Cardiac hypertrophy is a pathological manifestation in heart under chronic stress and a significant factor in the pathogenesis of heart failure. In the past decade or so, numerous intracellular signaling pathways and molecular mechanisms have been discovered in the pathological hypertrophy, from transcriptional regulation to signaling cascades, from epigenomic modulation to redox homeostasis. It is clear that the regulatory network contributing to cardiac hypertrophy involves complex interactions between positive and negative regulators with different molecular and mechanistic characteristics.
Earlier studies, particularly from Dr. Sadoshima’s group, have revealed the importance of thioredoxins (Trxs) mediated regulation of oxidative stress in cardiac hypertrophy, particularly through targeted modification of HDACII and ASK 1–7. These studies have contributed to the current understanding that oxidative injury is a major underlying mechanism and contributor of pathological hypertrophy in diseased heart and established the mechanistic basis for the anti-hypertrophic function of Trx-1. In this issue of Circulation Research, Yang et al from the same group report the identification and functional characterization a microRNA, miR-98 as one potentially important downstream effector of Trx-1 in suppressing cardiac hypertrophy8, thus representing yet another excellent showcase for the intricate molecular network in cardiac hypertrophy regulation spanning from redox regulation, epigenomic modulation to gene regulation.
Since small non-coding RNAs (miRNAs) enter the realm of gene regulation in cardiomyocytes, their impact has been nothing but small. There is an explosion of reports on the discovery and characterization of different miRNA species and their involvement in almost every aspect of cardiac biology and diseases, establishing an exciting new dimension in gene regulation network for cardiac development and pathogenesis9, 10. In early studies, miRNA profiling identified scores of miRNA species that were up-and down-regulated during cardiac hypertrophy11–14. Subsequent characterizations establish that some of these miRNA have the ability to induce or suppress cardiomyocyte hypertrophy. In addition to its critical role in cardiomyocyte differentiation and normal development15, miR-1 also negatively regulates myocyte sizes and endothelin-1 or isoproterenol induced hypertrophy16. Similar role as a negative regulator of hypertrophy was also demonstrated for miR-13317. On the other hand, miR-23, miR-195 and miR-208 were reported to promote cardiac hypertrophy. Numerous downstream targets have been established for these miRNAs. Not surprisingly, some of the targets include well established key transcription factors for cardiac hypertrophy, such as Gata4, Mef-2a and NFATc416, 18. In addition, other hypertrophy related genes have been targeted by miRNAs as well, including muscle specific ring finger protein 1 (MuRF1), GTPase-activating protein (RasGAP), cyclin-dependent kinase 9 (Cdk9), fibronectin, and Ras homolog enriched in brain (Rheb), RhoA, Cdc42, thyroid hormone receptor associated protein 1 and myostatin, just to name a few13, 19–21.
