Abstract
Obesity is a risk factor for development of certain cancers but the basis for this risk is unclear. In this study, we developed a novel mouse model that demonstrates directly how lipogenic phenotypes commonly associated with diet induced-metabolic syndromes can influence hepatic cancer development. Activated AKT and β-catenin (AKT/CAT) genes were hydrodynamically co-delivered using the Sleeping Beauty transposon to initiate liver tumorigenesis. AKT/CAT and MET/CAT combination induced microscopic tumor foci by 4 weeks while no tumorigenesis resulted from delivery of AKT, MET or CAT alone. Primary AKT/CAT tumor cells were steatotic (fatty) hepatocellular adenomas which progressed to hepatocellular carcinomas (HCCs) upon in vivo passage, whereas primary MET/CAT tumors emerged directly as frank HCC. Conversion of AKT/CAT tumor cells to frank HCC during passage was associated with induction of the human HCC marker alpha-fetoprotein and the stem cell marker CD133. Using hierarchical clustering and gene set enrichment analysis, we compared the primary murine AKT/CAT and MET/CAT tumors to a panel of 53 human HCCs and determined these two mouse models could be stratified as distinct subtypes associated in humans with poor clinical prognosis. The chief molecular networks identified in primary and passaged AKT/CAT tumors were steatosis and lipid metabolic pathways, respectively. Our findings show how co-activation of the AKT and CAT pathways in hepatocytes can efficiently model development of a lipogenic tumor phenotype. Further, we believe our approach could speed the dissection of microenvironmental factors responsible for driving steatotic-neoplastic transformation to frank carcinoma, through genetic modification of existing immunodefined transgenic models.
Keywords: hepatocellular, lipogenic, transposon, tumor, AKT
Introduction
Knowledge of the genetic lesions associated with hepatocellular carcinoma (HCC) development has exploded in recent years with the advent of genome scale analysis and functional genomics (1). MET, beta-catenin (CAT) and AKT are among the onco-proteins and signaling pathways frequently dysregulated in HCC. In human cancers including HCC, oncogenic MET is a regulator of metastasis, angiogenesis and vascular invasion. The MET gene signature in HCC is correlated with very poor prognosis (2). Mutations in beta-catenin occur in up to 50% of all HCC. Many HCCs also demonstrate the activation of AKT and it has been reported that both hepatitis B and C can activate PI3K/AKT signaling (3). It is well established that AKT plays a key role in tumorigenesis by stimulating cell proliferation and inhibiting apoptosis. Phospho-AKT is also correlated with early disease reoccurrence and poor prognosis (4). It has been shown in several mouse models that driving hepatic AKT through either constitutively activated AKT or PTEN knockout or E2f1 overexpression, results in lipogenic hepatocytes and steatohepatitis (5–7).
Molecular and epidemiological studies show chronic infection with hepatitis virus B and C or exposure to environmental toxins is strongly associated with approximately 70% of HCC development. The remaining 30% of HCC is correlated with metabolic stress resulting from dietary factors and obesity (8). A common link between these diversely initiated etiological pathways is the dependence upon inflammation. The initial hepatic transcriptional response to metabolic stress is induction of inflammation-associated genes which are eventually replaced with lipogenic genes resulting in hepatosteatosis with a clear cell appearance (9). Chronic dietary exposure results in a sustained inflammatory response that is further exacerbated by the fatty liver microenvironment resulting in steatohepatitis. Furthermore, a transcriptional regulatory link between lipogenesis, inflammation and transformation has been recently established demonstrating genes involved in these three networks are functionally interdependent in mediating oncogenesis (10).
A better understanding of the pathologic ability of pre-neoplastic precursor cells to perceive inflammatory mediators as tumorigenic growth signals may allow us to discover new opportunities for inhibiting or treating inflammation-driven hepatic cancers. As an initial step in determining what collaborative microenvironmental/extracellular signals predispose hepatocytes to tumorigenesis, we have developed and characterized an AKT/CAT driven hepatic tumor model that accurately portrays the lipogenic phenotype seen in some human liver cancers. This model can rapidly be applied to any strain, displays progression, and is serially transplantable into syngeneic immunocompetent or transgenically immunodefined hosts (11).
Materials and Methods
Vector Construction
pT3-EF5-flox, pT3-EF5-hMET, and HSB2 were provided by Dr. Xin Chen (UCSF, San Francisco, CA). A Δ90Nbeta-catenin (first 90 amino acids deleted) was PCR amplified from RCAS-βCATS37AHA (Addgene (12)) using the following primers: 5’-CACCACAATGGCTCAGAGGGTACGAGCT-3’, 5’-TTACAGGTCAGTATCAAACC-3’. The Myr-AKT insert was PCR amplified from RCAS-myrAKT (Addgene, (13)) using the following primers: 5’-CACCCTTATGGGGAGCAGCAAG-3’, 5’-TCAGGCTGTGCCACTGGCTGA-3’. These PCR products where cloned into pENTR (Invitrogen) and subsequently transferred to pT3-EF5-flox using the manufacturers protocol to produce pT3CAT and pT3AKT (Supplemental Figure 1).
Hydrodynamic Injection and Monitoring
Hydrodynamic delivery and monitoring using strains FVB/n and C57/BL6 as previously described (11, 14). All mice were handled, fed, and housed in accordance with an approved NCI Frederick Institutional Animal Care and Use protocol.
Orthotopic Tumor Passage
Tumor cell suspensions were made by physical dissociation of liver-tumor in HBSS followed by filtration though 100 µm sieves. Cells (3×105 cells/0.5 ml) were administered as previously described (15).
Tissue Collection and Histology
Tumor bearing livers were removed and each sub-divided identically (by lobe) for subsequent analysis. Distinction between carcinoma and adenoma is based on trabeculation, differentiation, cell plate and invasion (16).
Quantitative PCR
RNA was prepared from flash frozen tissue using the RNA miniprep kit (Qiagen) and AllPrep DNA/RNA miniprep kit (Qiagen). Analysis was performed according to ABI’s manufacturer’s protocol using the following primer sets (ABI identifier): keratin 19 (Mm00492980_m1), keratin 7 (Mm00466676_m1), tumor-associated calcium signal transducer 1 (Mm00493214_m1), alpha fetoprotein (Mm00431715_m1), delta-like 1 homolog (Drosophila) (Mm00494477_m1), prominin 1 (Mm00477115_m1), dickkopf homolog 1 (Xenopus laevis) (Mm00438422_m1), BMP and activin membrane-bound inhibitor homolog (Xenopus laevis) (Mm03024088_g1), and apolipoprotein C-I (Mm01319370_g1). mRNA expression levels were normalized to endogenous beta-actin and expressed relative to normal livers. Data were log transformed and analyzed by one-way ANOVA and Tukey post test to determine statistical significance as indicated.
Transposon Replication Assay
100ng DNA was subjected to DpnI (10 units) digestion overnight at 37°C. Quantitative PCR was performed using the Taqman kit (ABI) with the following primers designed to a pT3 promoter region that harbors two DpnI sites:
| T3Pr3’In.200F | 5’-TGGAATTTGCCCTTTTTGAG-3’ |
| T3Pr3’Ot/In.200/228R | 5’-TCTGCAGAATTCCACCACAC-3’ |
Immunohistochemistry
Formalin-fixed slides were subjected to hematoxylin and eosin, Masson trichrome, Periodic acid Schiff (PAS), tunnel (APO) and immunohistochemical (IHC) staining. The indirect immunoperoxidase procedure using ABC reagent kit (DAB as chromagen), Vector Laboratories, Burlingame, CA was used and the slides were stained with antibodies including: rabbit anti-human AFP (DakoCytomation, Carpinteria, CA, 1:500 dilution), CD45 (BD Biosciences, 1:100 dilution), LFABP (Santa Cruz, 1:500 dilution), beta-catenin (Abcam 1:100 dilution), and Ki67 (Abcam 1:500 dilution). Formalin-fixed tissues were also cryopreserved and used for oil red-O staining.
