Abstract
Background
The basis for increased susceptibility of atopic dermatitis (AD) patients to develop disseminated viral skin infections such as eczema herpeticum (ADEH+) is poorly understood.
Objective
We sought to determine whether atopic dermatitis subjects prone to disseminated viral skin infections have defects in their interferon responses.
Methods
GeneChip profiling was used to identify differences in gene expression of peripheral blood mononuclear cells (PBMC) from patients with a history of ADEH+ as compared to ADEH− and non-atopic controls. Key differences in protein expression were verified by ELISPOT and/or ELISA. Clinical relevance was further demonstrated by a mouse model of disseminated viral skin infection and genetic association analysis for genetic variants in IFNG and IFNGR1 and ADEH among 435 cases and controls.
Results
We demonstrate by global gene expression analysis selective transcriptomic changes within the interferon (IFN) superfamily of PBMCs from ADEH+ subjects reflecting low IFNγ and IFNγ receptor gene expression. IFNγ protein production was also significantly lower in ADEH+ (N=24) compared to ADEH− (N=20) and non-atopic (NA; N=20) controls. IFNγ receptor knockout (KO) mice developed disseminated viral skin infection after epicutaneous challenge with vaccinia virus (VV). Genetic variants in IFNG and IFNGR1 SNPs were significantly associated with ADEH (112 cases, 166 controls) and IFNγ production: a 2-SNP (A–G) IFNGR1 haplotype (rs10457655 and rs7749390) showed the strongest association with a reduced risk of ADEH+ ((13.2% ADEH+ vs 25.5% ADEH−, P = 0.00057).
Conclusions
ADEH+ patients have reduced IFNγ production, and IFNG and IFNGR1 SNPs are significantly associated with ADEH+ and may contribute to an impaired immune response to herpes simplex virus (HSV).
Clinical Implications
Atopic dermatitis subjects prone to disseminated viral skin infections have defects in their interferon responses.
Capsule summary
Using genomic, immunologic and genetic approaches, these investigators demonstrated that atopic dermatitis subjects prone to disseminated viral skin infections have defects in their interferon responses.
Keywords: atopic dermatitis; infection; eczema herpeticum; IFNG, IFNGR1
INTRODUCTION
Atopic dermatitis (AD) is a complex chronic skin disease affecting up to 30% of children, often persisting into adulthood.1,2 A small subset of AD patients suffer from disseminated viral infections, i.e. eczema herpeticum (ADEH+), after herpes simplex virus (HSV) infection3 or eczema vaccinatum (EV) after smallpox vaccination4with vaccinia virus (VV). As a result, it is recommended that all AD patients avoid routine smallpox vaccination. Smallpox vaccination is also not advised for family members who have close contact with AD patients since life threatening or disfiguring EV has been reported in such individuals.5 This is a major impediment to mass vaccination since the conventional smallpox vaccine is withheld from AD patients not prone to viral infections (ADEH−).6,7 To address this major health care problem, the National Institutes of Health/National Institute of Allergy and Infectious Diseases (NIH/NIAID) formed the Atopic Dermatitis Vaccinia Network (ADVN) to identify biomarkers and gene variants, which could lead to early identification of AD patients prone to disseminated viral infections.
The primary goal of the current study was to determine whether ADEH+ subjects have identifiable defects that reduce their ability to control HSV and VV viral skin infections. We evaluated global transcriptional differences in peripheral blood mononuclear cells (PBMCs) from ADEH+ patients, compared to ADEH− and non-atopic control (NA) subjects. Expression analysis of 38,500 genes demonstrated significant association of ADEH+ with transcriptomics of the interferon (IFN) superfamily. Most noteworthy, type II IFNγ and its receptor were downregulated in ADEH+, but not in ADEH−, patients. Indeed, we confirmed that IFNγ protein production in PBMCs from ADEH+ subjects was significantly lower than ADEH− and NA control subjects. The biological and clinical significance of this defect in IFNγ generation were further explored by examining the response of mice genetically deficient in IFNγ receptor expression to viral skin inoculation, and by evaluation of IFNG and IFNG receptor gene variants in ADEH+ versus ADEH− and NA control subjects.
METHODS
Subjects
Twenty-four ADEH+, 20 ADEH− and 20 non-atopic subjects were evaluated for CMI as described in Table EI of this article’s Online Repository at www.jacionline.org. A subset of these subjects (N=27) were selected for geneChip profiling studies. Additional subjects were evaluated in genetic association studies as described in Table E II. ADEH+ subjects (N=112) were defined as subjects with AD who had at least one EH episode as described in reference 3. HSV infection was laboratory confirmed. ADEH− subjects (N=166) were defined as subjects with AD with no history of EH. Non-atopic, healthy controls (N=157) were defined as having no personal history of chronic disease including atopy. All study participants were further evaluated by a detailed history and physical examination, as well as a questionnaire to assess history of cutaneous viral infections and concomitant medication use.
GeneChip profiling experiments
RNA was hybridized to an Affymetrix GeneChip U133_Plus2 (54613 probe sets for 38,500 genes) and hybridization signals were measured using an Agilent Gene Array Scanner. The global ADEH+ transcriptional response was evaluated by MAPPFinder with 1.5 fold change cutoff, and q value 10% as described previously.8 The MAPPFinder compatible files were prepared using the GenMAPP converting tool and significant bioprocesses were selected by choosing gene oncology (GO) terms containing greater than 50% of the total number of ADEH+-associated genes and a Z-score >2.
ELISPOT assay
PBMCs were isolated, cryopreserved and stored.9 Cells were resuspended at 1 × 107 cells/mL in RPMI with 10% human serum AB in 10 mM Hepes. Fifty microliters of the cell suspension and 50 µL of HSV or mock antigens prepared were added to microtiter wells pre-coated with anti-human IFNγ mAb at 5µg/ml (Endogen). After overnight incubation, plates were washed and bound IFNγ was revealed with biotin-labeled mAb anti-human IFNγ (Endogen), streptavidin-AP (Pierce) and 1-Step NBT/BCIP (Pierce). Spot forming cells (SFC) were then counted.
Virus preparation
The Western Reserve (VR1354; ATCC, Manassas, VA) strains of VV were propagated in HeLa S3 (ATCC#CCK-2.2) human adenocarcinoma cells.10
Mice
IFNγ R−/−, B6.129S7-Ifngr1tm1Agt/J and control C57BL/6J mice were purchased from The Jackson Laboratory (Bar Harbor, ME). All protocols were approved by the Institutional Animal Care and Use Committee at National Jewish Health.
Inoculation of VV into murine skin
The dorsal thoracic of mice were clipped and treated with Nair® to remove all hair. Seventy-two hours following hair removal, mice were anesthetized and inoculated with 1 × 107 PFU of Western Reserve VV by scarification in the dorsal thorax region. Each animal received 15 pricks with a bifurcated needle. Mice were monitored daily for the formation of satellite lesions. After the exposure period, total RNA was isolated from skin biopsies and blood lysates by chloroform:phenol extraction and isopropanol precipitation according to manufacturer’s guidelines (Molecular Research Center, Inc.).
