Abstract
microRNAs (miRNAs) are a family of 21-23 nucleotide endogenous non-coding RNAs that post-transcriptionally regulate gene expression in a sequence-specific manner. Typically, miRNAs downregulate target genes by recognizing and recruiting protein complexes to 3′UTRs, followed by translation repression or mRNA degradation. miR-92 is a well-studied oncogene in mammalian systems. Here, using zebrafish as a model system, we uncovered a novel tissue-inductive role for miR-92 during early vertebrate development. Overexpression resulted in reduced endoderm formation during gastrulation with consequent cardia and viscera bifida. By contrast, depletion of miR-92 increased endoderm formation, which led to abnormal Kupffer's vesicle development and left-right patterning defects. Using target prediction algorithms and reporter constructs, we show that gata5 is a target of miR-92. Alteration of gata5 levels reciprocally mirrored the effects of gain and loss of function of miR-92. Moreover, genetic epistasis experiments showed that miR-92-mediated defects could be substantially suppressed by modulating gata5 levels. We propose that miR-92 is a critical regulator of endoderm formation and left-right asymmetry during early zebrafish development and provide the first evidence for a regulatory function for gata5 in the formation of Kupffer's vesicle and left-right patterning.
Keywords: Kupffer's vesicle, gata5, Left-right asymmetry, miR-92 (mir92), Zebrafish
INTRODUCTION
microRNAs (miRNAs) are short non-coding RNAs that post-transcriptionally downregulate gene expression by pairing with complementary sequences in the 3′UTR of target mRNAs (Bartel, 2004; He and Hannon, 2004; Kim et al., 2009). In animals, the pairings are usually imperfect and typically result in translation inhibition and/or mRNA destabilization (Chekulaeva and Filipowicz, 2009; Filipowicz et al., 2008). miRNAs play crucial roles in differentiation, cell specification, proliferation and the response to stress, but the exact function of many individual miRNAs remains to be elucidated (Ambros, 2004; Flynt et al., 2007; Flynt et al., 2009; Hatfield et al., 2005; Liu and Olson, 2010; Shkumatava et al., 2009; Takacs and Giraldez, 2010). In part, this is because of the imperfect pairing with target mRNAs, which complicates the identification of bona fide targets given the large number of possible pairing interactions. Nucleotides 2-7 at the 5′ end of miRNAs constitute the seed region, which plays a crucial role in target pairing (Lewis et al., 2003). However, the seed sequence is not the only determinant, as other nucleotides of the miRNA as well as the sequence context within the 3′UTR also play important roles (Grimson et al., 2007; Li et al., 2008).
Previous work has shown that miR-92 functions as an oncogene (reviewed by Croce, 2009). In humans, two different miR-92 loci, MIR-92A and MIR-92B, are encoded in the polycistronic MIR-17-92 and MIR-106A-363 clusters, respectively (reviewed by Petrocca et al., 2008a). Amplification of the MIR-17-92 cluster is frequently observed in hematopoietic malignancies and other solid tumors (Ota et al., 2004; Petrocca et al., 2008b; Volinia et al., 2006). Also, both clusters are common insertion sites in multiple types of retrovirally induced murine leukemias (Cui et al., 2007; Joosten et al., 2002; Landais et al., 2007; Lund et al., 2002; Mikkers et al., 2002; Suzuki et al., 2002; Wang et al., 2006). In a mouse B-cell lymphoma model, enforced overexpression of the miR-17-92 cluster significantly accelerated disease onset and progression (He et al., 2005). Consistent with a role in cancer, transcriptional activation of the miR-17-92 cluster is directly regulated by c-Myc and E2F3 (O'Donnell et al., 2005; Sylvestre et al., 2007; Woods et al., 2007). Multiple downstream targets of the miR-17-92 cluster have been identified that contribute to its tumorigenic function, including E2F1 (Lazzerini Denchi and Helin, 2005; O'Donnell et al., 2005; Woods et al., 2007), p21/CDKN1A (Ivanovska et al., 2008; Petrocca et al., 2008b), Bim/BCL2L11, Pten (Koralov et al., 2008; Petrocca et al., 2008b; Ventura et al., 2008; Xiao et al., 2008), TSP1 (thrombospondin 1) and CTGF (connective tissue growth factor) (Dews et al., 2006). Control of these factors illustrates the important role that miRNAs can play in differentiated cells and during tumorigenesis. By contrast, the exact role of these and other miRNAs during development is less well understood (Fontana et al., 2007; Koralov et al., 2008; Takacs and Giraldez, 2010; Ventura et al., 2008; Xiao et al., 2008). Here, we focus on the significant issue of normal miRNA function in controlling differentiation and patterning during early vertebrate embryogenesis.
During early vertebrate development, the formation of embryonic germ layers and of body axes are the result of the complex integration and precise regulation of multiple signaling pathways (Hamada et al., 2002; Zorn and Wells, 2009). In zebrafish, Nodal signaling is both necessary and sufficient for mesoendoderm specification (the common progenitors of both endoderm and mesoderm) (Rodaway et al., 1999; Schier et al., 1997). The transcription factor Gata5 (Evans et al., 1988) apparently acts directly downstream of Nodal signaling to specify endodermal fate via the activation of another transcription factor, sox32 (casanova), which induces expression of the essential endodermal marker sox17 (Alexander et al., 1999; Warga and Nusslein-Volhard, 1999). Depletion of Gata5 in mutants and morphants impairs endoderm formation, whereas excess Gata5 causes an expansion of endoderm (Reiter et al., 1999; Reiter et al., 2001). Here, we reveal a central role for miR-92 as a regulator of endoderm specification via control of gata5. Further, we show that loss of miR-92 inhibits the formation and function of Kupffer's vesicle, a transient organ equivalent to the embryonic node in amniotes that plays a crucial role in establishing left-right asymmetry (Bakkers et al., 2009; Essner et al., 2002; Fliegauf et al., 2007; McGrath et al., 2003; Nonaka et al., 1998; Nonaka et al., 2005; Raya and Izpisua Belmonte, 2006; Tabin and Vogan, 2003; Tanaka et al., 2005). In contrast to the large number of studies that have examined a role for miR-92 in cancer, our results reveal a novel and essential role for miR-92 during early vertebrate development.
MATERIALS AND METHODS
Zebrafish lines and maintenance
Wild-type AB and sox17:gfp (Sakaguchi et al., 2006) lines of zebrafish (Danio rerio) were used. Embryos were grown at 28°C in egg water and staged by morphology (Kimmel et al., 1995) and age [hours post-fertilization (hpf)].
Microinjection
Zebrafish embryos were injected at the one-cell stage with miRNAs, morpholinos (MOs) or mRNA reporters. miR-92a, miR-92b and control (ctrl) miRNAs were prepared by annealing single-stranded RNAs [synthesized by Integrated DNA Technologies (IDT)]:
miR-92a sense, 5′-UAUUGCACUUGUCCCGGCCUGUUU-3′;
miR-92a antisense, 5′-ACAGGCCGGGACAAGUGCAAUAUU-3′;
miR-92b sense, 5′-UAUUGCACUCGUCCCGGCCUCCUU-3′;
miR-92b antisense, 5′-GGAGGCCGGGACGAGUGCAAUAUU-3′;
ctrl mRNA sense, 5′-CUCUAGGUUAAACUCCUGGUU-3′; and
ctrl miRNA antisense, 5′-UUGAGAUCCAAUUUGAGGACC-3.
Annealing was performed by mixing equal amounts of sense and antisense strands, heating to 65°C for 5 minutes, and gradually cooling to room temperature. Annealed duplexes were aliquoted and stored at −80°C. Unless otherwise indicated, 0.5 ng of miR-92a and miR-92b were injected. Control miRNAs were injected at 1 ng per embryo. In Fig. 5G, 0.2 ng of miR-92a and miR-92b were used to avoid general toxicity caused by injection of large amounts of RNA.