Microarray
A total of 200ng RNA was amplified and incubated for 16h at 37°C (Ambion). The efficiency of the in vitro transcription was quantified using a NanoDrop ND1000 (Thermo Scientific). Array hybridization, washing, Cy3-streptavidin labeling (Amersham Biosciences), and scanning was performed on an iSCAN using reagents and protocols supplied by the manufacturer (illumina). 750ng biotinylated cRNA was hybridized to mouseRef-8v3 beadchips for 18h at 58°C. Image analysis and data extraction was automated using iSCAN Control Software (Illumina).
Statistical analysis
Data collection was performed using GenomeStudio (Illumina). The detection score for genes was computed from the z-value relative to that of negative controls. The technical error was estimated by iterative robust least squares fit and the data set normalized using quantile algorithm and background subtraction. FDR-adjusted p-values were calculated using the Benjamini-Hochberg procedure. Normal liver samples injected with empty T3 transposon were used as reference. All data was log2 transformed and significant differentially expressed genes between the surrounding liver and tumor were identified by Bootstrap-t (p≤0.001, 5,000 repetitions). Integration of human HCC and mouse data sets was performed by z-transformation. The probability of overall survival was estimated according to Kaplan-Meier and Mantel-Cox statistics (GraphPad Prism5.01). Hierarchical cluster analysis based on Euclidian distance was performed with Cluster 3, and results were visualized with TreeView v1.6. Analysis of network connectivity was completed using Ingenuity Pathway Analysis (IPA7.1). The significance of each network and the connectivity was estimated in IPA. Nonparametric gene set enrichment analysis (GSEA) was performed with the GSEA tool developed by the Broad Institute.
Results
AKT cooperates with beta-catenin to induce hepatocellular adenoma
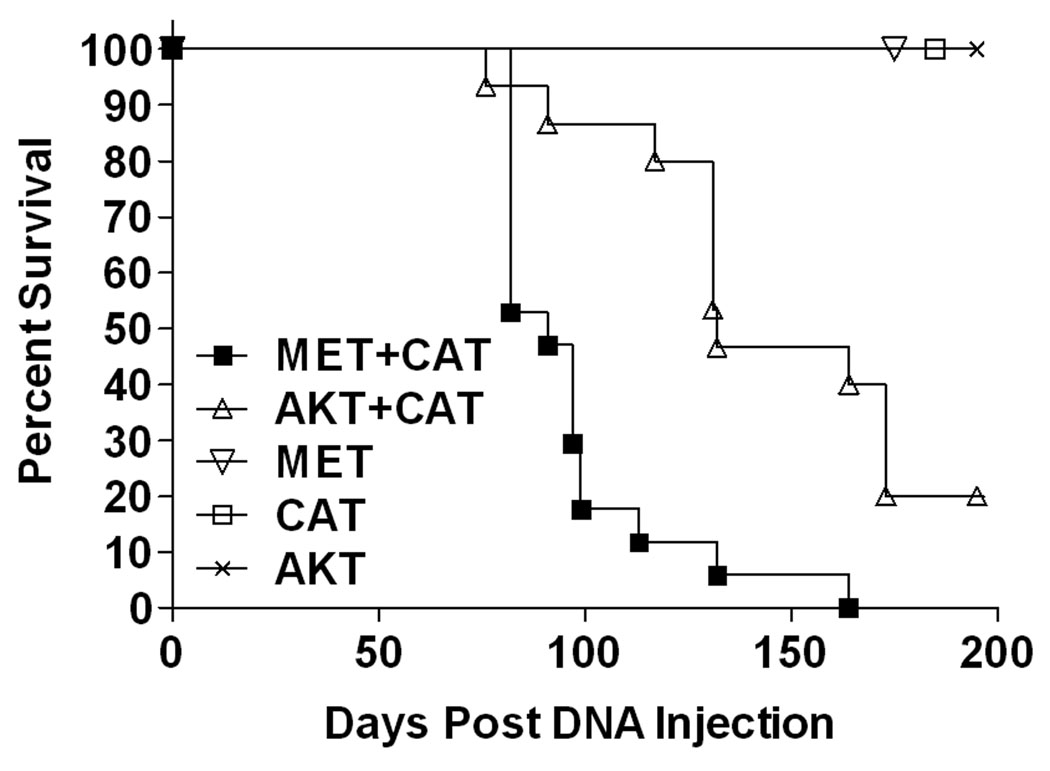
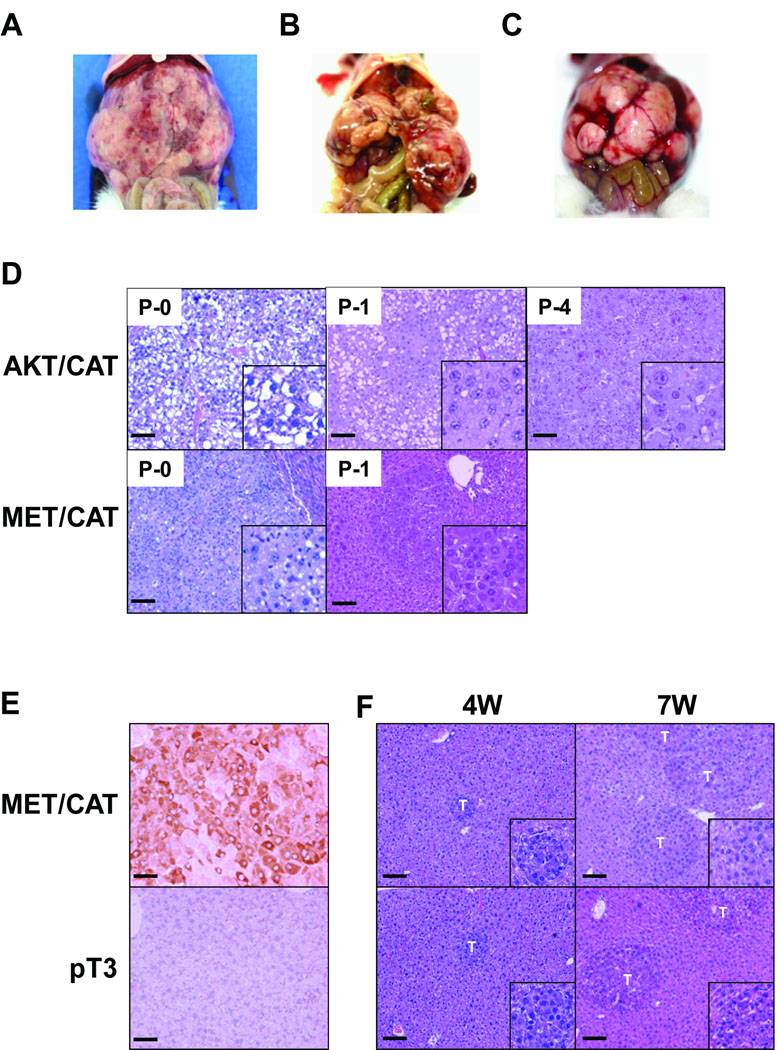
The SB transposon hydrodynamic injection system was selected to deliver transforming oncogenes to hepatocytes in vivo (Supplemental Figure 1). We hypothesized that the co-delivery of activated AKT and CAT in the liver would drive both steatogenesis and tumorigenesis. We compared the combination of activated AKT and CAT to MET and CAT which had been previously shown to induce HCCs by this method (11). Survival studies using FVB/n mice with either AKT or MET combined with CAT resulted in a median perimoribund liver tumor burden of 132 and 91 days, respectively (Figure 1). No pathology was observed in single oncogene groups, demonstrating that oncogene cooperation is required for tumorigenesis using these specific oncogenes. The gross characteristics of the resulting tumors were dependent upon the genes delivered (Figure 2A–C). The tumors were multifocal with greater than 100 nodules per liver. MET/CAT tumor was composed of tissue resembling normal liver and showing areas of intratumoral hemorrhage and bile-filled vesicles (Figure 2A). The AKT/CAT tumors were pale, likely due to lipid accumulation, with intratumoral hemorrhage (Figure 2B). Tumors were induced in both C57/BL6 and FVB/n mice with similar kinetics and penetrance. Histopathology identified foci of hyperplasia/neo-hepatocyte atypia along with hepatocellular carcinoma composed of cells with comparatively smaller cytoplasmic to nuclear volumes relative to adjacent hepatocytes in MET/CAT liver samples (Figure 2D, MET/CAT P-0). Histopathology of AKT/CAT liver tumor samples revealed the presence of foci composed of vacuolated cells with large cytoplasm to nuclear volumes consistent with hepatocellular adenoma (HCA) and hepatosteatosis (Figure 2D, AKT/CAT P-0). Histochemical staining with oil red-O and PAS confirmed these vacuoles consisted of lipid and not glycogen (Supplemental Figure 2, panels A and B). Immunohistochemical staining for alpha-fetoprotein (AFP), a common HCC marker, showed areas of reactive cells only in MET/CAT but not in vector control (pT3) or AKT/CAT –transfected liver (Figure 2E and data not shown).
