Abstract
Clear cell renal cell carcinoma (CCRCC) evolves due to mutations in the von Hippel-Lindau (VHL) tumor suppressor gene. Although the loss of VHL enables the survival and proliferation of CCRCC cells, it is also expected to introduce vulnerabilities which may be exploited for therapeutics discovery. To this end, we developed a high throughput screen to identify small molecules derived from plants, microorganisms and marine organisms to which CCRCC cells are sensitive. Screening over 8000 compounds using this approach, we report here the identification of the microbially-derived compound carminomycin I (CA) as an effective inhibitor of VHL-defective (VHL−/−) CCRCC cell proliferation. CA also induced apoptosis in CCRCC cells by a mechanism independent of p53 or hypoxia-inducible factor HIF2. We found that P-glycoprotein (P-gp) sequestered CA within the Golgi complex. Interestingly, Golgi sequestration was critical for the antiproliferative effects of CA and P-gp inhibitors abrogated this activity. Furthermore, CA induced cleavage of the Golgi protein p115 and the translocation of its C-terminal fragment to the nucleus. Finally, examination of the activity of the VHL-interacting Golgi protein ERGIC-53 showed that VHL could mediate protection from CA in CCRCC cells. Our natural products-based screening approach has revealed the P-gp mediated localization of anticancer compounds within the Golgi in CCRCC cells as a potential strategy of targeting VHL-deficient CCRCC cells.
Introduction
Clear cell renal cell carcinoma (CCRCC) is the most commonly found form of kidney cancer. It occurs in hereditary and sporadic forms both of which arise due to structural alterations in the von Hippel-Lindau (VHL) gene (1, 2). The most studied function of the VHL gene product (pVHL) is the regulation of levels of the hypoxia inducible factor (HIF)-α family of transcription factors (3). However, the increasing number of proteins with which it interacts as well as genotype-phenotype correlations in VHL disease all strongly suggest that pVHL possesses functions unrelated to HIF that may contribute to the genesis and progression of CCRCC. How HIF-independent functions contribute to tumor development after pVHL inactivation, however, remains unclear. Elucidation of these functions should lead to enhanced understanding of renal carcinogenesis and should enable the identification of novel targets that may be exploited in therapeutic discovery.
Natural products (i.e., plant, microbial and marine organism-derived compounds) have in the past proved a fertile source of bioactive compounds that have led to both the development of newer chemotherapeutic interventions and a better understanding of the underlying disease biology. In the area of anticancer compounds, natural products either represent actual drugs (such as taxol, vincristin, campthotecin, etc.) or have served as the basis for the development of a large number of currently clinically approved drugs. We, therefore, set out to address the challenges of identifying small molecules selectively targeting VHL−/− CCRCC cells using a forward chemical genetics approach employing natural products-based high throughput screening.
Materials and Methods
Cells and cell culture
The 786-O cell line was obtained from the repository of the Developmental Therapeutics Program at the National Cancer Institute. UOK-121, and UOK-127 cell lines were obtained from the National Cancer Institutes’s Urologic Oncology Branch Renal Cell Carcinoma Cell Line Repository. The cell lines have been characterized at the repositories using, among others, single nucleotide polymorphism arrays and by oligonucleotide-base HLA typing. They have also been subjected to spectral karyotyping and have been characterized for variations in short tandem repeats (4, 5). All cell lines were placed under cryostorage after they were acquired from the repositories and used within 6 months of thawing fresh vials. Routinely, cells were grown in DMEM supplemented with 10% fetal bovine serum and incubated at 37°C in 5% CO2. Stably transduced 786-OVHL, UOK-121VHL and UOK-127 VHL as well 786-O, UOK-121, and UOK-127 cell lines were generated using retroviral gene delivery with or without VHL as described previously (6). During this time, cell morphology, growth curve and possible mycoplasma contamination were regularly checked to ensure absence of contamination.
High-throughput screening
During the screening campaign, 5000 cells of both 786-O and 786-OVHL cell lines were plated per well in different 384-well plates and plates were incubated overnight. After this period, test compounds were added at a final concentration of 10 μmol/L. Reduction of 2,3-bis[2-methoxy-4-nitro-5-sulfophenyl]-2H-tetrazolium-5-carboxanilide (XTT) by metabolically active cells to a colored product was used to assess effect of compounds on cells after 24 h. A total of 8,326 compounds were tested using this screening assay.
Analysis of Cell Death
Cells were treated with serial dilutions of either the vehicle control or hit compounds in a dose-response format and were incubated for 24 h. Cell viability was assessed using the above XTT assay. Caspase activity in treated cells was determined using Caspase-Glo 3/7, 8, and 9 assays according to the manufacturer’s instructions in 384-well plates (Promega). Lactate dehydrogenase (LDH) release from necrotic cells into the extracellular fluid was determined using the CytoTox-One membrane integrity assay kit following the manufacturer’s protocol (Promega).