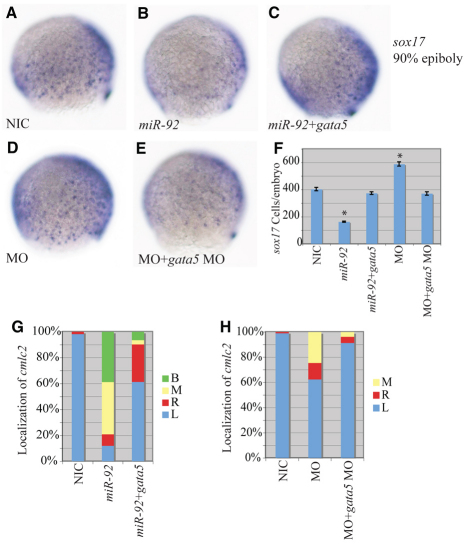
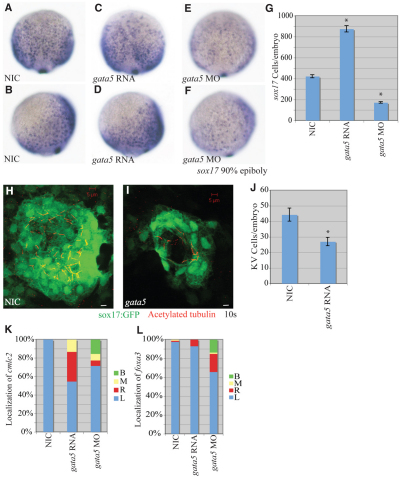
Fig. 5.
Epistatic interaction between miR-92 and gata5. (A-E) Localization of sox17-expressing cells in wild-type and variously injected zebrafish embryos at 90% epiboly. MO refers to miR-92 MO. Views are lateral with dorsal to the right. (F) Numbers of sox17-expressing cells in NICs (n=4), miR-92 gain-of-function (n=4), miR-92 and gata5 co-injected (n=7), miR-92 loss-of-function (n=4) and miR-92 MO and gata5 MO co-injected (n=7) embryos. Error bars represent s.e.m. *, P<0.01 between injected embryos and NIC; Student's t-test. (G) Percentages of left (L), right (R), midline (M) and bilateral (B) localized cmlc2 in NIC (n=112), miR-92 gain-of-function (n=67) and miR-92 and gata5 co-injected (n=31) embryos. (H) Percentages of left, right, midline and bilateral localized cmlc2 in NIC (n=97), miR-92 loss-of-function (n=61) and miR-92 MO and gata5 MO co-injected (n=81) embryos.
All MOs were obtained from Gene Tools:
MO92a1, 5′-ACAGGCCGGGACAAGTGCAATA-3′;
MO92a2, 5′-CACACAGCATTGCTACCAATCCCAA-3′;
MO92a2′, 5′-CACAGAGCATTGCGGCCGATCCCAA-3′;
MO92b1, 5′-GGAGGCCGGGACGAGTGCAATA-3′; and
MO92b2, 5′-TGAACAACACTGCACAACATCCCAC-3′.
Unless otherwise indicated, 1 ng of MO92a1 and MO92b1 were injected per embryo and are jointly referred to as MO1; 3 ng of MO92a2, MO92a2′ and MO92b2 were injected per embryo and are referred to jointly as MO2. Standard control MOs (GeneTools) were injected at 10 ng per embryo; 1 pmol of a gata5 translation blocker MO (5′-AAGATAAAGCCAGGCTCGAATACAT-3′) was injected per embryo (Holtzinger and Evans, 2007). In vitro transcribed, capped GFP reporter mRNAs and gata5 mRNAs were injected at 25 pg and 20 pg per embryo, respectively.
Dorsal forerunner cell (DFC) injections were performed as described (Amack and Yost, 2004). For delivery into DFCs, embryos were injected with 1 ng of a miR-92a and miR-92b mixture at 3 hpf.
Molecular cloning
The gata5 (NM_131235.2) 3′UTR was amplified by RT-PCR using forward (5′-CCACCGAATTCTGATCCGAGACC-3′) and reverse (5′-GGAGGCTCGAGAAACGATATAATTCC-3′) primers. The resulting cDNA was cloned downstream of the GFP open reading frame in the pCS2+ vector (Rupp et al., 1994). Deletion of both MREs was created by reverse PCR (Coolidge and Patton, 1995) using the following primers: gata5 D1F, 5′-TCCACCAAAAATATGGTGGATG-3′; gata5 D1R, 5′-ACATCATAGATATGCCACCATAAATCA-3′; gata5 D2F, 5′-GACCCGCGCCGCTT-3′; and gata5 D2R, 5′-GGAATACAATACAACATTGACAGAGTC-3′. All clones were verified by DNA sequencing.
In situ hybridization
Embryos were fixed in 4% paraformaldehyde (PFA) in 1×PBS. Digoxygenin-labeled RNA probes were synthesized using a Roche DIG RNA Labeling Kit. cDNA templates included foxa3 (Field et al., 2003), cmlc2 (Yelon et al., 1999), sox17 (Amack and Yost, 2004), ntl (Schulte-Merker et al., 1994), lrdr (Essner et al., 2005), foxj1 (Neugebauer et al., 2009) and fgf8 (Yamauchi et al., 2009). Whole-mount in situ hybridization was performed as described (Thisse and Thisse, 2008). Embryos were mounted in 100% glycerol and images were obtained using a Zeiss Axiophot camera. The number of sox17-expressing cells was manually counted over a large number of images, as indicated in the relevant figure legends.
Northern blotting
Total RNA from zebrafish embryos was separated on 12% polyacrylamide gels and electroblotted to positively charged nylon membranes. DNA oligonucleotides complementary to miR-92 were labeled with [α32P]dATP using StarFire Labeling Kits (IDT). Hybridizations were carried out in 7% SDS and 0.2 M NaPO4 (pH 7.2) for 16 hours followed by washes in 2×SSPE containing 0.1% SDS.
Quantitative PCR
Total RNA from embryos at 90% epiboly was isolated using TRI reagent (Molecular Research Center). cDNAs were prepared from 20 ng total RNA and quantitative PCR was performed using the Power SYBR Green PCR Master Mix (Applied Biosystems) on an iCycler iQ Multicolor machine (Bio-Rad) using primers 5′-GGTGTGGGCGAAAGATGAGC-3′ (forward) and 5′-CTCGTAGACGTTCGGCCTCC-3′ (reverse), with annealing at 60°C.
Immunoblotting
Proteins were extracted from deyolked embryos at 1 day post-fertilization (dpf) in lysis buffer [25 mM HEPES (pH 7.5), 5 mM MgCl2, 300 mM NaCl, 1 mM EDTA, 0.2 mM EGTA, 1 mM DTT, 10% glycerol, 1.0% Triton X-100, 1 mM PMSF]. Then, 20 μg total protein was separated on 10% SDS-PAGE gels and transferred to PVDF-plus membranes (GE Osmonics). Rabbit polyclonal antibodies against GFP (Torrey Pines Biolabs) and α-tubulin (Abcam) were used at 1:1000 and 1:500 dilution, respectively. Anti-rabbit HRP-conjugated secondary antibodies (GE Healthcare) were then used for visualization with ECL reagents (Perkin Elmer). For quantification, GFP levels were normalized to α-tubulin control levels, after which the ratio of GFP in the presence versus absence of miR-92 was determined.
Immunohistochemistry
Embryos were fixed in 4% PFA in 1×PBS, permeabilized in 0.5% Triton X-100 in 1×PBS for 1 hour, then incubated in blocking buffer (5% donkey serum, 5 mg/ml BSA, 1% DMSO, 0.1% Tween 20 in 1×PBS) at room temperature for 2 hours. Mouse polyclonal primary antibodies against acetylated tubulin (Sigma) were diluted 1:800. Cy3-conjugated secondary antibodies against mouse IgG (Jackson ImmunoResearch) were used at 1:100. Embryos were mounted in GVA mount (Invitrogen). Samples were imaged on a Zeiss LSM 510META confocal microscope.
RESULTS
Overexpression of miR-92 results in partial viscera and cardia bifida
To characterize the function of miR-92 during early vertebrate development, we performed both gain- and loss-of-function experiments. miR-92 is among the earliest expressed miRNAs detected during zebrafish development following analysis by deep sequencing (C. Wei and J.G.P., unpublished). It localizes to the developing gut, liver and heart by 2-3 dpf (Wienholds et al., 2005). Gain-of-function experiments were performed by injecting miR-92 into zebrafish embryos at the one-cell stage, followed by assessment of the effects on gut and liver tissues by in situ hybridization with the pan-endodermal marker forkhead box a3 (foxa3) (Reiter et al., 1999). In non-injected controls (NICs), foxa3 localized to the developing gut tube, liver and pancreas primordia at 50 hpf, with liver on the left and pancreas on the right-hand side of the midline (Fig. 1A,B). Strikingly, miR-92 injection caused over 70% of the embryos to display aberrant foxa3 localization, showing a bifurcated gut tube with duplication of liver primordia (Fig. 1A,B). A similar phenotype has been observed previously and is referred to as viscera bifida (Nair and Schilling, 2008). For the remaining embryos, 20% showed localization of foxa3 along the midline, indicating a lack of gut looping, and another 3% had undetectable levels of foxa3, suggesting possible defects in endodermal specification.
