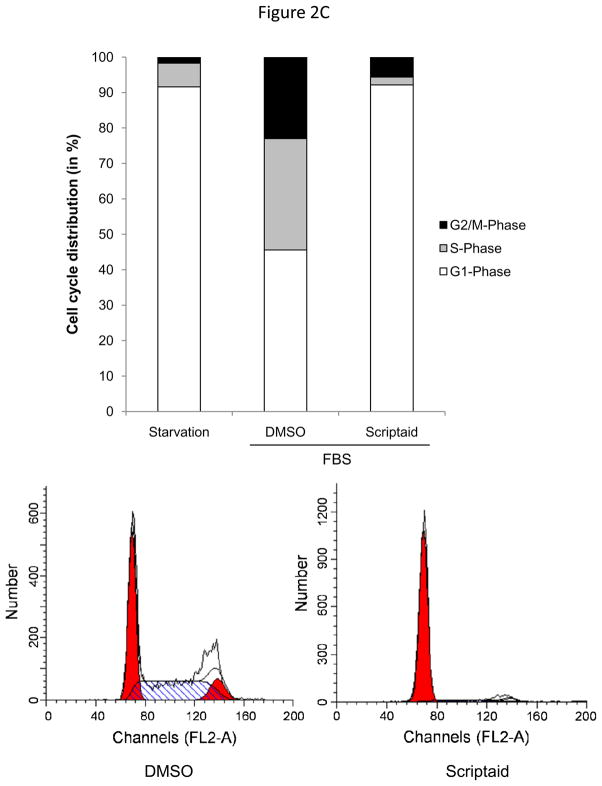
Figure 2.
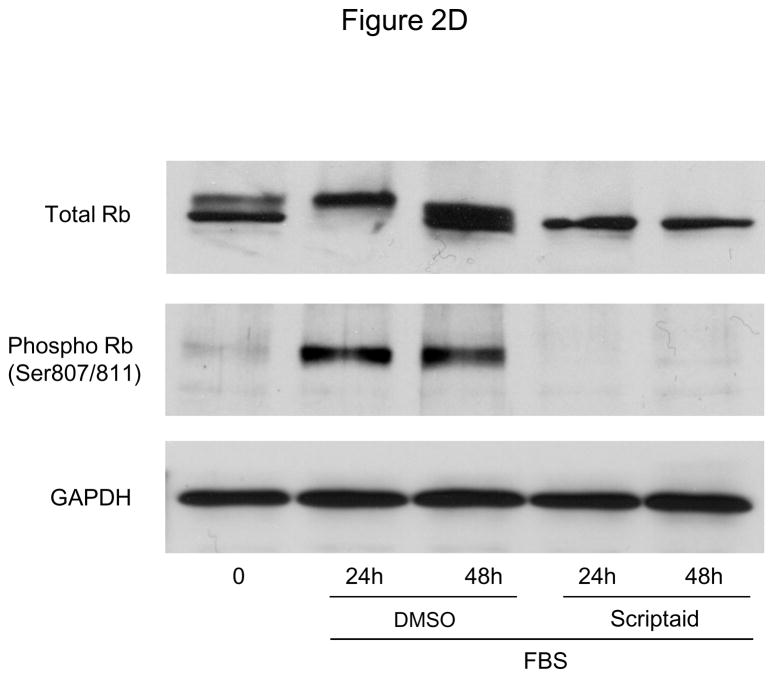
The HDAC inhibitor Scriptaid prevents mitogen-induced SMC proliferation. A: Serum-deprived SMC were pretreated with DMSO or 6 μM Scriptaid, and stimulated with 10% FBS. Cells were counted at indicated time points using a hemocytometer. B: CFSE labeled SMC were starved for 24 h, treated with DMSO or different doses of Scriptaid, and stimulated with 10% FBS. After 48 h cells were analyzed by FACS. C: SMC were treated as described in A. Cell cycle distribution was assessed at baseline and 24 h after FBS stimulation using FACS analysis. D: Cells were stimulated as described in A. Whole cell lysate was collected at the indicated time points and analyzed for protein expression of total Rb, phosphorylated Rb and GAPDH. The autoradiograms shown are representative of three independently performed experiments using different cell preparations. Data is presented as mean ± SEM (* p < 0.05 vs. DMSO).
HDAC activity is required for G1→S phase progression of the cell cycle
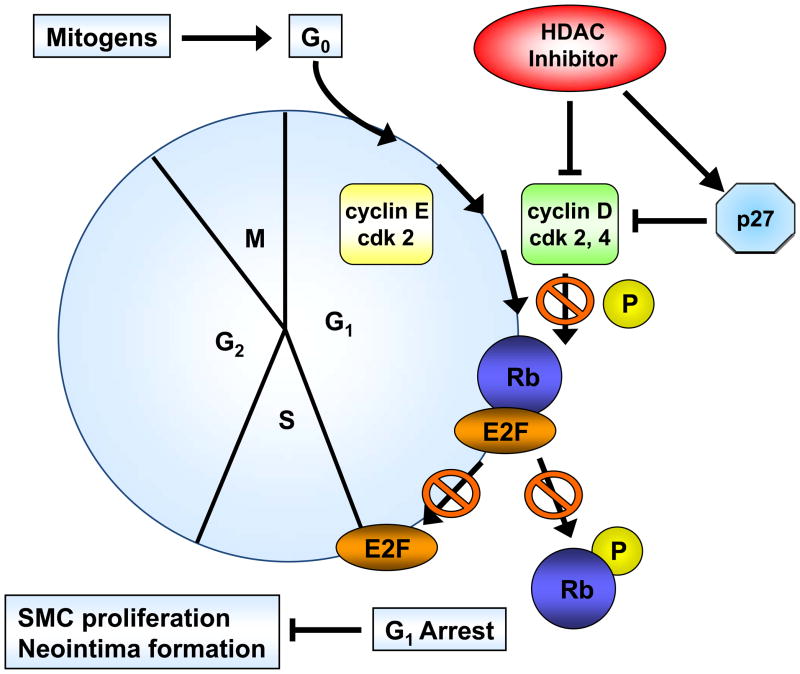
To further determine the mechanisms by which HDAC inhibition prevents SMC proliferation, we next determined cell cycle distribution using flow cytometry. As depicted in Figure 2C, treatment with Scriptaid inhibited mitogen-induced G1→S progression of SMC. A similar G1-arrest was observed following mitogenic stimulation with platelet-derived growth factor (PDGF, Supplemental Figure V). Cell cycle progression through the G1→S phase checkpoint is governed by the retinoblastoma protein (Rb), which is phosphorylated in response to mitogenic stimulation by cyclin-dependent kinase (CDK)/cyclin complexes allowing cells to enter S phase.16 Consistent with the G0/G1 arrest induced by HDAC inhibition, Scriptaid treatment completely prevented mitogen-induced phosphorylation of Rb (Figure 2D). Collectively, these findings demonstrate that Scriptaid inhibits SMC proliferation by preventing mitogen-induced Rb phosphorylation and subsequent cell cycle progression.