Western blotting
After compound treatment, cells were trypsinized, neutralized and pelleted. Cell pellets were washed twice with ice-cold PBS. Pellets were then lysed with NP-40 lysis buffer (Sigma) containing protease inhibitor cocktail (Roche) 1 tablet per 5 mL of lysis buffer and 1.02 μmol/L phenylmethylsulfonyl fluoride. Total protein concentration was determined using a BCA kit (Pierce) with bovine serum albumin (Sigma) as the standard. Cellular protein (10-50 μg) was resolved by SDS–PAGE and electrophoretically transferred onto polyvinylidene difluoride membranes (Invitrogen). Membranes were blocked with Odyssey Blocking Buffer (Li-Cor) overnight at 4 °C and then incubated with specific primary antibodies. All antibodies except the following were obtained from Santa Cruz Biotechnology and used at 1:200 to 1:1000 dilution: VHL, PARP, cleaved PARP (Cell Signaling); A1AT, Bak (Abcam); and α-tubulin and β-actin (Sigma). GAPDH, β-actin or α-tubulin was used as a loading control and changes in protein levels shown were found to be significant relative to loading controls. Following incubation with primary antibody, membranes were washed and incubated with anti-mouse IRDye 680 and anti-rabbit IRDye 800 secondary antibodies (Li-Cor). Blots were scanned using an Odyssey infrared imaging system and processed using its software (Li-Cor). Band intensities were determined using the Odyssey band quantitation software after background subtraction using the median top-bottom method. Intensities of bands of interest were normalized to the signals from the corresponding loading control bands.
Cell cycle analysis
Following treatment with 0.1 μmol/L CA for 16 h, 0.5-1.0×106/ml live 786-O and 786-OVHL cells were harvested and stained with propidium iodide using Krishan’s buffer (0.1% sodium citrate, 0.02 mg/mL RNase A, 0.3% NP-40 and 50 μg/mL propidium iodide) at a pH of 7.4.
RNAi and quantitative reverse transcription-PCR
HIF-2α targeted siRNA and control siRNA termed Mock were kind gifts from Dr. Olga Aprelikova of the Laboratory of Tumor and Stem Cell Biology at the National Cancer Institute and have been described previously. The sequence for the HIF-2α siRNA used was 5′-GGUGUGCUGUUUGGAGUUCdTdT-3′. A control siRNA termed Mock (target sequence 5′-GCGCGCTTTGTAGGATTCG-3′) was purchased from Qiagen and had no significant homology to any known human sequence. 786-O cells (1–2 × 105/well) were transfected with siRNA oligos (30–60 nM final concentration) in 6-well plates, using Lipofectamine (Invitrogen) following the manufacturer’s protocol. Forty-eight hours later, the transfected RCC cells were washed and used for subsequent experiments. Transduction of 786-O cells via lentiviral P-gp shRNA was carried out using commercially available lentiviral supernatants (Santa Cruz) following the manufacturer’s protocol. Total RNA was extracted and purified using the RNeasy mini kit (Qiagen), and 2 μg total RNA was reverse transcribed using ImProm-II reverse transcription reagent (Promega). Primers and probes for human P-gp, HIF-2α, AIAT, and β–actin were purchased from Applied Biosystems (Assays-on-demand).
P-glycoprotein ATPase assay
The assay was performed using the Pgp-Glo Assay kit (Promega) following the manufacturer’s protocol.
α1-antitrypsin (A1AT) ELISA
Cells were grown in T25-flasks to ~80% confluence and conditioned cell culture medium was harvested after 24 h. For ELISA, 100 μL undiluted conditioned medium was used. ELISA was conducted according to manufacturer’s instructions (Alpco).
Immunocytochemistry
Cells grown on cover slips or seeded on glass chips with an O- or L-shaped fibronectin micropatterns (Cytoo) were fixed with 4% fresh paraformaldehyde and permeabilized with either 0.1% Triton X-100 in PBS or cold methanol. Samples were blocked using Odessey blocking buffer overnight at 4°C. Anti-p115 targeting the p115 C-terminus (1:100, Novus Biologicals), anti-ERGIC-53 (1;1000, Alexis), anti-P-gp (1:100, SantaCruz or 1:100 Abcam), and anti-GM130 (1:100, Epitomics) monoclonal antibodies were incubated with samples overnight at 4°C. Finally, samples were washed and incubated for 45 min with the appropriate secondary antibody from among the following: Alexa Fluor 488 goat anti-rabbit; Alexa Fluor 488 chicken anti-goat and/or Alexa Fluor 594 goat anti-mouse (Invitrogen). Counterstaining was done where shown using Prolong Gold antifade reagent with DAPI (Invitrogen) as the mounting medium. In some double labeled experiments, 786-O cells were first transduced with Organelle Lights Golgi-RFP (Invitrogen) as a marker for the Golgi. Images were acquired using Zeiss LSM 510 microscopes at 63x magnification (Zeiss).
Image Analysis
Images were processed with the program ImageJ (http://www.uhnres.utoronto.ca/facilities/wcif/imagej/). Quantitative analysis of colocalization was performed using the plugin “colocalization threshold” of ImageJ. Details of the image analysis protocol can be found in the Supplemental Data.
Results
High throughput screen (HTS)
To identify small molecule inhibitors of VHL−/− cell proliferation, an HTS assay was developed using the paired cell lines 786-O and 786-OVHL run in parallel in separate 384-well plates. The CCRCC cell line 786-O was selected for assay development based on its properties such as tolerance to up to 3% DMSO, a doubling time of about 24 hours in microplates, compatibility with automation in cell plating and liquid handling, as well as ABC transporter profile. VHL+/+ 786-O cells (786-OVHL) were then generated by transduction using a VHL expression construct. Expression of the VHL gene product (pVHL) in stably transduced cells was confirmed by western blotting (Supplementary Fig. S1A). Similarly, other VHL+/+ cell lines were also generated using the VHL−/− CCRCC cell lines UOK-121 and UOK-127. Consistency and reproducibility of the HTS assay was assessed using a set of 352 compounds as a validation library. A very high correlation coefficient (0.954; R2 = 0.89–0.91) was obtained from three independent experiments. In addition, how well the assay performed during the screen was also routinely evaluated via determination of the Z-factor (7). The Z-factor determined during the screen for all plates with 786-O cells using actinomycin D at 10 μmol/L as the positive control and 1% DMSO as the negative control was found to be 0.68 ± 0.082 indicating that the assay performed well during HTS. In the primary screen, 56 compounds inhibited cell proliferation by more than two-fold in 786-O cells compared to that in 786-OVHL cells with these compounds also showing at least 50% inhibition in 786-O cells.