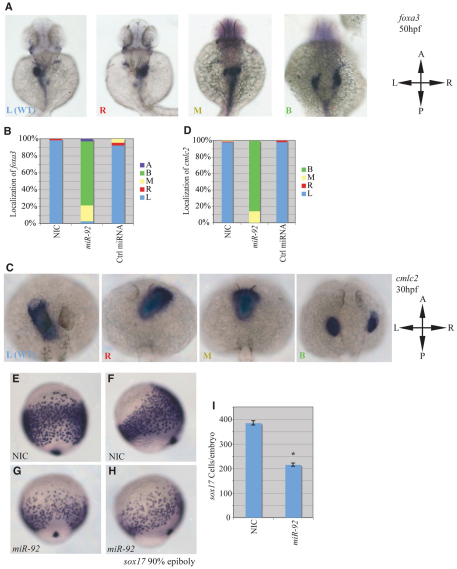
Fig. 1.
miR-92 gain of function. Gain-of-function experiments were performed by injection of single-cell zebrafish embryos with miR-92 followed by localization of specific markers, as indicated. (A) Localization of foxa3 in wild-type embryos and those injected with miR-92 at 50 hpf. Views are dorsal with anterior to the top. Images are of representative embryos with liver primordia localized to either the left (L), right (R), midline (M) or bilateral (B) positions. (B) Percentages of left, right, midline or bilateral localization of foxa3 in non-injected control (NIC) (n=188), miR-92-injected (n=37) and control miRNA-injected (n=90) embryos. In rare cases, no expression of foxa3 was detected (A, absent). (C) Localization of cmlc2 in wild type and in embryos injected with miR-92 at 30 hpf. Views are dorsal with anterior to the top. Images are of representative embryos with cardiac primordia localized to the left, right, midline or bilateral positions. (D) Percentages of left, right, midline and bilateral localization of cmcl2 in NIC (n=103), miR-92-injected (n=98) and control miRNA-injected (n=53) embryos. (E-H) Localization of sox17-expressing cells in wild-type embryos and miR-92-overexpressing embryos at 90% epiboly. (E,G) Dorsal views with anterior to the top. (F,H) Lateral views with dorsal to the right. (I) Numbers of sox17-expressing cells in NIC (n=16) and miR-92-injected embryos (n=11). Error bars represent s.e.m. *, P<0.01; Student's t-test. A, anterior; P, posterior; L, left; R, right.
The developing heart is also enriched for miR-92 and its development depends indirectly on the proper establishment of endodermal fates (Wienholds et al., 2005; Reiter et al., 1999; Alexander et al., 1999; Kikuchi et al., 2000; Schier et al., 1997). Thus, we also tested the effects of miR-92 overexpression on heart development by examining the localization of the cardiac marker cmlc2 (cardiac myosin light chain 2; myl7 – Zebrafish Information Network) (Reiter et al., 1999). In NICs, cmlc2 localized to cardiac primordia on the left side of the embryo at 30 hpf, as expected (Fig. 1C,D). By contrast, injection of miR-92 caused over 85% of the embryos to display bilateral cmlc2 expression, indicating a failure of heart fusion that is referred to as cardia bifida (Reiter et al., 1999; Reiter et al., 2001; Holtzinger and Evans, 2007; Stainier, 1996). The remaining 15% of embryos showed cmlc2 along the midline, indicating an inability of the heart tube to undergo normal looping. The specificity of these gain-of-function experiments was supported by an absence of changes in foxa3 or cmlc2 expression upon injection of unrelated and control miRNAs (Fig. 1B,D). Also, the effects of miR-92 were dose dependent (see Table S1 in the supplementary material).
Overexpression of miR-92 results in a reduction of endoderm
Viscera and cardia bifida result from the failure to coalesce the relevant mesendodermal organ progenitors at the midline during the segmentation stages and may be secondary to a variety of earlier defects in endoderm or mesoderm formation (Alexander et al., 1999; Kikuchi et al., 2000; Nair and Schilling, 2008; Ober et al., 2004; Reiter et al., 1999; Reiter et al., 2001; Schier et al., 1997). To address this issue, we examined the effects of miR-92 gain of function on the expression of genes that act early in the specification and determination of endoderm and mesoderm. No significant defects were detected in mesoderm formation with miR-92 injection, as indicated by the normal expression pattern of ntl (see Fig. S1I-P in the supplementary material). By contrast, miR-92 injection caused a dramatic decrease in the specification of early endoderm cells, as indicated by the decreased numbers of sox17- and sox32-expressing cells at the end of gastrulation (Fig. 1E-I and see Fig. S1A-H in the supplementary material). This suggests a selective impairment in the formation of endodermal cells with no effect on the formation of the mesodermal germ layer.
Depletion of miR-92 results in aberrant left-right patterning of internal organs
We next performed loss-of-function experiments using antisense morpholinos (MOs) to block miR-92 activity during early development (see Figs S2 and S3 in the supplementary material). In NICs, the localization of foxa3 to the developing liver was primarily on the left side at 50 hpf (Fig. 2A). However, injection of MOs against miR-92 resulted in 20-40% of the embryos displaying abnormal left-right localization of foxa3 in the developing liver. Similarly, 40% of the morphants showed localization of cmcl2 to either the right or the middle, as compared with normal left-sided heart patterning in NICs (Fig. 2B). Thus, loss of miR-92 resulted in a significant incidence of aberrant left-right patterning.
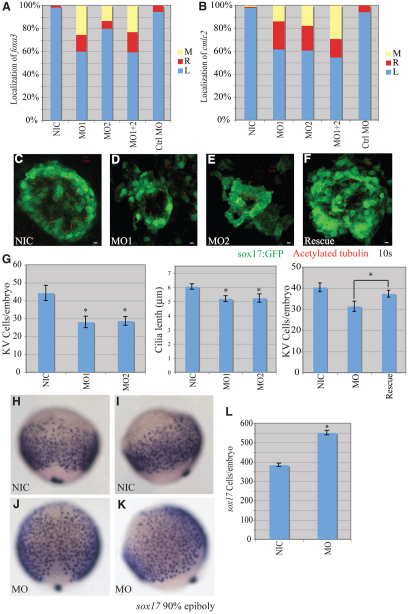
Fig. 2.
miR-92 loss of function. Loss-of-function experiments were performed by injection of antisense MOs against miR-92 into single-cell zebrafish embryos followed by localization of markers, as indicated. (A) Percentages of left (L), right (R) and midline (M) localized foxa3 in NIC (n=188), miR-92 MO1-injected (n=55), miR-92 MO2-injected (n=15), MO1+2-injected (n=126) and control MO-injected (n=56) embryos. (B) Percentages of left, right and midline localized cmcl2 in NIC (n=103), MO1-injected (n=102), MO2-injected (n=97), MO1+2-injected (n=62) and control MO-injected (n=53) embryos. (C-F) Confocal z-stacks of Kupffer's vesicle (KV) in NICs, MO1-injected and MO2-injected embryos and in miR-92 morphants co-injected with miR-92 RNA into the dorsal forerunner cells (DFCs) (Rescue) at the10-somite stage using a sox17:gfp transgenic line that labels KV cells green. Motile cilia were identified by immunohistochemistry with antibodies against acetylated tubulin (red). Scale bars: 5 μm. (G) GFP-positive KV cells were counted and cilia length measured in the indicated embryos. Error bars represent s.e.m. *, P<0.01 for KV cell number between miR-92 morphants and NICs, P<0.05 for cilia length and P<0.05 for KV cell number between miR-92 morphants and rescue embryos; Student's t-test. MO refers to miR-92 MO. (H-K) Localization of sox17-expressing cells in wild-type embryos (NIC) and miR-92 morphants (MO) at 90% epiboly. (H,J) Dorsal views with anterior to the top. (I,K) Lateral views with dorsal to the right. (L) Numbers of sox17-expressing cells in NICs (n=16) and miR-92 morphants (MO; n=13). Error bars represent s.e.m. *, P<0.01; Student's t-test.
Several lines of evidence suggest that the observed left-right asymmetry defects are specific to depletion of miR-92. First, two independent MOs (MO1 and MO2) yielded identical results (Fig. 2A,B). Second, synergistic and similar effects were observed upon co-injection of both MOs at much lower doses than the individual injections. Third, the defects were dose dependent (see Table S2 in the supplementary material). Fourth, injection of control MO did not affect the left-right localization of either marker (Fig. 2A,B).