HDAC inhibition modifies the expression of cyclin-dependent kinase inhibitors
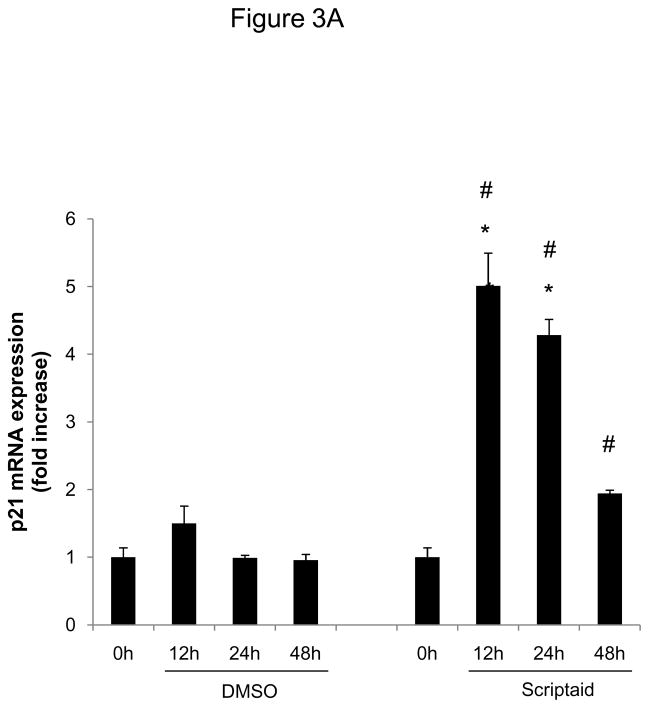
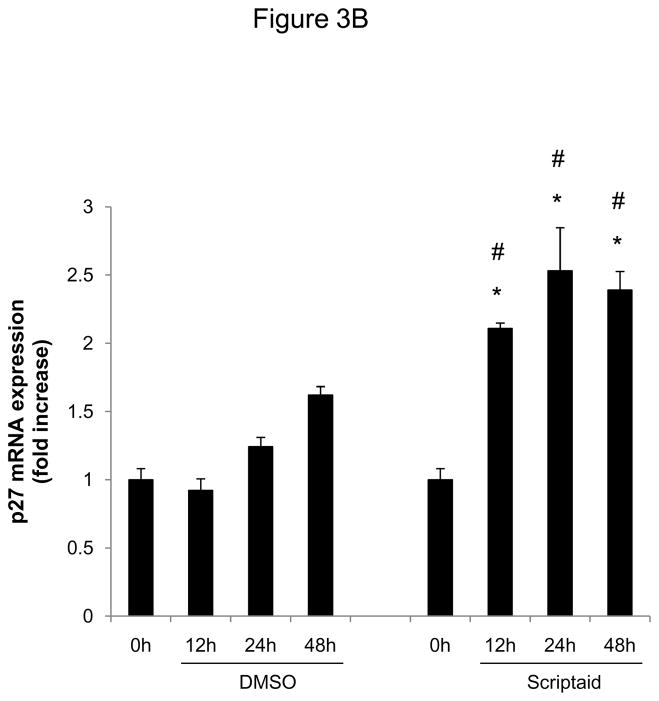
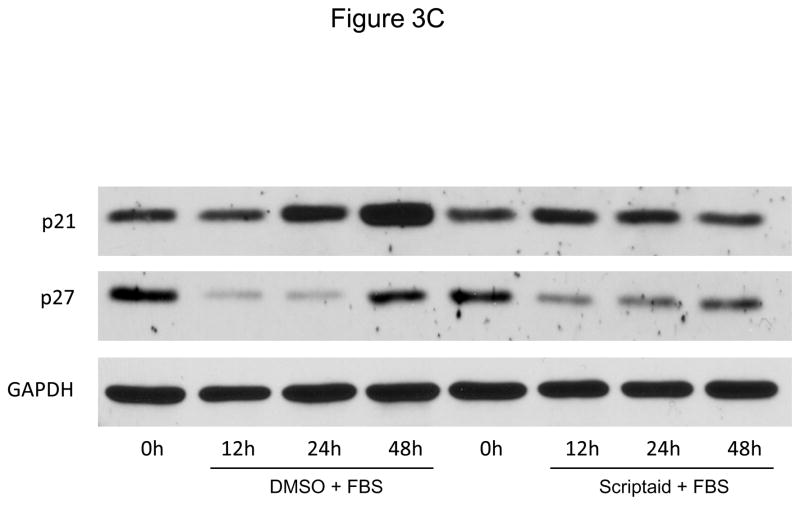
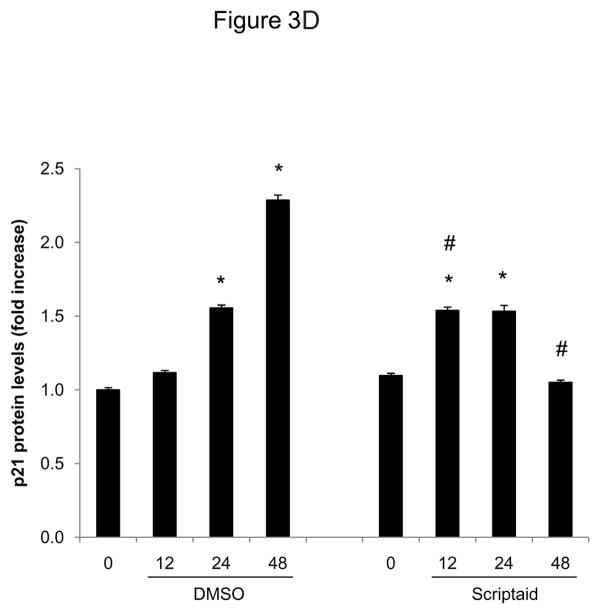
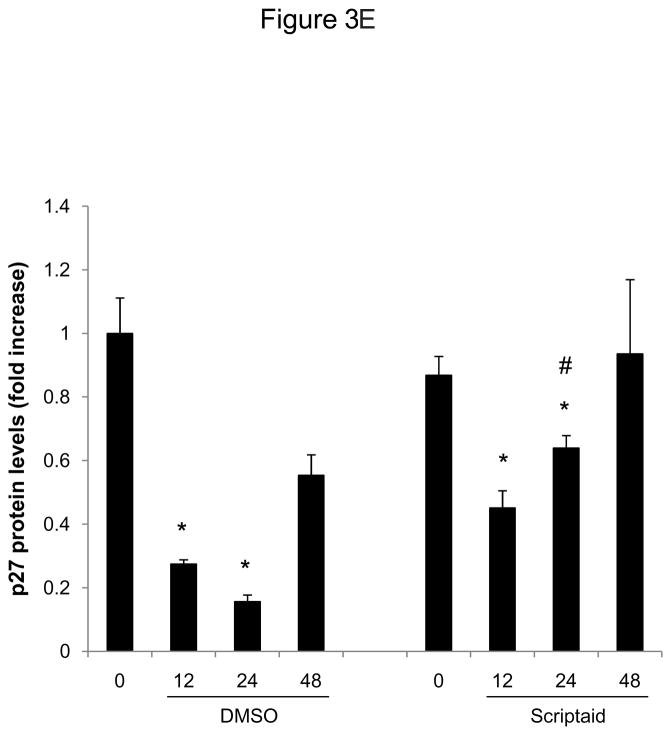
Considering the profound inhibition of Rb phosphorylation by Scriptaid in SMC, we next investigated whether HDAC inhibition modulates the expression of cyclin-dependent kinase inhibitors (CDKI) p21Cip1 and p27Kip1, which negatively regulate the activity of CDK/cyclin complexes and exhibit growth-inhibitory effects on SMC.17 Analysis of gene expression profiles revealed that Scriptaid treatment increased mRNA expression levels of p21Cip1 and p27Kip (Figure 3A and B). Albeit this transcriptional induction, HDAC inhibition attenuated the increase in p21Cip1 protein levels observed in response to mitogenic stimulation (Figure 3C). Furthermore, mitogen-induced downregulation of p27Kip1 protein levels was slightly altered in SMC treated with Scriptaid (Figure 3C). Collectively, these observations demonstrate increased transcript levels of the CDKI p21Cip1 and p27Kip in response to Scriptaid treatment and point to a previously unrecognized translational block of p21Cip1 transcription by HDAC inhibition.
