Summary
Kinetochores are proteinaceous scaffolds implicated in the formation of load-bearing attachments of chromosomes to microtubules during mitosis. Kinetochores contain distinct chromatin- and microtubule-binding interfaces, generally defined as inner and outer kinetochore, respectively [reviewed in 1]. The constitutive centromere-associated network (CCAN) and the Knl1-Mis12-Ndc80 complexes (KMN) network are the main multi-subunit protein assemblies in the inner and outer kinetochore, respectively. The point of contact between the CCAN and the KMN network is unknown. Cenp-C is a conserved CCAN component whose central and C-terminal regions have been implicated in chromatin binding and dimerization [2–10]. Here, we show that a conserved motif in the N-terminal region of Cenp-C binds directly and with high affinity to the Mis12 complex. Expression in HeLa cells of the isolated N-terminal motif of Cenp-C prevents outer kinetochore assembly, causing chromosome mis-segregation. The KMN network is also responsible for kinetochore recruitment of the components of the spindle assembly checkpoint, and we observe checkpoint impairment in cells expressing the Cenp-C N-terminal segment. Our studies unveil a crucial and likely universal link between the inner and outer kinetochore.
Keywords: Kinetochore, centromere, CENP-C, Mis12, Mtw1, Ndc80, Knl1, Dsn1, CENP-A
Results and Discussion
The KMN network is a supra-molecular assembly of the 4-subunit Mis12 complex (abbreviated as Mis12C and containing Dsn1, Nnf1, Nsl1 and Mis12), the 4-subunit Ndc80 complex (Ndc80C, containing Ndc80, Nuf2, Spc24 and Spc25), and the 2-subunit Knl1 complex (Knl1C, containing Knl1 and Zwint). The inner core of the kinetochore is built on specialized chromatin containing the histone H3 variant Cenp-A as well as neighbouring H3-containing nucleosomes (reviewed in reference [1]). A group of proteins, including Cenp-C, Cenp-H, Cenp-I, Cenp-K to Cenp-U, Cenp-W and Cenp-X, comprises the CCAN (also known as NAC/CAD) and are closely associated with centromeric chromatin (reviewed in reference [1]). Within the CCAN, at least two subunits, Cenp-N and Cenp-C, contact Cenp-A directly [3, 5, 11].
Among the first kinetochore subunits to be cloned, Cenp-C (whose predicted domains are shown in Figure 1A) is enriched in affinity purifications of tagged subunits of the KMN network and is required for kinetochore recruitment of a subset of outer kinetochore proteins in different species [12–23]. Thus, Cenp-C might provide an attachment site for the recruitment of outer kinetochore components and their associated partners, including the components of the spindle assembly checkpoint. Here, we set out to formally test this hypothesis and describe an interaction responsible for these properties of Cenp-C.
