Abstract
IL-1F6, IL-1F8 and IL-1F9 and the IL-1R6(RP2) receptor antagonist IL-1F5 constitute a novel IL-1 signaling system that is poorly characterized in skin. To further characterize these cytokines in healthy and inflamed skin, we studied their expression in healthy control (NN), uninvolved psoriasis (PN) and psoriasis plaque (PP) skin using QRT-PCR and immunohistochemistry. Expression of IL-1F5, -1F6, -1F8, and -1F9 were increased 2-3 orders of magnitude in PP versus PN skin, which was supported immunohistologically. Moreover, treatment of psoriasis with etanercept led to significantly decreased IL-1F5, -1F6, -1F8 and -1F9 mRNAs, concomitant with clinical improvement. Similarly increased expression of IL-1F5, -1F6, -1F8 and -1F9 was seen in the involved skin of two mouse models of psoriasis. Suggestive of their importance in inflamed epithelia, IL-1α and TNF-α induced IL-1F5, -1F6, -1F8, and -1F9 transcript expression by normal human keratinocytes. Microarray analysis revealed that these cytokines induce the expression of anti-microbial peptides and matrix metalloproteins by reconstituted human epidermis. In particular, IL-1F8 increased mRNA expression of HBD2, HBD3 and CAMP and protein secretion of HBD2 and HBD3. Collectively, our data suggest important roles for these novel cytokines in inflammatory skin diseases and identify these peptides as potential targets for antipsoriatic therapies.
Keywords: Skin, inflammation, cytokine, IL-1, psoriasis, anti-microbial peptides
INTRODUCTION
The IL-1 family of cytokines is a set of structurally related molecules that are primary mediators of inflammation (1). This family now consists of 11 cytokines and 9 cell surface receptors (reviewed in (2, 3)), the best characterized of which are IL-1α, IL-1β, IL-1ra and IL-18. Seven novel members were recently identified from DNA databases almost simultaneously by several different groups (4-7) and to reduce confusion, a systematic nomenclature was proposed (8). IL-1F6(9), IL-1F8(10, 11) and IL-1F9 (10, 12) all have reported pro-inflammatory properties. IL-1F5 shares 52% amino acid similarity with IL-1ra (1) and as such antagonizes IL-1F6 (9) and IL-1F9 (12) at the IL-1R3(RAcP)-IL-1R6(Rrp2) receptor complex (10). IL-1F7 (6) acts as an attenuator of IL-18 activity (13) and may act as an inhibitor of innate immune signaling. IL-1F10 (14) has been reported to bind soluble IL-1R1 to unknown effect. Finally, IL-1F11 (IL-33), has been reported to be a Th2-inducing cytokine (15) having important roles in the stimulation of mast cells, eosinophils, basophils and Th2 T cells, but also has a role in boosting IFN-γ production by NK cells (16).
Many of these cytokines are overexpressed in inflammatory disorders such as psoriasis (9, 12, 17, 18), which is a T-cell-mediated skin disease characterized by red, scaly and well-demarcated plaques on the skin, nail abnormalities, and, in some patients, a destructive arthritis (19). Importantly, overexpression of IL-1F6 in mouse basal keratinocytes leads to skin inflammation (9) that is exacerbated on an IL-1F5 deficient background, resulting in psoriasiform skin lesions. IL-1F5 (9), -1F6(9, 12) and -1F9 (12) mRNA have been shown to be elevated in psoriasis plaques, and keratinocytes were identified as the predominant source (12). Taken together these data indicate that IL-1F5, -1F6, -1F8, -1F9 and IL-1R6 constitute an distinct IL-1 signaling system analogous to IL-1α, -1β, -1ra and IL-1R1 that is present and active in epithelia.
The regulation and function of these new IL-1 family members in the skin is poorly understood. To better define the role of this family in skin inflammation, we acquired biopsies of healthy control, uninvolved and lesional psoriasis skin and assessed the expression of IL-1F5, -1F6, -1F8, -1F9 and their receptors by real time quantitative reverse transcription PCR (qRT-PCR), which we then confirmed by immunohistochemical evaluation. Next we examined their mRNA expression levels in lesional skin during treatment with the anti-TNF-α biologic etanercept (Enbrel®). To gain further insight into the role of these cytokines in T-cell-mediated skin inflammation we assessed how their expression was modulated by the inflammatory cytokines IL-1α, TNF-α, IL-17A and IL-22. IL-1F5 and -1F9 were efficiently induced by IL-1α, IL-17A and TNF-α. However, although IL-1F9 mRNA and intracellular protein could be induced by these cytokines, secretion of IL-1F9 by keratinocytes required application of extracellular ATP. In terms of effector functions, these newly-described cytokines induced the expression of a wide swath of epithelial defense proteins, with IL-1F8 being a particularly efficient inducer of human β-defensin (HBD) secretion. Taken together, our data suggest important roles for these novel cytokines in the pathogenesis of psoriasis and identify these peptides as potential targets for antipsoriatic therapies.
MATERIALS AND METHODS
Study Population
Twenty individuals with chronic plaque psoriasis and twenty normal controls were enrolled (age range 18-75 years). Entry criteria were the manifestation of one or more well-demarcated, scaly, erythematous psoriatic plaques that were not limited to the scalp and no systemic anti-psoriatic treatments for 2 weeks before biopsy. Biopsies sites varied between patients depending on site of active plaques whereas biopsies of uninvolved and control skin were from the buttocks.
Biopsies were also obtained from uninvolved and lesional skin of individuals responding to treatment for chronic plaque psoriasis with the biologic agent etanercept (Enbrel®). Thirty subjects were enrolled based on criteria previously published (20). Inclusion criteria included age greater than 18 years and stable plaque-type psoriasis involving at least 10% body surface area. Exclusion criteria included use of systemic psoriasis therapy within 4 weeks, topical therapy within 2 weeks, or severe co-morbid diseases. For 12 weeks, subjects received etanercept 50mg twice a week subcutaneously (open-label). At baseline, 6 mm punch biopsies were obtained under local anesthesia (lidocaine) from uninvolved skin and a target plaque. Subsequent biopsies were taken on days 1, 6, 14, 21 and 28 of therapy from the same target plaque.
Informed consent was obtained from all subjects, under protocols approved by the Institutional Review Board of the University of Michigan. This study was conducted in compliance with good clinical practice and according to the Declaration of Helsinki Principles.
Real Time Quantitative Reverse Transcription PCR (qRT-PCR)
After removal from the skin, biopsies were snap-frozen in liquid nitrogen and stored at −80°C until use. Biopsies were pulverized with a hammer while still frozen, dissolved in RLT buffer (Qiagen, Chatsworth, CA), homogenized using glass beads (Biospec Products Inc, Bartlesville, OK) and total RNA was isolated (RNeasy Mini kit, Qiagen). 200ng RNA was reverse transcribed (High Capacity cDNA Transcription kit, Applied Biosystems Inc., Foster City, CA) and transcripts quantified using a 7900HT Fast Real-time PCR system (Applied Biosystems) using Taqman primer sets purchased from Applied Biosystems (IL1A Hs00174092_m1, IL-1R8 Hs00990788_m1, IL1B Hs00174097_m1, IL-1R9 Hs00213600_m1, IL1RN (IL1F3) Hs00174099_m1, DEFB103 Hs00218678_m1, IL18 (IL1F4) Hs00155517_m1, DEFB104 Hs00414476_m1, IL1F5 Hs00202179_m1, CAMP Hs00189038_m1, IL1F6 Hs00205367_m1, CCL20 Hs00355476_m1, IL1F8 Hs00205359_m1, SERPINB1 Hs00961948_m1, IL1F9 Hs00219742_m1, CXCL8 (IL-8) Hs00174103_m1, IL-1R1 Hs00168392_m1, PI3 (elafin) Hs00268204_m1, IL-1R2 Hs00174759_m1, MMP1 Hs00233958_m1, IL1RAP (R3) Hs00370506_m1, MMP9 Hs00234579_m1, IL-1R4 Hs01073300_m1, MMP10 Hs00233987_m1, IL-1R5 Hs00175381_m1, MMP19 Hs00275699_m1, IL1rL2 (R6) Hs00909276_m1, IL-1R7 Hs00187256_m1, RPLP0 Hs99999902_m1). All values were normalized to the expression of the housekeeping gene ribosomal protein, large, P0 (RPLP0). Mouse skin mRNA was extracted, reverse transcribed and transcripts quantified as above using primers for murine IL-1 family genes (IL-1A Mm99999060_m1, IL-1F5 Mm00497802_m1, IL-1F6 Mm00457645_m1, IL-1F8 Mm01337545_m1, IL-1F9 Mm00463327_m1) and 18S rRNA housekeeping gene (Hs99999901_s1). Data were tested for normality and statistical significance calculated using Student’s t-test or Mann Whitney test as appropriate using GraphPad Prism version 5.00 for Windows (GraphPad Software, San Diego, CA).