Figure 1. Survival after hydrodynamic delivery of oncogene expressing SB transposons.
FVB/n mice (6–8 weeks) were administered 25 µg of the indicated oncogene by hydrodynamic tail vein injection. Mice were scored when peri-moribund with tumor burden. MET/CAT: n=17, AKT/CAT: n=15, MET: n=5, CAT: n=5, AKT: n=5.
Figure 2. Gross Morphology and histology of AKT- and MET- CAT tumor models.
Panels A–C: Hydrodynamic oncogene expressing SB tumors, (A) MET/CAT, (B) AKT/CAT, (C) serially passaged AKT/CAT tumors. Panel D: Histology (H&E) (100X, bar =100um; inset is 200X) of AKT- and MET- CAT tumors, top panels (P-0, -1, -4) are AKT/CAT tumor imaged at indicated passage progressing from clear cytoplasm (steatosis; HCA) to dense cytoplasm with trabeculation (HCC). The lower panel shows MET/CAT tumor (P-0, -1) which does not show alteration upon passage. Panel E: AFP immunohistochemistry (200X, bar =50um). Seven week MET/CAT and vector control (pT3) transfected livers were stained for AFP protein. No staining was observed in vector control or in AKT/CAT transfected livers (data not shown). Panel F: Microscopic (100X, bar =100um; inset is 200X) tumor growth in MET/CAT transfection can be detected as early as four weeks. “T” indicates tumor focus.
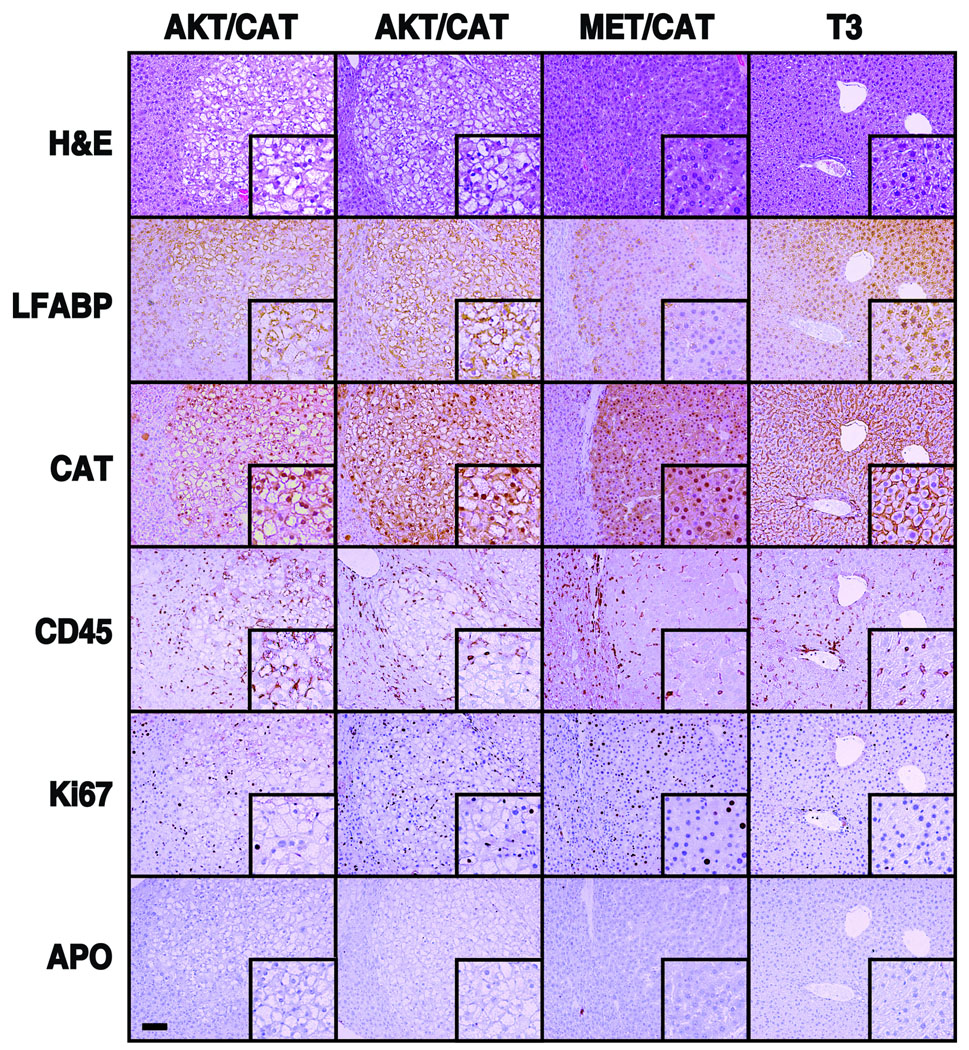
Immunohistochemical stains characteristic of specific HCA subtypes and HCC-associated pathogenesis were also carried out on AKT/CAT and MET/CAT models (Figure 3)(17). Liver FABP loss, associated with the human HNF1-alpha deficient HCA subtype, did not occur in either AKT/CAT or MET/CAT models. Staining for beta-catenin demonstrated tumor cell cytoplasmic and nuclear staining in both models. This is consistent with stabilized beta-catenin transposition-dependent tumorigenesis and is a common feature of both HCC and the beta-catenin subtype of HCA(17). An inflammation-associated increase in leukocyte infiltration was not evident by comparison of CD45 stained tumor and empty-vector. However staining for proliferation antigen Ki67 demonstrates the presence of morphologically smaller and elongated nuclei scattered throughout the tumor bearing livers consistent with leukocyte proliferation and an inflammatory response in both tumor models. Tumor cell proliferation was more apparent in MET/CAT compared to AKT/CAT. Staining for apoptosis showed no evidence for model-associated cell death. Finally, Masson trichrome staining did not support the co-induction of fibrosis in these tumor models however isolated incidences of intra-tumoral connective tissue was evident in some higher passage AKT/CAT tumors (Supplemental Figure 3 and data not shown).
Figure 3. Immunohistochemical analysis of: HCA/HCC markers, apoptosis, leukocyte infiltration, and proliferation associated with AKT/CAT, MET/CAT and control (T3) liver.