Depletion of miR-92 alters Kupffer's vesicle function
Specification of left-right patterning is highly regulated and propagated through several stages of embryogenesis in a complex genetically controlled program (Bakkers et al., 2009; Hamada et al., 2002; Raya and Izpisua Belmonte, 2006). In zebrafish, motile cilia in Kupffer's vesicle (KV) appear to be crucial for left-right asymmetry, analogous to their counterparts in the node in amniotes (Amack et al., 2007; Amack and Yost, 2004). Evidence gathered from both zebrafish and mouse embryos suggests a strong connection between node/vesicle structures and proper left-right patterning, especially with regard to the flow-generating function of primary cilia (Amack et al., 2007; Amack and Yost, 2004; Kreiling et al., 2008; Marszalek et al., 1999; Murcia et al., 2000; Nonaka et al., 1998; Okada et al., 1999; Schneider et al., 2008; Supp et al., 1999; Takeda et al., 1999). We therefore tested for KV defects in the miR-92 morphants. To visualize KV, we used a sox17:gfp transgenic line to label KV cells with GFP. A substantial reduction in cell numbers within KV was observed in the miR-92 morphants (Fig. 2C-E,G). To evaluate further the structural deficits in KV function in the miR-92 morphants, we examined the number and length of monocilia within KV by immunohistochemistry with antibodies against acetylated tubulin (Essner et al., 2005). A significant reduction in cilia number and a modest, but significant, decrease in their length were observed in miR-92 morphants (Fig. 2G and see Fig. S4 in the supplementary material). These results suggest that miR-92 is required for the proper development of KV and cilia within KV.
To ensure that the KV defects were specific to the loss of miR-92, we performed rescue experiments in which exogenous miR-92 was expressed in dorsal forerunner cells (DFCs) (Amack and Yost, 2004). DFCs are the progenitor cells that form KV, and if loss of miR-92 from these cells is responsible for the observed defects, we should be able to rescue such defects by targeted miR-92 gain of function in the DFCs of morphant embryos. Indeed, specific restoration of miR-92 expression in DFCs was able to partially, but significantly, rescue KV size in morphant embryos (Fig. 2F,G). In addition, expression of miR-92 in DFCs also rescued left-right patterning defects when examined using the cmlc2 marker (see Fig. S5 in the supplementary material). These results indicate that the observed KV defects are specific to miR-92 depletion in DFCs.
Depletion of miR-92 results in increased endoderm
Normal KV development from DFCs requires both endodermal and mesodermal signals, in addition to genes involved in general ciliogenesis (Alexander et al., 1999; Amack et al., 2007; Amack and Yost, 2004; Neugebauer et al., 2009; Oteiza et al., 2008). We therefore examined the expression and localization of markers involved in both general ciliogenesis and the formation of endoderm and mesoderm. No significant defects in general ciliogenesis or mesoderm formation were detected, as indicated by normal expression of lrdr (dnah9 – Zebrafish Information Network), foxj1, fgf8 and ntl, as well as the normal development of motile cilia in the pronephros and inner ear (see Figs S1 and S6 in the supplementary material). By contrast, loss of miR-92 caused a dramatic increase in the number of sox17-expressing endodermal cells and these cells were much more spread out at the end of gastrulation, indicating defects in endoderm formation (Fig. 2H-L). Consistent with increased cell number, there was an overall 1.6-fold increase in sox17 mRNAs in the miR-92 morphants, as detected by quantitative PCR. This increase was suppressed by co-injection of miR-92 (see Fig. S7 in the supplementary material). Taken together, these results suggest that miR-92 is required for proper endoderm specification and for the development of structures that are crucial for controlling fundamental aspects of organogenesis and overall body patterning.
gata5 is a target of miR-92
Online target prediction algorithms [TargetScan (http://www.targetscan.org/), MicroCosm Targets (http://www.ebi.ac.uk/enright-srv/microcosm/htdocs/targets/v5/) and PicTar (http://www.pictar.org/)] were used to search for potential targets of miR-92. The zinc-finger transcription factor gata-binding protein 5 (gata5) was identified as a potential target as it contains two potential miRNA recognition elements (MREs) in its 3′UTR (Fig. 3A). Gata5 is a critical regulator of vertebrate endoderm development, as overexpression of Gata5 increases endodermal cell numbers, whereas reduced Gata5 expression causes a reduction in endodermal cell numbers (Reiter et al., 1999; Reiter et al., 2001). Also, gata5 localizes to DFCs during early zebrafish development (data not shown). Hence, gata5 is a compelling miR-92 target during early zebrafish development.
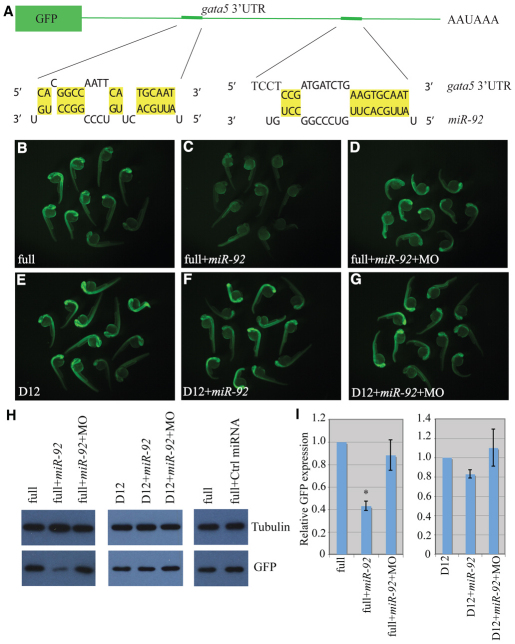
Fig. 3.
gata5 is a target of miR-92. (A) The GFP reporter fused to the 3′UTR of gata5. Base pairing between miR-92 and two miRNA recognition elements (MREs) is shown. (B-G) Single-cell zebrafish embryos were injected with mRNAs encoding GFP reporters and fluorescence was monitored at 1 dpf. Embryos were injected with reporters containing either the full-length gata5 3′UTR (full) or a construct in which both MREs were deleted (D12). MO refers to miR-92 MO. (H) Western blot of lysates from embryos injected as above were performed with antibodies against GFP or α-tubulin as a loading control. (I) Quantification of relative GFP expression. Error bars represent s.e.m. *, P<0.01, between indicated injection and control embryos (n>3); Student's t-test.
To determine whether gata5 is a bona fide target of miR-92, we analyzed the interaction between the gata5 3′UTR and miR-92 using GFP reporter assays. The full-length gata5 3′UTR and a deletion construct lacking both MREs were cloned downstream of the GFP open reading frame. RNA transcripts from these constructs were then injected into single-cell zebrafish embryos in the presence or absence of miR-92. The following day, GFP expression levels were monitored by fluorescence microscopy and by western blotting with antibodies against GFP. In both assays, GFP levels were reduced by miR-92 co-injection, and this was dependent on the presence of intact miR-92 MREs (Fig. 3). Importantly, co-injection of miR-92 antisense MOs rescued expression of GFP in the presence of miR-92 (Fig. 3D,G-I). Injection of a control miRNA had no effect on GFP levels (Fig. 3H). These results suggest that gata5 is a bona fide target of miR-92. Consistent with this, an increase in gata5 transcript levels was observed in miR-92 morphants as compared with NICs (data not shown).
Regulation of gata5 by miR-92 during early zebrafish development
The hypothesis that miR-92 targets gata5 is not only supported by the reporter assays but also by previous work that showed that altered levels of gata5 control endoderm formation, as monitored by changes in sox17-expressing cell numbers (Reiter et al., 1999; Reiter et al., 2001). Also, loss of gata5 has been shown to cause cardia and viscera bifida (Reiter et al., 1999; Reiter et al., 2001). However, no prior reports have implicated gata5 in KV defects or consequent left-right asymmetry defects. Thus, we sought to test directly whether miR-92 control of gata5 would induce KV defects and changes in body plan. If gata5 were regulated by miR-92 during early zebrafish development, then altered expression of Gata5 should reciprocally mirror the effects caused by gain and loss of miR-92, which would then enable powerful genetic epistasis experiments to verify the interaction between miR-92 and gata5.
First, we confirmed that knockdown of gata5 causes a reduction of sox17-expressing cells at the end of gastrulation and that injection of gata5 mRNA causes an increase in sox17-expressing cells (Fig. 4A-G) (Reiter et al., 1999; Reiter et al., 2001). Second, we confirmed that gata5 knockdown causes cardia and viscera bifida defects, as observed in gata5 mutants (faust) and gata5 morphants (Holtzinger and Evans, 2007; Reiter et al., 1999; Reiter et al., 2001) (Fig. 4K,L). After confirming that our experimental system recapitulated earlier work, we then tested the effects of excess Gata5 expression on organogenesis and left-right patterning. When we injected gata5 mRNA and determined the localization of foxa3 and cmcl2, we found that 45% and 7% of embryos displayed altered left-right patterning of cardia and viscera primordia, respectively (Fig. 4K,L). These results show that the increase or reduction of endodermal cell numbers caused by raising or lowering Gata5 levels, together with the finding that increased and decreased levels of miR-92 have converse effects that can be offset by altered gata5 expression, support the idea that the miR-92-gata5 regulatory interaction is involved in allocating correct endodermal cell numbers and maintaining proper left-right patterning.