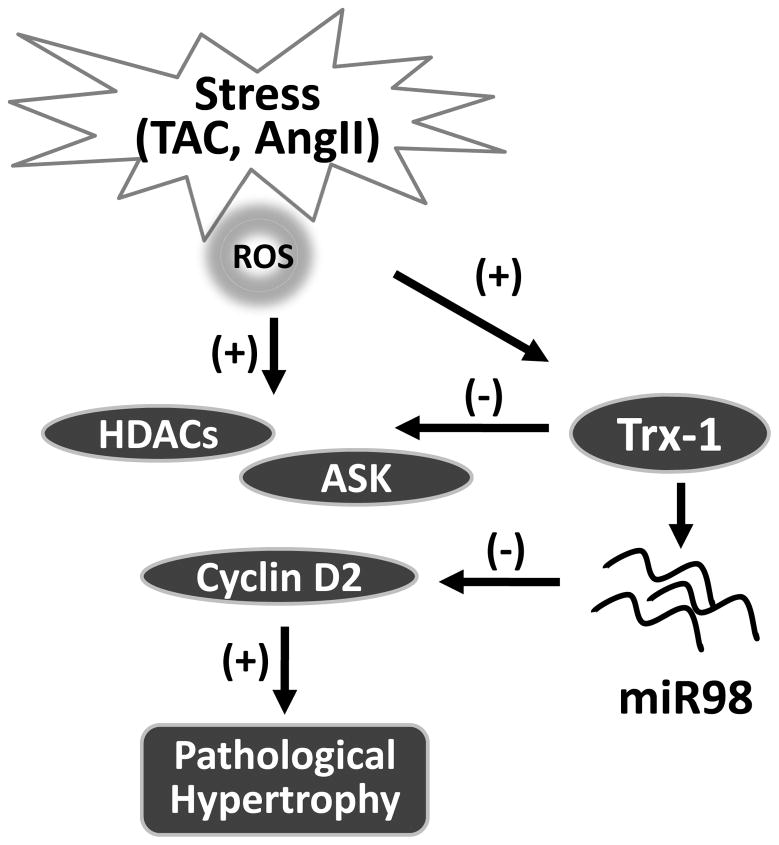
MiR-98 is a member of the let-7 family. Let7 family members are expressed in many tissues and at high levels in heart22, 23. It has been shown that the expression of let-7 family members were upregualated by ischemia/reperfusion in mouse heart24 and in human failing heart25–27. However, their function in hypertrophy regulation was never reported. In the current study8, Yang et al found that miR-98 was also upregulated in pressure overloaded or Ang II treated hearts and isolated cardiomyocytes. One of the most intriguing findings from this report, though, is that miR98/let-7 appears to be a downstream effector of Trx1. First, they found that Trx1 overexpression was sufficient to induce the expression of several members of let-7 miRNA family, including miR-98 in cardiomyocytes. Further, they demonstrated that Trx-1 expression was required for Ang II-induced miR-98 expression. More importantly, without miR98, Trx1 failed to suppress Ang II induced hypertrophy, suggesting that miR98 is indispensible for Trx1’s anti-hypertrophic function. These experiments provide a compelling argument that miR-98 is downstream of Trx1 and critical to Trx1 mediated anti-hypertrophy function in cardiac hypertrophy. Therefore, in addition to modulating the oxidative activation of stress kinases, class II HDACs and transcription factor, a non-protein player at epigenomic level was added into this complex negative feedback scheme of hypertrophy regulation (Figure 1).
Figure 1.
miR-98 and Trx-1 mediated negative feedback loop in cardiac hypertrophy.
The authors went further to characterize cyclin D2 as a functional target of miR98. They showed that miR-98 regulated AngII induced cyclin D2 expression via a specific binding motif in cyclin D2 3’-UTR and Trx1 overexpression decreased the level of cyclin D2 in a miR-98 dependent manner. They also showed that knockdown of cyclin D2 attenuated Ang II induced hypertrophy and miR98 mediated suppression of hypertrophy was attenuated by overexpression of cyclin D2. This is consistent with the conclusion that miR98 functions as a key player in an intricate negative feedback loop for hypertrophy regulation. However, a major caveat in this conclusion is that most of the experimental data is established in cultured myocytes or acutely treated hearts via adenovirus vectors, thus the exact role of miR98/cyclin-D2 axis in cardiac hypertrophy needs further examination with better genetic tools. In fact, miR-133 was shown to suppress cardiac hypertrophy in vitro and in vivo using acute knockdown approaches but had no significant impact on cardiac hypertrophy in genetic knockout model 28. Therefore, possibility of off-target effects and compensatory changes may complicate the observation and conclusions.