Quantitative real time RT-PCR
RNeasy Mini Kits (Qiagen, Valencia, CA) were used according to the manufacturer’s protocol. RNA was reverse transcribed into cDNA and analyzed by real-time RT-PCR using an ABI Prism 7300 sequence detector (Applied Biosystems, Foster City, CA) as previously described.10,11 Primers and probes for mouse GAPDH were purchased from Applied Biosystems. The primer and probe sequences for VV recognize a subunit of a DNA-directed RNA polymerase expressed within two hours of viral entry.12 To allow for comparisons between samples and groups, quantities of all targets in test samples were normalized to the corresponding GAPDH or total cDNA levels.13
Gene association studies
Eight IFNG SNPs and six IFNGR1 SNPs were genotyped (See Table EIII), which included previously reported functional SNPs (2 in IFNG, 2 in IFNGR1) in addition to highly tagging SNPs selected from the HapMap database (http://www.hapmap.org/). SNP genotyping was performed using TaqMan Allelic discrimination Assays on the 7900HT Sequence Detection System (Applied Biosystems).
Statistical analysis
ELISPOT values were log10 transformed to satisfy statistical assumptions. Diagnostic group comparisons employed analysis of covariance (ANCOVA) models adjusting for age (mock-stimulated endpoints) or adjusting for age and mock-stimulated values (HSV-stimulated endpoints) using SAS version 9.1. All P-values were considered descriptive and exploratory, and P-values < 0.05 were considered noteworthy. All statistical analyses for mouse experiments were conducted using Graph Pad Prism, version 4.01 and SAS version 9.1. Data were analyzed using a student’s t test or one-way analysis of variance (ANOVA) followed by a Tukey-Kramer post hoc test.14 Genetic association analysis was performed using the Cochran–Armitage trend test under an additive model using PLINK software.15 Haplotype analyses were performed using sliding windows of 2–4 SNPs where empiric P-values for haplotype frequency differences were generated over 10,000 permutations. Associations with SFC were performed using a linear regression analysis adjusted for confounding variables (age and gender).
RESULTS
Screening geneChip profiling studies
Candidate targets to identify susceptibility to ADEH were evaluated by Affymetrix GeneChip profiling.8 Total mRNA was isolated from sham-treated and VV-treated PBMCs from ADEH+ (n=5), ADEH− (n=11), and healthy NA (n=11) individuals. The global pathway analysis identified the IFN superfamily as having the most altered genes in VV-challenged ADEH+ PBMCs (Table I). The detailed analysis of expression of individual interferon superfamily genes revealed that the overwhelming majority of IFN-related genes were upregulated, including type 1 IFN-α,-β,-ε,-ω subfamilies (Table II). Increased expression of the Type I IFNs, known to have potent anti-viral effects, would not account for the increased propensity of ADEH+ subjects toward disseminated viral infections. Their lack of effect may be related to the observed downregulation of interferon (alpha, beta, and omega) receptor 1 (Table II). Most importantly, the combination of IFNγ (type II interferon) and IFNγ receptor were significantly downregulated in PBMCs from ADEH+ patients. Moreover, the downregulation of these genes was not detected in PBMCs from ADEH− patients. These data demonstrate that decreased VV-induced expression of IFNγ and its receptor is ADEH+ specific.
Table I.
PBMC biological processes significantly associated with ADEH+1
| GOID | GO Name | Changed* genes |
Measured genes |
% changed genes |
Z Score |
|---|---|---|---|---|---|
| 5132 | Interferon-alpha/beta receptor binding | 5 | 5 | 100.00 | 4.730 |
| 51047 | Positive regulation of secretion | 10 | 18 | 55.56 | 4.097 |
| 42036 | Negative regulation of cytokine biosynthesis | 8 | 13 | 61.54 | 4.040 |
| 48041 | Focal adhesion formation | 7 | 11 | 63.64 | 3.896 |
| 30004 | Monovalent inorganic cation homeostasis | 7 | 11 | 63.64 | 3.896 |
| 46328 | Regulation of JNK cascade | 10 | 19 | 52.63 | 3.880 |
| 45576 | Mast cell activation | 5 | 7 | 71.43 | 3.641 |
| 46330 | Positive regulation of JNK cascade | 7 | 12 | 58.33 | 3.594 |
| 45072 | Regulation of interferon-gamma biosynthesis | 8 | 15 | 53.33 | 3.517 |
| 51291 | Protein hetero-oligomerization | 6 | 10 | 60.00 | 3.417 |
| 5248 | Voltage-gated sodium channel activity | 5 | 8 | 62.50 | 3.239 |
| 45073 | Regulation of chemokine biosynthetic process | 5 | 8 | 62.50 | 3.239 |
| 51181 | Cofactor transport | 5 | 8 | 62.50 | 3.239 |
| 30641 | Hydrogen ion homeostasis | 5 | 8 | 62.50 | 3.239 |
| 16805 | Dipeptidase activity | 5 | 8 | 62.50 | 3.239 |
| 8354 | Germ cell migration | 5 | 8 | 62.50 | 3.239 |
| 6878 | Copper ion homeostasis | 5 | 8 | 62.50 | 3.239 |
| 17015 | Regulation of TGF beta receptor signaling pathway | 6 | 11 | 54.55 | 3.115 |
| 32395 | MHC class II receptor activity | 6 | 11 | 54.55 | 3.115 |
| 3711 | Transcriptional elongation regulator activity | 6 | 11 | 54.55 | 3.115 |
| 50798 | Activated T cell proliferation | 6 | 11 | 54.55 | 3.115 |
| 5375 | Copper ion transporter activity | 5 | 9 | 55.56 | 2.896 |
| 32602 | Chemokine production | 5 | 9 | 55.56 | 2.896 |
| 50755 | Chemokine metabolic process | 5 | 9 | 55.56 | 2.896 |
| 6825 | Copper ion transport | 5 | 9 | 55.56 | 2.896 |
| 42033 | Chemokine biosynthetic process | 5 | 9 | 55.56 | 2.896 |
| 7202 | Phospholipase C activation | 5 | 9 | 55.56 | 2.896 |
| 18149 | Peptide cross-linking | 5 | 9 | 55.56 | 2.896 |
considering that genes driving a bioprocess can be either up or down regulated dependent on a gene function (i.e. inhibitors or activators), all genes were counted independently of the direction of a change.
Bioprocesses with Z-score < 2.0 and %-change >50 were considered significantly affected by ADEH+.
Table II.