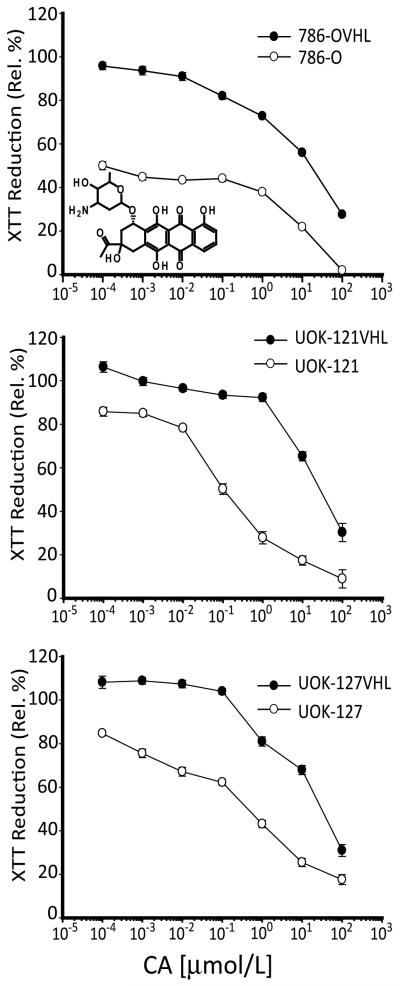
Hit compounds identified in the primary screen were serially diluted and subjected to retesting in a dose-response format. Of the 56 compounds identified as hits in the primary screen, 15 were found to show attenuation of cell proliferation in 786-O cells in a dose dependent manner. To enable a broader evaluation of effect of hit compounds on VHL−/− cell proliferation, hits were also evaluated in paired VHL+/+ and VHL−/− UOK-121 and UOK-127 cells. All 15 hit compounds tested also showed differential inhibition of cell proliferation in paired UOK-121 and UOK-121VHL cells as well as in UOK-127 and UOK-127VHL cells (Fig. 1). Because of its fluorescent nature, we chose one of the hit compounds, carminomycin I (CA), as a model to illustrate the utility of natural products in providing a novel perspective on targeting cancer cells in general and CCRCC cells in particular. Below, we present our findings from the follow-up work done using CA.
Figure 1.
Effect of CA on proliferation of VHL−/− and VHL+/+ CCRCC cells. Cells were treated with serial dilutions of CA for 24 h and data shown is expressed as percentage of XTT reduction relative to DMSO treated cells. Ecah point represents mean ± SEM from two independent experiments with a point in an experiment averaged from 8 replicates.
CA induces apoptosis in 786-O cells
The most studied function of pVHL is the negative regulation of HIF via ubiquitin-mediated degradation. Therefore, we examined whether the observed cytotoxicity in 786-O cells was HIF mediated and whether the observed resistance to CA in 786-OVHL cells was dependent on HIF downregulation. We found, however, that CA exerts its effects on 786-O cells through HIF independent mechanisms (Supplementary Fig. S2). Absence of lactate dehydrogenase release into the grwoth medium and the lack of vesicles that stained positive for Beclin-1, Atg12 or LC3B also allowed the determination that CA did not curtail the viability of 786-O cells through either necrosis or autophagy. Measurement of caspase activity over a 24 h period after CA treatment, on the other hand, revealed increased caspase-3 activity as early as 2 h after treatment. More pronounced differential casapase-3 activity between 786-O and 786-OVHL cells became apparent after 24 h at all CA concentrations tested- 0.1 nmol/L to 100 μmol/L (Supplementary Fig. S3A). Significant differential caspase-8, and to a lesser extent caspase-9, activity in 786-O and 786-OVHL cells was observable only 24 h after treatment at concentrations between 0.1 and 10 μmol/L (Supplementary Fig. 3). This suggested that apoptosis mediated by caspase-3 activation may contribute to the observed effect of CA on cell proliferation. Caspase-3 activation at lower CA test concentrations also appears not to directly correlate with activation of either caspase-8 or -9 suggesting that an alternative initiation pathway may play a role.
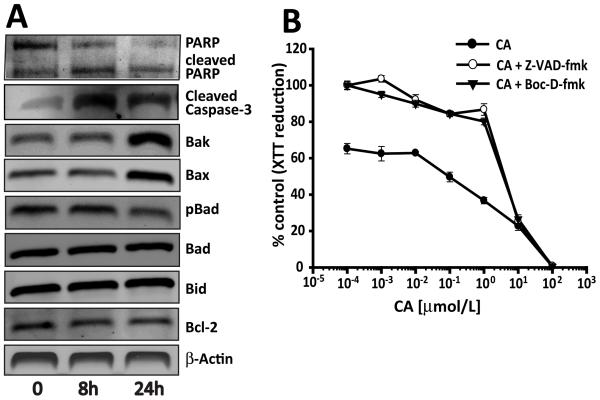
Real-time monitoring of effects of CA treatment on 786-O cells led to the observation of blebbing and changes to the cell membrane characteristic of apoptosis (Supplementary Fig. S3B). Genomic DNA extracted from CA treated 786-O cells also exhibited characteristic internucleosomal laddering of genomic DNA fragments in agarose gel electrophoresis (Supplementary Fig. S3C). Induction of apoptosis by CA in 786-O cells was further supported through monitoring PARP and caspase-3 cleavage as well as through changes in the levels of a panel of proteins commonly involved in apoptotic signaling via western blotting (Fig. 2A). Finally, treatment of 786-O cells with the pan caspase inhibitors Z-VAD-fmk or Boc-D-fmk resulted in reversal of effects of CA on cell proliferation (Fig. 2B). Taken together, these results indicate that CA curtailed proliferation of 786-O cells through induction of apoptosis.