Comprehensive as a single report, this study also opens many more questions that need to be addressed. The underlying mechanisms of Trx1 mediated miR98 induction are unclear. Is this a general response to Trx1’s anti-oxidative effect or through other Trx1 dependent pathway? Since several members of the let-7 family other than miR-98 are also up-regulated by Trx1, do they share similar function in hypertrophy regulation? In addition to cyclinD2, other downstream targets of miR98 might also contribute to its anti-hypertrophic effect. The authors noted that miR-98 inhibits NFAT signaling pathway in cardiomyocytes, but limited information was provided. More importantly, this report focuses mostly on the effect of hypertrophy in terms of myocyte size while other associated pathological changes from contractility, metabolism to electrophysiology are not fully addressed. It needs to be established whether miR98 is involved solely in reducing myocyte hypertrophy or also directly or indirectly attenuates other aspects of cardiac pathologies. Finally, the expression pattern and the potential role of miR98 in human hypertrophic cardiomyopathy is unknown.
Nevertheless, this study offers a real opportunity for translation into a targeted therapy for hypertrophy. The functional relevance of miR98 in hypertrophy as demonstrated from this study by both loss-of-function and gain-of-function approaches in intact hearts provided a proof of concept that targeted expression of miR98 may attenuate pathological hypertrophy and improve the clinic outcome. Although Trx1 is a potent suppressor of pathological hypertrophy, it would be a major challenge to manipulate the Trx1 gene expression or protein function in intact hearts via gene or protein therapy1. In contrast, long-term administration of miRNAs or antagomers can now be achieved via systemic delivery29, 30. The discovery of miR-98 as a downstream mediator of Trx1 should provide another valuable target of intervention for cardiac hypertrophy. The study is yet another piece of evidence that the impact of small RNA in cardiac biology and therapy is only getting bigger.
Acknowledgments
Source of funding: YW is supported by NIH Grants (HL70079, HL103205, HL098954, HL080111, HL088640). HS is a supported by11SDG5570029 from American Heart Association.
List of Abbreviations
- Ang II
Angiotensin II
- miRNA
MicroRNA
- miR-98
MicroRNA-98
- NRCMs /CMs
Neonatal rat cardiomyocytes
- Trx1
Thioredoxin 1, 3’UTR, 3 prime untranslated region
Footnotes
Disclosures: None.
References
- 1.Matsushima S, Zablocki D, Sadoshima J. Application of recombinant thioredoxin1 for treatment of heart disease. J Mol Cell Cardiol. 2010 doi: 10.1016/j.yjmcc.2010.09.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Oka S, Ago T, Kitazono T, Zablocki D, Sadoshima J. The role of redox modulation of class ii histone deacetylases in mediating pathological cardiac hypertrophy. J Mol Med. 2009;87:785–791. doi: 10.1007/s00109-009-0471-2. [DOI] [PubMed] [Google Scholar]
- 3.Ago T, Liu T, Zhai P, Chen W, Li H, Molkentin JD, Vatner SF, Sadoshima J. A redox-dependent pathway for regulating class ii hdacs and cardiac hypertrophy. Cell. 2008;133:978–993. doi: 10.1016/j.cell.2008.04.041. [DOI] [PubMed] [Google Scholar]
- 4.Ago T, Sadoshima J. Thioredoxin1 as a negative regulator of cardiac hypertrophy. Antioxid Redox Signal. 2007;9:679–687. doi: 10.1089/ars.2007.1529. [DOI] [PubMed] [Google Scholar]