Stains are indicated in the left column and transposons are listed across the top. Two separate nodules from different livers were used for AKT/CAT. All slides are generated from serial sections (100X, bar =100um; inset is 200X) and framed to show uninvolved liver tissue at the left margin for reference. LFABP staining was not altered in either model. Beta-catenin staining (DAB) demonstrated tumor specific nuclear accumulation in both models compared to adjacent liver tissue and control (T3). CD45 staining shows little evidence of tumor infiltration however there is an increased incidence of CD45 staining in surrounding liver tissue. This coincides with Ki67 staining of small or elongated nuclei consistent with leukocyte proliferation. Ki67 staining indicates MET/CAT is proliferating more than AKT/CAT. There is no evidence of apoptosis in either model.
Serial passage of AKT/CAT tumors correlates with increased malignancy
The ability to serially transplant tumors was tested by intrasplenic injection of 3×105 dispersed tumor cells. This approach assures hepatic dispersion of single cell suspension throughout the liver combined with pooling many primary tumors to further minimize the likelihood of clonal dominance or competition. Surprisingly the MET/CAT tumors had a very long latency (Table 1), averaging 146 days compared to 91 days for the initial DNA driven tumor. Histology (Figure 2D MET/CAT P-1) shows little or no change in pathology. The second passage was carried out for more than 200 days with no evidence of tumor. AKT/CAT tumor cells gave rise to perimoribund liver tumor masses (Figure 2C) with latencies of 64 through 69 days for at least 4 serial passages (Table 1). Histochemical analysis showed the degree of steatosis declined with passage resulting in denser cytoplasms, possible glycogen granules and accompanying trabeculation (Figure 2D, AKT/CAT P-1, P-4). The incidence of invasiveness increased with passage reflected by the increase in proportion of carcinoma foci per passage (Table 1). This increase in carcinoma is consistent with a progressive transition from adenoma to carcinoma and supported by histology showing the dense cytoplasm phenotype within or attached to foci of clear cell appearance (Figure 2D AKT/CAT P-1, P-4). No evidence of extra-hepatic metastatic or primary growth was detected using these hydrodynamically delivered oncogene combinations or by intrasplenic passage.
Table 1.
Median time to peri-moribund (days) and malignant composition as percentage carcinoma content from histological classification (carcinoma/adenoma) upon serial passage of tumors generated by hydrodynamic transfection.
| AKT/CAT | P-0 | P-1 | P-2 | P-3 | P-4 |
|---|---|---|---|---|---|
| survival | 132, n=15 range=97 |
68, n=13 range=154 |
67, n=51 range=100 |
64, n=35 range=117 |
69, n=11 range=49 |
| malignancy | 12.5%, n=4 | 22.4%, n=12 | 57.6%, n=38 | 84.6%, n=25 | 90.0%,n=10 |
| MET/CAT | P-0 | P-1 | |||
| survival | 91, n=17 range=82 |
146, n=6 range=93 |
- | - | - |
| malignancy | 25.4%, n=5 | 58.3%, n=6 | - | - | - |
Expression patterns of hepatocyte and hepatic progenitor markers
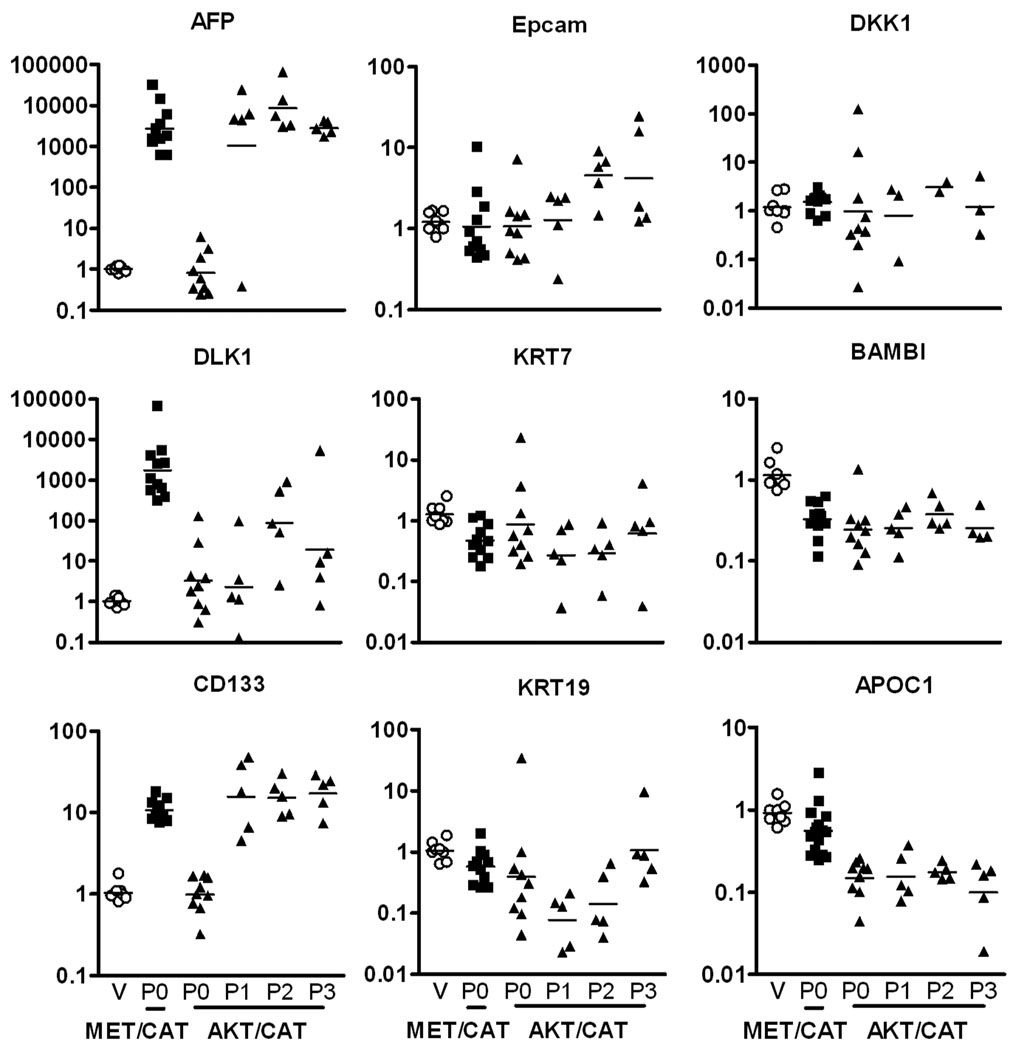
In order to ascertain the relationship of these two tumor models to human hepatocellular carcinoma subtypes, a panel of human HCC prognostic signatures for hepatocyte and hepatic progenitor markers was employed (18). The MET/CAT model showed significant increases in hepatoblast markers AFP, DLK1 and CD133 relative to vector control (pT3) control-transfected liver (Figure 4; p<0.05). Primary AKT/CAT displayed no induction of AFP or CD133 and sporadic induction of DLK1. Upon passage, AKT/CAT tumors induced expression of AFP and CD133 to levels equal to the MET/CAT model with DLK1 remaining unchanged. Serial passages of AKT/CAT are stable with respect to AFP, CD133, KRT7, DKK1, BAMBI, and APOC1, whereas DLK1, Epcam, and KRT19 are highly variable consistent with clonal selection. Applying the human HCC prognostic outcomes predicted by this panel as described previously (18), the resulting expression patterns stratify MET/CAT as a hepatocyte progenitor-like HCC (AFP+, Epcam−) with poor prognosis and primary AKT/CAT as mature hepatocyte-like HCC (AFP−, Epcam−) with fair prognosis. Upon serial passage of the AKT/CAT model, upregulation of AFP places it into the same hepatocyte progenitor-like/poor prognosis group as MET/CAT.