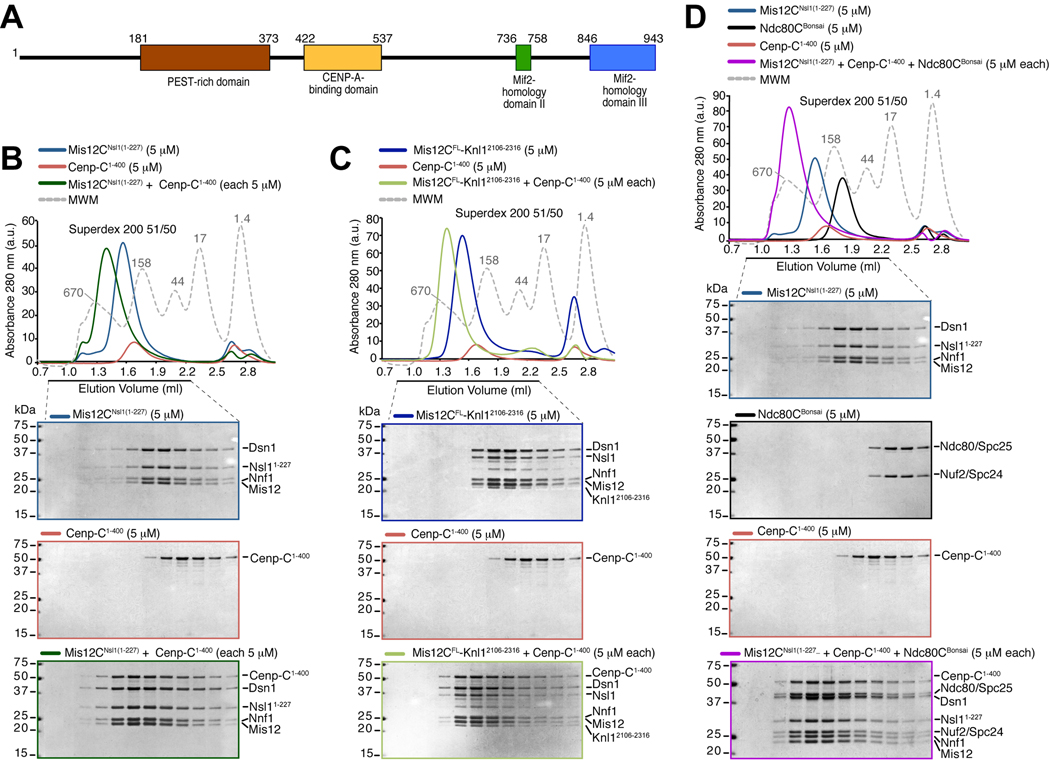
Figure 1. Cenp-C1–400 binds Mis12C.
(A) Schematic depiction of the domain organization of human Cenp-C. (B) Size-exclusion chromatography elution profiles and SDS-PAGE analysis of recombinant Mis12CNsl1(1–227) (upper panel), recombinant Cenp-C1–400 (middle panel), and their stoichiometric combination (lower panel). Complex formation is indicated by a shift in the elution profile of Cenp-C1–400 and Mis12CNsl1(1–227) and their appearance, in stoichiometric amounts, in early elution volumes. (C) As in B, but with Mis12CFL-Knl12106–2316 instead of Mis12CNsl1(1–227). The middle panel is the same as in B. (D) Incorporation of the Ndc80Bonsai complex in the Cenp-C1–400-Mis12CNsl1(1–227) complex. The upper panel and lower middle panel are the same as in B.
The N-terminal region of Cenp-C binds directly to the Mis12 complex
A Cenp-C deletion construct retaining only the C-terminal half of Cenp-C localizes normally to kinetochores but fails to recruit outer kinetochore components [18]. Thus, we initially tested whether a recombinant segment of Cenp-C encompassing residues 1–400 (Cenp-C1–400) bound directly to recombinant KMN constituents reconstituted as recently described [24, 25]. When analyzed by size-exclusion chromatography (SEC, which separates based on size and shape), stoichiometric combinations of Cenp-C1–400 and Ndc80CBonsai (an engineered Ndc80C retaining microtubule-binding and kinetochore-binding domains [24]) did not interact (Figure S1A). Conversely, Cenp-C1–400 interacted tightly with an engineered version of Mis12C in what appeared to be a 1:1 complex (Figure 1B). The Mis12C utilized in these experiments, Mis12CNsl1(1–227), bears a 50-residue deletion in the C-terminal tail of the Nsl1 subunit that prevents the interaction of Nsl1 with Knl1 [25]. Additional deletion mutants of Mis12C, including Mis12CNsl1(1-258), Mis12CMicro and Mis12CMini (the latter two bear additional deletions in the Nsl1 and Dsn1 subunit, see Methods), also interacted with Cenp-C1–400 (Figure S1B and data not shown). Cenp-C1–400 also interacted with the Mis12C-Knl12106–2316 complex, a complex of full-length Mis12C and the C-terminal region of Knl1 [25] (Figure 1C). Although Cenp-C was unable to bind Ndc80CBonsai or full length Ndc80C, a ternary complex was formed when Mis12C was also present (Figure 1D and Figure S1C–D). Overall, these results indicate that Cenp-C binds directly to Mis12C, and that the interaction is compatible with additional interactions of Mis12C with Ndc80C and Knl1. The Cenp-C-Mis12C interaction appears stoichiometric, in agreement with a recent analysis of kinetochore subunit copy number [26].