- 5.Ago T, Yeh I, Yamamoto M, Schinke-Braun M, Brown JA, Tian B, Sadoshima J. Thioredoxin1 upregulates mitochondrial proteins related to oxidative phosphorylation and tca cycle in the heart. Antioxid Redox Signal. 2006;8:1635–1650. doi: 10.1089/ars.2006.8.1635. [DOI] [PubMed] [Google Scholar]
- 6.Yoshioka J, Schulze PC, Cupesi M, Sylvan JD, MacGillivray C, Gannon J, Huang H, Lee RT. Thioredoxin-interacting protein controls cardiac hypertrophy through regulation of thioredoxin activity. Circulation. 2004;109:2581–2586. doi: 10.1161/01.CIR.0000129771.32215.44. [DOI] [PubMed] [Google Scholar]
- 7.Yamamoto M, Yang G, Hong C, Liu J, Holle E, Yu X, Wagner T, Vatner SF, Sadoshima J. Inhibition of endogenous thioredoxin in the heart increases oxidative stress and cardiac hypertrophy. J Clin Invest. 2003;112:1395–1406. doi: 10.1172/JCI17700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yanfei Yang TA, Zhai Peiyong, Abdellatif Maha, Sadoshima Junichi. Thioredoxin 1 negatively regulates angiotensin ii-induced cardiac hypertrophy through upregulation of mir-98/let-7. Circ Res. 2011 doi: 10.1161/CIRCRESAHA.110.228437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Condorelli G, Latronico MVG, Dorn GW. Micrornas in heart disease: Putative novel therapeutic targets? European Heart Journal. 2010;31:649–658. doi: 10.1093/eurheartj/ehp573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Small EM, Frost RJA, Olson EN. Micrornas add a new dimension to cardiovascular disease. Circulation. 2010;121:1022–1032. doi: 10.1161/CIRCULATIONAHA.109.889048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.van Rooij E, Sutherland LB, Liu N, Williams AH, McAnally J, Gerard RD, Richardson JA, Olson EN. A signature pattern of stress-responsive micrornas that can evoke cardiac hypertrophy and heart failure. Proceedings of the National Academy of Sciences. 2006;103:18255–18260. doi: 10.1073/pnas.0608791103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cheng Y, Ji R, Yue J, Yang J, Liu X, Chen H, Dean DB, Zhang C. Micrornas are aberrantly expressed in hypertrophic heart: Do they play a role in cardiac hypertrophy? Am J Pathol. 2007;170:1831–1840. doi: 10.2353/ajpath.2007.061170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Sayed D, Hong C, Chen I-Y, Lypowy J, Abdellatif M. Micrornas play an essential role in the development of cardiac hypertrophy. Circ Res. 2007;100:416–424. doi: 10.1161/01.RES.0000257913.42552.23. [DOI] [PubMed] [Google Scholar]
- 14.Tatsuguchi M, Seok HY, Callis TE, Thomson JM, Chen J-F, Newman M, Rojas M, Hammond SM, Wang D-Z. Expression of micrornas is dynamically regulated during cardiomyocyte hypertrophy. Journal of Molecular and Cellular Cardiology. 2007;42:1137–1141. doi: 10.1016/j.yjmcc.2007.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhao Y, Ransom JF, Li A, Vedantham V, von Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ, Srivastava D. Dysregulation of cardiogenesis, cardiac conduction, and cell cycle in mice lacking mirna-1–2. Cell. 2007;129:303–317. doi: 10.1016/j.cell.2007.03.030. [DOI] [PubMed] [Google Scholar]
- 16.Ikeda S, He A, Kong SW, Lu J, Bejar R, Bodyak N, Lee K-H, Ma Q, Kang PM, Golub TR, Pu WT. Microrna-1 negatively regulates expression of the hypertrophy-associated calmodulin and mef2a genes. Mol Cell Biol. 2009;29:2193–2204. doi: 10.1128/MCB.01222-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Care A, Catalucci D, Felicetti F, Bonci D, Addario A, Gallo P, Bang M-L, Segnalini P, Gu Y, Dalton ND, Elia L, Latronico MVG, Hoydal M, Autore C, Russo MA, Dorn GW, Ellingsen O, Ruiz-Lozano P, Peterson KL, Croce CM, Peschle C, Condorelli G. Microrna-133 controls cardiac hypertrophy. Nat Med. 2007;13:613–618. doi: 10.1038/nm1582. [DOI] [PubMed] [Google Scholar]