Fig. 4.
gata5 effects on early zebrafish development. (A-F) Localization of sox17-expressing cells in wild-type, gata5 gain-of-function and gata5 loss-of-function embryos at 90% epiboly. (A,C,E) Dorsal views with anterior to the top. (B,D,F) Lateral views with dorsal to the right. (G) Numbers of sox17-expressing cells in NICs (n=4), gata5 gain-of-function (n=7) and gata5 loss-of-function (n=8) embryos. Error bars represent s.e.m. *, P<0.01 between injected embryos and NIC; Student's t-test. (H,I) Confocal z-stacks of KV in NIC and gata5-injected embryos at the 10-somite stage. KVs were labeled using a sox17:gfp (green) transgenic line as above. Motile cilia were labeled with antibodies against acetylated tubulin (red). Scale bars: 5 μm. (J) KV cell number was counted as above. Error bars represent s.e.m. *, P<0.01 between NIC and gata5 gain of function; Student's t-test. (K) Percentages of left (L), right (R), midline (M) and bilateral (B) localized cmcl2 in NIC (n=49), gata5 gain-of-function (n=75) and gata5 loss-of-function (n=84) embryos. (L) Percentages of left, right, midline and bilateral localized foxa3 in NIC (n=83), gata5 gain-of-function (n=142) and gata5 loss-of-function (n=73) embryos.
Because we discovered a link between miR-92 expression in DFCs and KV formation, it was important to determine whether the gata5-induced left-right asymmetry defects could be explained by defective KV formation. Using the sox17:gfp transgenic line that allows visualization of KV formation, we found a dramatic reduction in KV cell number after gata5 overexpression (Fig. 4H-J and see Fig. S4 in the supplementary material).
miR-92-mediated defects can be partially suppressed by modulation of Gata5
Genetic epistasis experiments were performed to test the hypothesis that miR-92 regulates gata5. If miR-92 acts as a negative regulator of gata5, co-injection of gata5 mRNA with miR-92 should suppress the miR-92 gain-of-function defects. Likewise, blocking gata5 function should suppress the miR-92 loss-of-function defects. As shown in Fig. 5, the reduction in the number of sox17-expressing cells caused by overexpression of miR-92 could be suppressed by co-injection of gata5 RNA. Similarly, co-injection of gata5 MOs with miR-92 MOs suppressed the increase in sox17-expressing cells (Fig. 5A-F).
For cardiac morphogenesis, we observed a partial, but significant, rescue of cmlc2 localization upon co-injection of miR-92 and gata5 RNAs that resulted in a reduction of cardia bifida from 40% to 6% (Fig. 5G). Similarly, blockage of gata5 function in miR-92 morphants resulted in a significant suppression (from 40% to 10%) of the left-right patterning defects (Fig. 5H). Thus, the effects of altered miR-92 levels can be partially suppressed by modulating gata5 levels. These results strongly support the hypothesis that miR-92 regulates endoderm formation and left-right asymmetry by controlling gata5 expression.
DISCUSSION
Our data suggest a model in which miR-92 acts as a critical regulator of early zebrafish development by precisely controlling gata5 expression. By repressing gata5 expression, excess miR-92 causes a reduction in endoderm during the blastula and gastrula stages and, at later stages, cardia/viscera bifida. By contrast, reduced levels of miR-92 cause increased endoderm formation, defects in KV development and, at later stages, abnormal left-right patterning of internal organs. We have uncovered a novel function for miR-92 in controlling endoderm formation and left-right asymmetry by modulating gata5.
Function of miR-92 during early development
The miR-92 family is among the earliest expressed miRNAs during zebrafish development, beginning at the mid-blastula stage. Spatially, miR-92 is expressed somewhat broadly during early development but with increased expression in DFCs and KV (data not shown). Only a limited number of miRNAs have been analyzed for their roles during these early stages when the germ layers develop and the body axes are patterned (Choi et al., 2007; Martello et al., 2007; Rosa et al., 2009). Here, we have shown a novel role for miR-92 in early zebrafish development in endoderm formation, KV function and left-right patterning via the control of gata5. To our knowledge, this is the first time that a miRNA has been identified as a regulator of endoderm formation, vertebrate left-right asymmetry and KV development.
Regulation of gata5 by miR-92
The precise regulation of Gata5 expression is not well understood. During zebrafish endoderm formation, gata5 is transcriptionally upregulated by phosphorylated Smad2 (Shivdasani, 2002). Hypermethylation of the GATA5 promoter region has been reported in various human cancer cell lines, concomitant with downregulation of GATA5 transcription (Akiyama et al., 2003; Guo et al., 2004; Guo et al., 2006; Hellebrekers et al., 2009). Our results suggest that gata5 is subject to post-transcriptional regulation by miR-92. Three lines of evidence support this hypothesis: (1) miR-92 silencing of GFP reporter expression is dependent on an intact gata5 3′UTR (Fig. 3); (2) the effects of gain and loss of function with gata5 are opposite to those observed with miR-92 (Fig. 4); and (3) miR-92-induced defects could be substantially suppressed by changing concentrations of gata5 in epistasis experiments (Fig. 5).
As a downstream effector of Nodal signaling and a regulator of endoderm specification, Gata5 needs to be precisely controlled, both temporally and spatially. In zebrafish, gata5 mRNA is found within five cell diameters of the blastoderm margin at the onset of gastrulation and overlaps with definitive endodermal cells during gastrulation (Reiter et al., 2001; Warga and Nusslein-Volhard, 1999). Excess expression or depletion of Gata5 alters endodermal cell numbers at the end of gastrulation (Reiter et al., 2001). We hypothesize that miR-92 contributes to the establishment of the proper gata5 expression patterns during the blastula and gastrula stages that guide the differentiation and allocation of proper endodermal cell numbers during gastrulation.
Interestingly, the MIR-17-92 cluster is upregulated in lung cancer cell lines, whereas GATA5 is downregulated (Guo et al., 2004; Volinia et al., 2006). It is tempting to hypothesize that both transcriptional regulation via DNA methylation and post-transcriptional regulation via MIR-92 contribute to the silencing of GATA5 in lung cancer. It will be interesting to determine whether gata5 is a general target of miR-92 in other types of tumor.
Misregulation of Gata5
We observed a decrease in sox17-expressing cell numbers upon miR-92 overexpression or gata5 knockdown (Figs 1 and 4). We also observed both cardia and viscera bifida in these embryos. The mechanisms underlying cardia and viscera bifida are not fully understood. It will be interesting to elucidate how endoderm defects cause cardia/viscera bifida. An increase in endoderm formation was observed upon miR-92 knockdown or gata5 overexpression (Figs 2 and 4), but it remains unclear whether endoderm cell numbers increase at the expense of reduced mesoderm cell numbers. Endoderm and mesoderm share common progenitors; endodermal cells become specified from mesodermal cells as a result of stronger Nodal signaling (Schier et al., 1997; Warga and Nusslein-Volhard, 1999). Perhaps miR-92 regulation of gata5 affects endoderm versus mesoderm fate decisions or reflects control of overall cell numbers within the endodermal lineage.
Examining the localization of cmlc2 and foxa3 surprisingly revealed left-right patterning defects in internal organs upon loss of miR-92 or overexpression of gata5. This is the first evidence, to our knowledge, for a regulatory function for gata5 in left-right asymmetry and also the first data to demonstrate defects in viscera morphogenesis upon gata5 overexpression. Previously, overexpression of gata5 was found to cause the expansion and ectopic development of cardiac tissue (Holtzinger and Evans, 2007; Reiter et al., 1999). Here, we did not observe significant expansion of cmlc2 expression nor ectopic localization, except for on the right-hand side or midline in both the gata5 gain-of-function and miR-92 loss-of-function experiments. A possible explanation for these differences could be in the use of different dosages for gata5 RNA injection in the two studies.