ADEH+ candidate genes in the interferon superfamily
| AFFY | Gene Title | Gene Symbol |
Fold Change* ADEH+ vs Normal |
q-value(%) ADEH+ vs Normal† |
Fold Change ADEH− vs Normal |
q-value(%) ADEH− vs Normal |
|---|---|---|---|---|---|---|
| Upregulated | ||||||
| 208259_x_at | Interferon, alpha 7¶ | IFNA7 | 5.33 | <1.0 | 2.59 | <0.1 |
| 211405_x_at | Interferon, alpha 17 | IFNA17 | 4.76 | <1.0 | 2.15 | <0.1 |
| 211145_x_at | Interferon, alpha 21 | IFNA21 | 4.59 | <1.0 | 1.55 | 0.554 |
| 207932_at | Interferon, alpha 8 | IFNA8 | 4.40 | <1.0 | 2.22 | 0.554 |
| 208344_x_at | Interferon, alpha 1 | IFNA1 | 4.17 | <1.0 | 2.34 | <0.1 |
| 207817_at | Interferon, omega 1 | IFNW1 | 3.97 | <1.0 | 3.47 | <0.1 |
| 207964_x_at | Interferon, alpha 4 | IFNA4 | 3.89 | <1.0 | 2.10 | 0.059 |
| 208261_x_at | Interferon, alpha 10 | IFNA10 | 3.58 | <1.0 | 2.39 | <0.1 |
| 208173_at | Interferon, beta 1, fibroblast | IFNB1 | 3.51 | <1.0 | 2.30 | <0.1 |
| 208182_x_at | Interferon, alpha 14 | IFNA14 | 3.40 | <1.0 | 2.44 | <0.1 |
| 211338_at | Interferon, alpha 2 | IFNA2 | 3.18 | <1.0 | 2.12 | <0.1 |
| 1552917_at | Interleukin 29 (interferon, lambda 1) | IL29 | 2.88 | <1.0 | 2.50 | <0.1 |
| 1553574_at | Interferon epsilon 1 | IFNE1* | 2.60 | <1.0 | 1.45 | 44.938 |
| 208448_x_at | Interferon, alpha 16 | IFNA16 | 2.56 | <1.0 | 2.42 | <0.1 |
| 1555464_at | Interferon induced with helicase C domain 1 | IFIH1* | 2.21 | <1.0 | 1.31 | 0.059 |
| 216502_at | Interferon stimulated exonuclease gene 20kda-like 2 | ISG20L2 | 1.55 | 2.608 | 1.76 | <0.1 |
| Downregulated | ||||||
| 211676_s_at | Interferon gamma receptor 1 | IFNGR1* | −1.63 | 3.738 | −1.21 | 87.248 |
| 225669_at | Interferon (alpha, beta and omega) receptor 1 | IFNAR1* | −1.59 | 2.608 | −1.14 | 87.911 |
| 210354_at | Interferon, gamma | IFNG* | −1.52 | 8.401 | 1.12 | 87.911 |
fold changes were generated by SAM 2.0 software applying multiple comparisons of 5 ADEH+ or 9 ADEH− samples versus 9 non-atopic controls (the two lowest signals among 11 ADEH− and 11 nonatopic (NA) samples were trimmed to increase the reliability of hybridization signals). Resulting fold changes are represented by bolded values. The symbols of genes with the significant (q <0.1) difference in expression during direct comparison of ADEH+ and ADEH− are marked with *.
interferons alpha, beta, epsilon, and omega belong to type I; and interferon gamma to type II interferons.
genes with significantly different expression in ADEH+ compared to ADEH−.
q-value represent the best false discovery rate seen for all of the possible gene lists a gene can be a part of, q <10% was considered significant.
IFNγ response of ADEH+ vs ADEH− subjects
Because IFNγ producing cells are known to be important cytotoxic effectors of immune protection against viruses, gene profiling was evaluated in subjects with ADEH+ (n=24), ADEH− (n= 20), or NA controls (n=20). Clinical characteristics of these participants are shown in Table EI of this article’s Online Repository at www.jacionline.org. We found a selective reduction of the IFNγ gene in ADEH+ subjects when IFNγ production was assessed post-mock stimulation or post-HSV stimulation using enzyme-linked immunosorbent spot (ELISPOT) technology to measure the frequency of HSV-specific IFNγ producing PBMCs.9
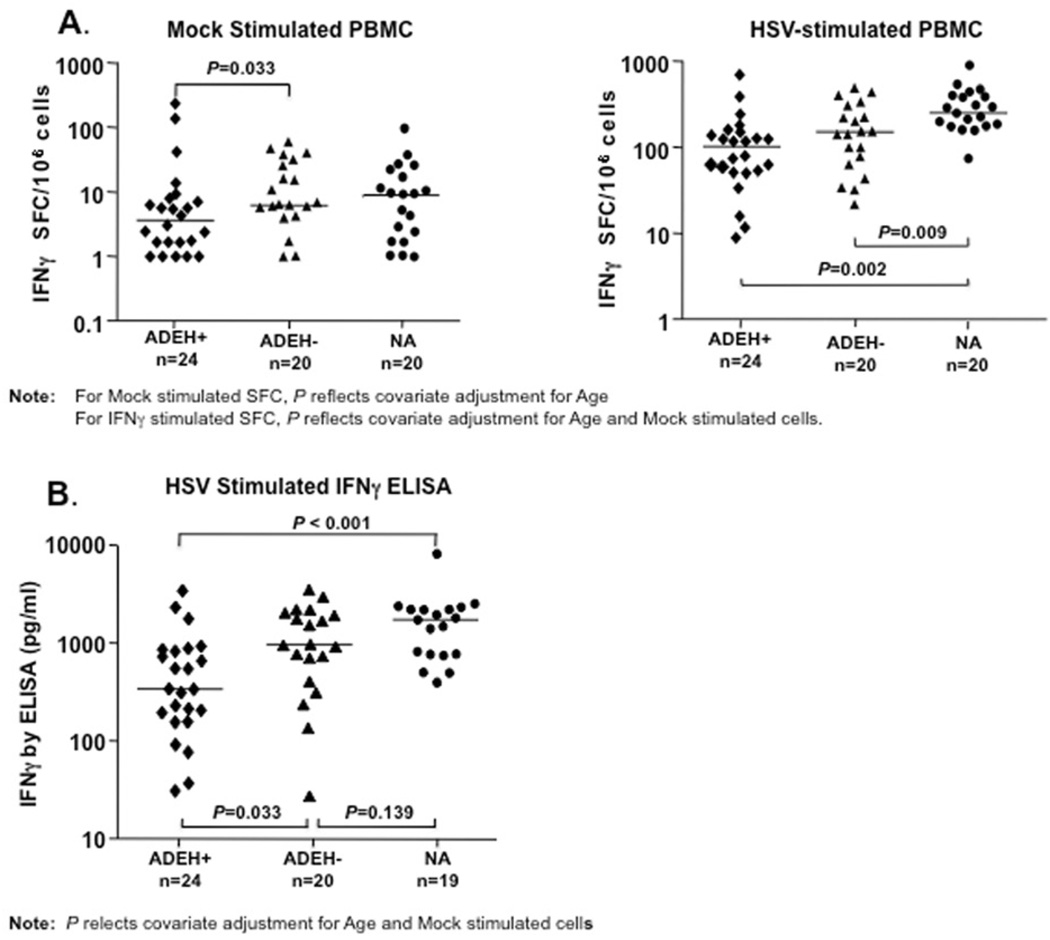
As shown in Figure 1A, IFNγ protein production was lower in PBMCs from ADEH+ subjects compared to PBMCs from the ADEH− and non-atopic control groups. Of note, after mock stimulation, IFNγ spot forming cells (SFCs) were lower (P=0.033) in ADEH+ subjects compared to ADEH− subjects, identifying a potential immune biomarker, which distinguishes propensity to disseminated viral skin infection in ADEH+ subjects. Furthermore, after HSV stimulation, IFNγ SFCs were significantly lower in both AD groups compared to NA; however, IFNγ SFCs were lowest in the ADEH+ group. Importantly, the lower levels of frequency of IFNγ producing cells observed in ADEH+ individuals was further confirmed by ELISA analysis of HSV-stimulated PBMC culture supernatants (Fig 1B), which showed lower levels (P=0.033) of IFNγ secretion in PBMC from ADEH+, as compared to ADEH− subjects. Lower levels of IFNγ secretion were also observed between ADEH+ compared to NA controls (P<0.001).