Figure 2.
Apoptosis following treatment with CA. (A) 786-O cells were treated with 0.1 μmol/L CA and protein lysates prepared at shown time points. These protein extracts were subjected to western blotting analysis. No significant changes in the levels of these proteins were seen in 786-OVHL cells. (B) 786-O cells were treated with Z-VAD-fmk (20 μmol/L) or Boc-D-fmk (20 μmol/L) prior to incubation with CA for 24 h. Cell viability was determined by XTT reduction assay. Values are expressed as a percentage of control. Ecah point represents mean ± SEM from two independent experiments with a point in an experiment averaged from 8 replicates.
CA induces survivin and p53 downregulation in 786-O cells
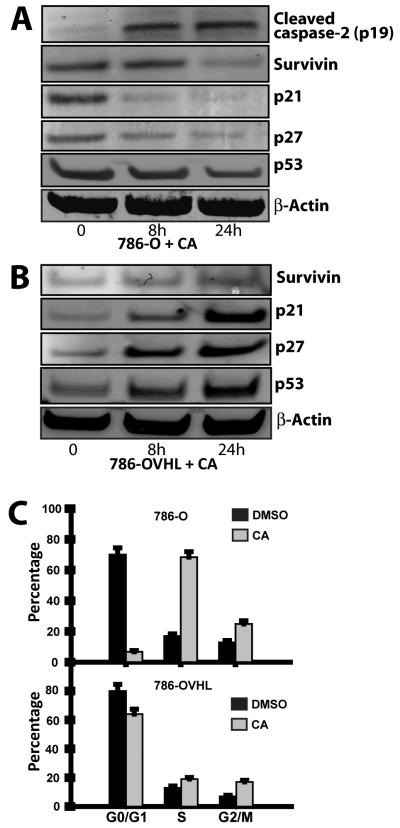
Resistance to cytokine-induced apoptosis in the 786-O cell line has been attributed to expression of high levels of apoptosis-inhibiting molecules (8). We thus examined, the effect of CA on the levels of expression of the inhibitor of apoptosis protein survivin. Western blotting of lysates from CA treated cells revealed a decrease in survivin levels in 786-O cells while its levels in 786-OVHL cells increased suggesting that CA owes its apoptotic effects in 786-O cells, in part, to its effects on survivin (Fig. 3). The activation of caspase-2 has been reported to repress the transcription of the survivin (9). Western blotting also revealed that CA treatment enhanced capsase-2 cleavage (Fig. 3A). Next, since upregulation of survivin in malignant cells has also been associated with accelerated S-phase shift, resistance to G1 arrest and activation of Cdk2/Cyclin E complex leading to Rb phosphorylation (10), we looked at effects of CA on the cell cycle and cell cycle checkpoint signaling pathway proteins. First, the distribution of live 786-O cells in each phase of the cell cycle was determined using flow cytometry following incubation with CA. The number of 786-O cells in S-phase significantly increased consistent with S-phase arrest (Fig. 3C). We also found that CA treatment in 786-O cells was accompanied by a decrease in the levels of phosphorylated Rb and an increase in levels of cyclins D3 and E with no significant change in Cdk2 levels (supplementary Fig. S4). Interestingly, cyclin D3 is known to induce the activation of caspase-2 (11) suggesting that caspase-2 mediated proapoptotic mitochondrial effects and downregulation of survivin both play a role in mediating the effects of CA on proliferation of 786-O cells.
Figure 3.
Effects of CA on p53 and related proteins. 786-O (A) and 786-OVHL (B) cells were treated with 0.1 μmol/L CA over a 24 h period. Protein extracts were prepared and subjected to western blot analysis. C) Effects of CA on cell cycle distribution in 786-O cells. Fluorescence-activated cell sorter (FACS) analysis of live 786-O cells without or with carminomycin (0.1 μmol/L) treatment. All cells were harvested 16 h after treatment and stained with propidium iodide in Krishan’s Buffer before analysis.
In order to better understand the molecular mechanisms that result in the differential activity seen between 786-O and 786-OVHL cells in response to CA treatment, we also examined the role of factors commonly involved in regulating apoptotic response. Activation of p53 has been shown to induce apoptosis in 786-O cells treated with either indomethacin (12) or UV-radiation (13). As a result, we first examined changes in p53 levels as well as changes in the levels of p53 downstream targets in both 786-O and 786-OVHL cells following CA treatment. Western blotting of 786-O protein lysates from treated cells showed that p53 levels in fact decreased in response to CA in 786-O cells (Fig. 3A). In contrast, p53 levels in 786-OVHL cells increased in response to CA. We also looked at changes in the level of p53 targets such as p21 and p27. Both p21 and p27 levels decreased in response to treatment with CA in 786-O cells, suggesting that apoptosis may not be mediated through activation of the p53 pathway in 786-O cells.