Figure 3.
HDAC inhibition modifies the expression of cyclin-dependent kinase inhibitors. A–C: Serum-deprived SMC were treated with DMSO or 6 μM Scriptaid, and stimulated with 10% FBS. Cells were harvested at the indicated time points and analyzed for mRNA and protein expression of p21Cip1 and p27Kip1 by real-time RT-PCR (A–B) and Western blotting (C). Quantification of the Western blotting experiments was performed by densitometry and normalization to GAPDH from three independently performed experiments. Data is presented as mean ± SEM (* p < 0.05 vs. baseline, # p < 0.05 vs. DMSO).
HDAC inhibition represses cyclin D1 transcription
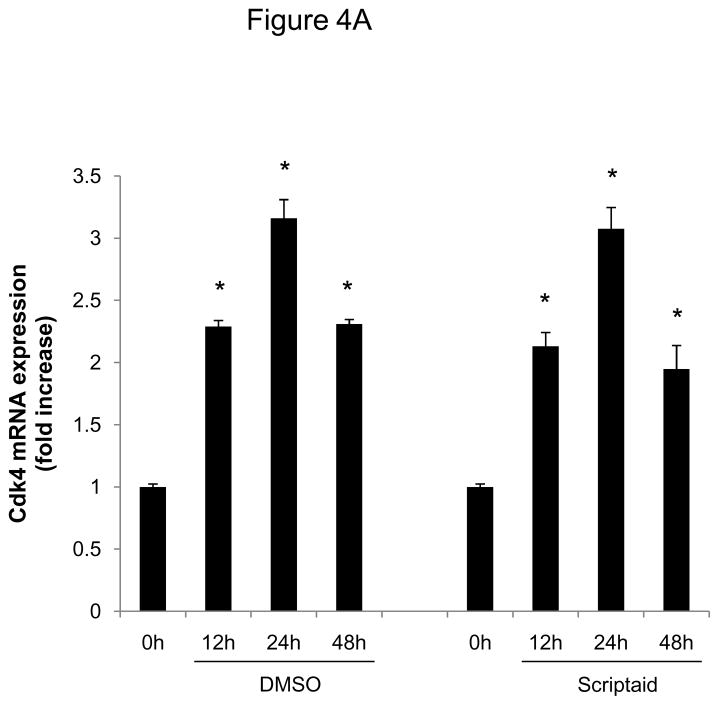
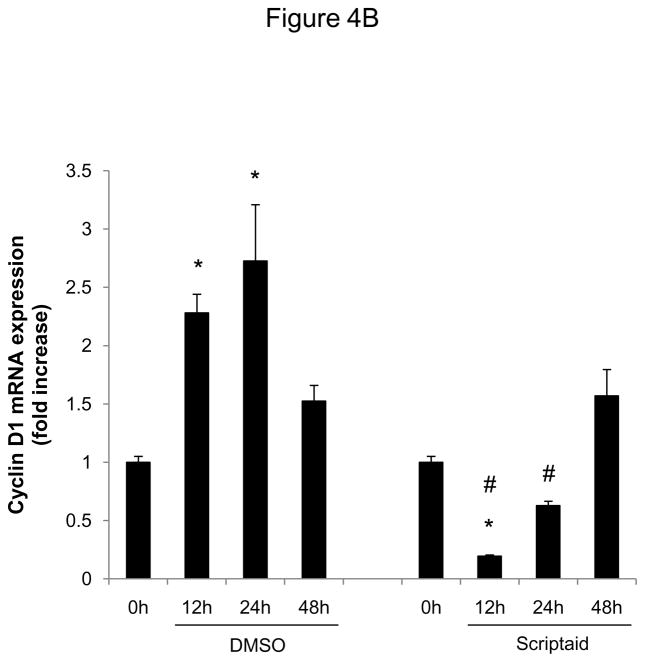
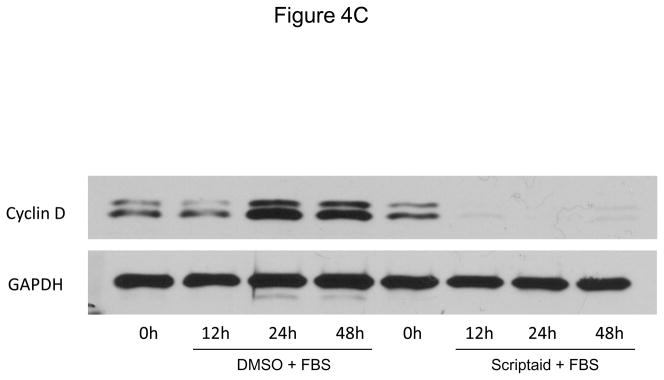
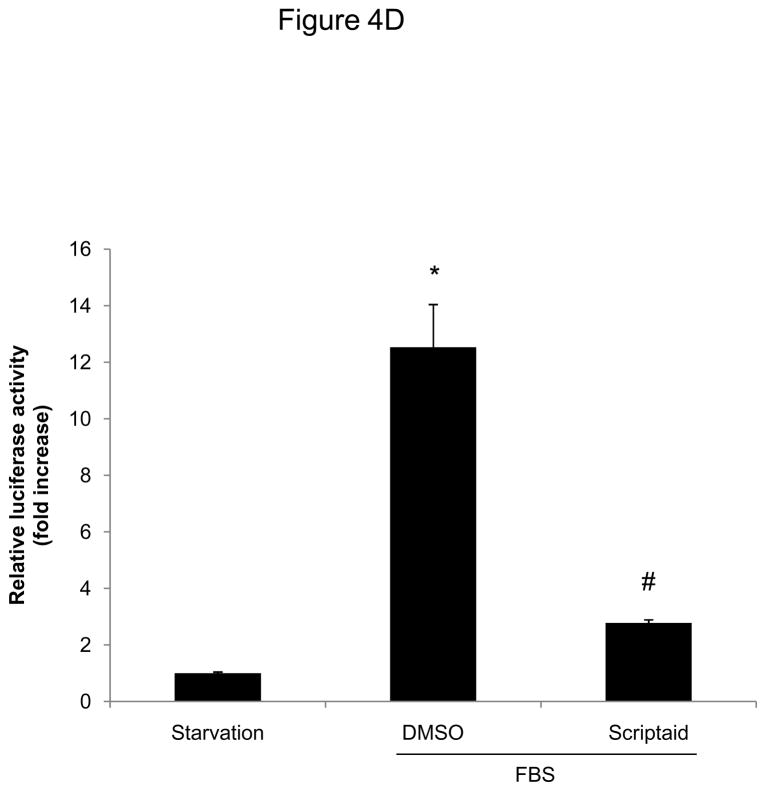
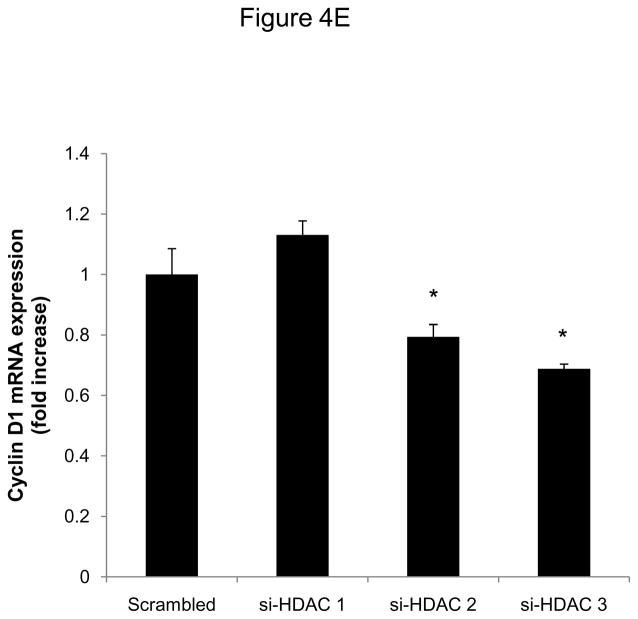
We next focused our analysis on the regulation of the CDK4/cyclin D1 complex by Scriptaid, the critical complex phosphorylating Rb. The induction of CDK4 mRNA expression in response to mitogenic stimulation was not modulated by HDAC inhibition (Figure 4A). In contrast, Scriptaid treatment profoundly repressed mitogen-induced cyclin D1 mRNA expression (Figure 4B and Supplemental Figure VI), despite a considerable increase in lysine 9 histone H3 acetylation at the cyclin D1 promoter (Supplemental Figure VII). This inhibition of cyclin D1 mRNA expression by Scriptaid translated into a complete prevention of mitogen-induced cyclin D1 protein expression by HDAC inhibition (Figure 4C). Transient transfection experiments further confirmed a solid inhibition of mitogen-induced cyclin D1 promoter activity in response to Scriptaid (Figure 4D). To investigate the specific contributions of class I HDAC to the observed inhibition of cyclin D1 transcription, SMC were transfected with siRNA against HDAC 1–3. As depicted in Figure 4E, knock down of HDAC 2 and 3 significantly reduced cyclin D1 mRNA levels. In concert, these data indicate that HDAC inhibition silences cyclin D1 expression through a transcriptional mechanism acting on the gene promoter, an effect that results in decreased Rb phosphorylation and cell cycle progression.