Immunohistochemistry
Human skin biopsies were snap frozen in OCT (Tissue-Tek, Sakura), sectioned at 5μm and stained with mouse-anti human IL-1F5 (5μg/ml, R&D Systems, Minneapolis, MN, USA), IL-1F8 (5μg/ml, R&D), goat-anti human IL-1F6 (15μg/ml, R&D) or rat anti-human IL-1F9 (10μg/ml, Santa Cruz Biotechnology). Subsequently the appropriate species of secondary detection kit was used (Vectorstain ABC, Vector Labs) followed by visualization with 3,3′-DAB substrate (BD Pharmingen, San Diego, CA) and counterstaining with CAT hematoxylin (Biocare Medical).The appropriate isotype control antibodies were utilized at the same concentrations as the primary detection antibodies and gave no discernable staining.
Adult mice were euthanized; their hair shaved and skin from the back was processed for paraffin sectioning. For paraffin sectioning, skin was placed in 10% buffered formalin (Surgipath Medical Industries, Richmond, IL), overnight at 4°C prior to dehydration and embedding (Sakura Finetech, Torrance, CA). Immunohistochemistry was performed on 5μm thick paraffin sections using high temperature antigen retrieval solution (DAKO, Carpinteria, CA) for anti-mouse IL-1F6/FIL1Є (1:100 dilution, R&D Systems), or matched goat IgG isotype control (R&D Systems). The antibody and isotype control were detected using anti-goat IgG ABC kits (VectorLabs) and were visualized with 3,3′-DAB substrate (Vector Labs). Slides were counterstained with hematoxylin. For each animal, 1 image was taken for an n=6.
Keratinocyte culture
Normal human keratinocyte (NHK) cultures were established from sun-protected adult human skin as described (21) in serum-free medium optimized for high-density keratinocyte growth (Medium 154, Invitrogen/Cascade Biologics, Portland, OR). NHKs were used for experiments in the second or third passage. All cells were plated at 5000 cells/cm2 and maintained to 4-days post-confluency. Cultures were then starved of growth factors in unsupplemented medium M154 for 24 hours before use. Experiments were carried out under low calcium (0.1mM) conditions. Cultures were stimulated with recombinant human cytokines from R&D Systems: TNF-α (0.1 – 10ng/ml), IL-1α (10 ng/ml), IL-17A (2 and 10 ng/ml), IL-22 (10ng/ml).
Reconstituted human epidermal (RHE) cultures were obtained from MatTek (EPI-200, MatTek Inc, Ashland, MA). After overnight equilibration in 5ml normal maintenance medium (MatTek), medium was refreshed and supplemented with recombinant human cytokines as indicated. Medium was sampled at 24 and 48h. In addition, 4mm diameter discs were punched from RHE cultures and formalin fixed, paraffin embedded, sectioned at 5μm and stained for HBD-2 expression (1:200 dilution, goat anti-BD-2, Peprotech, Rocky Hill, NJ) or IL-1F9 expression (10μg/ml, Santa Cruz Biotechnology) and visualized with 3,3′-DAB (BD Pharmingen).
RNA processing and microarray hybridization
RHE cultures were removed from their support filters and dissolved in RLT buffer (Qiagen), homogenized using glass beads (Biospec) and total RNA was isolated (RNeasy Mini kit, Qiagen. RNA quantity and quality were measured using an Agilent 2100 Bioanalyzer (Agilent Technologies, Palo Alto, CA). Only samples yielding intact 18S and 28S ribosomal RNA profiles were used. cDNA and in vitro transcription for probe biotinylation were performed using 5μg of total RNA according to the manufacturer’s protocols (Ambion). Samples were run on Human Gene ST 1.0 arrays to query the expression 28,869 genes with 764,885 distinct probes (Affymetrix, Foster City, CA). The Robust Multichip Average method (22) was used to process the raw data. Raw microarray data has been deposited in the NCBI Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo) and is accessible through GEO Series accession number GSE25400.
ELISAs
Estimation of HBD-2 was carried out as previously described (23). IL-8 ELISA was performed as recommended (R&D Systems). Secreted HBD-3 was measured using maxisorb microtiter plates (NUNC) coated overnight at 4°C with 50μl/well rabbit anti-human BD-3 Ab (Peprotech) diluted to 3μg/ml in PBS. Plates were blocked with 200ul 1%BSA (in PBS), for 1h at RT. After washing 4 times with 300μl PBS-0.05%Tween-20, 50ul/well of samples and standards were added in duplicate and incubated for 2h at RT. After washing, 50μl/well of biotinylated rabbit anti-human BD-3 Ab (Peprotech) was added at 0.25μg/ml in PBS-0.1%BSA-0.05% Tween20 for 2h at RT. After a further 4 washes, 50μl/well streptavidin-HRP (R&D Systems, 1/200 dilution in PBS-0.1%Tween-20) was added for 30 min at RT. Following which, plates were washed again 4 times and 100μl SureBlue TMB substrate solution (KPL, Gaithersberg, MD, USA) was added to each well and incubated for up to 20 min at RT protected from light. The color reaction was stopped by addition of 50μl/well of 2N sulfuric acid and absorbance read at 450 and 620nm.
Secreted IL-1F9 was measured by ELISA. Maxisorp microtiter plates (NUNC) were coated with 100μl/well of monoclonal rat anti-human IL-1F9 antibody (R&D Systems, 2μg/ml in PBS) overnight at 4°C. After washing 3 times with 0.05% BSA in PBS, 50μl sample or recombinant IL-1F9 (R&D Systems) was added to each well in duplicate for 2h at RT. After washing 3 times, biotinylated polyclonal goat anti-human IL-1F9 detection antibody (RnD Systems, 2μg/ml in 0.05% BSA in PBS) was added for 2h at RT. After a further 3 washes, 50μl/well streptavidin-HRP (R&D Systems, 1/200 dilution in PBS-0.05%Tween-20) was added for 30 min at RT. Following which, plates were washed again 3 times and 100μl SureBlue TMB substrate solution (KPL) was added to each well and incubated for up to 20 min at RT protected from light. The color reaction was stopped by addition of 50μl/well of 2N sulfuric acid and absorbance read at 450 and 620nm.
RESULTS
Expression of IL-1F5, -1F6, -1F8 and -1F9 is increased in plaque psoriasis skin
Given the pro-inflammatory nature of IL-1α, the identification of new members of the IL-1 family raised intriguing possibilities about their involvement in epithelial inflammation. Thus, we examined the expression of these IL-1 family cytokines and their receptors using real-time quantitative RT-PCR on snap-frozen biopsies of healthy, non-lesional psoriasis and plaque psoriasis skin from 20 volunteers.