The exact causes of the observed KV defects upon altered miR-92 and gata5 levels remain to be determined. Development of KV from DFCs is a complex procedure that is not fully understood (Oteiza et al., 2008). Both cell-autonomous and non-autonomous signals contribute to the induction, migration, proliferation and polarization of DFCs (Amack et al., 2007; Amack and Yost, 2004; Choi et al., 2007; Oteiza et al., 2008). We observed impaired KV development upon miR-92 loss of function that could be suppressed by restoration of miR-92 expression specifically within DFCs of miR-92 morphants. This suggests that the requirement for miR-92 during KV development is cell-autonomous, although the involvement of other signaling pathways or mechanisms cannot be excluded. Previously, diminished KVs were found in embryos with decreased endodermal cell numbers (Alexander et al., 1999). Here, both the miR-92 loss-of-function and gata5 gain-of-function experiments resulted in an increase in endodermal cell numbers and smaller KVs, but it remains unclear at what stage of development the KV defects arise.
miR-92 targets
One explanation for the partial suppression of miR-92 defects in epistasis experiments with gata5 is that gata5 is not the only target of miR-92. Other genes important for endoderm formation and left-right asymmetry might also be subject to regulation by miR-92. Many genes involved in cell cycle control, proliferation or apoptosis are predicted to be targets of miR-92 by online target prediction algorithms. During normal gastrulation movements, endodermal cells migrate dorsally in a characteristic pattern. Besides an increase in endodermal cell number, depletion of miR-92 caused endodermal cells to spread out more during migration, which might indicate additional defects in such as cell adhesion. This phenotype was not observed in gata5-overexpressing embryos, suggesting that miR-92 might regulate genes involved in cell adhesion or migration. If true, this could be another way in which miR-92 contributes to cancer by controlling cell migration and metastasis.
Supplementary Material
Acknowledgements
We thank the Solnica-Krezel and Zhong labs at Vanderbilt University and the Yost lab at the University of Utah School of Medicine for their generous sharing of probes for in situ hybridization, antibodies for immunohistochemistry and gata5 MOs. We thank Chris Wright and Lilianna Solnica-Krezel for critical comments on the manuscript. This work was supported by NIH grant GM 075790 to J.G.P. and a Vanderbilt University Dissertation Enhancement Grant to N.L. Deposited in PMC for release after 12 months.
Footnotes
Competing interests statement
The authors declare no competing financial interests.
Supplementary material
Supplementary material for this article is available at http://dev.biologists.org/lookup/suppl/doi:10.1242/dev.056697/-/DC1
References
- Akiyama Y., Watkins N., Suzuki H., Jair K. W., van Engeland M., Esteller M., Sakai H., Ren C. Y., Yuasa Y., Herman J. G., et al. (2003). GATA-4 and GATA-5 transcription factor genes and potential downstream antitumor target genes are epigenetically silenced in colorectal and gastric cancer. Mol. Cell. Biol. 23, 8429-8439 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alexander J., Rothenberg M., Henry G. L., Stainier D. Y. (1999). casanova plays an early and essential role in endoderm formation in zebrafish. Dev. Biol. 215, 343-357 [DOI] [PubMed] [Google Scholar]
- Amack J. D., Yost H. J. (2004). The T box transcription factor no tail in ciliated cells controls zebrafish left-right asymmetry. Curr. Biol. 14, 685-690 [DOI] [PubMed] [Google Scholar]
- Amack J. D., Wang X., Yost H. J. (2007). Two T-box genes play independent and cooperative roles to regulate morphogenesis of ciliated Kupffer's vesicle in zebrafish. Dev. Biol. 310, 196-210 [DOI] [PubMed] [Google Scholar]
- Ambros V. (2004). The functions of animal microRNAs. Nature 431, 350-355 [DOI] [PubMed] [Google Scholar]
- Bakkers J., Verhoeven M. C., Abdelilah-Seyfried S. (2009). Shaping the zebrafish heart: from left-right axis specification to epithelial tissue morphogenesis. Dev. Biol. 330, 213-220 [DOI] [PubMed] [Google Scholar]
- Bartel D. P. (2004). MicroRNAs: genomics, biogenesis, mechanism, and function. Cell 116, 281-297 [DOI] [PubMed] [Google Scholar]
- Chekulaeva M., Filipowicz W. (2009). Mechanisms of miRNA-mediated post-transcriptional regulation in animal cells. Curr. Opin. Cell Biol. 21, 452-460 [DOI] [PubMed] [Google Scholar]
- Choi W. Y., Giraldez A. J., Schier A. F. (2007). Target protectors reveal dampening and balancing of Nodal agonist and antagonist by miR-430. Science 318, 271-274 [DOI] [PubMed] [Google Scholar]
- Coolidge C. J., Patton J. G. (1995). Run-around PCR: a novel way to create duplications using polymerase chain reaction. Biotechniques 18, 763-764 [PubMed] [Google Scholar]
- Croce C. M. (2009). Causes and consequences of microRNA dysregulation in cancer. Nat. Rev. Genet. 10, 704-714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cui J. W., Li Y. J., Sarkar A., Brown J., Tan Y. H., Premyslova M., Michaud C., Iscove N., Wang G. J., Ben-David Y. (2007). Retroviral insertional activation of the Fli-3 locus in erythroleukemias encoding a cluster of microRNAs that convert Epo-induced differentiation to proliferation. Blood 110, 2631-2640 [DOI] [PubMed] [Google Scholar]
- Dews M., Homayouni A., Yu D., Murphy D., Sevignani C., Wentzel E., Furth E. E., Lee W. M., Enders G. H., Mendell J. T., et al. (2006). Augmentation of tumor angiogenesis by a Myc-activated microRNA cluster. Nat. Genet. 38, 1060-1065 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Essner J. J., Vogan K. J., Wagner M. K., Tabin C. J., Yost H. J., Brueckner M. (2002). Conserved function for embryonic nodal cilia. Nature 418, 37-38 [DOI] [PubMed] [Google Scholar]
- Essner J. J., Amack J. D., Nyholm M. K., Harris E. B., Yost H. J. (2005). Kupffer's vesicle is a ciliated organ of asymmetry in the zebrafish embryo that initiates left-right development of the brain, heart and gut. Development 132, 1247-1260 [DOI] [PubMed] [Google Scholar]
- Evans T., Reitman M., Felsenfeld G. (1988). An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc. Natl. Acad. Sci. USA 85, 5976-5980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Field H. A., Ober E. A., Roeser T., Stainier D. Y. (2003). Formation of the digestive system in zebrafish. I. Liver morphogenesis. Dev. Biol. 253, 279-290 [DOI] [PubMed] [Google Scholar]
- Filipowicz W., Bhattacharyya S. N., Sonenberg N. (2008). Mechanisms of post-transcriptional regulation by microRNAs: are the answers in sight? Nat. Rev. Genet. 9, 102-114 [DOI] [PubMed] [Google Scholar]
- Fliegauf M., Benzing T., Omran H. (2007). When cilia go bad: cilia defects and ciliopathies. Nat. Rev. Mol. Cell Biol. 8, 880-893 [DOI] [PubMed] [Google Scholar]
- Flynt A. S., Li N., Thatcher E. J., Solnica-Krezel L., Patton J. G. (2007). Zebrafish miR-214 modulates Hedgehog signaling to specify muscle cell fate. Nat. Genet. 39, 259-263 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Flynt A. S., Thatcher E. J., Burkewitz K., Li N., Liu Y., Patton J. G. (2009). miR-8 microRNAs regulate the response to osmotic stress in zebrafish embryos. J. Cell Biol. 185, 115-127 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fontana L., Pelosi E., Greco P., Racanicchi S., Testa U., Liuzzi F., Croce C. M., Brunetti E., Grignani F., Peschle C. (2007). MicroRNAs 17-5p-20a-106a control monocytopoiesis through AML1 targeting and M-CSF receptor upregulation. Nat. Cell Biol. 9, 775-787 [DOI] [PubMed] [Google Scholar]