Figure 4.
HDAC inhibition represses cyclin D1 transcription. A and B: Serum-deprived SMC were treated with DMSO or 6 μM Scriptaid, and stimulated with 10% FBS. Cells were harvested at indicated time points and analyzed for mRNA expression of Cdk4 (A) and cyclin D1 (B). C: Cells were treated as described in A. Whole cell lysate was collected and analyzed for protein expression of cyclin D1. D: SMC were transfected with a cyclin D1 promoter luciferase reporter plasmid. Following transfection, cells were treated with DMSO or 6 μM Scriptaid in 10% FBS. Luciferase activity was assayed after 24 h. Transfection efficiency was adjusted by normalizing firefly luciferase activities to renilla luciferase activities generated by cotransfection with pRL-CMV. E: Cells were transfected twice with siRNA against HDAC 1, 2, 3 and serum-deprived for 24 h. Following this starvation period, synchronized cells were stimulated with 50 ng/ml PDGF and mRNA expression of cyclin D1 was analyzed after 6h. Data is presented as mean ± SEM (* p < 0.05 vs. baseline or scrambled siRNA, # p < 0.05 vs. DMSO).
Inhibition of HDAC activity alters E2F target gene expression
Phosphorylation of Rb releases its repressive function on the transcription factor E2F and allows transactivation of gene promoters that drive cells into S phase.16 To validate the notion that histone deacetylase activity operates to promote this transcriptional cascade at the G1→S transition, we next analyzed downstream Rb/E2F target gene expression, including cyclin A and MCM6. As predicted, the inhibition of Rb phosphorylation in response to HDAC inhibition resulted in a solid silencing of the downstream bona fide E2F target genes MCM6 and cyclin A (Supplemental Figure VIII and IX). These data indicate that the inhibition of Rb phosphorylation by Scriptaid translates into a downstream stable repression of E2F target genes.