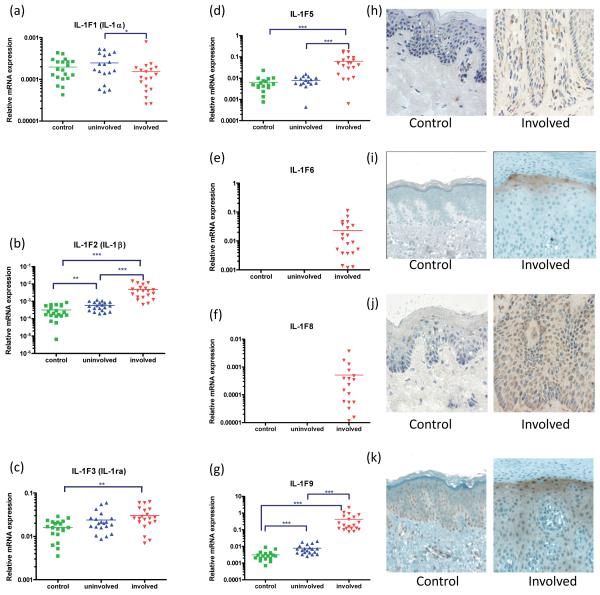
The expression of several IL-1 family members was significantly increased in plaque psoriasis skin (Figure 1). IL-1β (p<0.001), IL-1F5 (p<0.001), IL-1F6 (p<0.001), IL-1F8 (p<0.001), IL-1F9 (p<0.001) mRNA were all significantly increased compared with symptomless psoriasis skin and healthy control skin. As assessed in terms of fold-change relative to its negligible expression in normal skin, IL-1F6 mRNA was up-regulated to a much greater degree than its antagonist cytokine IL-1F5 (Figure 1d, e). This upregulation in psoriasis plaques was also evident in terms of protein expression, where IL-1F5 (Figure 1h), -1F6 (i), -1F8 (j) and -1F9 (k) proteins were all up-regulated in the keratinocytes of lesional skin compared with healthy control skin as assessed immunohistochemically. In addition, IL-1F5, -1F8 and -1F9 were also detected in the mononuclear cell infiltrate in lesional skin. We also assessed the expression of all IL-1 receptor transcripts in healthy, non-lesional and plaque skin. Of note, IL-1R2 (the IL-1α/β decoy receptor) was significantly downregulated (p<0.001) in plaque skin, as was IL-1R4 (p<0.001), the receptor for IL-1F11 (IL-33) and IL-1R8 and IL-1R9, whose ligands and functions in skin are currently unknown. However, neither IL-1R3 (IL-1RAcP) nor IL-1R6 (IL-1Rrp2) was differentially expressed in these tissues (Supplemental Figure 1).
Figure 1. The IL-1 family members IL-1F5, -1F6, -1F8 and -1F9 are over-expressed in plaque psoriasis skin.
Quantitative real-time RT-PCR revealed significant over-expression of IL-1β, IL-1ra, IL-1F5, -1F6, -1F8 and -1F9 mRNA in psoriasis skin plaques (a-g). This was confirmed by immunohistochemical detection showing diffuse cytoplasmic expression of IL-1F5 (h), -1F6 (i), heavy peri-nuclear staining of IL-1F8 (j) and heavy up-regulation of IL-1F9 in the upper spinous layers (k) of psoriasis plaques compared with healthy control skin. RT-PCR values expressed relative to the housekeeping gene RPLP0 (36B4). Statistical significance indicated * p<0.05, ** p<0.01, *** p<0.001 (n=18-20, 2-tailed t-test or Mann Whitney test as appropriate). Images 600x final magnification.
Biologic treatment of psoriasis decreases IL-1F5, -1F6, -1F8 and -1F9 expression in skin
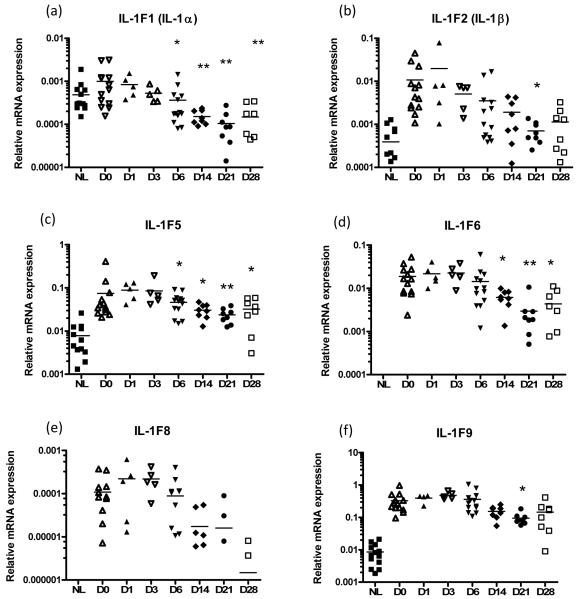
Therapies targeting TNF-α such as etanercept, a human TNF receptor fusion protein, have been shown to have high efficacy in psoriasis (24). To appreciate how expression of the IL-1 family of cytokines is modulated during disease remission, we obtained skin biopsies from patients responding to etanercept therapy for chronic plaque psoriasis. Biopsies were taken on days 1, 6, 14, 21 and 28 of treatment and processed for mRNA analysis, revealing that etanercept treatment lead to significantly decreased expression of IL-1α (p<0.001, ANOVA), IL-1β (p<0.001), IL-1F5 (p<0.01), -1F6 (p<0.001), -1F8 (p<0.001) and -1F9 (p<0.001) (Figure 2).
FIGURE 2. Treatment of chronic plaque psoriasis with the TNF-α scavenger etanercept (a human TNF receptor fusion protein) leads to a significant decrease in IL-1α (p<0.001), IL-1β (p<0.001), IL-1F5 (p<0.01), -1F6 (p<0.001), -1F8 (p<0.001) and -1F9 (p<0.001) mRNA expression in lesional skin (a-f, 1-way ANOVA with Dunnett’s test).
These changes in IL-1 family expression co-ordinate with decreases in disease severity. NL, non-lesional skin, D0-D28, day 0 - day 28 post-treatment lesional skin biopsy.
IL-1F5, -1F6, -1F8 and -1F9 are over-expressed in two mouse models of psoriasis
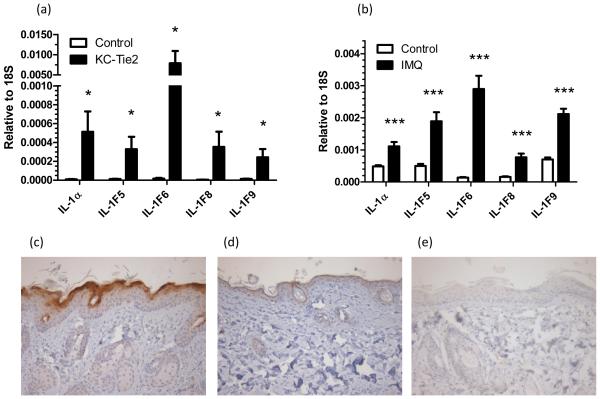
Given the dramatic over-expression of IL-1F5, -1F6, -1F8 and -1F9 in involved psoriasis skin, we went on to examine whether these cytokines were over-expressed in two established mouse models of psoriasis. We analyzed mRNA from lesional and unaffected control skin from the KC-Tie2 (Tg(KRT5-tTA)1Dmt x Tg(TetOS-Tek)1Dmt) transgenic model (25, 26) and the imiquimod-induced mouse model (27) of psoriasis (Figure 3). In both of these mouse models, the inflamed skin showed striking and significant over-expression of IL-1α, IL-1F5, -1F6, -1F8 and -1F9 transcripts compared with skin from mono-transgenic control littermates (Figure 3a) or non-imiquimod-treated skin (Figure 3b), respectively. Moreover, immunohistochemistry revealed robust expression of IL-1F6 in the lesional skin of KC-Tie2 mice (Figure 3c) compared with mono-transgenic littermates (Figure 3d).