- Grimson A., Farh K. K., Johnston W. K., Garrett-Engele P., Lim L. P., Bartel D. P. (2007). MicroRNA targeting specificity in mammals: determinants beyond seed pairing. Mol. Cell 27, 91-105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo M., Akiyama Y., House M. G., Hooker C. M., Heath E., Gabrielson E., Yang S. C., Han Y., Baylin S. B., Herman J. G., et al. (2004). Hypermethylation of the GATA genes in lung cancer. Clin. Cancer Res. 10, 7917-7924 [DOI] [PubMed] [Google Scholar]
- Guo M., House M. G., Akiyama Y., Qi Y., Capagna D., Harmon J., Baylin S. B., Brock M. V., Herman J. G. (2006). Hypermethylation of the GATA gene family in esophageal cancer. Int. J. Cancer 119, 2078-2083 [DOI] [PubMed] [Google Scholar]
- Hamada H., Meno C., Watanabe D., Saijoh Y. (2002). Establishment of vertebrate left-right asymmetry. Nat. Rev. Genet. 3, 103-113 [DOI] [PubMed] [Google Scholar]
- Hatfield S. D., Shcherbata H. R., Fischer K. A., Nakahara K., Carthew R. W., Ruohola-Baker H. (2005). Stem cell division is regulated by the microRNA pathway. Nature 435, 974 [DOI] [PubMed] [Google Scholar]
- He L., Hannon G. J. (2004). MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet. 5, 522-531 [DOI] [PubMed] [Google Scholar]
- He L., Thomson J. M., Hemann M. T., Hernando-Monge E., Mu D., Goodson S., Powers S., Cordon-Cardo C., Lowe S. W., Hannon G. J., et al. (2005). A microRNA polycistron as a potential human oncogene. Nature 435, 828-833 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hellebrekers D. M., Lentjes M. H., van den Bosch S. M., Melotte V., Wouters K. A., Daenen K. L., Smits K. M., Akiyama Y., Yuasa Y., Sanduleanu S., et al. (2009). GATA4 and GATA5 are potential tumor suppressors and biomarkers in colorectal cancer. Clin. Cancer Res. 15, 3990-3997 [DOI] [PubMed] [Google Scholar]
- Holtzinger A., Evans T. (2007). Gata5 and Gata6 are functionally redundant in zebrafish for specification of cardiomyocytes. Dev. Biol. 312, 613-622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ivanovska I., Ball A. S., Diaz R. L., Magnus J. F., Kibukawa M., Schelter J. M., Kobayashi S. V., Lim L., Burchard J., Jackson A. L., et al. (2008). MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol. 28, 2167-2174 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joosten M., Vankan-Berkhoudt Y., Tas M., Lunghi M., Jenniskens Y., Parganas E., Valk P. J., Lowenberg B., van den Akker E., Delwel R. (2002). Large-scale identification of novel potential disease loci in mouse leukemia applying an improved strategy for cloning common virus integration sites. Oncogene 21, 7247-7255 [DOI] [PubMed] [Google Scholar]
- Kikuchi Y., Trinh L. A., Reiter J. F., Alexander J., Yelon D., Stainier D. Y. (2000). The zebrafish bonnie and clyde gene encodes a Mix family homeodomain protein that regulates the generation of endodermal precursors. Genes Dev. 14, 1279-1289 [PMC free article] [PubMed] [Google Scholar]
- Kim V. N., Han J., Siomi M. C. (2009). Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell Biol. 10, 126-139 [DOI] [PubMed] [Google Scholar]
- Kimmel C. B., Ballard W. W., Kimmel S. R., Ullmann B., Schilling T. F. (1995). Stages of embryonic development of the zebrafish. Dev. Dyn. 203, 253-310 [DOI] [PubMed] [Google Scholar]
- Koralov S. B., Muljo S. A., Galler G. R., Krek A., Chakraborty T., Kanellopoulou C., Jensen K., Cobb B. S., Merkenschlager M., Rajewsky N., et al. (2008). Dicer ablation affects antibody diversity and cell survival in the B lymphocyte lineage. Cell 132, 860-874 [DOI] [PubMed] [Google Scholar]
- Kreiling J. A., Balantac Z. L., Crawford A. R., Ren Y., Toure J., Zchut S., Kochilas L., Creton R. (2008). Suppression of the endoplasmic reticulum calcium pump during zebrafish gastrulation affects left-right asymmetry of the heart and brain. Mech. Dev. 125, 396-410 [DOI] [PubMed] [Google Scholar]
- Landais S., Landry S., Legault P., Rassart E. (2007). Oncogenic potential of the miR-106-363 cluster and its implication in human T-cell leukemia. Cancer Res. 67, 5699-5707 [DOI] [PubMed] [Google Scholar]
- Lazzerini Denchi E., Helin K. (2005). E2F1 is crucial for E2F-dependent apoptosis. EMBO Rep. 6, 661-668 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lewis B. P., Shih I. H., Jones-Rhoades M. W., Bartel D. P., Burge C. B. (2003). Prediction of mammalian microRNA targets. Cell 115, 787-798 [DOI] [PubMed] [Google Scholar]
- Li N., Flynt A. S., Kim H. R., Solnica-Krezel L., Patton J. G. (2008). Dispatched Homolog 2 is targeted by miR-214 through a combination of three weak microRNA recognition sites. Nucleic Acids Res. 36, 4277-4285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu N., Olson E. N. (2010). MicroRNA regulatory networks in cardiovascular development. Dev. Cell 18, 510-525 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund A. H., Turner G., Trubetskoy A., Verhoeven E., Wientjens E., Hulsman D., Russell R., DePinho R. A., Lenz J., van Lohuizen M. (2002). Genome-wide retroviral insertional tagging of genes involved in cancer in Cdkn2a-deficient mice. Nat. Genet. 32, 160-165 [DOI] [PubMed] [Google Scholar]
- Marszalek J. R., Ruiz-Lozano P., Roberts E., Chien K. R., Goldstein L. S. (1999). Situs inversus and embryonic ciliary morphogenesis defects in mouse mutants lacking the KIF3A subunit of kinesin-II. Proc. Natl. Acad. Sci. USA 96, 5043-5048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martello G., Zacchigna L., Inui M., Montagner M., Adorno M., Mamidi A., Morsut L., Soligo S., Tran U., Dupont S., et al. (2007). MicroRNA control of Nodal signalling. Nature 449, 183-188 [DOI] [PubMed] [Google Scholar]
- McGrath J., Somlo S., Makova S., Tian X., Brueckner M. (2003). Two populations of node monocilia initiate left-right asymmetry in the mouse. Cell 114, 61-73 [DOI] [PubMed] [Google Scholar]
- Mikkers H., Allen J., Knipscheer P., Romeijn L., Hart A., Vink E., Berns A. (2002). High-throughput retroviral tagging to identify components of specific signaling pathways in cancer. Nat. Genet. 32, 153-159 [DOI] [PubMed] [Google Scholar]
- Murcia N. S., Richards W. G., Yoder B. K., Mucenski M. L., Dunlap J. R., Woychik R. P. (2000). The Oak Ridge Polycystic Kidney (orpk) disease gene is required for left-right axis determination. Development 127, 2347-2355 [DOI] [PubMed] [Google Scholar]
- Nair S., Schilling T. F. (2008). Chemokine signaling controls endodermal migration during zebrafish gastrulation. Science 322, 89-92 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neugebauer J. M., Amack J. D., Peterson A. G., Bisgrove B. W., Yost H. J. (2009). FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature 458, 651-654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nonaka S., Tanaka Y., Okada Y., Takeda S., Harada A., Kanai Y., Kido M., Hirokawa N. (1998). Randomization of left-right asymmetry due to loss of nodal cilia generating leftward flow of extraembryonic fluid in mice lacking KIF3B motor protein. Cell 95, 829-837 [DOI] [PubMed] [Google Scholar]
- Nonaka S., Yoshiba S., Watanabe D., Ikeuchi S., Goto T., Marshall W. F., Hamada H. (2005). De novo formation of left-right asymmetry by posterior tilt of nodal cilia. PLoS Biol. 3, e268 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Donnell K. A., Wentzel E. A., Zeller K. I., Dang C. V., Mendell J. T. (2005). c-Myc-regulated microRNAs modulate E2F1 expression. Nature 435, 839-843 [DOI] [PubMed] [Google Scholar]
- Ober E. A., Olofsson B., Makinen T., Jin S. W., Shoji W., Koh G. Y., Alitalo K., Stainier D. Y. (2004). Vegfc is required for vascular development and endoderm morphogenesis in zebrafish. EMBO Rep. 5, 78-84 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okada Y., Nonaka S., Tanaka Y., Saijoh Y., Hamada H., Hirokawa N. (1999). Abnormal nodal flow precedes situs inversus in iv and inv mice. Mol. Cell 4, 459-468 [DOI] [PubMed] [Google Scholar]
- Ota A., Tagawa H., Karnan S., Tsuzuki S., Karpas A., Kira S., Yoshida Y., Seto M. (2004). Identification and characterization of a novel gene, C13orf25, as a target for 13q31-q32 amplification in malignant lymphoma. Cancer Res. 64, 3087-3095 [DOI] [PubMed] [Google Scholar]