FIG 1.
IFNγ production is decreased in PBMC from ADEH+ as compared to ADEH− and NA human subjects. Panel A) shows IFNγ SFC after mock (left panel) or HSV stimulation (right panel) ex vivo. Panel B) shows IFNγ protein measurements in culture supernatants of HSV stimulated PBMC. PBMC from each subject group were stimulated for six days with mock or HSV antigens, then culture supernatants removed and analyzed for IFNγ secretion by ELISA. Datapoints represent the difference between IFNγ secretion in HSV vs mock stimulated PBMCs for each subject; P-values reflect adjusted (ANCOVA derived) comparisons. Horizontal lines represent median values.
Response of mice deficient in IFNγ receptor to VV
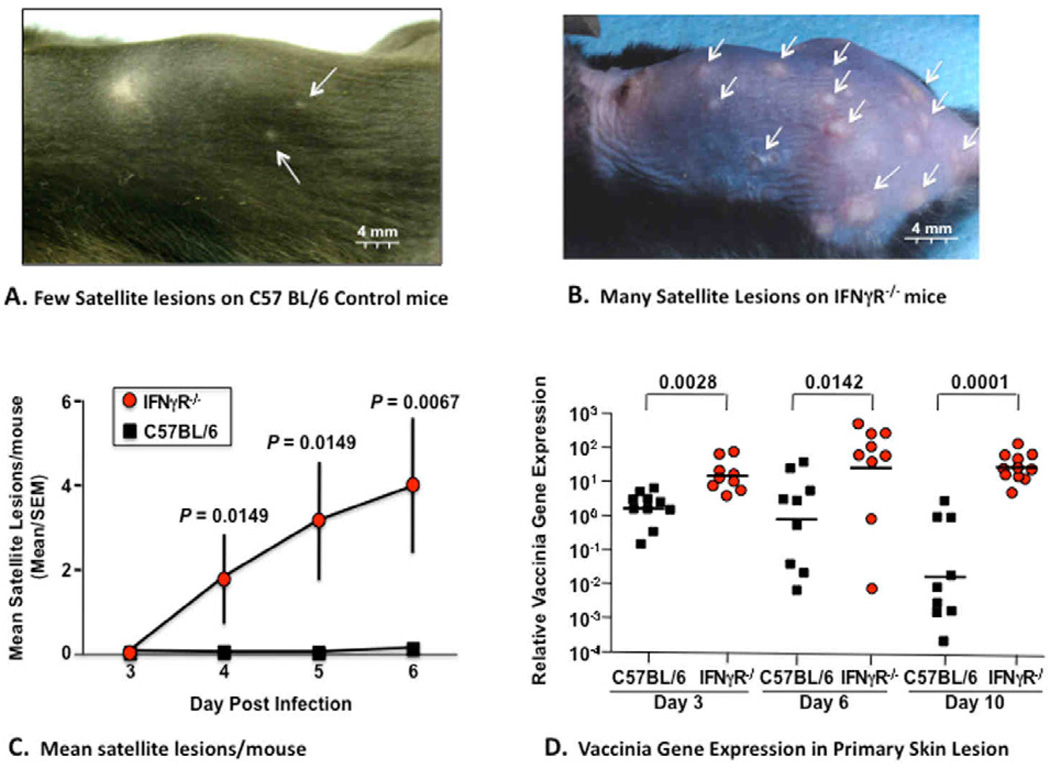
It is well established that IFNγ is highly effective at inhibiting HSV and VV replication in vitro in cell lines or in vivo after systemic infection or ocular injection.16–19 However, the role of IFNγ in controlling skin infection has not been previously reported. Since the primary objective of the current study was to identify the critical immune responses required to control VV replication in the skin after smallpox vaccination and gene profiling demonstrated reduced IFNγ and IFNγ receptor expression in ADEH+ subjects (Table II), we assessed the response of IFNγ receptor gene knockout (IFNγ R−/−) versus control mice with the same genetic background (C57 BL6) after inoculation with VV by scarification in a manner identical to the technique used for administration of smallpox vaccines to human skin. Following inoculation of VV into the skin, a significantly greater number of VV containing satellite lesions (Fig 2A–C) appeared on the skin of IFNγ R−/−, as compared to control mice by day four. The satellite lesions were also larger in diameter (Fig 2A, B). IFNγ R−/− mice also had significantly greater weight loss (P<0.001; data not shown), reduced survival (P=0.0025; data not shown), and increased viral load (P<0.01) in their primary inoculation sites (Fig 2D) as compared to control mice.
FIG 2.
Disseminated viral skin infection in IFNγ receptor KO mice after inoculation with vaccinia virus (VV). Clinical appearance of satellite lesions are shown after epicutaneous inoculation of C57 Bl/6 control mice (Panel A) and IFNRγ−/− mice (Panel B) with VV. Panel C demonstrates that the mean number of satellite lesions was significantly greater in IFNRγ−/− mice than control mice. Panel D shows data points of relative vaccinia gene expression (measured by real time PCR) was significantly greater at all timepoints in IFNRγ−/− mice than control mice.
IFNG and IFNG receptor (IFNGR1) gene variants are associated with ADEH+
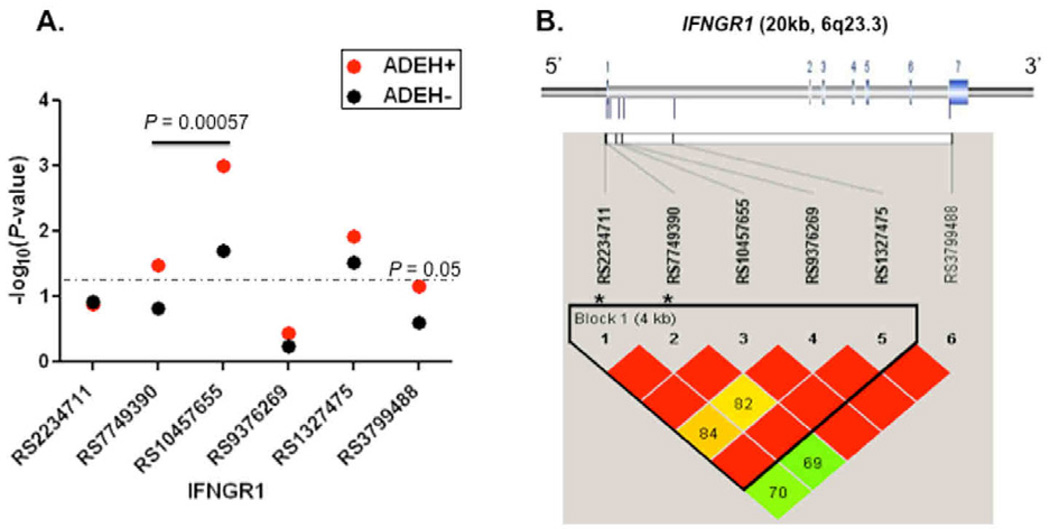
Because there were significantly lower numbers of IFNγ SFCs in the ADEH+ group compared to the ADEH− group even after mock stimulation, we explored the association between genetic variants in the IFNG gene and its related gene, IFNGR1, and risk of ADEH+ phenotype. Using both tagging and functional SNPs, we tested for association between SNPs and ADEH status. Detailed information on the participants in the ADVN has been previously described.20 We present here with updated information (See Table EII). As shown in Fig 3A, significant associations were observed for ADEH+ and three SNPs in IFNGR1 (rs1327475, P = 0.012; rs10457655, P = 0.001; rs7749390, P = 0.033). Associations with ADEH+ were further enhanced by haplotype analysis; a 2-SNP (A–G) haplotype spanning a region of <1.0kb in intron 1 of IFNGR1 (rs10457655 and rs7749390) within one single linkage disequilibrium (LD) block (Fig 3B) showed the strongest inverse association with ADEH+ (13.2% ADEH+ vs 25.5% ADEH−, P = 0.00057). Although none of the individual IFNG SNPs were significantly associated with risk of ADEH, significant associations were observed in haplotype analyses. Specifically, a four-marker haplotype spanning <4.3 kb (rs2069727, rs2069718, rs2430561, rs2069705) showed the strongest association of the possible haplotypes (P = 0.0027) and was less common in patients without ADEH+ (i.e. was a risk haplotype; ADEH+ vs ADEH−, 5.4% vs 1.1%, Table III and Fig E1).