FIGURE 3. IL-1F5, -1F6, -1F8 and -1F9 are over-expressed in the involved skin of 2 mouse models of psoriasis.
mRNA was isolated from the involved skin of the of bi-transgenic KC-Tie2 mice and mono-transgenic controls (a, n=10), and imiquimod (IMQ)-treated C57BL/6 mice and untreated controls (b, n=6). Transcripts for IL-1F5, -1F6, -1F8 and -1F9 were all significantly increased in the involved skin of both mice compared with their corresponding controls. Statistical significance indicated * p<0.05, ** p<0.01, *** p<0.001 (2-tailed t-test or Mann Whitney test as appropriate). IL-1F6 protein was heavily expressed in the involved skin of the of bi-transgenic KC-Tie2 mice (c) compared with mono-transgenic controls (d) as assessed by immunohistochemistry. No staining was detected in the corresponding isotype control (e). Representative photomicrographs of n=6 bi-transgenic and mono-transgenic mice.
Psoriasis-associated cytokines induce IL-1F5, -1F8 and -1F9 expression by keratinocytes
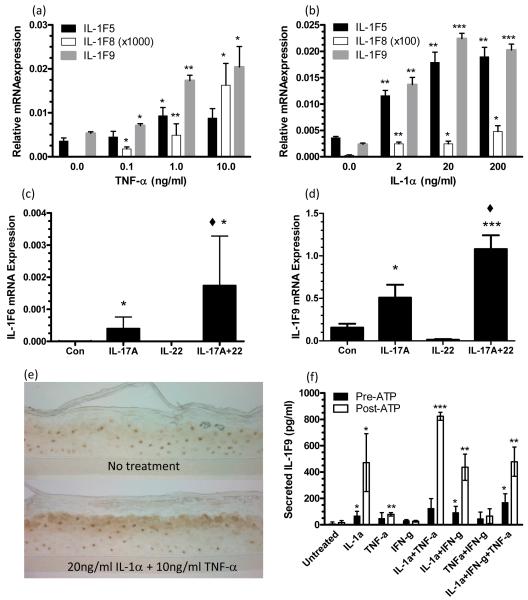
To begin to understand the induction of IL-1 cytokines in inflamed skin, we treated normal NHK with the psoriasis-associated cytokines TNF-α, IL-1α, IL-17A, and IL-22. Stimulation of NHK with TNF-α dose-dependently induced IL-1F5, -1F8 and -1F9 mRNAs (Figure 4a). IL-1α treatment lead to dose-dependent increased expression of IL-1F5, -1F8, -1F9 (Figure 4b). IL-17A induced IL-1F6 and -1F9 expression (Figure 4c,d) and while IL-22 alone was ineffective, it potentiated the effects of IL-17A for the induction of IL-1F6 (p<0.05) and IL-1F9 (p<0.001) (Figure 4c,d).
FIGURE 4. The inflammatory cytokines IL-1a, TNF-a and IL-17A induce expression of IL-1 family members by cultured keratinocytes.
Post-confluent NHK were treated as indicated for 24h and qRT-PCR analyses revealed induction of IL-1F5, -1F8, -1F9 by TNF-a (a) and IL-1a treatment (b). IL-1F6 (c) and -1F9 (d) transcripts were induced by IL-17A which could be augmented by IL-22. IL-1α and TNF-α promote keratinocyte IL-1F9 expression (e) but extracellular ATP (1mM, 2h) is required for optimal secretion of IL-1F9 into the culture medium as analyzed by IL-1F9 ELISA (f). IL-1F9 was expressed in the upper spinous layers of cultures of reconstituted human epidermis as manifested by immunohistochemical staining with antibodies against IL-1F9 (e). Bars show mean ± SD (n=3). Statistical significance indicated * p<0.05, ** p<0.01, *** p<0.001 versus no treatment, and ◆ p<0.05 for IL17A+IL-22 versus IL-17A alone.
The induction of IL-1F9 protein was confirmed in IL-1α and TNF-α-treated RHE cultures (Figure 4e). Individually, both IL-1α (4f) and TNF-α (4f) could induce keratinocyte expression of IL-1F9, however, a combination of the two cytokines was most effective (Figure 4e, f). Although this stimulation was particularly efficient at inducing IL-1F9 protein expression in tissue as detected immunohistochemically, little secreted IL-1F9 was detected in conditioned culture medium. Given that like IL-1α IL-1F5 – IL-1F9 do not have leader sequences that target their secretion (7), and IL-1α is likely secreted by an ATP-dependent alternative pathway (28), we stimulated NHK with exogenous ATP for 2h following 24h of cytokine stimulation (Figure 4f). Application of ATP markedly induced secretion of IL-1F9 from the keratinocyte cultures, as measured by ELISA of conditioned medium at 2h (Figure 4f).
IL-1F8 functions as an inducer of antimicrobial peptide and MMP expression by keratinocytes
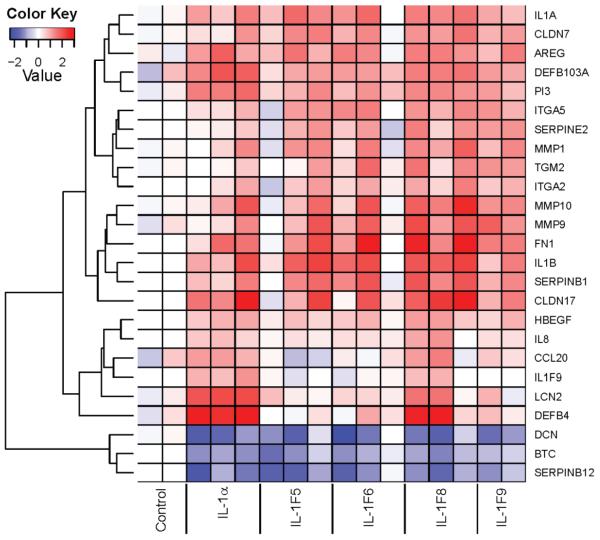
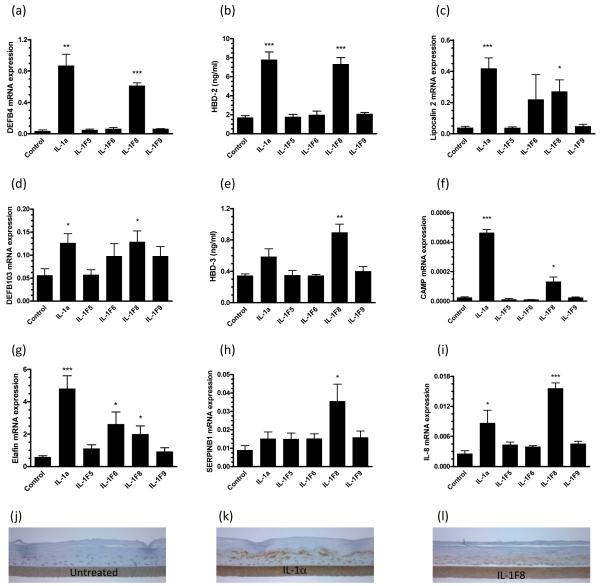
Although induction of NF-κB activation by IL-1F6 (10), -1F8 (10) and -1F9 (10, 12) has been demonstrated, no function has been assigned to these cytokines. Thus we treated RHE cultures with IL-1F5, -1F6, -1F8 and -1F9 and performed microarray analyses using the Affymetrix Human Gene ST 1.0 platform. IL-1F5, -1F6, -1F8, -1F9 upregulated (>2-fold) 52, 69, 77 and 37 transcripts and down-regulated (>2-fold) 246, 54, 20 and 11 transcripts respectively. A heat map illustrating a selection of psoriasis and inflammation-associated transcripts is shown in Figure 5. Subsequent QRT-PCR analysis confirmed that several antimicrobial peptides are significantly induced by IL-1F8, including HBD-2 (DEFB4, Figure 6a), lipocalin2 (6c), HBD-3 (6d), CAMP (6f), elafin (6g), serpinB1 (6h) and IL-8 (6i). IL-1F8-induced secretion of the defensins HBD-2 and HBD-3 into the conditioned cell culture medium was detected by ELISA (Figure 6b,e). The upregulation of HBD-2 protein expression by IL-1α and IL-1F8 treated RHE cultures could also be detected immunohistochemically as shown in Figure 6j-l. Collectively, these data illustrate for the first time the induction of antimicrobial peptides by IL-1F8.