Sensitivity to CA in 786-O cells is mediated through its active localization within the Golgi
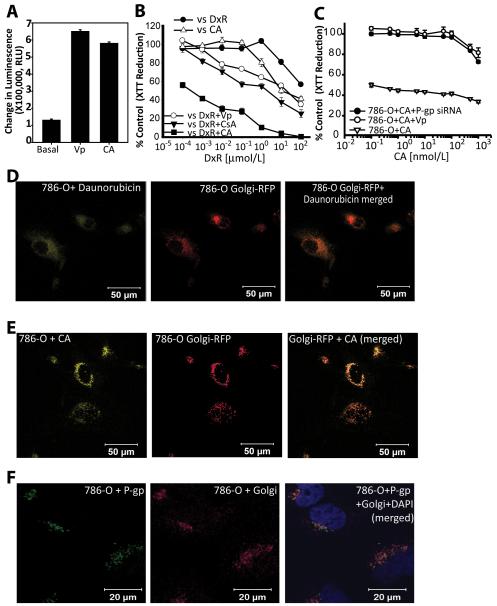
Resistance to anthracycline compounds like CA is known to be mediated by the ATP-binding cassette transporter superfamily of membrane proteins. More pertinently, sequestration of the CA related anthracycline daunorubicin within intracellular organelles has been reported to be mediated by p-glycoprotein (P-gp) (14). We, therefore, first looked at whether CA potentially was a substrate for P-gp. The results showed that CA stimulated basal P-gp ATPase activity by more than 4-fold suggesting that it is a substrate for transport by P-gp like other anthracycline antibiotics (Fig. 4A). Since P-gp mediated resistance to the anthracycline doxorubicin in OVCAR-3 cells is reversible by other P-gp substrates, we looked at whether this resistance could also be reversed by CA. Pretreatment of OVCAR-3 cells with 0.1 μmol/L CA resulted in an enhanced sensitivity to doxorubicin compared to pretreatement with the P-gp inhibitors verapamil or cyclosporin A (Fig. 4B). Next, 786-O cells were pretreated with verapamil at 50 μmol/L for 30 min and then incubated with varying concentrations of CA. It was found that 786-O cells became resistant to CA (Fig. 4C). To further confirm involvement of P-gp, siRNA was used to knock-down P-gp in 786-O cells (786-O Pgp-KD). Compared to 786-O cells treated with a GFP control siRNA, 786-O Pgp-KD cells displayed increased resistance to CA (Fig. 4C).
Fig. 4.
CA is sequestered within the Golgi. (A) CA stimulates P-gp ATPase activity. The difference in luminescence between 100 μmol/L Na3VO4− treated and DMSO (control) treated P-gp in cell membrane fractions constituted Basal activity. The decrease in luminescence as a result of CA treatment compared to Na3VO4− treatment represents CA stimulated P-gp ATPase activity. Determination of unconsumed ATP after stopping P-gp ATPase activity was made using luciferase and luminescence measured is presented in relative light units (RLU). Verapamil was used as a positive control. Results shown are averages of three independent experiments performed in triplicate ± SEM. (B) Resistance to doxorubicin in the P-gp expressing cell line OVCAR-3, is significantly reversed via cotreatment with CA (0.1μmol/L) compared to the P-gp inhibitors verapamil (Vp, 50μmol/L) and cyclosporine A (CsA, 6μmol/L). (C) The P-gp inhibitor verapamil and P-gp siRNA reverse the sensitivity of 786-O cells to CA. (D) The known P-gp substrate and CA analog daunorubicin (DxR) exhibits perinuclear localization in 786-O cells. (E) Subcellular localization of CA in the Golgi in 786-O cells transduced with an RFP-Golgi marker and CA (10 μmol/L/2 h). (F) Determination of presence of P-gp in the Golgi was made in fixed and permeabilized 786-O cells co-stained with antibodies against P-gp (green) and the Golgi marker GM130 (red). Colocalization analysis was undertaken as described in the supplemetal information online.
Following this, we examined whether CA, like the anthracycline and P-gp substrate daunorubicin, was also sequestered within 786-O cells. Because CA is a fluorescent compound, we used confocal fluorescence microscopy of CA treated 786-O cells and determined that like daunorubicin (Fig. 4D), CA localized within the perinuclear region in vesicles (Fig. 4E). This observation appears to exclude the known nuclear effect on DNA of anthracyclines as a potential mechanism of cytotoxicity (15). Since the perinuclear distribution of CA appeared consistent with localization in either the endoplasmic reticulum (ER) or the Golgi, 786-O cells transfected with either a Golgi-localizing RFP (786-O Golgi-RFP) or ER-localizing dsRed reporter (786-O ER-dsRed) were generated to enable a comparison of the localization pattern of CA in 786-O cells. Comparison of the fluorescence images of 786-O Golgi-RFP, 786-O ER-dsRed and 786-O cells treated with CA suggested that CA might localize in the Golgi. Further support for this was obtained from colocalization studies done in 786-O Golgi-RFP cells using quantitative analysis of confocal images which revealed significant colocalization. 74%±0.4% (median±MAD) of CA signals were found within the Golgi and 54%±0.1% of the Golgi compartment showed CA signals (n=30). Similarly, to confirm the presence of P-gp in the Golgi, we also carried out a colocalization experiment using P-gp immunofluorescence while using GM130 as a Golgi marker in fixed 786-O cells grown on glass coverlips. Here, 46%±13% (median±MAD) of P-gp signals were found within the Golgi and 67%±9% of the Golgi compartment showed P-gp signal (n=30) indicating a significant P-gp presence in the Golgi (Fig. 4F). 786-O cells grown on O- and L-shaped fibronectin micropatterns on glass chips (Supplementary Fig. S5) to normalize cell position, shape, polarity and internal organelle positioning also confirmed the presence of P-gp in the Golgi in addition to the cell membrane (16).