HDAC inhibition reduces neointima formation following vascular injury
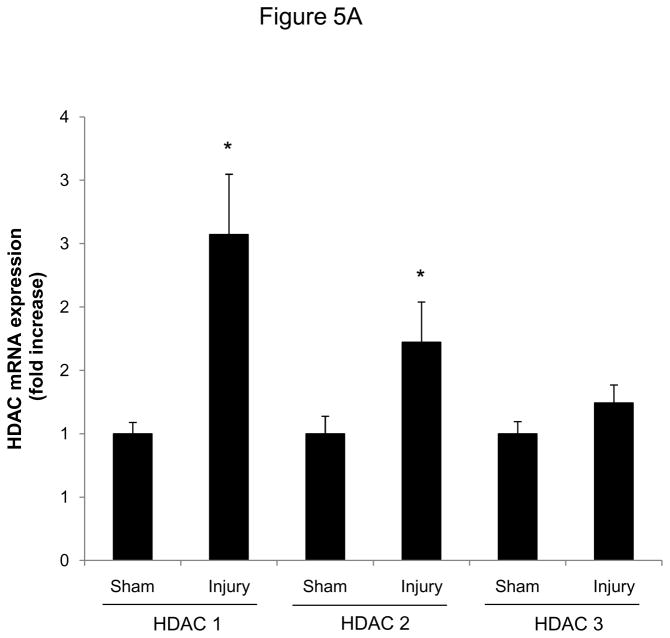
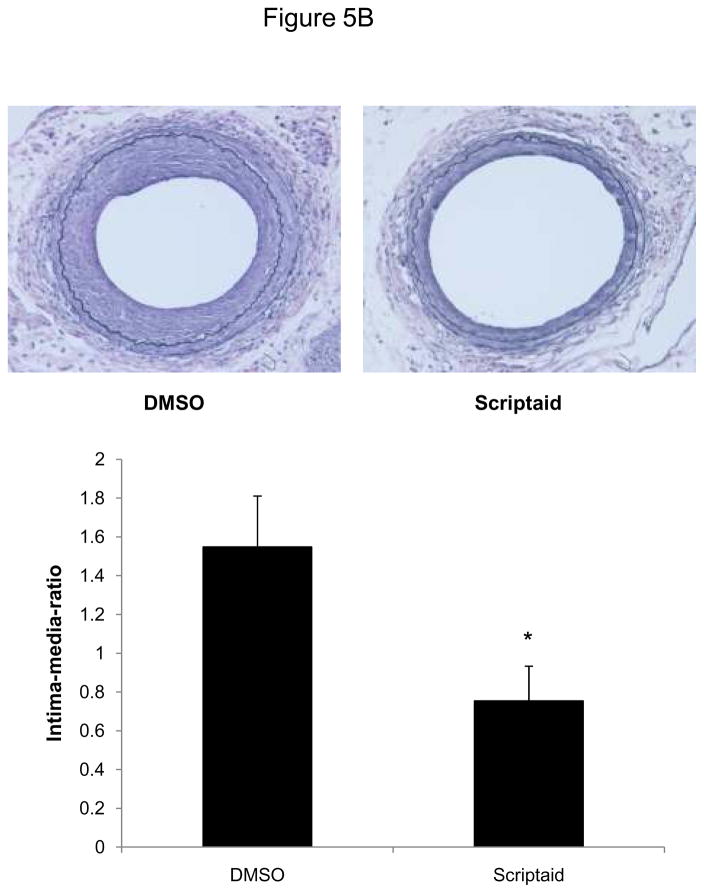
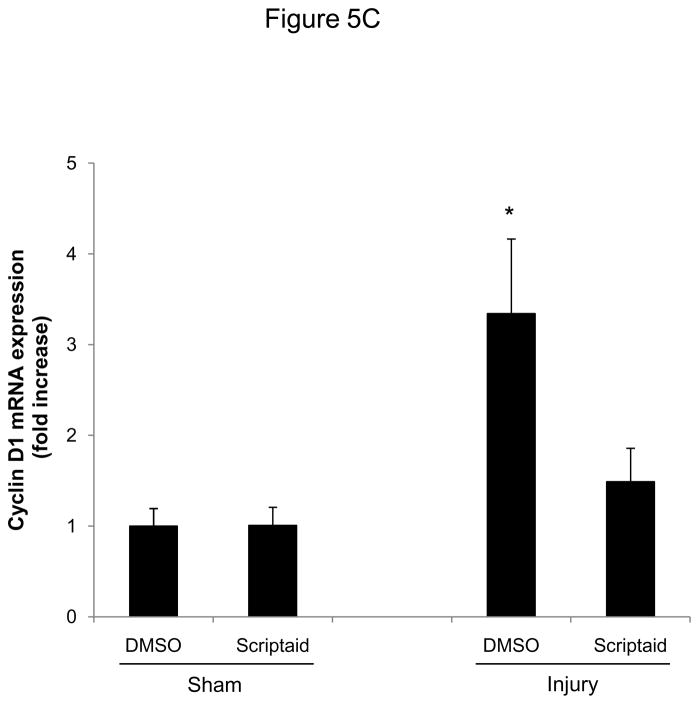
Finally, we analyzed HDAC expression in mice following vascular injury and tested whether the observed attenuation of SMC proliferation by HDAC inhibition is applicable in vivo. Consistent with our in vitro studies, expression of class I HDAC increased in femoral arteries following vascular injury (Figure 5A). Next, we treated mice with Scriptaid and performed an endothelial denudation injury. The treatment dose of Scriptaid in these experiments was based on prior studies in mice and results in a solid inhibition of HDAC in vivo without toxicity.14–15 As depicted in Figure 5B, endovascular injury of the mouse femoral artery resulted in a concentric neointima formation. In contrast, treatment with Scriptaid considerably attenuated neointima formation, and morphometric quantification revealed an approximately 60% reduction in the intima-to-media ratio following HDAC inhibition (Figure 5B and Supplemental Table I). To recapitulate our in vitro findings, we analyzed cyclin D1 expression in femoral arteries of DMSO and Scriptaid-treated mice following vascular injury. As demonstrated in Figure 5C, HDAC inhibition significantly decreased injury-induced cyclin D1 mRNA expression. Finally, to assess SMC proliferation in vivo we performed immunohistochemical staining of PCNA. As depicted in representative stainings in Figure 5D, Scriptaid treatment reduced neointimal PCNA expression following vascular injury. These findings establish that HDAC inhibition in mice prevents proliferative remodeling in response to endovascular injury.
Figure 5.
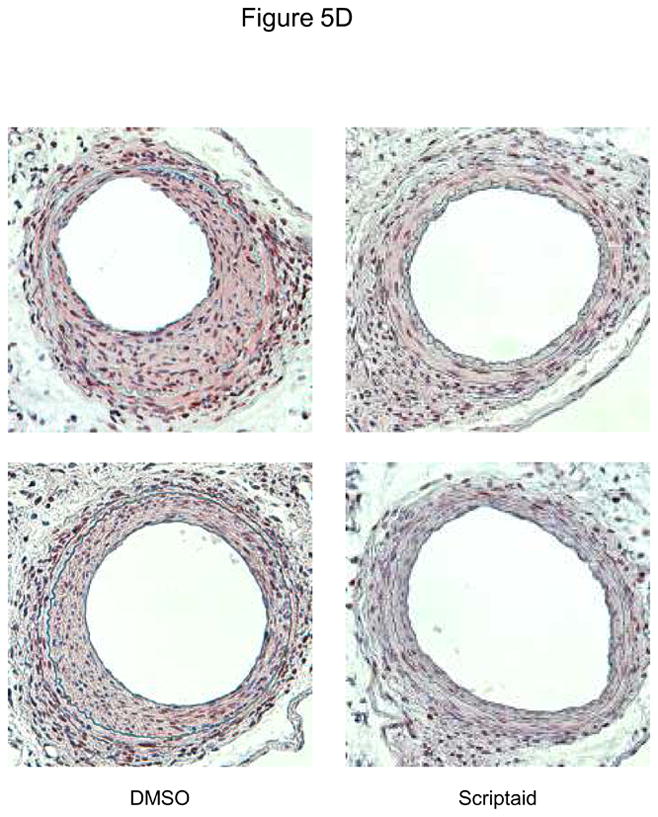
HDAC inhibition reduces neointima formation following vascular injury. A: Guide wire-induced endothelial denudation injuries were performed on the left femoral artery of C57BL/6J mice (n = 4). mRNA expression of HDAC 1–3 was analyzed after 48 h. B: Following endothelial denudation injuries, mice were treated with Scriptaid or DMSO for 28 days (n = 9 each group). Tissues were harvested and elastic Verhoeff-van Gieson stain was performed to visualize the internal and external elastic lamina. The ratio of intima to media was calculated as the intimal area divided by medial area. C: Cyclin D1 mRNA expression was analyzed 48 after endothelial denudation injuries (each group n = 4) or sham surgery (each group n = 3) and treatment with Scriptaid or DMSO. D: Representative immunohistochemical stainings of PCNA expression. Tissues were harvested after 28 days of Scriptaid or DMSO treatment. Data is presented as mean ± SEM (* p < 0.05 vs. baseline)
Discussion
Epigenetic control of gene expression constitutes a conserved regulatory layer of transcriptional control that orchestrates important cellular processes, including proliferation, differentiation, and inflammation.9–11 However, whether epigenetic modifications of chromatin accessibility operate to modulate SMC proliferation remains elusive. In the present study, we report that HDAC expression is increased in response to mitogenic stimulation of SMC and required for proliferation by regulating G1→S phase progression of the cell cycle. Furthermore, our in vivo experiments characterize histone acetylation as a previously unrecognized critical event for neointima formation in response to vascular injury.