FIGURE 5. Microarray analysis revealed the induction of anti-microbial peptides and molecules involved in the remodeling and barrier function of the epidermis.
Reconstituted human epidermal cultures were stimulated for 24h with 25ng/ml recombinant IL-1a, 5μg/ml IL-1F5, -1F6, -1F8 or -1F9 and analyzed using Affymetrix Human Gene ST 1.0 microarrays. X-axis contains data from replicate RHE cultures. Color key: Dark red, 3-fold increased expression; dark blue 2.5-fold decreased expression compared with the average expression values of the untreated (control) cultures.
FIGURE 6. IL-1F8 induces expression of epithelial defense proteins by cultured human keratinocytes.
Reconstituted human epidermal cultures were stimulated for 24h with 25ng/ml recombinant IL-1α, 5μg/ml IL-1F5, -1F6, -1F8 or -1F9. QRT-PCR shows IL-1F8 increased expression of the anti-microbial peptides DEFB4 (a), lipocalin 2 (c) DEFB103 (d), CAMP (f), elafin (g), serpin B1 (h) and IL-8 (i). ELISA of the conditioned culture medium revealed respective 4-fold and 2.6-fold increases in secreted HBD-2 (b) and HBD-3 peptides (e) in response to IL-1F8. Induction of HBD-2 could also be visualized immunohistochemically (j-l). Bars, mean ± S.D (n=6). Statistical significance indicated * p<0.05, ** p<0.01, *** p<0.001.
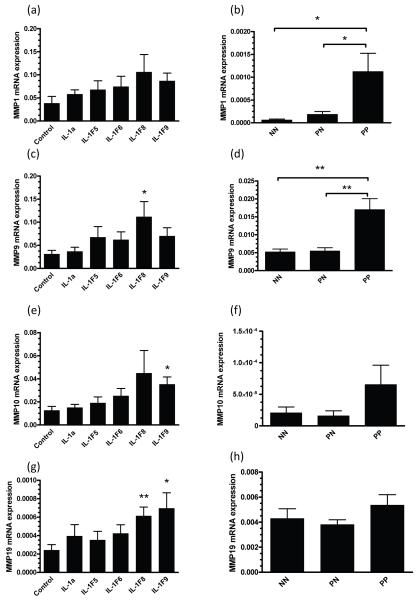
Our microarray analysis also highlighted a number of matrix metalloproteinases (MMP) as targets of IL-1F8 and IL-1F9 treatment of RHE cultures (Figure 5) which is of interest as a number of MMPs are over-expressed in lesional psoriasis skin (29, 30). Using QRT-PCR we demonstrate that IL-1F8 could significantly induce 3.6-fold and 2.6-fold increases in MMP9 and MMP19 mRNA respectively (Figure 7c, g) and IL-1F9 induced 2.8-fold and 2.9-fold increases in MMP10 and MMP19 transcript expression respectively (Figure 7e, g). Interestingly, MMP1 (18-fold, p<0.05) and MMP9 (2.8-fold, p<0.01) were significantly elevated in lesional psoriasis skin (Figure 7).
FIGURE 7. IL-1F8 induces MMP expression by reconstituted human epidermal (RHE) cultures.
Reconstituted human epidermal cultures were stimulated for 24h with 25ng/ml recombinant IL-1a or 5μg/ml IL-1F5, -1F6, -1F8 or -1F9. QRT-PCR revealed significant increases in MMP9 (3.6-fold, c), MMP10 (3.6-fold, e) and MMP19 (2.6-fold, g) transcripts in response to IL-1F8, while IL-1F9 induced 2.8-fold and 2.9-fold increases in MMP10 (e) and MMP9 (g) respectively. MMP1 (b) and MMP9 (d) were both significantly over-expressed in psoriasis lesions (PP) compared with symptomless skin (NN or PN). Bars, mean ± S.D (n=9). Statistical significance indicated * p<0.05, ** p<0.01, *** p<0.001.
DISCUSSION
Epidermal inflammation is driven by the coordinated action of a number of cytokines and growth factors, including the IL-1 family. With the discovery of IL-1F5 (4-7, 31), IL-1F6 (7), IL-1F8 (5, 7) and IL-1F9 (6), these cytokines together with their receptors IL-1R6(RP2) and IL-1R3(RAcP) form a novel and distinct IL-1 signaling system. Since over-expression of IL-1F6 in the basal keratinocytes of an IL-1F5-deficient mouse lead to psoriasiform hyperplasia (9), we proceeded to characterize this cytokine family in human psoriasis skin. Expression of IL-1F5, -1F6, -1F8, and -1F9 transcripts were increased 2-3 orders of magnitude in PP versus PN skin (Figure 1) which was supported immunohistologically and is in accord with the earlier findings that IL-1β (32), IL-18 (33), IL-1F5 (17), IL-1F6 (9) and IL-1F9 (9, 17) are all markedly elevated in lesional psoriasis skin. IL-1α mRNA and protein have been previously shown to be decreased in psoriasis plaques (32) which is consistent with the data we present here (Figure 1a). Interestingly neither receptor for these cytokines was differentially expressed (IL-1R3 and IL-1R6, Supplementary Figure 1). We also found significantly decreased expression of IL-1α, IL-1β, IL-1F5, IL-1F6, IL-1F8 and IL-1F9 mRNAs in skin during treatment of psoriasis with etanercept (Figure 2), which is consistent the earlier findings that two other IL-1 family members, IL-1β (34) and IL-18 (35), are downregulated during effective treatment of psoriasis, and with studies in a mouse-human psoriasis xenograft system which show markedly decreased IL-1F6 expression in the skin lesions during effective treatment with etanercept (36).
Given the marked expression of these cytokines in psoriasis lesions and that the overexpression of IL-1F6 leads to psoriasiform inflammation in the mouse (9) we examined two mouse models of psoriasis (Figure 3) and determined that IL-1F5, -1F6, -1F8 and -1F9 were all dramatically elevated in the lesional skin of both KC-Tie2 and imiquimod-treated mice. We also demonstrate that IL-1F6 protein was robustly expressed in the lesional skin of KC-Tie2 mice (Figure 3c). It is particularly striking that in the KC-Tie2 mice, IL-1F6 mRNA is expressed 25-times more abundantly than that of its receptor antagonist IL-1F5 (Figure 3a).
Although IL-1F6, -1F8 and -1F9 have been shown to activate NF-κB (10, 12), JNK and/or ERK1/2-mediated pathways in cell lines (10), no function has yet been assigned to these molecules in skin. Thus we treated reconstituted epidermal cultures with recombinant IL-1F6, -1F8 and -1F9 and performed microarray analysis. Strikingly, these cytokines induced a broad pattern of increased anti-microbial peptide (AMP), matrix metalloprotein (MMP) and growth factor expression (Figure 5). In particular IL-1F8 was effective at inducing keratinocytes to express HBD-2 and HBD-3, the secretion of which was readily detectable in conditioned culture medium (Figure 6). This is an interesting observation in the light of the fact that the mononuclear cell infiltrate in psoriasis plaques is positive for IL-1F6, -1F8 and -1F9 immunoreactivity (Figure 1h-k) and that the expression of these cytokines can be induced in blood monocytes in culture (A. Johnston, unpublished observation) and thus may contribute to AMP and MMP expression in inflamed skin. In addition, the mutual induction of IL-1α, IL-1β and IL-1F9 may act as a positive feedback loop driving AMP expression by the epidermis. It is interesting to note that the increases in MMP expression in PP vs PN or NN skin tend to be larger than those induced by cytokine treatment of RHE cultures (Figure 7). These differences may relate to the fact that RHE cultures continue to express the “regenerative maturation” program of epidermal differentiation that is also characteristic of psoriasis (37) though they are capable of reverting to a more normal differentiation profile after grafting to immunocompromised mice (38).