FIG 3.
Summary of genetic associations for six IFNGR1 SNPs. Panel A) the −log10-transformed P-values for IFNGR1 SNPs and ADEH+ and ADEH−. The dashed gray line represents a P value of 0.05. The thick line represents a 2-SNP haplotype. Panel B) Gene structure and pattern of LD (D') in European American healthy controls, with red to green reflecting higher to lower D' values. Seven exons (color in blue) in IFNGR1 were presented (upper panel).*Functional SNPs.
Table III.
Strongest association of IFNG haplotype with ADEH+ among European Americans
| rs2069727* | rs2069718 | rs2069716 | rs2430561* | Haplotype ADEH+ |
Frequency ADEH− |
P-value |
|---|---|---|---|---|---|---|
| C | G | C | A | 0.05 | 0.04 | 0.60 |
| C | G | T | A | 0.36 | 0.40 | 0.337 |
| T | G | T | A | 0.05 | 0.01 | 0.0027 |
| T | A | T | T | 0.41 | 0.39 | 0.664 |
| T | G | T | T | 0.12 | 0.15 | 0.332 |
| OMNIBUS, P = 0.033 | ||||||
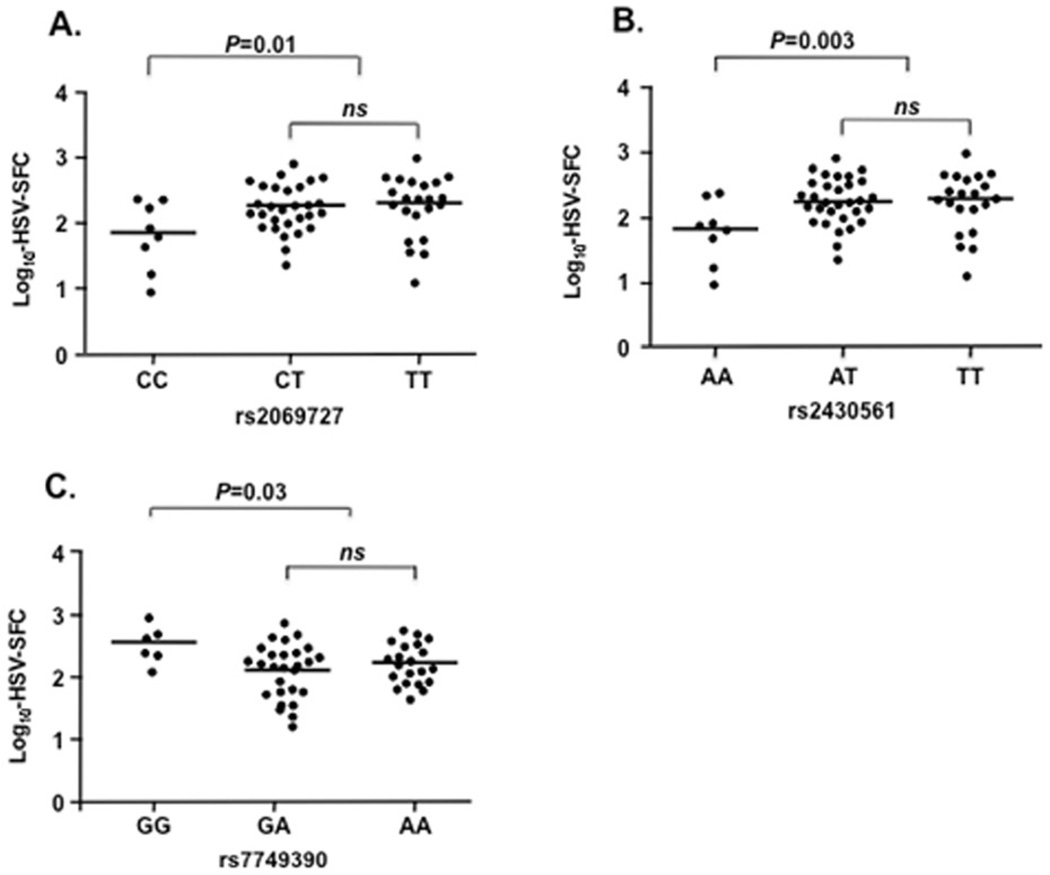
Marker associated with IFNγ production
We also tested whether genetic variants in IFNG and IFNGR1 were associated with different immune responses to HSV exposure as defined by HSV-induced SFCs in PBMCs from the subgroup of ADVN subjects (n=64) tested for CMI in Fig 1. We observed significant associations for two IFNG markers (rs2069727, rs2430561) and reduced IFNγ production (P=0.01 and 0.003, respectively, Figs. 4A and 4B). Marker rs2430561, in perfect LD (D'=1) with rs2069727, is located within a putative nuclear factor κB (NF-κB) binding site and has been reported to be associated with low IFNγ production.21–23
FIG 4.
Association of IFNG and IFNGR1SNPs with IFNγ production as determined by the log10-transformed mean SFC/106 cells. IFNG SNPs rs2069727 CC genotype and rs2430561 AA were significantly associated with reduced IFNγ production (P = 0.01[CC vs CT+TT] and P = 0.003 [AA vs AT+TT], respectively, Panel A and Panel B). IFNGR1SNP rs7749390 GG genotype was significantly associated with increased IFNγ production (P = 0.03 [GG vs GA+AA]), Panel C). ns: not significant.
We found that two functional SNPs in IFNGR1 were associated with an increase in IFNγ production (rs223471 [−56C/T], 24 P = 0.026 [data not shown]; rs7749390,25 P = 0.030, Fig 4C). The SNP rs7749390 was also significantly associated with a reduced risk of the ADEH+ phenotype (35.6%ADEH+ vs 45.1% ADEH−, P = 0.03, Fig 3A). Collectively, these data suggest that these variants are involved in gene regulation and directly affecting the levels of IFNγ expression.