During the course of atherosclerotic lesion formation, pathways activated by growth factors and cytokines ultimately converge to the transcriptional modulation of cell cycle-regulatory genes promoting proliferation of SMC.1 In our study, we illustrate that class I HDAC represent a group of mitogen-responsive genes in SMC that are upregulated at a transcriptional level. Functional experiments established that siRNA-mediated knock-down of class I HDAC 1–3 reduces SMC proliferation, illustrating a novel mitogenic function for HDAC activity in SMC proliferation. These observations are consistent with previous reports documenting an induction of HDAC 1 in different murine cell types in response to growth factors,18 and reduced proliferation of tumor cells and embryonic stem cells following knock-down or genetic knockout of class I HDAC.19–21
Inhibitors of HDAC stabilize the acetylation of histone proteins and have been primarily studied in cancer cells leading to their investigation in clinical trials.22 One of the best described events following HDAC inhibitor treatment is the induction of growth arrest in G1- and/or G2/M-phase of the cell cycle.8, 23 In our study, treatment of quiescent SMC with the HDAC inhibitor Scriptaid induced G1 arrest following mitogenic stimulation. The molecular mechanisms by which histone acetylation serves its mitogenic function are likely multifactorial and involve transcriptional events orchestrating the activation of a mitogenic gene expression program. In SMC, phosphorylation of Rb by CDK/cyclin complexes was completely abolished by HDAC inhibition resulting in decreased expression of downstream E2F target genes. Considering the complete prevention of mitogen-induced Rb phosphorylation by HDAC inhibition, we focused our initial mechanistic studies on the upstream CDKI and CDK/cyclin complexes, which mediate the phosphorylation of Rb during cell cycle progression. Transcript levels of p21CIP1, the CDKI upregulated by HDAC inhibitors in most cell types,8, 24 were induced by Scriptaid treatment in SMC. However, this transcriptional induction of p21CIP1 mRNA did not translate to increased protein levels, pointing to previously unrecognized translational or posttranslational regulation of p21CIP1 protein by HDAC inhibition in SMC. Considering that induced p21CIP1 limits SMC proliferation,25 this finding indicates that the inhibition of SMC proliferation by histone acetylation likely operates independent of p21CIP1. In several prior studies the growth arrest induced by HDAC inhibition has been attributed to the induction of p21CIP1.8, 24 Similarly, Okamoto et al. have noted an induction of p21CIP1 protein in rat SMC by TSA treatment and suggested a p21CIP1-dependent growth arrest.26 However, our data is more consistent with a recent study by Wilting et al. using a genetic model of HDAC 1 and HDAC 2 deficiency.27 In this compelling study, it was confirmed that the growth arrest induced by histone acetylation occurs independently of p21CIP1.27 A potential explanation for this discrepancy might be differences in experimental design and, more likely, the less potent inhibition of HDAC by the frequently used TSA in these studies.13 Finally, the CDKI p27Kip was mildly upregulated at a transcriptional level and its decrement following mitogenic stimulation was modestly prevented in response to HDAC inhibition in SMC. Since p27Kip has previously been described to be upregulated by HDAC inhibition and to inhibit SMC proliferation, a contribution of p27Kip to the growth arrest induced by HDAC inhibition in SMC may be conceivable.17, 28 However, collectively, neither the moderate induction of p27Kip by Scriptaid nor the block of mitogen-induced p21CIP protein expression can explain the profound inhibition of SMC proliferation observed by HDAC inhibition in our experiments.
The CDK4/cyclin D1 complex initiates the phosphorylation-dependent inactivation of the retinoblastoma protein allowing S phase entry and cell proliferation.29 As expected, HDAC inhibition resulted in an increase of lysine 9 histone H3 acetylation at the cyclin D1 promoter. This histone modification has been shown to correlate with activation of gene expression.7 However, in SMC HDAC inhibition completely abolished the transcriptional induction of cyclin D1 in the early G1 phase. Transient transfection experiments suggest that HDAC inhibition silences cyclin D1 expression through a transcriptional mechanism. Consistent with this observation in SMC, an inhibition of cyclin D1 mRNA and protein expression by HDAC inhibitor treatment has been observed in cancer cells and certain somatic cell types.30–32 The transcriptional repression of cyclin D1 transcription in cancer cell types appears to be stimulus-dependent and involves an inhibition of either a NF-κB or AP-1-dependent activation of the cyclin D1 promoter.30–31 HDAC inhibition impairs the binding of NF-κB through post-translational modifications including the acetylation of the NF-κB subunit p52.31 In contrast AP-1-dependent activation of the cyclin D1 promoter is inhibited through impaired recruitment of the preinitiation complex to the c-jun promoter resulting in reduced AP-1 protein levels.30 Following siRNA-mediated knock down of class I HDAC, we observed the largest decrease in cyclin D1 expression using siRNA against HDAC 3. Interestingly, the repression of c-jun transcription by HDAC inhibition was also conferred through HDAC 3.30 Both the NF-κB and AP-1 transcriptional signaling pathways are activated during the mitogenic response of SMC, and CDK4/cyclin D1 activity is induced during neointima formation.33–35 Considering further that overexpression of cyclin D1 induces SMC proliferation,36 we would infer that the silencing of cyclin D1 expression by histone acetylation contributes at least in part to the inhibition of SMC proliferation by the HDAC inhibitor Scriptaid.
Using a murine model of endovascular endothelial denudation,37 HDAC inhibition reduced cyclin D1 and PCNA expression and neointima formation in response to vascular injury. This observation confirms that the inhibition of SMC proliferation by histone acetylation noted in SMC culture systems is applicable to proliferative vascular remodeling in vivo. Furthermore, this anti-proliferative activity of HDAC inhibition in the vascular wall is consistent with the well-established induction of growth arrest and associated reduction in tumor growth in various species treated with HDAC inhibitors.38–39 Interestingly, in further support of the reduction of neointima formation by HDAC inhibition in the present study, the HDAC inhibitor tributyrin was previously found to inhibit SMC migration,40 a critical early event for pathological vascular remodeling.1 Therefore, in addition to an inhibition of mitogen-induced proliferation, the HDAC inhibitor Scriptaid may prevent neointima formation by limiting SMC migratory responses.
In summary, we demonstrate in the present study that class I HDAC serve a mitogenic role in SMC. HDAC inhibition induces cell cycle arrest by preventing mitogen-induced Rb phosphorylation. This inhibition of Rb phosphorylation occurs through a differential regulation of cell cycle regulatory genes orchestrating the G1→S phase transition checkpoint, including a transcriptional repression of cyclin D1. Finally, we establish that HDAC inhibition limits the proliferative response underlying neointima formation. These findings identify HDAC as a critical component of a transcriptional cascade regulating SMC proliferation and suggest that HDAC might play a pivotal role in the development of proliferative vascular diseases, including atherosclerosis and in-stent restenosis.