Decorin (DCN) and betacellulin (BTC) were both consistently down-regulated by the IL-1 family cytokines in our microarray experiments (Figure 5) and both of these transcripts are heavily down-regulated in lesional psoriasis skin (23, 39). BTC is the only epidermal growth factor receptor (EGFR) ligand which is decreased in psoriasis skin (23, 40). Its role in skin biology is largely unknown, although transgenic overexpression of the extracellular domain of BTC in the mouse leads to alterations in hair follicle development and cycling, and increased angiogenesis at experimental wounding sites (41). Decorin has been reported to interact with transforming growth factor (TGF)-β (42), and via this interaction it is thought to participate in cell cycle control (43).
While others have shown that IL-1F9 is induced in NHK by treatment with IL-1α and TNF-α (5), here we demonstrate for the first time that treatment with exogenous ATP is required for secretion of IL-1F9 into the medium (Figure 4e,f). The same phenomenon was recently described for IL-1F6 release from transfected murine bone marrow-derived macrophages (44). IL-1F6, -1F8 and -1F9, like IL-1α and -1β, lack signal sequences for conventional secretion (7) via the ER/Golgi pathway, and are secreted via a non-classical pathway involving multivesicular bodies and exosomes (28) in a process triggered by extracellular ATP. Interestingly, like IL-1α (45), one role of IL-1F5, IL-1F6, IL-1F8 and/or IL-1F9 may involve acting as a transcription factor and as such would not require secretion. To that end, both IL-1F7 (46) and IL-1F11 (IL-33) (47) translocate to the nucleus where they function as a transcription factors.
We typically required microgram/ml quantities of IL-1F5, -1F6, -1F8 and -1F9 for in vitro activity, which is consistent with earlier findings (10, 12). Thus, we must consider the possibility that these cytokines require some form of post-translational modification that is not included in the commercial recombinant preparations we have used, such as glycosylation (48) and / or trimming of N-terminal amino acid residues (36). It will be important to focus on understanding how these molecules might function intracellularly as transcription factors and to further characterize how they are processed within the cell for secretion, as both modes of action may be important in tissue homeostasis and defense.
IL-1 was the first cytokine discovered in skin (49) and with the discovery that IL-1 is crucial for T-cell differentiation (50, 51), IL-1 immunobiology is undergoing a renaissance. In the light of the present study, we now have a better understanding of the broader IL-1 family in the pathobiology of psoriasis. Further, given that we have shown that these new IL-1 family members are able to drive antimicrobial peptide expression in skin and generate a psoriasis-like gene expression profile, it is possible that the use of an IL-1R6 antagonist, analogous to IL-1ra/anakinra in the cryopyrin-associated periodic syndromes (52, 53) and rheumatoid arthritis (54), may prove to be therapeutically beneficial in psoriasis.
Supplementary Material
Acknowledgments
This work was supported in part by a National Psoriasis Foundation/USA Discovery Grant (AJ) and a Dermatology Foundation Research Grant (AJ), the National Institute for Arthritis, Musculoskeletal and Skin Disease (NIAMS), National Institutes of Health (R01 AR 052889 to JTE), American Skin Association and Dermatology Foundation (JEG). JTE is supported by the Ann Arbor Veterans Affairs Hospital.
Abbreviations used in this paper
- NHK
normal human keratinocyte
- qRT-PCR
quantitative reverse transcription polymerase chain reaction
- RHE
reconstituted human epidermis
- HBD
human β-defensin
- MMP
matrix metalloproteinase
- AMP
anti-microbial peptide
REFERENCES
- 1.Dunn E, Sims JE, Nicklin MJ, O’Neill LA. Annotating genes with potential roles in the immune system: six new members of the IL-1 family. Trends Immunol. 2001;22:533–536. doi: 10.1016/s1471-4906(01)02034-8. [DOI] [PubMed] [Google Scholar]
- 2.Dinarello CA. Immunological and inflammatory functions of the interleukin-1 family. Annu Rev Immunol. 2009;27:519–550. doi: 10.1146/annurev.immunol.021908.132612. [DOI] [PubMed] [Google Scholar]
- 3.Sims JE, Smith DE. The IL-1 family: regulators of immunity. Nat Rev Immunol. 2010;10:89–102. doi: 10.1038/nri2691. [DOI] [PubMed] [Google Scholar]
- 4.Mulero JJ, Pace AM, Nelken ST, Loeb DB, Correa TR, Drmanac R, Ford JE. IL1HY1: A novel interleukin-1 receptor antagonist gene. Biochem Biophys Res Commun. 1999;263:702–706. doi: 10.1006/bbrc.1999.1440. [DOI] [PubMed] [Google Scholar]
- 5.Kumar S, McDonnell PC, Lehr R, Tierney L, Tzimas MN, Griswold DE, Capper EA, Tal-Singer R, Wells GI, Doyle ML, Young PR. Identification and initial characterization of four novel members of the interleukin-1 family. J Biol Chem. 2000;275:10308–10314. doi: 10.1074/jbc.275.14.10308. [DOI] [PubMed] [Google Scholar]
- 6.Busfield SJ, Comrack CA, Yu G, Chickering TW, Smutko JS, Zhou H, Leiby KR, Holmgren LM, Gearing DP, Pan Y. Identification and gene organization of three novel members of the IL-1 family on human chromosome 2. Genomics. 2000;66:213–216. doi: 10.1006/geno.2000.6184. [DOI] [PubMed] [Google Scholar]
- 7.Smith DE, Renshaw BR, Ketchem RR, Kubin M, Garka KE, Sims JE. Four new members expand the interleukin-1 superfamily. J Biol Chem. 2000;275:1169–1175. doi: 10.1074/jbc.275.2.1169. [DOI] [PubMed] [Google Scholar]
- 8.Sims JE, Nicklin MJ, Bazan JF, Barton JL, Busfield SJ, Ford JE, Kastelein RA, Kumar S, Lin H, Mulero JJ, Pan J, Pan Y, Smith DE, Young PR. A new nomenclature for IL-1-family genes. Trends Immunol. 2001;22:536–537. doi: 10.1016/s1471-4906(01)02040-3. [DOI] [PubMed] [Google Scholar]
- 9.Blumberg H, Dinh H, Trueblood ES, Pretorius J, Kugler D, Weng N, Kanaly ST, Towne JE, Willis CR, Kuechle MK, Sims JE, Peschon JJ. Opposing activities of two novel members of the IL-1 ligand family regulate skin inflammation. J Exp Med. 2007;204:2603–2614. doi: 10.1084/jem.20070157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Towne JE, Garka KE, Renshaw BR, Virca GD, Sims JE. Interleukin (IL)-1F6, IL-1F8, and IL-1F9 signal through IL-1Rrp2 and IL-1RAcP to activate the pathway leading to NF-kappaB and MAPKs. J Biol Chem. 2004;279:13677–13688. doi: 10.1074/jbc.M400117200. [DOI] [PubMed] [Google Scholar]