Figure 4. Quantitative PCR of HCC subtype associated and Epcam coregulated gene markers.
The gene expression patterns generated by the following hepatic progenitor and differentiation markers can differentiate four subtypes of HCC (18). AFP, DLK1, and CD133 are stem/hepatoblast markers; Epcam, KRT7, and KRT19 are stem/epithelial markers; DKK1 and BAMBI are Wnt signaling markers; and APOC1 is a mature hepatocyte marker. Liver transfected with pT3 (empty vector, column V, open circle), MET/CAT P-0 tumors (column underscored P0, filled square) and AKT/CAT P-0 and serially passaged tumors (columns underscored: P0 through P3, filled triangle). Each symbol represents the QPCR signal normalized to beta-actin and relative to the mean pT3 signal (set to one) for the respective marker to generate fold induction. The bar designates the geometric mean value of each group.
Development of an assay to selectively detect replication of integrated oncogene-transposons as a sensitive surrogate marker of early tumorigenesis
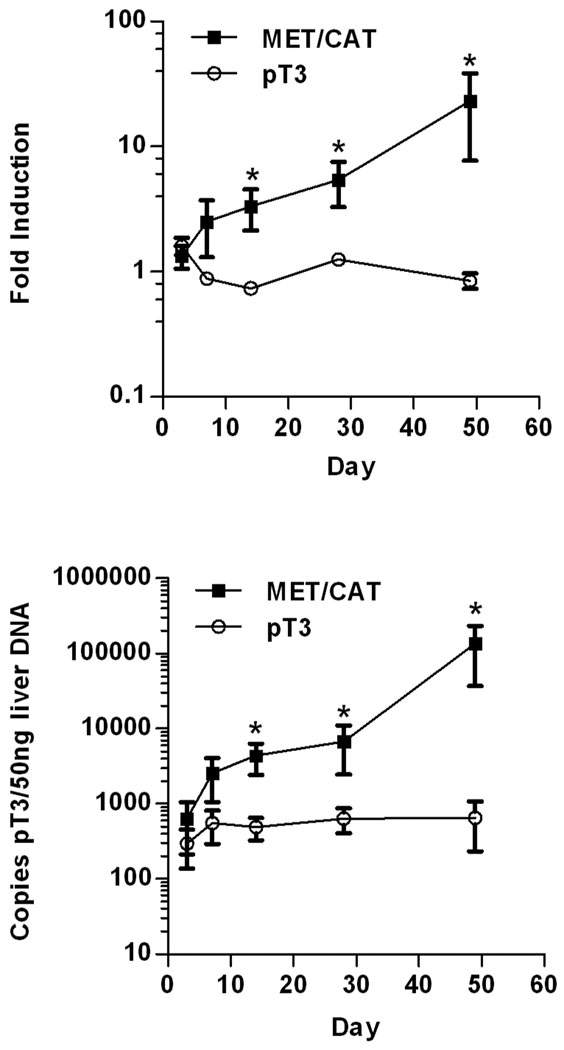
To detect the earliest signs of tumor induction in this model, a trait or marker linked to the process of tumorigenesis was sought since histological analysis indicated that liver tumors were difficult to observe prior to 4 weeks (Figure 2F). We surmised AFP is a likely candidate since it is a common marker of human HCC and it is upregulated several thousand fold in end-stage MET/CAT tumors (Figure 4). Additionally, it has been shown by immunohistochemistry that AFP is not detectable in CAT- or MET- alone transfected livers-but is robustly induced when these oncogenes are co-delivered (11). By 3 days post injection, AFP mRNA levels were not significantly different between MET/CAT and pT3 control vectors (Figure 5A). An upward trend of 3 fold AFP message in oncogene DNA injected liver was evident at 1 week. At 2 weeks, AFP increased to a significant difference of 5 fold that was maintained through four weeks. Between four and seven weeks, an average of 27–fold induction of AFP occurred.
Figure 5. Detection of early tumorigenesis.
Panel A: Quantitative PCR for AFP message. This graph shows a 7 week time course of AFP mRNA expression in MET/CAT (10 µg, filled square) and vector control (pT3, 10 µg, open circle) transfected livers. Asterisks indicate statistically significant (p<0.05) data points. Days 3 and 7, n=5; days 14, 28, and 49, n=10. Symbols represent mean and brackets standard error of the mean (SEM).
Panel B: Transposon replication assay. Graph shows time course analysis of integrated transposon DNA replication (DpnI digested, QPCR) for MET/CAT (10 ug, filled square) and vector control (pT3, 10 ug, open circle) transfected livers. Asterisks indicate statistically significant (p<0.05) data points. Days 3 and 7, n=5; days 14, 28, and 49, n=10.
Since different markers are unique to different collaborating oncogene pairs (no AFP in AKT/CAT, Figure 4), a universal approach was developed that selectively quantitated integrated-replicated transposon delivery vectors using DpnI and QPCR. This assay measured the replication of transfected MET/CAT transposons in the same tumor samples used for AFP mRNA QPCR (Figure 5B). The increase in transposon DNA occurred with similar kinetics to the increase in AFP message in the MET/CAT model. The replication assay shows a two-fold greater sensitivity over AFP QPCR with respect to the empty vector control from three days to 4 weeks although the first statistically significant difference matches with AFP QPCR at 14 days. Between 4 and 7 weeks this difference increases from 10 to over 200-fold (relative to pT3 vector control) consistent with an increase in tumor growth but without a proportional increase in AFP message. Similar trends were observed with AKT/CAT (data not shown). Therefore, the replication assay is more sensitive than AFP QPCR and can be applied to any pT3-transposon based model.