DISCUSSION
Using immunologic, genomic and genetic approaches, we demonstrate that low IFNγ expression contributes to the increased susceptibility of a subset of AD patients to develop severe viral infections. Global gene expression profiling of VV-stimulated PBMCs from ADEH+, ADEH−, and healthy NA subjects revealed a significant association of genes that code for IFNs with the ADEH+ phenotype (Table I). Further analysis of the IFN family revealed significant (q value <0.1) associations of genes that code for type I and II IFNs, with an aberrant response to stimulation with VV (Table II). Interestingly, gene expression of type I IFNs was significantly increased in ADEH+ subjects, suggesting an abortive effort to mount an anti-viral response in these patients. This may be due to the observed downregulation of interferon alpha, beta, and omega receptor 1. In contrast, type II IFNγ was significantly downregulated. Although type I IFNs have a direct anti-viral effect in vitro, which can be reproduced in vivo at pharmacologic doses, IFNγ plays a crucial role in the initiation and propagation of the anti-viral cell-mediated defense. In vivo studies in mice have demonstrated a non-redundant synergistic effect of type I and type II IFNs to mount an effective host anti-viral response.26
Importantly, our gene profiling studies also revealed a significant downregulation of the IFNγ receptor gene in ADEH+ subjects, but not in ADEH− patients. Low IFNγ receptor gene expression in combination with low IFNγ expression would be expected to reinforce the poor anti-viral response in these patients. The biological significance of this observation was supported by our finding of disseminated VV infection after scarification of VV into the skin of IFNγ receptor knockout mice. The clinical phenotype in these IFNγ receptor knockout mice of multiple VV induced satellite lesions (Fig 2), increased viremia and reduced survival is similar to clinical outcomes of EV in humans.4,5
Because there were significantly less IFNγ secretion (Fig 1B) and lower numbers of IFNγ SFCs after HSV stimulation of PBMC in the ADEH+ group compared to the ADEH− group, even after mock stimulation (Fig 1A), we tested whether genetic variants in the gene encoding IFNG and its receptor, IFNGR1, were associated with risk of ADEH+ and contributed to the reduced production of IFNγ. In this study, we observed significant associations for IFNGR1 SNPs (rs1327475, rs10457655, and rs7749390) and risk of ADEH+ phenotype, and this association was strengthened by haplotype analysis. The strongest association was for a protective haplotype AG (rs10457655 and rs7749390; P = 5.7×10−4) that reduced risk of ADEH+ phenotype by over half (OR=0.44) and was relatively common in this European American sample, with a prevalence of 13.2% in ADEH+ patients compared to 25.5% in ADEH− patients. Interestingly, this haplotype includes the functional SNPs rs7749390, a SNP located on the exon/intron splicing site of IFNGR1.25 This SNP has been associated with increased levels of IFNγ production and reduced risk of ADEH+, suggesting that this SNP may protect against ADEH+ by regulating IFNγ secretion after exposure to HSV. It is possible that this SNP breaks a consensus splicing site sequence, resulting in the intron remaining in mature mRNA and subsequently the production of aberrant proteins.27 Although there are several other known IFNGR1 mutations (V61E, Y66C, C77F) that have been associated with partial or complete IFNGR1 deficiency,28 we didn't genotype those because of low minor allele frequencies (MAF<5%) and the limited power in the existing sample to detect associations; however, future studies in an expanded dataset should include these known variants. In addition, we observed associations for ADEH− and two IFNGR1 SNPs (rs1327475 and rs10457655, Fig 3A), but the associations were stronger for ADEH+, suggesting that these SNPs may confer an increased susceptibility of atopic dermatitis (AD) patients to develop disseminated viral skin infections (ADEH+).
Significant associations were also observed for IFNG variants and risk of ADEH. In particular, a four-SNP haplotype showed the strongest association with increased risk of ADEH (P=0.0027). Of interest, this haplotype includes two SNPs (rs2069727 and rs2430561, IFNG+874T/A) that were significantly associated with reduced IFNγ production ex vivo in response to HSV exposure. SNP rs2430561 has previously been associated with low IFNγ production in psoriasis vulgaris21 and tuberculosis22 as well as risk of atopic asthma, AD, and allergic rhinitis.23 Although none of the individual SNPs were associated with ADEH per se, the significant associations observed between disease and haplotypes comprised of these SNPs suggest that as of yet untyped variants within or around the four-SNP haplotype locus are causally related to risk of ADEH. In this study, we selected eight IFNG SNPs which included two previously described functional variants in IFNG (rs2069709, rs2430561) and six tagging SNPs spanning a 13.1 kb region on chromosome 12q14 (Fig E1).
Although the genetics portion of the current study was not designed to interrogate the mechanism(s) by which the IFNG and IFNGR functional variants are associated with ADEH and altered IFNγ production, this constitutes an obvious next step. We also acknowledge that other variants in these two genes may also contribute to risk of ADEH. As indicated above, we purposely did not genotype SNPs of low MAF given our current limitations of power; however, it is recognized that a considerable proportion of heritable disease risk in complex traits is in fact associated with rare variants12 [usually defined as those with a frequency of less than 5%−1% or lower.29 Testing for association between rare variants and rare traits is inherently problematic given the challenges of patient recruitment for a robustly powered discovery sample and the further difficulties of access to similarly robust replication samples. Based on our registry enrollment, we estimate that ADEH affects less than 3% of AD subjects,3 and can possibly be defined as a rare trait. Rare diseases are typically characterized as being highly penetrant and caused by polymorphisms of large effect that are often rare. Rare variants of large effects, combined with multiple other rare mutations, together can explain a large proportion of the genetic basis for complex diseases.30 On the other hand, there is evidence that common alleles with modest to large effect underlie risk for certain rare but complex traits (i.e., cleft palate31). Our own recent studies demonstrated that a relatively uncommon null mutation in the gene encoding filaggrin (FLG; R501X) was three times more prevalent in ADEH+ patients compared to ADEH− patients (24% vs. 8%, respectively), and the relative risk for ADEH+ disease was nearly doubled (OR=11.8 vs. 6.2; P=0.0008). Thus, although we recognize the potential for association between ADEH and as of yet untyped markers in IFNG and IFNGR1, we believe our tagging approach combined with a focus on known functional variants is a comprehensive first step for detecting causal loci of large effects in ADEH.
Although the genetic association studies including IFNG and IFNGR1 SNPs are supportive of our primary observation in this study - that IFNγ responses are diminished in both murine and human conditions associated with disseminated viral infection - it will ultimately be critical to seek replication of these associations in independent populations of ADEH+ patients. The primary sample used for the tests for association with risk of ADEH+ was the group of European American patients and healthy controls from the NIAID-supported ADVN. We cannot speculate at this time whether or not similar associations would be observed in populations of different ancestry. However, our studies on associations between IFNG and IFNGR1 variants and IFNγ production were conducted in both European American and African Americans, and we observed no difference in the levels of IFNγ between these two racial groups (Fig E2). We believe these findings suggest that the relevance of the functional IFNG (+874T-A) and IFNGR1 (−56C/T and +95T/C) SNPs in regulating IFNγ production is not ethnic- or race-specific, and subsequently contributes to a reduced risk of ADEH.