Supplementary Material
Figure 6.
HDAC inhibition targets cell cycle progression. Cell cycle progression is dependent on the orchestrated expression, activation and holoenzyme formation of cyclins and cyclin-dependent kinases (CDKs). HDAC inhibition blocks cell cycle progression through repression of cyclin D1. Impaired activation of cyclin–CDK complexes inhibits mitogen-induced Rb phosphorylation and downstream activation of E2F-regulated genes, resulting in G1 arrest and reduced SMC proliferation.
Acknowledgments
none
Sources of Funding - These studies were supported in part by the National Institutes of Health/National Center for Research Resources Centers of Biomedical Research Excellence in Obesity and Cardiovascular Disease Grant P20 RR021954.
Footnotes
Disclosure - The authors have nothing to disclose that could be perceived as real or apparent conflict(s) of interest.
References
- 1.Dzau VJ, Braun-Dullaeus RC, Sedding DG. Vascular proliferation and atherosclerosis: New perspectives and therapeutic strategies. Nat Med. 2002;8:1249–1256. doi: 10.1038/nm1102-1249. [DOI] [PubMed] [Google Scholar]
- 2.Doran AC, Meller N, McNamara CA. Role of smooth muscle cells in the initiation and early progression of atherosclerosis. Arterioscler Thromb Vasc Biol. 2008;28:812–819. doi: 10.1161/ATVBAHA.107.159327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moses JW, Leon MB, Popma JJ, Fitzgerald PJ, Holmes DR, O’Shaughnessy C, Caputo RP, Kereiakes DJ, Williams DO, Teirstein PS, Jaeger JL, Kuntz RE the SIRIUS Investigators. Sirolimus-eluting stents versus standard stents in patients with stenosis in a native coronary artery. N Engl J Med. 2003;349:1315–1323. doi: 10.1056/NEJMoa035071. [DOI] [PubMed] [Google Scholar]
- 4.Zhang QJ, Goddard M, Shanahan C, Shapiro L, Bennett M. Differential gene expression in vascular smooth muscle cells in primary atherosclerosis and in stent stenosis in humans. Arterioscler Thromb Vasc Biol. 2002;22:2030–2036. doi: 10.1161/01.atv.0000042206.98651.15. [DOI] [PubMed] [Google Scholar]
- 5.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 6.Bernstein BE, Meissner A, Lander ES. The mammalian epigenome. Cell. 2007;128:669–681. doi: 10.1016/j.cell.2007.01.033. [DOI] [PubMed] [Google Scholar]
- 7.Jenuwein T, Allis CD. Translating the histone code. Science. 2001;293:1074–1080. doi: 10.1126/science.1063127. [DOI] [PubMed] [Google Scholar]
- 8.Sambucetti LC, Fischer DD, Zabludoff S, Kwon PO, Chamberlin H, Trogani N, Xu H, Cohen D. Histone deacetylase inhibition selectively alters the activity and expression of cell cycle proteins leading to specific chromatin acetylation and antiproliferative effects. J Biol Chem. 1999;274:34940–34947. doi: 10.1074/jbc.274.49.34940. [DOI] [PubMed] [Google Scholar]
- 9.McDonald OG, Wamhoff BR, Hoofnagle MH, Owens GK. Control of srf binding to carg box chromatin regulates smooth muscle gene expression in vivo. J Clin Invest. 2006;116:36–48. doi: 10.1172/JCI26505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Margariti A, Zampetaki A, Xiao Q, Zhou B, Karamariti E, Martin D, Yin X, Mayr M, Li H, Zhang Z, De Falco E, Hu Y, Cockerill G, Xu Q, Zeng L. Histone deacetylase 7 controls endothelial cell growth through modulation of {beta}-catenin. Circ Res. 106:1202–1211. doi: 10.1161/CIRCRESAHA.109.213165. [DOI] [PubMed] [Google Scholar]
- 11.Miao F, Gonzalo IG, Lanting L, Natarajan R. In vivo chromatin remodeling events leading to inflammatory gene transcription under diabetic conditions. J Biol Chem. 2004;279:18091–18097. doi: 10.1074/jbc.M311786200. [DOI] [PubMed] [Google Scholar]
- 12.Nomiyama T, Zhao Y, Gizard F, Findeisen HM, Heywood EB, Jones KL, Conneely OM, Bruemmer D. Deficiency of the nr4a neuron-derived orphan receptor-1 attenuates neointima formation after vascular injury. Circulation. 2009;119:577–586. doi: 10.1161/CIRCULATIONAHA.108.822056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Su GH, Sohn TA, Ryu B, Kern SE. A novel histone deacetylase inhibitor identified by high-throughput transcriptional screening of a compound library. Cancer Res. 2000;60:3137–3142. [PubMed] [Google Scholar]
- 14.Keen JC, Yan L, Mack KM, Pettit C, Smith D, Sharma D, Davidson NE. A novel histone deacetylase inhibitor, scriptaid, enhances expression of functional estrogen receptor alpha (er) in er negative human breast cancer cells in combination with 5-aza 2′-deoxycytidine. Breast Cancer Res Treat. 2003;81:177–186. doi: 10.1023/A:1026146524737. [DOI] [PubMed] [Google Scholar]
- 15.Kong Y, Tannous P, Lu G, Berenji K, Rothermel BA, Olson EN, Hill JA. Suppression of class i and ii histone deacetylases blunts pressure-overload cardiac hypertrophy. Circulation. 2006;113:2579–2588. doi: 10.1161/CIRCULATIONAHA.106.625467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Weinberg RA. E2f and cell proliferation: A world turned upside down. Cell. 1996;85:457–459. doi: 10.1016/s0092-8674(00)81244-1. [DOI] [PubMed] [Google Scholar]
- 17.Tanner FC, Boehm M, Akyurek LM, San H, Yang Z-Y, Tashiro J, Nabel GJ, Nabel EG. Differential effects of the cyclin-dependent kinase inhibitors p27kip1, p21cip1, and p16ink4 on vascular smooth muscle cell proliferation. Circulation. 2000;101:2022–2025. doi: 10.1161/01.cir.101.17.2022. [DOI] [PubMed] [Google Scholar]
- 18.Bartl S, Taplick J, Lagger G, Khier H, Kuchler K, Seiser C. Identification of mouse histone deacetylase 1 as a growth factor- inducible gene. Mol Cell Biol. 1997;17:5033–5043. doi: 10.1128/mcb.17.9.5033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Senese S, Zaragoza K, Minardi S, Muradore I, Ronzoni S, Passafaro A, Bernard L, Draetta GF, Alcalay M, Seiser C, Chiocca S. A role for histone deacetylase 1 in human tumor cell proliferation. Mol Cell Biol. 2007:MCB.00494–00407. doi: 10.1128/MCB.00494-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lagger G, O’Carroll D, Rembold M, Khier H, Tischler J, Weitzer G, Schuettengruber B, Hauser C, Brunmeir R, Jenuwein T, Seiser C. Essential function of histone deacetylase 1 in proliferation control and cdk inhibitor repression. Embo J. 2002;21:2672–2681. doi: 10.1093/emboj/21.11.2672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Glaser KB, Li J, Staver MJ, Wei RQ, Albert DH, Davidsen SK. Role of class I and class II histone deacetylases in carcinoma cells using sirna. Biochem Biophys Res Commun. 2003;310:529–536. doi: 10.1016/j.bbrc.2003.09.043. [DOI] [PubMed] [Google Scholar]