CA induces p115 cleavage
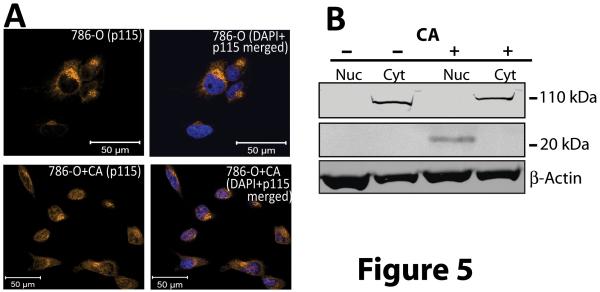
Because CA is sequestered within the Golgi, we next investigated whether effects on Golgi proteins played a role in its proapoptotic activity. Cleavage and import into the nucleus of the C-terminus of the Golgi protein p115 has been shown to promote apoptosis (17). Western blotting and immunofluorescence examination employing an antibody that recognizes the c-terminus of p115 in CA treated 786-O cells showed that this fragment was detectable in the nucleus of 786-O cells whereas none was seen in the nucleus of untreated controls (Fig. 5).
Figure 5.
Cleaved c-terminal fragment of p115 translocates to the nucleus on CA treatment. A) 786-O cells were treated with 0.01 μmol/L CA for 24 h and were then stained with an antibody targeting the C-terminus of p115 (golden). Nuclei were stained with DAPI. Note the accumulation of p115 immunoreactive material in the nuclei after CA treatment. B) Western blotting of fractionated Cytosolic (Cyt) and nuclear (Nuc) protein extracts of CA treated (+) and control (−) 786-O cells using an antibody recognizing the c-terminus of p115.
CA modulates ERGIC-53 activity
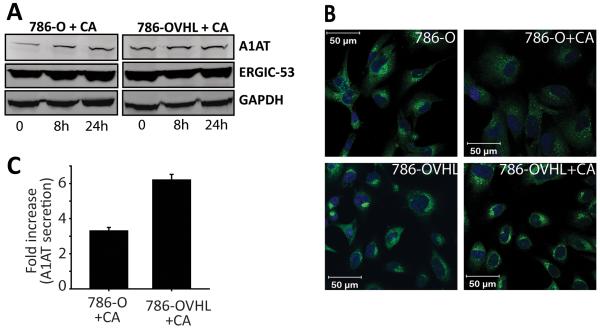
In order to better understand the differential response to CA treatment seen between 786-O and 786-OVHL cells, we asked whether the interaction of Golgi proteins with pVHL afforded 786-OVHL cells protection from the effects of CA. One of the well studied Golgi proteins known to interact with pVHL is the ER-Golgi intermediate compartment protein ERGIC-53 (also known as LMAN1) which constitutively cycles between the ER, ERGIC, and the cis-Golgi elements (18). CA treatment produced no significant changes in ERGIC-53 levels in both 786-O and 786-O VHL cells (Fig. 6A). We, therefore, looked into whether its anterograde and retrograde trafficking was impacted by CA treatment in 786-O cells and whether pVHL afforded protection from effects of CA on its trafficking. Immunofluorescence confocal experiments revealed that its trafficking was impacted in 786-O which showed significantly decreased characteristic accumulation in ERGIC and cis-Golgi elements that is seen in untreated cells (Fig. 6B). No such disruption was evident in CA treated 786-OVHL cells. We then looked at export of a well-known ERGIC-53 client protein, α1-antitrypsin (A1AT), as a way to assess impact of CA on ERGIC-53 activity (19). Western blotting revealed that intracellular A1AT levels increased in CA treated 786-O cells indicating that A1AT was increasingly being retained within treated cells while its levels remained unchanged in treated 786-OVHL cells (Fig. 6A). We also compared A1AT-secretion from 786-O and 786-OVHL cells after treatment with CA. Conditioned medium from CA or DMSO control treated 786-O and 786-OVHL cells was examined using an A1AT-specific ELISA. A 3.3-fold increase in secreted A1AT levels was measured in 786-O cells relative to untreated controls (5.01 μg/L/1000 cells) where as a 6.2-fold increase was observed in 786-OVHL cells relative to control cells (15.52 μg/L/1000 cells; Fig. 6C).
Fig. 6.
CA modulates ERGIC-53 activity but not its levels. (A) Intracellular levels of ERGIC-53 and its client A1AT were monitored over 24 h after treatment with 0.01 μmol/L CA via western blotting. (B) Effect of CA on distribution of ERGIC-53 in 786-O and 786-OVHL cells visualized by immunofluorescence microscopy. Cells were treated with either DMSO (control) or 0.01 μmol/L CA for 24 h. Note the diffuse staining for ERGIC-53 (green fluorescence) in the perinuclear area in CA treated 786-O cells. (C) A1AT levels in conditioned medium obtained from 786-O and 786-OVHL cells treated with 0.01 μmol/L CA were measured 24 h after treatment using ELISA. Values were then normalized to cell number and are reported relative to controls. Data represent the average of three experiments ± SEM.