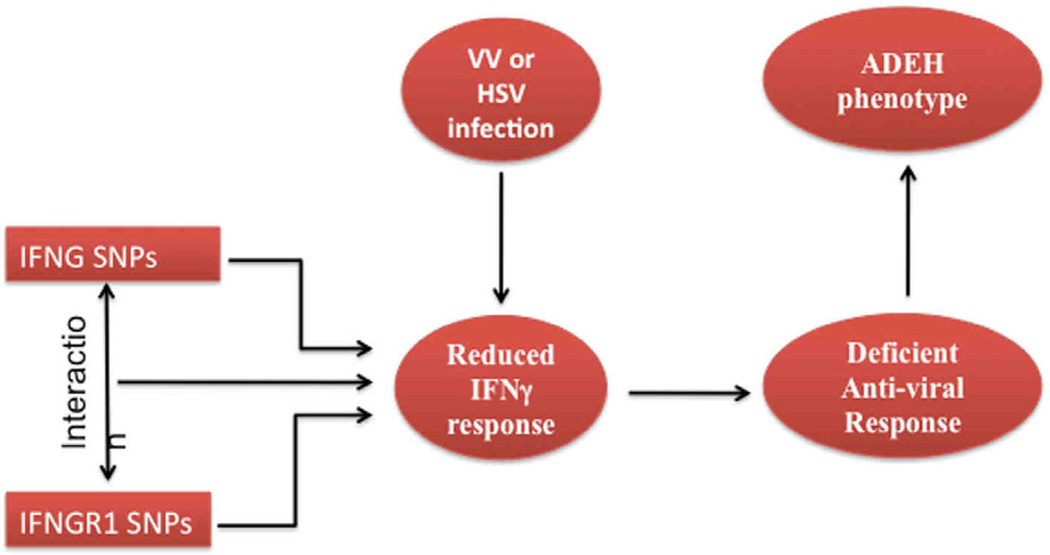
In summary, we used several different approaches to demonstrate a low IFNγ expression in patients with ADEH+ as well as association between IFNG and IFNGR1 SNPs and the ADEH+ phenotype and IFNγ production in a multicenter case-control study (see Figure 5). Thus, our data demonstrates, for the first time, that IFNG and IFNG receptor variants may impair the anti-viral response against viruses such as HSV and vaccinia and increase the risk of ADEH. The propensity of this small subset of AD patients to develop disseminated skin infections may be due to a combination of skin barrier defects facilitating skin viral penetration (reflected in their increased association with filaggrin null mutations) and a defective IFNγ systemic immune response. Of note, skin barrier defects are common in AD, even in ADEH− subjects. These skin barrier defects can relate to genetic null mutations or local Th2 mediated downregulation of the innate immune response.32 Of note, in this study we failed to find an association between enhanced Th2 systemic immune responses and ADEH (Table II). However, the current study found that IFNγ protein production were significantly lower in ADEH+, but not ADEH−, subjects suggesting that a defective systemic IFNγ immune response that fails to control viral replication plays a key role in the pathogenesis of ADEH. A defect in Th1 cytokines in ADEH may also contribute to the severe eczema and increased atopy associated with this form of AD.3, 33 A clear understanding of these risk factors may improve our ability to identify patients at greatest risk for ADEH+, and ultimately lead to early intervention to prevent this devastating complication of AD.
FIG 5.
Overview of Impaired Interferon Response Leading to Atopic Dermatitis Complicated by Eczema Herpeticum
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Maureen Sandoval for her assistance in the preparation of this manuscript and the nursing staff in the CTRC (Trudy Madigan RN and Janice Herrell RN) for their help in patient recruitment and sample collection. We also thank the ADVN coordinators (Trista Berry BS, Susan Tofte RN, Shahana Baig-Lewis, Peter Brown BS, Lisa Heughan BA, CCRC, Meggie Nguyen BS, Doru Alexandrescu MD, Lorianne Stubbs RC, Deborra James RN, CCRC, Reena Vaid MD, Diana Lee MD), ADVN regulatory advisors (Judy Lairsmith BA), laboratory and biotracking technicians (Jessica Scarpola, Muralidhar Bopparaju, Mary Bolognino MS, Lisa Latchney MS), NIAID-DAIT support (Marshall Plaut, MD and Joy Laurienzo Panza RN, BSN, CCRC), DACI Laboratory (Robert Hamilton PhD) and all of the patients who participated in this study. Special thanks to Gloria David PhD, Jamie Reese BS and Susan Lieff PhD at Rho, Inc., for coordination of the study.
This research was supported by the Clinical Translational Scientific Award from the National Center for Research Resources UL1 RR02580, The Atopic Dermatitis Vaccinia Network NIH/NIAID contracts N01 AI40029, N01 AI40030 and N01 AI40033. DYML was supported in part by NIAMS grant AR41256, and the Edelstein Family Foundation. KCB was supported in part by the Mary Beryl Patch Turnbull Scholar Program.
ABBREVIATIONS
- AD
Atopic dermatitis
- ADEH−
AD without a history of eczema herpeticum
- ADEH+
AD with a history of eczema herpeticum
- ADVN
Atopic Dermatitis Vaccinia Network
- CMI
Cell mediated immunity
- EA
European American
- EV
Eczema vaccinatum
- GO
Gene ontology
- HSV
Herpes simplex virus
- IFNγ R−/−
Interferon gamma receptor gene knockout
- NA
Non-atopics
- NF-κB
Nuclear factor kappa B
- NIH/NIAID
National Institutes of Health/National Institute of Allergy and Infectious Diseases
- PBMC
Peripheral blood mononuclear cell
- SAM 2.20
Significance Analysis of Microarrays
- SFCs
Spot-forming cells
- VV
Vaccinia virus (VV)
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
The authors have no conflict of interest to declare
REFERENCES
- 1.Odhiambo JA, Williams HC, Clayton TO, Robertson CF, Asher MI. Global variations in prevalence of eczema symptoms in children from ISAAC Phase Three. J Allergy Clin Immunol. 2009;124:1251–1258. doi: 10.1016/j.jaci.2009.10.009. [DOI] [PubMed] [Google Scholar]
- 2.Bieber T. Atopic dermatitis. N Engl J Med. 2008;358:1483–1494. doi: 10.1056/NEJMra074081. [DOI] [PubMed] [Google Scholar]
- 3.Beck LA, Boguniewicz M, Hata T, Schneider LC, Hanifin J, Gallo R, et al. Phenotype of atopic dermatitis subjects with a history of eczema herpeticum. J Allergy Clin Immunol. 2009;124:260–269. doi: 10.1016/j.jaci.2009.05.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Moses AE, Cohen-Poradosu R. Images in clinical medicine. Eczema vaccinatum--a timely reminder. N Engl J Med. 2002;346:1287. doi: 10.1056/NEJMicm010892. [DOI] [PubMed] [Google Scholar]
- 5.Vora S, Damon I, Fulginiti V, Weber SG, Kahana M, Stein SL, et al. Severe eczema vaccinatum in a household contact of a smallpox vaccinee. Clin Infect Dis. 2008;46:1555–1561. doi: 10.1086/587668. [DOI] [PubMed] [Google Scholar]
- 6.Fauci AS. Smallpox vaccination policy--the need for dialogue. N Engl J Med. 2002;346:1319–1320. doi: 10.1056/NEJM200204253461711. [DOI] [PubMed] [Google Scholar]
- 7.Lane JM, Ruben FL, Neff JM, Millar JD. Complications of smallpox vaccination, 1968: results of ten statewide surveys. J Infect Dis. 1970;122:303–309. doi: 10.1093/infdis/122.4.303. [DOI] [PubMed] [Google Scholar]