Comparative transcriptome analysis shows the AKT/CAT and MET/CAT relationship to human HCC
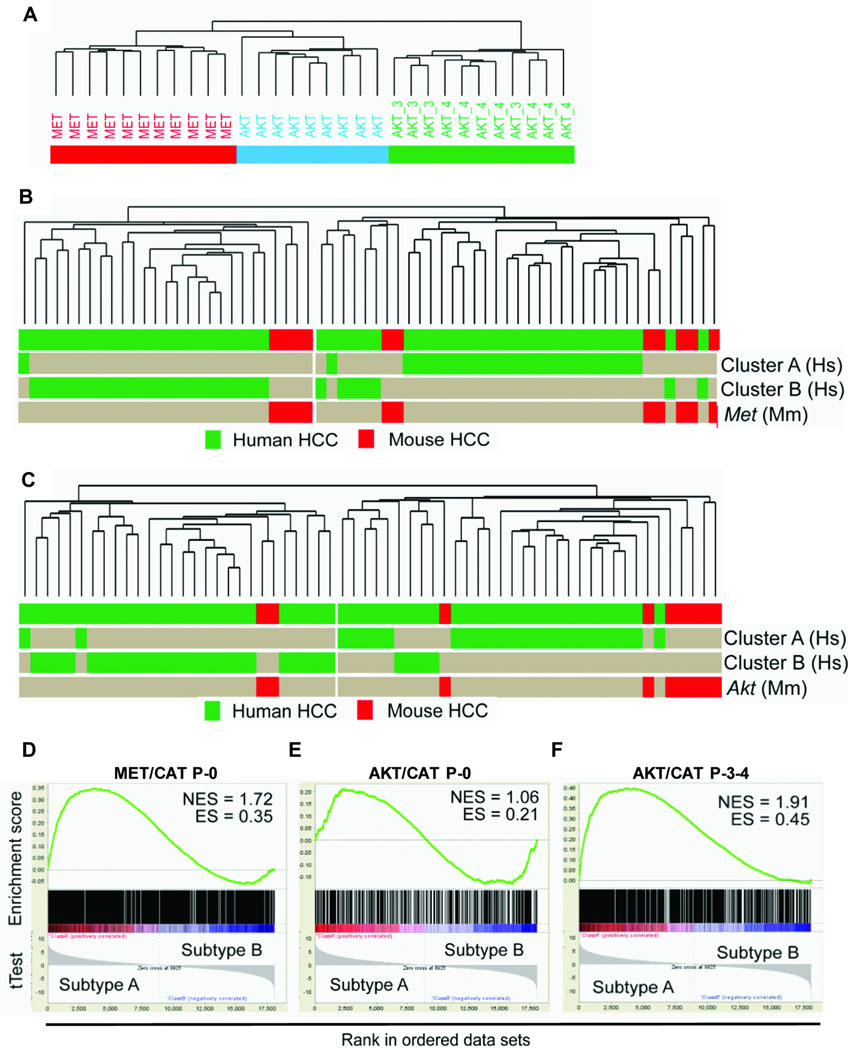
RNA was purified from a pool of 10–15 tumor nodules from each liver and subjected to expression analysis. This included end-point tumor harvested from initial MET/CAT (n=11) and AKT/CAT (n=9), and from serially passaged (P-3 and P-4, n=11) AKT/CAT. Age-matched normal liver transfected with empty pT3 transposon vector (n=8) was used as the control. Unsupervised hierarchical cluster analysis was carried out on a set of 1953 genes that showed a differential expression (tumor to normal) of at least 2 in 50% of the samples and separated the tumors into three clusters according to the transfected DNA and passage (Figure 6A). This confirmed that the modeling technique and the sampling method effectively squelched clonal variation and the resulting gene expression patterns were reproducible and specific to the transfected genes and not due to technical variability. Interestingly, P-0 AKT/CAT and P-0 MET/CAT are more similar than P-0 AKT/CAT is to passaged AKT/CAT (Figure 6A). This may reflect the early stage of transformation in P-0 tumors likely dominated by a parental hepatic gene signature.
Figure 6. Comparative genomic analysis.
Panels A: Hierarchical clustering of gene expression profiles from AKT/CAT P-0 (AKT), MET/CAT P-0 (MET), and serially passaged AKT/CAT (AKT -3, -4) tumors. The relatedness of the respective transcriptional profiles is graphically represented by their arrangement in the dendrogram.
Panel B–C: Dendrogram of Unsupervised hierarchical cluster analysis of integrated human (n=53), P-0 MET/CAT (Panel B, n=11) and P-0 AKT/CAT (Panel C, n=9) hepatocellular tumor transcriptomes, see Supplemental Figure 4 for hierarchical clustering. The horizontal rows delineate the distribution of different cohorts and prognostic subclasses. The length of the branches is proportionate to the degree of transcriptome similarity. Differently shaded bars represent human and mouse HCC tissues (labeled at right end), respectively. To derive the AKT/CAT signature, tumor and surrounding liver tissue was utilized for analysis. The MET/CAT signature was derived by comparison to empty vector control liver since non-transformed tissue was not available.
Panel D–F: Gene set enrichment analysis of gene signatures from: MET/CAT P-0 (Panel D), AKT/CAT P-0 (Panel E), and passaged AKT/CAT P-3-4 (Panel F). Enrichment-(ES) and normalized enrichment-(NES) scores are shown. The t-Test is graphed below each correlated gene in the ranked data set. Passaging of AKT/CAT enriches (NES=1.91) for human subtype A gene expression surpassing the association with MET/CAT signature (NES=1.72).
The association of the two tumor models and human HCC was further examined by cross-species comparison. The human HCC data set was generated from a cohort of 53 HCCs obtained from Caucasian and Chinese patients and hybridized to Illumina bead chips (19). Hierarchical cluster analysis (Supplemental Figure 4) was performed on an integrated set of 53 human HCC and 11 P-0 MET/AKT (Figure 6B) or 9 P-0 AKT/CAT (Figure 6C) derived murine tumors. A list of 2403 or 833 orthologous genes between the two species (MET- or AKT- CAT, respectively) represented on both platforms was identified. Integration and cluster analysis delineated clinically relevant subgroups of human HCC. Both the AKT/CAT and MET/CAT-responsive gene signatures clustered predominantly with poor prognosis human HCCs according to the previous described A (poor prognosis) and B (good prognosis) subclassification (Figure 6B and C) (20). Cumulative survival analysis of the human HCC cases classified by MET/CAT and AKT/CAT gene signature clustering (Supplemental Figure 5) verified this correlation (P<0.04). Gene set enrichment analysis (GSEA) of both the AKT/CAT and MET/CAT gene signatures demonstrated a significant positive correlation between upregulated genes and the gene expression signature of subclass A human HCC (Figure 6D and E). Furthermore, GSEA of serial passaged AKT/CAT tumors significantly increased the enrichment compared to P-0 AKT/CAT tumors, establishing a functional genomics correlation with serial passage and accompanying histological changes (Figure 6F and Figure 2D).
Since the histology of P-0 AKT/CAT was characteristically clear cell indicating a high degree of cytoplasmic lipid, we hypothesized that this model may be reflecting the human phenotype driven largely by obesity through molecular mechanisms that depend on inflammatory mediators. Ingenuity Pathway Analysis was used to identify statistically significant patterns of gene expression associated with known biological phenomena. Network comparison of AKT/CAT P-0 to MET/CAT P-0 gene expression revealed liver steatosis is selectively associated with AKT/CAT P-0 (p<0.004) and not MET/CAT P-0 (p<0.09) (Supplemental Figure 6A). Steatohepatitis-associated genes were also selectively expressed in AKT/CAT P-0 but below the threshold of p<0.05. Comparison of AKT/CAT primary to passaged tumor showed the most significant shared pathway in common was lipid metabolism (p<6.14×10−6 and p<3.02×10−11, respectively) (Supplemental Figure 6B). Ranking networks characterized as “molecular and cellular functions” ranked lipid metabolism as first and third of the five most significant pathways (Supplemental Table 1) in AKT/CAT P-3-4 and AKT/CAT P-0, respectively. This corresponds to an increase of 94 to 218 proteins to this network, constituted in part by an induction of lipid catabolic genes (Supplemental Tables 2A and 2B) coinciding with the decrease in histological steatosis (Figure 2D), and increased malignancy. These transcriptome analyses are consistent with AKT/CAT gene expression resulting in hepatocytes acquiring a steatotic feature due to aberrant lipid metabolism that increases with malignancy (passaged AKT/CAT) accompanied by inflammation (steatohepatitis).
Discussion
The combination of activated AKT and CAT rapidly produced steatotic liver tumors that favored adenoma pathology compared to carcinoma from MET and CAT (11). The AKT/CAT tumors were readily transplantable and became progressively malignant with increased passage in contrast to MET/CAT. This process was accompanied by an increase in cytoplasmic granularity and reduced lipid content. Microarray analysis confirmed that primary AKT/CAT tumors are steatogenic/lipogenic and that passage further expands the lipid metabolic network to include genes involved in lipid degradation and oxidation that could account for the concurrent reduction in lipid content.
The association of unique HCC tumor phenotypes with specific beta-catenin activating mutations have been recently reported (21). Although conflicting accounts of beta-catenin activation and prognostic outcomes exist, the activated beta-catenin (deletion mutation) models described herein align with poor prognostic outcome (22–28). Integration and comparison with a human HCC cohort shows passaged AKT/CAT and MET/CAT models are associated with the poor prognosis subgroup (subtype A (20)). Classification using a panel (18) of hepatocyte and hepatic progenitor markers derived from human HCC prognostic signatures stratify primary AKT/CAT tumor as mature hepatocyte-like HCC with fair prognosis contrasted to MET/CAT and serial passaged AKT/CAT as hepatocyte progenitor-like HCCs with poor prognosis.