Discussion
The main goal of the work presented above is the demonstration of the utility of a natural products based screening approach as a viable strategy for exploring newer ways of targeting CCRCC. To date, other than the modulation of HIF-2 activity, there have not been many viable molecular targets proposed for therapeutic discovery in CCRCC. In this regard, we discovered a compound that exploits other vulnerabilities created by genetic and epigenetic alterations that have taken place in CCRCC. The screen employed an unbiased cell-based assay that has led to the identification of carminomycin I (CA) as a selective inhibitor of CCRCC cell proliferation.
The similarity in the rate of change in XTT reduction of CA treated paired VHL−/− and VHL+/+ cells reveals that although CA impacts cell viability to a significant extent in VHL−/− cells, it did not impact the rate of change in viability to a similar extent compared to VHL+/+ cells. This suggests that the presence of pVHL impacts sensitivity to CA but not the rate of change in cell viability. We found that CA induced apoptosis in VHL−/− cells to a significant extent and that this activity was HIF and p53 independent. At low concentrations CA induced the activation of caspases 2 and 3 with little or no effect on the activation of caspases-8 or -9. This effect was accompanied by downregulation of the inhibitor of apoptosis protein survivin, a caspase-2 target. Although survivin levels appear to be steady at the 8 h time point while the level of caspase-2 is increasing in response to CA treatment, this does not necessarily imply that caspase-2 activation does not contribute to survivin down regulation. In fact, stress response to anthracyclines is known to result in the rapid induction of both mRNA and protein of inhibitor of apoptosis proteins including survivin which is followed by a gradual decrease of their expression (20). We also determined that CA was sequestered within the Golgi in CCRCC cells and that mediation by P-gp, which was also found within the Golgi, was required for its activity. However, whether CA is also sequestered within the Golgi of other P-gp expressing cell lines awaits further investigation. Its localization within the Golgi in CCRCC cells suggests that it might exert its action, at least in part, through effects on Golgi proteins. We, therefore, explored effects of CA on Golgi proteins and whether resistance to CA in VHL+/+ CCRCC cells was related to interaction of pVHL with Golgi proteins. We found that CA treatment resulted in the detection of the C-terminal fragment of p115 in the nucleus indicating that its apoptotic effects can in part be explained through its effects on p115 (21). Although CA had no effects on levels of the Golgi protein ERGIC-53, which is one of the few Golgi-proteins known to have a direct protein-protein interaction with pVHL, it impacted its distribution pattern between the ER and the Golgi. Our results also indicated that the secretion into the growth medium of A1AT, an ERGIC-53 client, was attenuated while intracellular A1AT levels increased in response to CA treatment. These are consistent with attenuation of function of ERGIC-53 (22).
CA is composed of an aglycone moiety and a monosaccharide residue. Since microsomes from mammalian tissues are known to metabolize anthracyclines to the superoxide anion of the aglycones and their monosaccharide residues through reductive glycosidic cleavage, it can be hypothesized that damage to Golgi proteins by the reactive oxygen species formed from the aglycone may be responsible for the effects of CA on proliferation in CCRCC cells (23-25). Alternatively, recognition of the monosaccharide residue in CA as a substrate may lead to attenuation of activity of Golgi proteins. Undoubtedly, however, CA also effects its activity through impact on additional pathways and targets. Demonstration of efficacy in xenograft models is pending preparative scale isolation of CA from the producing micro-organism.
In summary, our findings illustrate a potential novel approach for targeting VHL−/− clear cell renal carcinoma. Our findings also underscore the utility of natural products in yielding compounds that may be used either in therapeutic development or as bioprobes.
Supplementary Material
Acknowledgements
We thank Dr Olga Aprelikova for providing reagents. This project has been funded in whole or in part with federal funds from the National Cancer Institute, National Institutes of Health, under contract N01-CO-12400. The content of this publication does not necessarily reflect the views or policies of the Department of Health and Human Services, nor does mention of trade names, commercial products, or organizations imply endorsement by the U.S. Government. This Research was supported in part by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research.