- 8.Grigoryev DN, Finigan JH, Hassoun P, Garcia JG. Science review: searching for gene candidates in acute lung injury. Crit Care. 2004;8:440–447. doi: 10.1186/cc2901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Weinberg A, Song LY, Wilkening C, Sevin A, Blais B, Louzao R, et al. Optimization and limitations of use of cryopreserved peripheral blood mononuclear cells for functional and phenotypic T-cell characterization. Clin Vaccine Immunol. 2009;16:1176–1186. doi: 10.1128/CVI.00342-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Howell MD, Jones JF, Kisich KO, Streib JE, Gallo RL, Leung DY. Selective killing of vaccinia virus by LL-37: implications for eczema vaccinatum. J Immunol. 2004;172:1763–1767. doi: 10.4049/jimmunol.172.3.1763. [DOI] [PubMed] [Google Scholar]
- 11.Ong PY, Ohtake T, Brandt C, Strickland I, Boguniewicz M, Ganz T, et al. Endogenous antimicrobial peptides and skin infections in atopic dermatitis. N Engl J Med. 2002;347:1151–1160. doi: 10.1056/NEJMoa021481. [DOI] [PubMed] [Google Scholar]
- 12.Amegadzie BY, Ahn BY, Moss B. Identification, sequence, and expression of the gene encoding a Mr 35,000 subunit of the vaccinia virus DNA-dependent RNA polymerase. J Biol Chem. 1991;266:13712–13718. [PubMed] [Google Scholar]
- 13.Howell MD, Streib JE, Kim BE, Lesley LJ, Dunlap AP, Geng D, et al. Ceragenins: a class of antiviral compounds to treat orthopox infections. J Invest Dermatol. 2009;129:2668–2675. doi: 10.1038/jid.2009.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Tukey JW. Some thoughts on clinical trials, especially problems of multiplicity. Science. 1977;198:679–684. doi: 10.1126/science.333584. [DOI] [PubMed] [Google Scholar]
- 15.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dobbs ME, Strasser JE, Chu CF, Chalk C, Milligan GN. Clearance of herpes simplex virus type 2 by CD8+ T cells requires gamma interferon and either perforin- or Fas-mediated cytolytic mechanisms. J Virol. 2005;79:14546–14554. doi: 10.1128/JVI.79.23.14546-14554.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huang S, Hendriks W, Althage A, Hemmi S, Bluethmann H, Kamijo R, et al. Immune response in mice that lack the interferon-gamma receptor. Science. 1993;259:1742–1745. doi: 10.1126/science.8456301. [DOI] [PubMed] [Google Scholar]
- 18.Esteban M, Benavente J, Paez E. Effect of interferon on integrity of vaccinia virus and ribosomal RNA in infected cells. Virology. 1984;134:40–51. doi: 10.1016/0042-6822(84)90270-8. [DOI] [PubMed] [Google Scholar]
- 19.Melková Z, Esteban M. Interferon-gamma severely inhibits DNA synthesis of vaccinia virus in a macrophage cell line. Virology. 1994;198:731–735. doi: 10.1006/viro.1994.1087. [DOI] [PubMed] [Google Scholar]
- 20.Gao PS, Rafaels NM, Hand T, Murray T, Boguniewicz M, Hata T, et al. Filaggrin mutations that confer risk of atopic dermatitis confer greater risk for eczema herpeticum. J Allergy Clin Immunol. 2009;124:507–513. doi: 10.1016/j.jaci.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baran W, Szepietowski JC, Mazur G, Baran E. IFN-gamma promoter gene polymorphism in psoriasis vulgaris. Biomarkers. 2008;13:52–58. doi: 10.1080/13547500701610273. [DOI] [PubMed] [Google Scholar]
- 22.Sallakci N, Coskun M, Berber Z, Gurkan F, Kocamaz H, Uysal G, et al. Interferon-gamma gene+874T-A polymorphism is associated with tuberculosis and gamma interferon response. Tuberculosis (Edinb) 2007;87:225–230. doi: 10.1016/j.tube.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 23.Hussein YM, Ahmad AS, Ibrahem MM, El Tarhouny SA, Shalaby SM, Elshal AS, et al. Interferon gamma gene polymorphism as a biochemical marker in Egyptian atopic patients. J Investig Allergol Clin Immunol. 2009;19:292–298. [PubMed] [Google Scholar]
- 24.Matsuda A, Ebihara N, Kumagai N, Fukuda K, Ebe K, Hirano K, et al. Genetic polymorphisms in the promoter of the interferon gamma receptor 1 gene are associated with atopic cataracts. Invest Ophthalmol Vis Sci. 2007;48:583–589. doi: 10.1167/iovs.06-0991. [DOI] [PubMed] [Google Scholar]
- 25.He J, Wang J, Lei D, Ding S. Analysis of functional SNP in ifng/ifngr1 in Chinese Han population with tuberculosis. Scand J Immunol. 2010;71:452–458. doi: 10.1111/j.1365-3083.2010.02393.x. [DOI] [PubMed] [Google Scholar]
- 26.Klotzbucher A, Mittnacht S, Kirchner H, Jacobsen H. Different effects of IFN gamma and IFN alpha/beta on "immediate early" gene expression of HSV-1. Virology. 1990;179:487–491. doi: 10.1016/0042-6822(90)90322-i. [DOI] [PubMed] [Google Scholar]
- 27.Sierakowska H, Sambade MJ, Agrawal S, Kole R. Repair of thalassemic human beta-globin mRNA in mammalian cells by antisense oligonucleotides. Proc Natl Acad Sci USA. 1996;93:12840–12844. doi: 10.1073/pnas.93.23.12840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.van de Wetering D, de Paus RA, van Dissel JT, van de Vosse E. Functional analysis of naturally occurring amino acid substitutions in human IFN-gammaR1. Mol Immunol. 2010;47:1023–1030. doi: 10.1016/j.molimm.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 29.Bodmer W, Bonilla C. Common and rare variants in multifactorial susceptibility to common diseases. Nat Genet. 2008;40:695–701. doi: 10.1038/ng.f.136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Jugessur A, Rahimov F, Lie RT, Wilcox AJ, Gjessing HK, Nilsen RM, et al. Genetic variants in IRF6 and the risk of facial clefts: single-marker and haplotype-based analyses in a population-based case-control study of facial clefts in Norway. Genet Epidemiol. 2008;32:413–424. doi: 10.1002/gepi.20314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Grigoryev DN, Howell MD, Watkins TN, Chen YC, Cheadle C, Boguniewicz M, et al. Vaccinia virus-specific molecular signature in atopic dermatitis skin. J Allergy Clin Immunol. 2010;125:153–159. doi: 10.1016/j.jaci.2009.10.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Takahashi N, Akahoshi M, Matsuda A, Ebe K, Inomata N, Obara K, et al. Association of the IL12RB1 promoter polymorphisms with increased risk of atopic dermatitis and other allergic phenotypes. Hum Mol Genet. 2005;14:3149–3159. doi: 10.1093/hmg/ddi347. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.