Structural analysis of KMN-Cenp-C subcomplexes
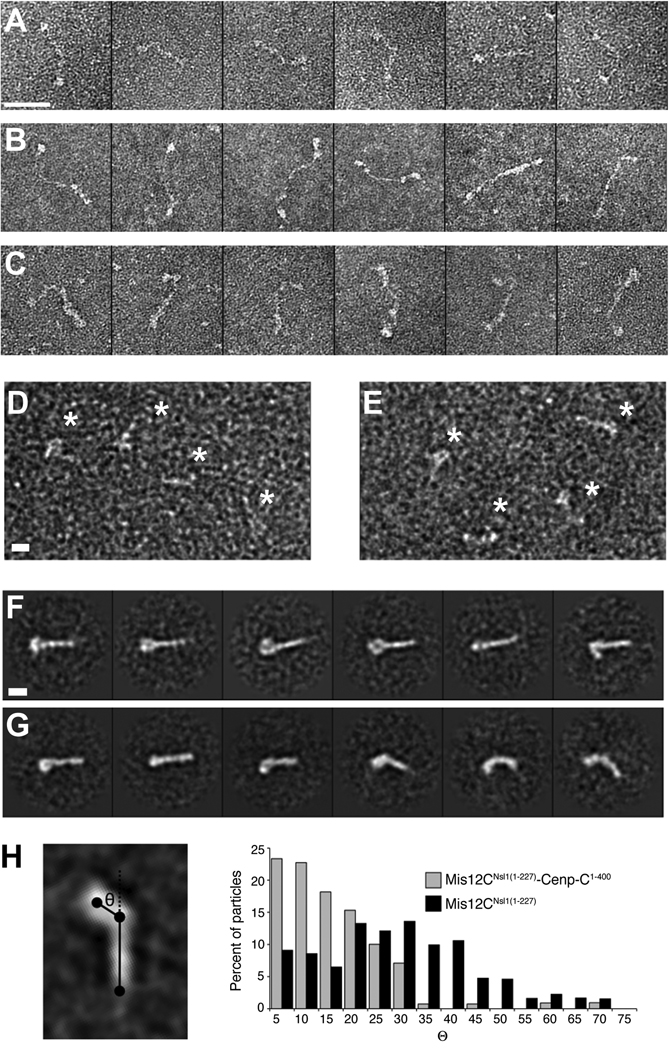
When imaged by negative-stain electron microscopy, the human Ndc80C appears as a 60-nm flexible rod (Figure 2A). When in complex, Ndc80C and Mis12C add their lengths in series, and appear as a ~80-nm whip, with a thicker “handle” corresponding to Mis12C (Figure 2B). This feature is emphasized in the complex of Ndc80C and Mis12C with Cenp-C1–400 (Figure 2C). Images of the Mis12C and Mis12C-Cenp-C1–400 complexes in the absence of Ndc80C (Figure 2D–E) were used for 2D single particle analysis (Figure 2F–G, Fig. S2). Class averages of the Mis12C-Cenp-C1–400 complex appear as a straight rod, generally with a globular domain visible at one end. In the absence of Cenp-C1–400, the averages are far more heterogeneous, showing both straight and bent conformations, as well as variability in the presence or absence of a visible globular head domain. When the two datasets were mixed, our classification procedure robustly sorted them into classes that recapture these essential features (Figure S2).