- 11.Magne D, Palmer G, Barton JL, Mezin F, Talabot-Ayer D, Bas S, Duffy T, Noger M, Guerne PA, Nicklin MJ, Gabay C. The new IL-1 family member IL-1F8 stimulates production of inflammatory mediators by synovial fibroblasts and articular chondrocytes. Arthritis Res Ther. 2006;8:R80. doi: 10.1186/ar1946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Debets R, Timans JC, Homey B, Zurawski S, Sana TR, Lo S, Wagner J, Edwards G, Clifford T, Menon S, Bazan JF, Kastelein RA. Two novel IL-1 family members, IL-1 delta and IL-1 epsilon, function as an antagonist and agonist of NF-kappa B activation through the orphan IL-1 receptor-related protein 2. J Immunol. 2001;167:1440–1446. doi: 10.4049/jimmunol.167.3.1440. [DOI] [PubMed] [Google Scholar]
- 13.Pan G, Risser P, Mao W, Baldwin DT, Zhong AW, Filvaroff E, Yansura D, Lewis L, Eigenbrot C, Henzel WJ, Vandlen R. IL-1H, an interleukin 1-related protein that binds IL-18 receptor/IL-1Rrp. Cytokine. 2001;13:1–7. doi: 10.1006/cyto.2000.0799. [DOI] [PubMed] [Google Scholar]
- 14.Lin H, Ho AS, Haley-Vicente D, Zhang J, Bernal-Fussell J, Pace AM, Hansen D, Schweighofer K, Mize NK, Ford JE. Cloning and characterization of IL-1HY2, a novel interleukin-1 family member. J Biol Chem. 2001;276:20597–20602. doi: 10.1074/jbc.M010095200. [DOI] [PubMed] [Google Scholar]
- 15.Schmitz J, Owyang A, Oldham E, Song Y, Murphy E, McClanahan TK, Zurawski G, Moshrefi M, Qin J, Li X, Gorman DM, Bazan JF, Kastelein RA. IL-33, an interleukin-1-like cytokine that signals via the IL-1 receptor-related protein ST2 and induces T helper type 2-associated cytokines. Immunity. 2005;23:479–490. doi: 10.1016/j.immuni.2005.09.015. [DOI] [PubMed] [Google Scholar]
- 16.Smithgall MD, Comeau MR, Yoon BR, Kaufman D, Armitage R, Smith DE. IL-33 amplifies both Th1- and Th2-type responses through its activity on human basophils, allergen-reactive Th2 cells, iNKT and NK cells. Int Immunol. 2008;20:1019–1030. doi: 10.1093/intimm/dxn060. [DOI] [PubMed] [Google Scholar]
- 17.Zhou X, Krueger JG, Kao MC, Lee E, Du F, Menter A, Wong WH, Bowcock AM. Novel mechanisms of T-cell and dendritic cell activation revealed by profiling of psoriasis on the 63,100-element oligonucleotide array. Physiol Genomics. 2003;13:69–78. doi: 10.1152/physiolgenomics.00157.2002. [DOI] [PubMed] [Google Scholar]
- 18.Theoharides TC, Zhang B, Kempuraj D, Tagen M, Vasiadi M, Angelidou A, Alysandratos KD, Kalogeromitros D, Asadi S, Stavrianeas N, Peterson E, Leeman S, Conti P. IL-33 augments substance P-induced VEGF secretion from human mast cells and is increased in psoriatic skin. Proc Natl Acad Sci U S A. 2010;107:4448–4453. doi: 10.1073/pnas.1000803107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Gudjonsson JE, Elder JT. Psoriasis. In: Wolff K, Goldsmith LA, Katz SI, Gilchrest BA, Paller AM, L. DJ, editors. Fitzpatrick’s Dermatology in General Medicine. McGraw-Hill; New York: 2007. pp. 169–194. [Google Scholar]
- 20.Moore A, Gordon KB, Kang S, Gottlieb A, Freundlich B, Xia HA, Stevens SR. A randomized, open-label trial of continuous versus interrupted etanercept therapy in the treatment of psoriasis. Journal of the American Academy of Dermatology. 2007;56:598–603. doi: 10.1016/j.jaad.2006.09.002. [DOI] [PubMed] [Google Scholar]
- 21.Elder JT, Fisher GJ, Zhang QY, Eisen D, Krust A, Kastner P, Chambon P, Voorhees JJ. Retinoic acid receptor gene expression in human skin. J Invest Dermatol. 1991;96:425–433. doi: 10.1111/1523-1747.ep12469889. [DOI] [PubMed] [Google Scholar]
- 22.Irizarry RA, Bolstad BM, Collin F, Cope LM, Hobbs B, Speed TP. Summaries of Affymetrix GeneChip probe level data. Nucleic Acids Res. 2003;31:e15. doi: 10.1093/nar/gng015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Johnston A, Gudjonsson JE, Aphale A, Guzman AM, Stoll SW, Elder JT. EGFR and IL-1 signaling synergistically promote keratinocyte antimicrobial defenses in a differentiation-dependent fashion. J Invest Dermatol. 2010 doi: 10.1038/jid.2010.313. In Press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mease PJ, Goffe BS, Metz J, VanderStoep A, Finck B, Burge DJ. Etanercept in the treatment of psoriatic arthritis and psoriasis: a randomised trial. Lancet. 2000;356:385–390. doi: 10.1016/S0140-6736(00)02530-7. [DOI] [PubMed] [Google Scholar]
- 25.Voskas D, Jones N, Van Slyke P, Sturk C, Chang W, Haninec A, Babichev YO, Tran J, Master Z, Chen S, Ward N, Cruz M, Jones J, Kerbel RS, Jothy S, Dagnino L, Arbiser J, Klement G, Dumont DJ. A cyclosporine-sensitive psoriasis-like disease produced in Tie2 transgenic mice. Am J Pathol. 2005;166:843–855. doi: 10.1016/S0002-9440(10)62305-X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wolfram JA, Diaconu D, Hatala DA, Rastegar J, Knutsen DA, Lowther A, Askew D, Gilliam AC, McCormick TS, Ward NL. Keratinocyte but not endothelial cell-specific overexpression of Tie2 leads to the development of psoriasis. Am J Pathol. 2009;174:1443–1458. doi: 10.2353/ajpath.2009.080858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.van der Fits L, Mourits S, Voerman JS, Kant M, Boon L, Laman JD, Cornelissen F, Mus AM, Florencia E, Prens EP, Lubberts E. Imiquimod-induced psoriasis-like skin inflammation in mice is mediated via the IL-23/IL-17 axis. J Immunol. 2009;182:5836–5845. doi: 10.4049/jimmunol.0802999. [DOI] [PubMed] [Google Scholar]
- 28.Qu Y, Franchi L, Nunez G, Dubyak GR. Nonclassical IL-1 beta secretion stimulated by P2X7 receptors is dependent on inflammasome activation and correlated with exosome release in murine macrophages. J Immunol. 2007;179:1913–1925. doi: 10.4049/jimmunol.179.3.1913. [DOI] [PubMed] [Google Scholar]
- 29.Suomela S, Kariniemi AL, Impola U, Karvonen SL, Snellman E, Uurasmaa T, Peltonen J, Saarialho-Kere U. Matrix metalloproteinase-19 is expressed by keratinocytes in psoriasis. Acta Derm Venereol. 2003;83:108–114. doi: 10.1080/00015550310007445. [DOI] [PubMed] [Google Scholar]
- 30.Cordiali-Fei P, Trento E, D’Agosto G, Bordignon V, Mussi A, Ardigo M, Mastroianni A, Vento A, Solivetti F, Berardesca E, Ensoli F. Decreased levels of metalloproteinase-9 and angiogenic factors in skin lesions of patients with psoriatic arthritis after therapy with anti-TNF-alpha. J Autoimmune Dis. 2006;3:5. doi: 10.1186/1740-2557-3-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Barton JL, Herbst R, Bosisio D, Higgins L, Nicklin MJ. A tissue specific IL-1 receptor antagonist homolog from the IL-1 cluster lacks IL-1, IL-1ra, IL-18 and IL-18 antagonist activities. Eur J Immunol. 2000;30:3299–3308. doi: 10.1002/1521-4141(200011)30:11<3299::AID-IMMU3299>3.0.CO;2-S. [DOI] [PubMed] [Google Scholar]
- 32.Cooper KD, Hammerberg C, Baadsgaard O, Elder JT, Chan LS, Sauder DN, Voorhees JJ, Fisher G. IL-1 activity is reduced in psoriatic skin. Decreased IL-1 alpha and increased nonfunctional IL-1 beta. J Immunol. 1990;144:4593–4603. [PubMed] [Google Scholar]