References
- 1.Gnarra JR, Tory K, Weng Y, et al. Mutations of the VHL tumor-suppressor gene in renal-carcinoma. Nat Genet. 1994;7:85–90. doi: 10.1038/ng0594-85. [DOI] [PubMed] [Google Scholar]
- 2.Latif F, Tory K, Gnarra J, et al. Identification of the vonHippel-Lindau disease tumor-suppressor gene. Science. 1993;260:1317–20. doi: 10.1126/science.8493574. [DOI] [PubMed] [Google Scholar]
- 3.Kaelin WG. Molecular basis of the VHL hereditary cancer syndrome. Nat Rev Cancer. 2002;2:673–82. doi: 10.1038/nrc885. [DOI] [PubMed] [Google Scholar]
- 4.National Institutes of Health . Developmental Therapeutics Program NCI; Maryland: dtp.nci.nih.gov [homepage on the Internet] [cited 2010 Aug 27]. Available from: http://dtp.nci.nih.gov/branches/btb/characterizationNCI60.html. [Google Scholar]
- 5.Yang Y, Padilla-Nash HM, Vira MA, et al. The UOK 257 cell line: a novel model for studies of the human Birt-Hogg-Dube gene pathway. Cancer Genetics and Cytogenetics. 2008;180:100–9. doi: 10.1016/j.cancergencyto.2007.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Woldemichael GM, Vasselli JR, Gardella RS, McKee TC, Linehan WM, McMahon JB. Development of a cell-based reporter assay for screening of inhibitors of hypoxia-inducible factor 2-induced gene expression. J Biomol Screen. 2006;11:678–87. doi: 10.1177/1087057106289234. [DOI] [PubMed] [Google Scholar]
- 7.Zhang JH, Chung TDY, Oldenburg KR. A simple statistical parameter for use in evaluation and validation of high throughput screening assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 8.Griffith TS, Fialkov JM, Scott DL, et al. Induction and regulation of tumor necrosis factor-related apoptosis-inducing ligand/Apo-2 ligand-mediated apoptosis in renal cell carcinoma. Cancer Res. 2002;62:3093–9. [PubMed] [Google Scholar]
- 9.Guha M, Xia F, Raskett CM, Altieri DC. Caspase 2-mediated tumor suppression involves survivin gene silencing. Oncogene. 2010;29 doi: 10.1038/onc.2009.428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Suzuki A, Hayashida M, Ito T, et al. Survivin initiates cell cycle entry by the competitive interaction with Cdk4/p16(INK4a) and Cdk2/Cyclin E complex activation. Oncogene. 2000;19:3225–34. doi: 10.1038/sj.onc.1203665. [DOI] [PubMed] [Google Scholar]
- 11.Mendelsohn AR, Hamer JD, Wang ZB, Brent R. Cyclin D3 activates Caspase 2, connecting cell proliferation with cell death. Proc Natl Acad Sci U S A. 2002;99:6871–6. doi: 10.1073/pnas.072290599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ou YC, Yang CR, Cheng CL, Raung SL, Hung YY, Chen CJ. Indomethacin induces apoptosis in 786-O renal cell carcinoma cells by activating mitogen-activated protein kinases and AKT. Eur J Pharmacol. 2007;563:49–60. doi: 10.1016/j.ejphar.2007.01.071. [DOI] [PubMed] [Google Scholar]
- 13.Schoenfeld AR, Parris T, Eisenberger A, et al. The von Hippel-Lindau tumor suppressor gene protects cells from UV-mediated apoptosis. Oncogene. 2000;19:5851–7. doi: 10.1038/sj.onc.1203985. [DOI] [PubMed] [Google Scholar]
- 14.Gong YP, Wang YT, Chen FY, et al. Identification of the subcellular localization of daunorubicin in multidrug-resistant K562 cell line. Leuk Res. 2000;24:769–74. doi: 10.1016/s0145-2126(00)00048-5. [DOI] [PubMed] [Google Scholar]
- 15.Duvernay VH, Pachter JA, Crooke ST. Molecular phamacological differences between carminomycin and its analog, carminomycin-11-methyl ether, and adriamycin. Cancer Res. 1980;40:387–94. [PubMed] [Google Scholar]
- 16.Thery M, Racine V, Piel M, et al. Anisotropy of cell adhesive microenvironment governs cell internal organization and orientation of polarity. Proc Nat Acad Sci. 2006;103:19771–6. doi: 10.1073/pnas.0609267103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mukherjee S, Shields D. Nuclear import is required for the pro-apoptotic function of the golgi protein p115. J Biol Chem. 2009;284:1709–17. doi: 10.1074/jbc.M807263200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hergovich A, Lisztwan J, Barry R, Ballschmieter P, Krek W. Regulation of microtubule stability by the von Hippel-Lindau tumour suppressor protein pVHL. Nat Cell Biol. 2003;5:64–70. doi: 10.1038/ncb899. [DOI] [PubMed] [Google Scholar]
- 19.Nyfeler B, Reiterer V, Wendeler MW, et al. Identification of ERGIC-53 as an intracellular transport receptor of alpha(1)-antitrypsin. J Cell Biol. 2008;180:705–12. doi: 10.1083/jcb.200709100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Abe S, Hasegawa M, Yamamoto K, et al. Rapid induction of IAP family proteins and Smac/DIABLO expression after proapoptotic stimulation with doxorubicin in RPMI 8226 multiple myeloma cells. Exp Mol Pathol. 2007;83:405–12. doi: 10.1016/j.yexmp.2007.04.001. [DOI] [PubMed] [Google Scholar]
- 21.Chiu R, Novikov L, Mukherjee S, Shields D. A caspase cleavage fragment of p115 induces fragmentation of the Golgi apparatus and apoptosis. J Cell Biol. 2002;159:637–48. doi: 10.1083/jcb.200208013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Appenzeller-Herzog C, Roche AC, Nufer O, Hauri HP. pH-induced conversion of the transport lectin ERGIC-53 triggers glycoprotein release. J Biol Chem. 2004;279:12943–50. doi: 10.1074/jbc.M313245200. [DOI] [PubMed] [Google Scholar]
- 23.Peters JH, Gordon GR, Kashiwase D, Lown JW, Yen SF, Plambeck JA. Redox activities of antitumor anthracyclines determined by microsomal oxygen-consumption and assays for superoxide anion and hydroxyl radical generation. Biochem Pharmacol. 1986;35:1309–23. doi: 10.1016/0006-2952(86)90276-5. [DOI] [PubMed] [Google Scholar]
- 24.Salvatorelli E, Guarnieri S, Menna P, et al. Defective one- or two-electron reduction of the anticancer anthracycline epirubicin in human heart - Relative importance of vesicular sequestration and impaired efficiency of electron addition. J Biol Chem. 2006;281:10990–1001. doi: 10.1074/jbc.M508343200. [DOI] [PubMed] [Google Scholar]
- 25.Asbell MA, Bullock FJ, Schwartz E, Yesair DW. Daunomycin and adriamycin metabolism via reductive glycosidic cleavage. J Pharmacol Exp Ther. 1972;182:63. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.