Figure 2. Electron microscopy analysis of the Cenp-C-Mis12C complex.
(A) Representative single particles of human Ndc80C (full length). Scale bar, 50 nm. (B) Representative single particles of Ndc80C-Mis12CNsl1(1–258). (C) Representative single particles of Ndc80C-Mis12CNsl1(1–258)-Cenp-C1–400 (D) Representative field of single particles of the Mis12CNsl1(1-258)-Cenp-C1–400 complex. Particles are indicated by asterisks. Scale bar, 10 nm. (E) Representative field of particles of Mis12CNsl1(1–258). Additional analyses on this complex were recently published [25, 39, 40]. (F) Selected reference-free class averages of the Mis12CNsl1(1–258)-Cenp-C1–400 complex. The complex appears as a linear rod, generally with a prominent bump at one end. Scale bar, 10 nm. (G) Selected reference-free class averages of Mis12CNsl1(1–258) alone. The complex adopts varying degrees of bending, and the globular lobe is often absent or diminished in size. (H) Bending angle analysis of the Mis12 complex in the presence and absence of Cenp-C1–400. The complex is substantially rigidified upon Cenp-C binding.
To investigate the complexes’ flexibility, we constructed a simplified 3-point model of each class average (Figure 2H), with points placed at the centre of the globular head, the hinge point, and the tip of the tail. We then measured the bending angle, θ, for each modelled class average. When weighted by the particle distribution across the classes, histograms of the bending angle distribution present in each sample population demonstrate far greater rigidity of the complex in the presence of Cenp-C1–400 (Figure 2H). Nevertheless, the Mis12 complex can access straight conformations in the absence of Cenp-C1–400, albeit infrequently relative to bent ones.
We applied chemical cross-linking methods to identify potential sites of contact between Cenp-C and the Mis12 complex, but the data, due to technical limitations, were not conclusive (not shown). The electron microscopy analysis also does not provide a firm localization of Cenp-C, for there is no strong additional density present in the Mis12C-Cenp-C1–400 complex averages that never appears in the averages of the Mis12C alone (Figure S2), despite the differences in the conformational landscapes of the two populations. This suggests that the majority of Cenp-C1–400 is flexible relative to Mis12C, in agreement with the small portion of Cenp-C1–400 which is sufficient for Mis12C binding (see below).
Mis12C binds a 20-residue motif in the N-terminal region of Cenp-C