- 33.Naik SM, Cannon G, Burbach GJ, Singh SR, Swerlick RA, Wilcox JN, Ansel JC, Caughman SW. Human keratinocytes constitutively express interleukin-18 and secrete biologically active interleukin-18 after treatment with pro-inflammatory mediators and dinitrochlorobenzene. J Invest Dermatol. 1999;113:766–772. doi: 10.1046/j.1523-1747.1999.00750.x. [DOI] [PubMed] [Google Scholar]
- 34.Gottlieb AB, Chamian F, Masud S, Cardinale I, Abello MV, Lowes MA, Chen F, Magliocco M, Krueger JG. TNF inhibition rapidly down-regulates multiple proinflammatory pathways in psoriasis plaques. J Immunol. 2005;175:2721–2729. doi: 10.4049/jimmunol.175.4.2721. [DOI] [PubMed] [Google Scholar]
- 35.Piskin G, Tursen U, Sylva-Steenland RM, Bos JD, Teunissen MB. Clinical improvement in chronic plaque-type psoriasis lesions after narrow-band UVB therapy is accompanied by a decrease in the expression of IFN-gamma inducers -- IL-12, IL-18 and IL-23. Exp Dermatol. 2004;13:764–772. doi: 10.1111/j.0906-6705.2004.00246.x. [DOI] [PubMed] [Google Scholar]
- 36.Blumberg H, Dinh H, Dean C, Jr., Trueblood ES, Bailey K, Shows D, Bhagavathula N, Aslam MN, Varani J, Towne JE, Sims JE. IL-1RL2 and Its Ligands Contribute to the Cytokine Network in Psoriasis. J Immunol. 2010;185:4354–4362. doi: 10.4049/jimmunol.1000313. [DOI] [PubMed] [Google Scholar]
- 37.Mansbridge JN, Knapp AM, Strefling AM. Evidence for an alternative pathway of keratinocyte maturation in psoriasis from an antigen found in psoriatic but not normal epidermis. J Invest Dermatol. 1984;83:296–301. doi: 10.1111/1523-1747.ep12340429. [DOI] [PubMed] [Google Scholar]
- 38.Smiley AK, Klingenberg JM, Boyce ST, Supp DM. Keratin expression in cultured skin substitutes suggests that the hyperproliferative phenotype observed in vitro is normalized after grafting. Burns. 2006;32:135–138. doi: 10.1016/j.burns.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 39.Gudjonsson JE, Ding J, Johnston A, Tejasvi T, Guzman AM, Nair RP, Voorhees JJ, Abecasis GR, Elder JT. Assessment of the psoriatic transcriptome in a large sample: additional regulated genes and comparisons with in vitro models. J Invest Dermatol. 2010;130:1829–1840. doi: 10.1038/jid.2010.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Piepkorn M, Predd H, Underwood R, Cook P. Proliferation-differentiation relationships in the expression of heparin-binding epidermal growth factor-related factors and erbB receptors by normal and psoriatic human keratinocytes. Arch Dermatol Res. 2003;295:93–101. doi: 10.1007/s00403-003-0391-x. [DOI] [PubMed] [Google Scholar]
- 41.Schneider MR, Antsiferova M, Feldmeyer L, Dahlhoff M, Bugnon P, Hasse S, Paus R, Wolf E, Werner S. Betacellulin regulates hair follicle development and hair cycle induction and enhances angiogenesis in wounded skin. J Invest Dermatol. 2008;128:1256–1265. doi: 10.1038/sj.jid.5701135. [DOI] [PubMed] [Google Scholar]
- 42.Hildebrand A, Romaris M, Rasmussen LM, Heinegard D, Twardzik DR, Border WA, Ruoslahti E. Interaction of the small interstitial proteoglycans biglycan, decorin and fibromodulin with transforming growth factor beta. Biochem J. 1994;302(Pt 2):527–534. doi: 10.1042/bj3020527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Stander M, Naumann U, Wick W, Weller M. Transforming growth factor-beta and p-21: multiple molecular targets of decorin-mediated suppression of neoplastic growth. Cell Tissue Res. 1999;296:221–227. doi: 10.1007/s004410051283. [DOI] [PubMed] [Google Scholar]
- 44.Martin U, Scholler J, Gurgel J, Renshaw B, Sims JE, Gabel CA. Externalization of the leaderless cytokine IL-1F6 occurs in response to lipopolysaccharide/ATP activation of transduced bone marrow macrophages. J Immunol. 2009;183:4021–4030. doi: 10.4049/jimmunol.0803301. [DOI] [PubMed] [Google Scholar]
- 45.Werman A, Werman-Venkert R, White R, Lee JK, Werman B, Krelin Y, Voronov E, Dinarello CA, Apte RN. The precursor form of IL-1alpha is an intracrine proinflammatory activator of transcription. Proc Natl Acad Sci U S A. 2004;101:2434–2439. doi: 10.1073/pnas.0308705101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sharma S, Kulk N, Nold MF, Graf R, Kim SH, Reinhardt D, Dinarello CA, Bufler P. The IL-1 family member 7b translocates to the nucleus and down-regulates proinflammatory cytokines. J Immunol. 2008;180:5477–5482. doi: 10.4049/jimmunol.180.8.5477. [DOI] [PubMed] [Google Scholar]
- 47.Carriere V, Roussel L, Ortega N, Lacorre DA, Americh L, Aguilar L, Bouche G, Girard JP. IL-33, the IL-1-like cytokine ligand for ST2 receptor, is a chromatin-associated nuclear factor in vivo. Proc Natl Acad Sci U S A. 2007;104:282–287. doi: 10.1073/pnas.0606854104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gudjonsson JE, Johnston A, Stoll SW, Riblett MB, Xing X, Kochkodan JJ, Ding J, Nair RP, Aphale A, Voorhees JJ, Elder JT. Evidence for altered Wnt signaling in psoriatic skin. J Invest Dermatol. 2010;130:1849–1859. doi: 10.1038/jid.2010.67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Luger TA, Stadler BM, Luger BM, Mathieson BJ, Mage M, Schmidt JA, Oppenheim JJ. Murine epidermal cell-derived thymocyte-activating factor resembles murine interleukin 1. J Immunol. 1982;128:2147–2152. [PubMed] [Google Scholar]
- 50.Veldhoen M, Hocking RJ, Atkins CJ, Locksley RM, Stockinger B. TGFbeta in the context of an inflammatory cytokine milieu supports de novo differentiation of IL-17-producing T cells. Immunity. 2006;24:179–189. doi: 10.1016/j.immuni.2006.01.001. [DOI] [PubMed] [Google Scholar]
- 51.Kryczek I, Wei S, Vatan L, Escara-Wilke J, Szeliga W, Keller ET, Zou W. Cutting edge: opposite effects of IL-1 and IL-2 on the regulation of IL-17+ T cell pool IL-1 subverts IL-2-mediated suppression. J Immunol. 2007;179:1423–1426. doi: 10.4049/jimmunol.179.3.1423. [DOI] [PubMed] [Google Scholar]
- 52.Hawkins PN, Lachmann HJ, McDermott MF. Interleukin-1-receptor antagonist in the Muckle-Wells syndrome. N Engl J Med. 2003;348:2583–2584. doi: 10.1056/NEJM200306193482523. [DOI] [PubMed] [Google Scholar]
- 53.Agostini L, Martinon F, Burns K, McDermott MF, Hawkins PN, Tschopp J. NALP3 forms an IL-1beta-processing inflammasome with increased activity in Muckle-Wells autoinflammatory disorder. Immunity. 2004;20:319–325. doi: 10.1016/s1074-7613(04)00046-9. [DOI] [PubMed] [Google Scholar]
- 54.Furst DE. Anakinra: Review of recombinant human interleukin-I receptor antagonist in the treatment of rheumatoid arthritis. Clinical Therapeutics. 2004;26:1960–1975. doi: 10.1016/j.clinthera.2004.12.019. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.