- Oteiza P., Koppen M., Concha M. L., Heisenberg C. P. (2008). Origin and shaping of the laterality organ in zebrafish. Development 135, 2807-2813 [DOI] [PubMed] [Google Scholar]
- Petrocca F., Vecchione A., Croce C. M. (2008a). Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res. 68, 8191-8194 [DOI] [PubMed] [Google Scholar]
- Petrocca F., Visone R., Onelli M. R., Shah M. H., Nicoloso M. S., de Martino I., Iliopoulos D., Pilozzi E., Liu C. G., Negrini M., et al. (2008b). E2F1-regulated microRNAs impair TGFbeta-dependent cell-cycle arrest and apoptosis in gastric cancer. Cancer Cell 13, 272-286 [DOI] [PubMed] [Google Scholar]
- Raya A., Izpisua Belmonte J. C. (2006). Left-right asymmetry in the vertebrate embryo: from early information to higher-level integration. Nat. Rev. Genet. 7, 283-293 [DOI] [PubMed] [Google Scholar]
- Reiter J. F., Alexander J., Rodaway A., Yelon D., Patient R., Holder N., Stainier D. Y. (1999). Gata5 is required for the development of the heart and endoderm in zebrafish. Genes Dev. 13, 2983-2995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reiter J. F., Kikuchi Y., Stainier D. Y. (2001). Multiple roles for Gata5 in zebrafish endoderm formation. Development 128, 125-135 [DOI] [PubMed] [Google Scholar]
- Rodaway A., Takeda H., Koshida S., Broadbent J., Price B., Smith J. C., Patient R., Holder N. (1999). Induction of the mesendoderm in the zebrafish germ ring by yolk cell-derived TGF-beta family signals and discrimination of mesoderm and endoderm by FGF. Development 126, 3067-3078 [DOI] [PubMed] [Google Scholar]
- Rosa A., Spagnoli F. M., Brivanlou A. H. (2009). The miR-430/427/302 family controls mesendodermal fate specification via species-specific target selection. Dev. Cell 16, 517-527 [DOI] [PubMed] [Google Scholar]
- Rupp R. A., Snider L., Weintraub H. (1994). Xenopus embryos regulate the nuclear localization of XMyoD. Genes Dev. 8, 1311-1323 [DOI] [PubMed] [Google Scholar]
- Sakaguchi T., Kikuchi Y., Kuroiwa A., Takeda H., Stainier D. Y. (2006). The yolk syncytial layer regulates myocardial migration by influencing extracellular matrix assembly in zebrafish. Development 133, 4063-4072 [DOI] [PubMed] [Google Scholar]
- Schier A. F., Neuhauss S. C., Helde K. A., Talbot W. S., Driever W. (1997). The one-eyed pinhead gene functions in mesoderm and endoderm formation in zebrafish and interacts with no tail. Development 124, 327-342 [DOI] [PubMed] [Google Scholar]
- Schneider I., Houston D. W., Rebagliati M. R., Slusarski D. C. (2008). Calcium fluxes in dorsal forerunner cells antagonize beta-catenin and alter left-right patterning. Development 135, 75-84 [DOI] [PubMed] [Google Scholar]
- Schulte-Merker S., Hammerschmidt M., Beuchle D., Cho K. W., De Robertis E. M., Nusslein-Volhard C. (1994). Expression of zebrafish goosecoid and no tail gene products in wild-type and mutant no tail embryos. Development 120, 843-852 [DOI] [PubMed] [Google Scholar]
- Shivdasani R. A. (2002). Molecular regulation of vertebrate early endoderm development. Dev. Biol. 249, 191-203 [DOI] [PubMed] [Google Scholar]
- Shkumatava A., Stark A., Sive H., Bartel D. P. (2009). Coherent but overlapping expression of microRNAs and their targets during vertebrate development. Genes Dev. 23, 466-481 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stainier D. Y., Fouquet B., Chen J.-N., Warren K. S., Weinstein B. M., Meiler S. E., Mohideen M. A., Neuhauss S. C., Solnica-Krezel L., Schier A. F., Zwartkruis F., Stemple D. L., Malicki J., Driever W., Fishman M. C. (1996). Mutations affecting the formation and function of the cardiovascular system in the zebrafish embryo. Development 123, 285-292 [DOI] [PubMed] [Google Scholar]
- Supp D. M., Brueckner M., Kuehn M. R., Witte D. P., Lowe L. A., McGrath J., Corrales J., Potter S. S. (1999). Targeted deletion of the ATP binding domain of left-right dynein confirms its role in specifying development of left-right asymmetries. Development 126, 5495-5504 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T., Shen H., Akagi K., Morse H. C., Malley J. D., Naiman D. Q., Jenkins N. A., Copeland N. G. (2002). New genes involved in cancer identified by retroviral tagging. Nat. Genet. 32, 166-174 [DOI] [PubMed] [Google Scholar]
- Sylvestre Y., De Guire V., Querido E., Mukhopadhyay U. K., Bourdeau V., Major F., Ferbeyre G., Chartrand P. (2007). An E2F/miR-20a autoregulatory feedback loop. J. Biol. Chem. 282, 2135-2143 [DOI] [PubMed] [Google Scholar]
- Tabin C. J., Vogan K. J. (2003). A two-cilia model for vertebrate left-right axis specification. Genes Dev. 17, 1-6 [DOI] [PubMed] [Google Scholar]
- Takacs C. M., Giraldez A. J. (2011). MicroRNAs as genetic sculptors: fishing for clues. Semin. Cell Dev. Biol. 21, 760-767 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takeda S., Yonekawa Y., Tanaka Y., Okada Y., Nonaka S., Hirokawa N. (1999). Left-right asymmetry and kinesin superfamily protein KIF3A: new insights in determination of laterality and mesoderm induction by kif3A−/− mice analysis. J. Cell Biol. 145, 825-836 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka Y., Okada Y., Hirokawa N. (2005). FGF-induced vesicular release of Sonic hedgehog and retinoic acid in leftward nodal flow is critical for left-right determination. Nature 435, 172-177 [DOI] [PubMed] [Google Scholar]
- Thisse C., Thisse B. (2008). High-resolution in situ hybridization to whole-mount zebrafish embryos. Nat. Protoc. 3, 59-69 [DOI] [PubMed] [Google Scholar]
- Ventura A., Young A. G., Winslow M. M., Lintault L., Meissner A., Erkeland S. J., Newman J., Bronson R. T., Crowley D., Stone J. R., et al. (2008). Targeted deletion reveals essential and overlapping functions of the miR-17 through 92 family of miRNA clusters. Cell 132, 875-886 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volinia S., Calin G. A., Liu C. G., Ambs S., Cimmino A., Petrocca F., Visone R., Iorio M., Roldo C., Ferracin M., et al. (2006). A microRNA expression signature of human solid tumors defines cancer gene targets. Proc. Natl. Acad. Sci. USA 103, 2257-2261 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang C. L., Wang B. B., Bartha G., Li L., Channa N., Klinger M., Killeen N., Wabl M. (2006). Activation of an oncogenic microRNA cistron by provirus integration. Proc. Natl. Acad. Sci. USA 103, 18680-18684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Warga R. M., Nusslein-Volhard C. (1999). Origin and development of the zebrafish endoderm. Development 126, 827-838 [DOI] [PubMed] [Google Scholar]
- Wienholds E., Kloosterman W. P., Miska E., Alvarez-Saavedra E., Berezikov E., de Bruijn E., Horvitz H. R., Kauppinen S., Plasterk R. H. A. (2005). MicroRNA expression in zebrafish embryonic development. Science 309, 310-311 [DOI] [PubMed] [Google Scholar]
- Woods K., Thomson J. M., Hammond S. M. (2007). Direct regulation of an oncogenic micro-RNA cluster by E2F transcription factors. J. Biol. Chem. 282, 2130-2134 [DOI] [PubMed] [Google Scholar]
- Xiao C., Srinivasan L., Calado D. P., Patterson H. C., Zhang B., Wang J., Henderson J. M., Kutok J. L., Rajewsky K. (2008). Lymphoproliferative disease and autoimmunity in mice with increased miR-17-92 expression in lymphocytes. Nat. Immunol. 9, 405-414 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamauchi H., Miyakawa N., Miyake A., Itoh N. (2009). Fgf4 is required for left-right patterning of visceral organs in zebrafish. Dev. Biol. 332, 177-185 [DOI] [PubMed] [Google Scholar]
- Yelon D., Horne S. A., Stainier D. Y. (1999). Restricted expression of cardiac myosin genes reveals regulated aspects of heart tube assembly in zebrafish. Dev. Biol. 214, 23-37 [DOI] [PubMed] [Google Scholar]
- Zorn A. M., Wells J. M. (2009). Vertebrate endoderm development and organ formation. Annu. Rev. Cell Dev. Biol. 25, 221-251 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.