Although histopathology and molecular analysis classify primary AKT/CAT tumors as adenoma, AKT/CAT tumor growth more resembles MET/CAT carcinoma and is much more penetrant and aggressive than the previously reported MET/dominant negative HNF1alpha adenoma model (11). This is consistent with the increased risk of malignant transformation observed in the activated beta-catenin subtype of human HCA (17). Given that progression from adenoma to carcinoma is rarely observed during human HCC development and mouse AKT/CAT adenoma progresses to carcinoma, the transient mouse adenoma intermediate is likely a fortunate consequence perhaps reflecting a temporal extension in carcinoma development. The lack of an equivalent pre-neoplastic microenvironmental cue such as fibrosis/cirrhosis may contribute to this difference between mouse and human HCC development. Therefore the primary AKT/CAT tumorigenesis model provides a useful bioassay for assaying host mediated contributions to this malignant progression in the context of lipogenesis (hepatosteatosis).
The collaboration of the PI3K/AKT/mTOR and Wnt/beta-catenin pathways in clear cell and hepatocellular carcinoma is supported by several lines of evidence. The PI3K/AKT/mTOR pathway is one of the most commonly utilized tumor signaling pathways able to stimulate proliferation, inhibit apoptosis and drive anaerobic glycolysis (29, 30). Expression of constitutive activated AKT in liver has been shown to lead to excessive glucose utilization, lipogenesis, and hyperplasia but not malignancy (6). Liver specific PTEN knockout results in AKT activation and produces very similar pathology including hepatomegally, steatosis, and steatohepatitis (7). PTEN knockouts can progress to HCC at 66–83% penetrance but required 44–78 weeks and produce tumors that are very similar to human non-alcoholic steatohepatitis (NASH) (31). The beta-catenin gene is the most commonly mutated locus associated with hepatocellular carcinoma and adenoma that is predisposed to malignancy (32). Wnt/Beta-catenin signaling is activated in clear cell –renal, -endometrial, and -ovarian cancers and can collaborate with the PTEN/AKT pathway in modeling mouse clear cell ovarian endometriod tumors (33, 34). Liver-specific expression of activated beta-catenin produces hepatomegally but not malignancy by itself. It has been shown to collaborate in hepatic tumorigenesis with MET, RAS, and Sprouty 2 utilizing the SB delivery system (35, 36). The resulting novel AKT/CAT model present here produces tumors more rapidly and with a higher penetrance than the existing transgenic models of hepatocellular carcinoma alone or in the context of hepatosteatosis (37).
Many lines of epidemiological, clinical, and pre-clinical data are converging upon the linkage between obesity, inflammation and oncogenesis. The liver is both an immunological and metabolic interface where dietary antigens and metabolic byproducts are processed. The liver also displays the greatest sensitivity to obesity linked cancer. The other types of cancers being linked to obesity include the clear cell subtypes of RCC and ovarian cancer (38, 39). The molecular pathological consequences of obesity in liver are steatosis and inflammation. Recently the ability of obesity to promote tumorigenesis has been molecularly linked to pathways mediated by inflammatory cytokines (40). The steatotic liver tumor model presented here is applicable to existing transgenic immunodefined hosts, reproducible, rapid, displays malignant progression and provides intrinsic molecular markers of early tumorigenesis. These attributes will accelerate our understanding of this linkage and perhaps suggests avenues of intervention into this form of liver cancer, as well as clear cell renal and epithelial –ovarian carcinomas.
Supplementary Material
Acknowledgement
This research was supported [in part] by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research. The authors would also like to thank Dr. John R. Ortaldo for assistance provided during the studies and suggestions made in the preparation of the manuscript.
This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under Contract No. HHSN261200800001E.
Footnotes
The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government.
References
- 1.Zender L, Villanueva A, Tovar V, Sia D, Chiang DY, Llovet JM. Cancer gene discovery in hepatocellular carcinoma. J Hepatol. 2010;52(6):921–929. doi: 10.1016/j.jhep.2009.12.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaposi-Novak P, Lee JS, Gomez-Quiroz L, Coulouarn C, Factor VM, Thorgeirsson SS. Met-regulated expression signature defines a subset of human hepatocellular carcinomas with poor prognosis and aggressive phenotype. J Clin Invest. 2006;116(6):1582–1595. doi: 10.1172/JCI27236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Vinciguerra M, Foti M. PTEN at the crossroad of metabolic diseases and cancer in the liver. Ann Hepatol. 2008;7(3):192–199. [PubMed] [Google Scholar]
- 4.Nakanishi K, Sakamoto M, Yamasaki S, Todo S, Hirohashi S. Akt phosphorylation is a risk factor for early disease recurrence and poor prognosis in hepatocellular carcinoma. Cancer. 2005;103(2):307–312. doi: 10.1002/cncr.20774. [DOI] [PubMed] [Google Scholar]
- 5.Coulouarn C, Gomez-Quiroz LE, Lee JS, et al. Oncogene-specific gene expression signatures at preneoplastic stage in mice define distinct mechanisms of hepatocarcinogenesis. Hepatology. 2006;44(4):1003–1011. doi: 10.1002/hep.21293. [DOI] [PubMed] [Google Scholar]
- 6.Ono H, Shimano H, Katagiri H, et al. Hepatic Akt activation induces marked hypoglycemia, hepatomegaly, and hypertriglyceridemia with sterol regulatory element binding protein involvement. Diabetes. 2003;52(12):2905–2913. doi: 10.2337/diabetes.52.12.2905. [DOI] [PubMed] [Google Scholar]
- 7.Watanabe S, Horie Y, Suzuki A. Hepatocyte-specific Pten-deficient mice as a novel model for nonalcoholic steatohepatitis and hepatocellular carcinoma. Hepatol Res. 2005;33(2):161–166. doi: 10.1016/j.hepres.2005.09.026. [DOI] [PubMed] [Google Scholar]
- 8.Parekh S, Anania FA. Abnormal lipid and glucose metabolism in obesity: implications for nonalcoholic fatty liver disease. Gastroenterology. 2007;132(6):2191–2207. doi: 10.1053/j.gastro.2007.03.055. [DOI] [PubMed] [Google Scholar]
- 9.Radonjic M, de Haan JR, van Erk MJ, et al. Genome-wide mRNA expression analysis of hepatic adaptation to high-fat diets reveals switch from an inflammatory to steatotic transcriptional program. PLoS One. 2009;4(8):e6646. doi: 10.1371/journal.pone.0006646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hirsch HA, Iliopoulos D, Joshi A, et al. A transcriptional signature and common gene networks link cancer with lipid metabolism and diverse human diseases. Cancer Cell. 2010;17(4):348–361. doi: 10.1016/j.ccr.2010.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tward AD, Jones KD, Yant S, et al. Distinct pathways of genomic progression to benign and malignant tumors of the liver. Proc Natl Acad Sci U S A. 2007;104(37):14771–14776. doi: 10.1073/pnas.0706578104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Orford K, Crockett C, Jensen JP, Weissman AM, Byers SW. Serine phosphorylation-regulated ubiquitination and degradation of beta-catenin. J Biol Chem. 1997;272(40):24735–24738. doi: 10.1074/jbc.272.40.24735. [DOI] [PubMed] [Google Scholar]