- 18.Li Q, Lin X, Yang X, Chang J. Nfatc4 is negatively regulated in mir-133a-mediated cardiomyocyte hypertrophic repression. American Journal of Physiology - Heart and Circulatory Physiology. 2010;298:H1340–H1347. doi: 10.1152/ajpheart.00592.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li P-F. Mir-23a functions downstream of nfatc3 to regulate cardiac hypertrophy. Proceedings of the National Academy of Sciences. 2009;106:12103–12108. doi: 10.1073/pnas.0811371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Rooij E, Sutherland LB, Qi X, Richardson JA, Hill J, Olson EN. Control of stress-dependent cardiac growth and gene expression by a microrna. Science. 2007;316:575–579. doi: 10.1126/science.1139089. [DOI] [PubMed] [Google Scholar]
- 21.Callis TE, Pandya K, Seok HY, Tang R-H, Tatsuguchi M, Huang Z-P, Chen J-F, Deng Z, Gunn B, Shumate J, Willis MS, Selzman CH, Wang D-Z. Microrna-208a is a regulator of cardiac hypertrophy and conduction in mice. The Journal of Clinical Investigation. 2009;119:2772–2786. doi: 10.1172/JCI36154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruvkun GB. The tiny rna world. Harvey Lect. 2003;99:1–21. [PubMed] [Google Scholar]
- 23.Boyerinas B, Park SM, Shomron N, Hedegaard MM, Vinther J, Andersen JS, Feig C, Xu J, Burge CB, Peter ME. Identification of let-7-regulated oncofetal genes. Cancer Res. 2008;68:2587–2591. doi: 10.1158/0008-5472.CAN-08-0264. [DOI] [PubMed] [Google Scholar]
- 24.van Rooij E, Sutherland LB, Thatcher JE, DiMaio JM, Naseem RH, Marshall WS, Hill JA, Olson EN. Dysregulation of micrornas after myocardial infarction reveals a role of mir-29 in cardiac fibrosis. Proceedings of the National Academy of Sciences. 2008;105:13027–13032. doi: 10.1073/pnas.0805038105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Thum T, Galuppo P, Wolf C, Fiedler J, Kneitz S, van Laake LW, Doevendans PA, Mummery CL, Borlak J, Haverich A, Gross C, Engelhardt S, Ertl G, Bauersachs J. Micrornas in the human heart: A clue to fetal gene reprogramming in heart failure. Circulation. 2007;116:258–267. doi: 10.1161/CIRCULATIONAHA.107.687947. [DOI] [PubMed] [Google Scholar]
- 26.Matkovich SJ, Van Booven DJ, Youker KA, Torre-Amione G, Diwan A, Eschenbacher WH, Dorn LE, Watson MA, Margulies KB, Dorn GW., II Reciprocal regulation of myocardial micrornas and messenger rna in human cardiomyopathy and reversal of the microrna signature by biomechanical support. Circulation. 2009;119:1263–1271. doi: 10.1161/CIRCULATIONAHA.108.813576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ikeda S, Kong SW, Lu J, Bisping E, Zhang H, Allen PD, Golub TR, Pieske B, Pu WT. Altered microrna expression in human heart disease. Physiological Genomics. 2007;31:367–373. doi: 10.1152/physiolgenomics.00144.2007. [DOI] [PubMed] [Google Scholar]
- 28.Liu N, Bezprozvannaya S, Williams AH, Qi X, Richardson JA, Bassel-Duby R, Olson EN. Microrna-133a regulates cardiomyocyte proliferation and suppresses smooth muscle gene expression in the heart. Genes & Development. 2008;22:3242–3254. doi: 10.1101/gad.1738708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.van Rooij E, Marshall WS, Olson EN. Toward microrna-based therapeutics for heart disease: The sense in antisense. Circ Res. 2008;103:919–928. doi: 10.1161/CIRCRESAHA.108.183426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hokaiwado N, Takeshita F, Banas A, Ochiya T. Rnai-based drug discovery and its application to therapeutics. IDrugs. 2008;11:274–278. [PubMed] [Google Scholar]