- 22.Ma X, Ezzeldin HH, Diasio RB. Histone deacetylase inhibitors: Current status and overview of recent clinical trials. Drugs. 2009;69:1911–1934. doi: 10.2165/11315680-000000000-00000. 1910.2165/11315680-000000000-000000000. [DOI] [PubMed] [Google Scholar]
- 23.Qiu L, Burgess A, Fairlie DP, Leonard H, Parsons PG, Gabrielli BG. Histone deacetylase inhibitors trigger a g2 checkpoint in normal cells that is defective in tumor cells. Mol Biol Cell. 2000;11:2069–2083. doi: 10.1091/mbc.11.6.2069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Richon VM, Sandhoff TW, Rifkind RA, Marks PA. Histone deacetylase inhibitor selectively induces p21waf1 expression and gene-associated histone acetylation. Proc Natl Acad Sci USA. 2000;97:10014–10019. doi: 10.1073/pnas.180316197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yang ZY, Simari RD, Perkins ND, San H, Gordon D, Nabel GJ, Nabel EG. Role of the p21 cyclin-dependent kinase inhibitor in limiting intimal cell proliferation in response to arterial injury. Proc Natl Acad Sci USA. 1996;93:7905–7910. doi: 10.1073/pnas.93.15.7905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Okamoto H, Fujioka Y, Takahashi A, Takahashi T, Taniguchi T, Ishikawa Y, Yokoyama M. Trichostatin a, an inhibitor of histone deacetylase, inhibits smooth muscle cell proliferation via induction of p21waf1. J Atheroscler Thromb. 2006;13:183–191. doi: 10.5551/jat.13.183. [DOI] [PubMed] [Google Scholar]
- 27.Wilting RH, Yanover E, Heideman MR, Jacobs H, Horner J, van der Torre J, Depinho RA, Dannenberg JH. Overlapping functions of hdac1 and hdac2 in cell cycle regulation and haematopoiesis. EMBO J. doi: 10.1038/emboj.2010.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chen JS, Faller DV. Histone deacetylase inhibition-mediated post-translational elevation of p27kip1 protein levels is required for g1 arrest in fibroblasts. J Cell Physiol. 2005;202:87–99. doi: 10.1002/jcp.20094. [DOI] [PubMed] [Google Scholar]
- 29.Baldin V, Lukas J, Marcote M, Pagano M, Draetta G. Cyclin d1 is a nuclear protein required for cell cycle progression in g1. Genes Dev. 1993;7:812–821. doi: 10.1101/gad.7.5.812. [DOI] [PubMed] [Google Scholar]
- 30.Yamaguchi K, Lantowski A, Dannenberg AJ, Subbaramaiah K. Histone deacetylase inhibitors suppress the induction of c-jun and its target genes including cox-2. J Biol Chem. 2005;280:32569–32577. doi: 10.1074/jbc.M503201200. [DOI] [PubMed] [Google Scholar]
- 31.Hu J, Colburn NH. Histone deacetylase inhibition down-regulates cyclin d1 transcription by inhibiting nuclear factor-kb/p65 DNA binding. Mol Cancer Res. 2005;3:100–109. doi: 10.1158/1541-7786.MCR-04-0070. [DOI] [PubMed] [Google Scholar]
- 32.Sharma P, Kumar S, Kundu GC. Transcriptional regulation of human osteopontin promoter by histone deacetylase inhibitor, trichostatin a in cervical cancer cells. Mol Cancer. 2010;9:178. doi: 10.1186/1476-4598-9-178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hu Y, Cheng L, Hochleitner B-W, Xu Q. Activation of mitogen-activated protein kinases (erk/jnk) and ap-1 transcription factor in rat carotid arteries after balloon injury. Arterioscler Thromb Vasc Biol. 1997;17:2808–2816. doi: 10.1161/01.atv.17.11.2808. [DOI] [PubMed] [Google Scholar]
- 34.Bellas RE, Lee JS, Sonenshein GE. Expression of a constitutive nf-kappa b-like activity is essential for proliferation of cultured bovine vascular smooth muscle cells. J Clin Invest. 1995;96:2521–2527. doi: 10.1172/JCI118313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Santiago FS, Ishii H, Shafi S, Khurana R, Kanellakis P, Bhindi R, Ramirez MJ, Bobik A, Martin JF, Chesterman CN, Zachary IC, Khachigian LM. Yin yang-1 inhibits vascular smooth muscle cell growth and intimal thickening by repressing p21waf1/cip1 transcription and p21waf1/cip1-cdk4-cyclin d1 assembly. Circ Res. 2007;101:146–155. doi: 10.1161/CIRCRESAHA.106.145235. [DOI] [PubMed] [Google Scholar]
- 36.Karpurapu M, Wang D, Van Quyen D, Kim T-K, Kundumani-Sridharan V, Pulusani S, Rao GN. Cyclin d1 is a bona fide target gene of nfatc1 and is sufficient in the mediation of injury-induced vascular wall remodeling. J Biol Chem. 2010;285:3510–3523. doi: 10.1074/jbc.M109.063727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Roque M, Fallon JT, Badimon JJ, Zhang WX, Taubman MB, Reis ED. Mouse model of femoral artery denudation injury associated with the rapid accumulation of adhesion molecules on the luminal surface and recruitment of neutrophils. Arterioscler Thromb Vasc Biol. 2000;20:335–342. doi: 10.1161/01.atv.20.2.335. [DOI] [PubMed] [Google Scholar]
- 38.Liu T, Tee AEL, Porro A, Smith SA, Dwarte T, Liu PY, Iraci N, Sekyere E, Haber M, Norris MD, Diolaiti D, Della Valle G, Perini G, Marshall GM. Activation of tissue transglutaminase transcription by histone deacetylase inhibition as a therapeutic approach for myc oncogenesis. Proc Natl Acad Sci USA. 2007;104:18682–18687. doi: 10.1073/pnas.0705524104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Imre G, Gekeler V, Leja A, Beckers T, Boehm M. Histone deacetylase inhibitors suppress the inducibility of nuclear factor-κb by tumor necrosis factor-α receptor-1 down-regulation. Cancer Res. 2006;66:5409–5418. doi: 10.1158/0008-5472.CAN-05-4225. [DOI] [PubMed] [Google Scholar]
- 40.Yan Z-Q, Yao Q-P, Zhang M-L, Qi Y-X, Guo Z-Y, Shen B-R, Jiang Z-L. Histone deacetylases modulate vascular smooth muscle cell migration induced by cyclic mechanical strain. J Biomech. 2009;42:945–948. doi: 10.1016/j.jbiomech.2009.01.012. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.