- 13.Orsulic S, Li Y, Soslow RA, Vitale-Cross LA, Gutkind JS, Varmus HE. Induction of ovarian cancer by defined multiple genetic changes in a mouse model system. Cancer Cell. 2002;1(1):53–62. doi: 10.1016/s1535-6108(01)00002-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Patil MA, Lee SA, Macias E, et al. Role of cyclin D1 as a mediator of c-Met- and beta-catenin-induced hepatocarcinogenesis. Cancer Res. 2009;69(1):253–261. doi: 10.1158/0008-5472.CAN-08-2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lee JK, Sayers TJ, Brooks AD, et al. IFN-gamma-dependent delay of in vivo tumor progression by Fas overexpression on murine renal cancer cells. J Immunol. 2000;164(1):231–239. doi: 10.4049/jimmunol.164.1.231. [DOI] [PubMed] [Google Scholar]
- 16.Mohr U, editor. WHO International Agency for Research on Cancer. New York: Springer-Verlag, Berlin; International Classification of Rodent Tumors The Mouse; pp. 64–68. [Google Scholar]
- 17.Bioulac-Sage P, Balabaud C, Zucman-Rossi J. Subtype classification of hepatocellular adenoma. Digestive surgery. 2010;27(1):39–45. doi: 10.1159/000268406. [DOI] [PubMed] [Google Scholar]
- 18.Yamashita T, Forgues M, Wang W, et al. EpCAM and alpha-fetoprotein expression defines novel prognostic subtypes of hepatocellular carcinoma. Cancer Res. 2008;68(5):1451–1461. doi: 10.1158/0008-5472.CAN-07-6013. [DOI] [PubMed] [Google Scholar]
- 19.Andersen JB, Loi R, Perra A, et al. Progenitor-derived hepatocellular carcinoma model in the rat. Hepatology. 2010;51(4):1401–1409. doi: 10.1002/hep.23488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lee JS, Chu IS, Heo J, et al. Classification and prediction of survival in hepatocellular carcinoma by gene expression profiling. Hepatology. 2004;40(3):667–676. doi: 10.1002/hep.20375. [DOI] [PubMed] [Google Scholar]
- 21.Cieply B, Zeng G, Proverbs-Singh T, Geller DA, Monga SP. Unique phenotype of hepatocellular cancers with exon-3 mutations in beta-catenin gene. Hepatology (Baltimore, Md) 2009;49(3):821–831. doi: 10.1002/hep.22695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hsu HC, Jeng YM, Mao TL, Chu JS, Lai PL, Peng SY. Beta-catenin mutations are associated with a subset of low-stage hepatocellular carcinoma negative for hepatitis B virus and with favorable prognosis. The American journal of pathology. 2000;157(3):763–770. doi: 10.1016/s0002-9440(10)64590-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wong CM, Fan ST, Ng IO. beta-Catenin mutation and overexpression in hepatocellular carcinoma: clinicopathologic and prognostic significance. Cancer. 2001;92(1):136–145. doi: 10.1002/1097-0142(20010701)92:1<136::aid-cncr1301>3.0.co;2-r. [DOI] [PubMed] [Google Scholar]
- 24.Fujito T, Sasaki Y, Iwao K, et al. Prognostic significance of beta-catenin nuclear expression in hepatocellular carcinoma. Hepato-gastroenterology. 2004;51(58):921–924. [PubMed] [Google Scholar]
- 25.Yamaoka H, Ohtsu K, Sueda T, Yokoyama T, Hiyama E. Diagnostic and prognostic impact of beta-catenin alterations in pediatric liver tumors. Oncology reports. 2006;15(3):551–556. [PubMed] [Google Scholar]
- 26.Cavard C, Colnot S, Audard V, et al. Wnt/beta-catenin pathway in hepatocellular carcinoma pathogenesis and liver physiology. Future oncology (London, England) 2008;4(5):647–660. doi: 10.2217/14796694.4.5.647. [DOI] [PubMed] [Google Scholar]
- 27.Yamashita T, Ji J, Budhu A, et al. EpCAM-positive hepatocellular carcinoma cells are tumor-initiating cells with stem/progenitor cell features. Gastroenterology. 2009;136(3):1012–1024. doi: 10.1053/j.gastro.2008.12.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Liu L, Zhu XD, Wang WQ, et al. Activation of beta-catenin by hypoxia in hepatocellular carcinoma contributes to enhanced metastatic potential and poor prognosis. Clinical cancer research : an official journal of the American Association for Cancer Research. 2010;16(10):2740–2750. doi: 10.1158/1078-0432.CCR-09-2610. [DOI] [PubMed] [Google Scholar]
- 29.Carnero A, Blanco-Aparicio C, Renner O, Link W, Leal JF. The PTEN/PI3K/AKT signalling pathway in cancer, therapeutic implications. Curr Cancer Drug Targets. 2008;8(3):187–198. doi: 10.2174/156800908784293659. [DOI] [PubMed] [Google Scholar]
- 30.Robey RB, Hay N. Is Akt the "Warburg kinase"?-Akt-energy metabolism interactions and oncogenesis. Semin Cancer Biol. 2009;19(1):25–31. doi: 10.1016/j.semcancer.2008.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Horie Y, Suzuki A, Kataoka E, et al. Hepatocyte-specific Pten deficiency results in steatohepatitis and hepatocellular carcinomas. J Clin Invest. 2004;113(12):1774–1783. doi: 10.1172/JCI20513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Imbeaud S, Ladeiro Y, Zucman-Rossi J. Identification of novel oncogenes and tumor suppressors in hepatocellular carcinoma. Semin Liver Dis. 2010;30(1):75–86. doi: 10.1055/s-0030-1247134. [DOI] [PubMed] [Google Scholar]
- 33.Zorn KK, Bonome T, Gangi L, et al. Gene expression profiles of serous, endometrioid, and clear cell subtypes of ovarian and endometrial cancer. Clin Cancer Res. 2005;11(18):6422–6430. doi: 10.1158/1078-0432.CCR-05-0508. [DOI] [PubMed] [Google Scholar]
- 34.Wu R, Hendrix-Lucas N, Kuick R, et al. Mouse model of human ovarian endometrioid adenocarcinoma based on somatic defects in the Wnt/beta-catenin and PI3K/Pten signaling pathways. Cancer Cell. 2007;11(4):321–333. doi: 10.1016/j.ccr.2007.02.016. [DOI] [PubMed] [Google Scholar]
- 35.Wangensteen KJ, Wilber A, Keng VW, et al. A facile method for somatic, lifelong manipulation of multiple genes in the mouse liver. Hepatology (Baltimore, Md) 2008;47(5):1714–1724. doi: 10.1002/hep.22195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lee SA, Ho C, Roy R, et al. Integration of genomic analysis and in vivo transfection to identify sprouty 2 as a candidate tumor suppressor in liver cancer. Hepatology. 2008;47(4):1200–1210. doi: 10.1002/hep.22169. [DOI] [PubMed] [Google Scholar]
- 37.Newell P, Villanueva A, Friedman SL, Koike K, Llovet JM. Experimental models of hepatocellular carcinoma. J Hepatol. 2008;48(5):858–879. doi: 10.1016/j.jhep.2008.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lowrance WT, Thompson RH, Yee DS, Kaag M, Donat SM, Russo P. Obesity is associated with a higher risk of clear-cell renal cell carcinoma than with other histologies. BJU Int. 2009;105(1):16–20. doi: 10.1111/j.1464-410X.2009.08706.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Olsen CM, Nagle CM, Whiteman DC, Purdie DM, Green AC, Webb PM. Body size and risk of epithelial ovarian and related cancers: a population-based case-control study. Int J Cancer. 2008;123(2):450–456. doi: 10.1002/ijc.23509. [DOI] [PubMed] [Google Scholar]
- 40.Park EJ, Lee JH, Yu GY, et al. Dietary and genetic obesity promote liver inflammation and tumorigenesis by enhancing IL-6 and TNF expression. Cell. 2010;140(2):197–208. doi: 10.1016/j.cell.2009.12.052. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.