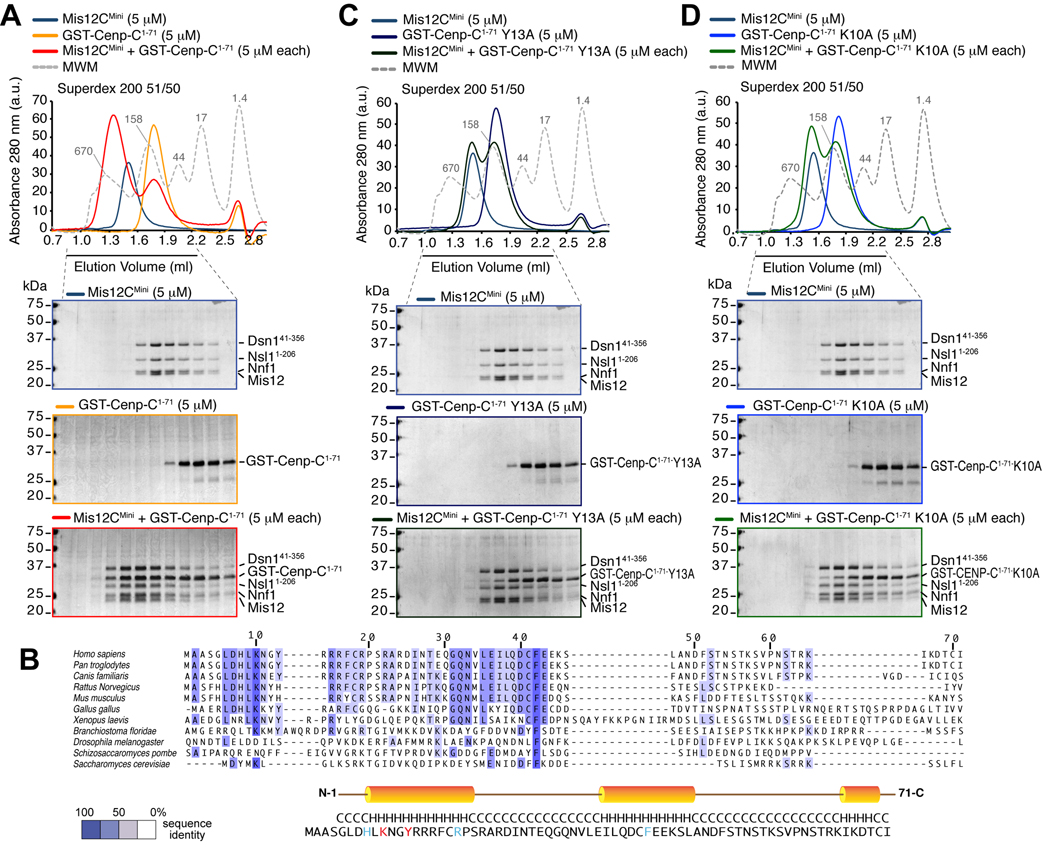
Next, we sought to dissect the Cenp-C region mediating the interaction with Mis12C. We created progressively shorter GST-fusions of the N-terminal region of Cenp-C and tested their ability to bind Mis12C (data not shown). GST-Cenp-C1–71 co-eluted with Mis12C from a size-exclusion column (Figure 3A), and the same was true for Cenp-C1–71 after removal of the GST moiety (Figure S3A).
Figure 3. Wild type but not mutant Cenp-C1–71 binds Mis12C.
(A) Size-exclusion chromatography elution profiles and SDS-PAGE analysis of recombinant Mis12CMini (upper panel), recombinant GST-Cenp-C1–71 (middle panel), and their stoichiometric combination (lower panel). Complex formation is indicated by a shift in the elution profile of GST-Cenp-C1–71 and Mis12CMini and their appearance, in stoichiometric amounts, in early elution volumes. (B) Multiple sequence alignment of the N-terminal region of Cenp-CV from the indicated species. The colouring scheme reports the degree of conservation. The position of three predicted helices is shown on the sequence of human Cenp-C. (C) As in A, but with a GST-Cenp-C1–71Y13A mutant. The upper panel is the same as in A. No shift in elution volume is observed, indicating that binding is impaired. (D) As in A, but with a GST-Cenp-C1–71K10A mutant. The upper panel is the same as in A. Also with this mutant no shift in elution volume is observed.
Cenp-C1–71 contains two short conserved motifs and is predicted to fold as three consecutive helices (Figure 3B). We tested the effects of several single alanine point mutations in conserved residues. Substitutions K10A and Y13A in predicted helix αA reduced the interaction between Cenp-C1–71 and the Mis12 complex (Figure 3C–D). On the other hand, introduction of alanine mutations in additional residues, including R19A and F42A in predicted helix αB did not affect Mis12C binding (Figure S3B–C). Essentially identical results were obtained when the chromatography experiments were performed with the isolated Cenp-C1–71 segment and related mutants after cleavage of the GST moiety (data not shown). Overall, these results indicate that the Mis12C-binding site of Cenp-C maps to a conserved (predicted) helix in the N-terminal region of Cenp-C. In agreement with this idea, further C-terminal deletions of Cenp-C, including Cenp-C1–46 and Cenp-C1–21, also co-eluted with Mis12C in SEC runs (Figure S3D–E).
Effects from impairing the Cenp-C:Mis12 interaction in cells
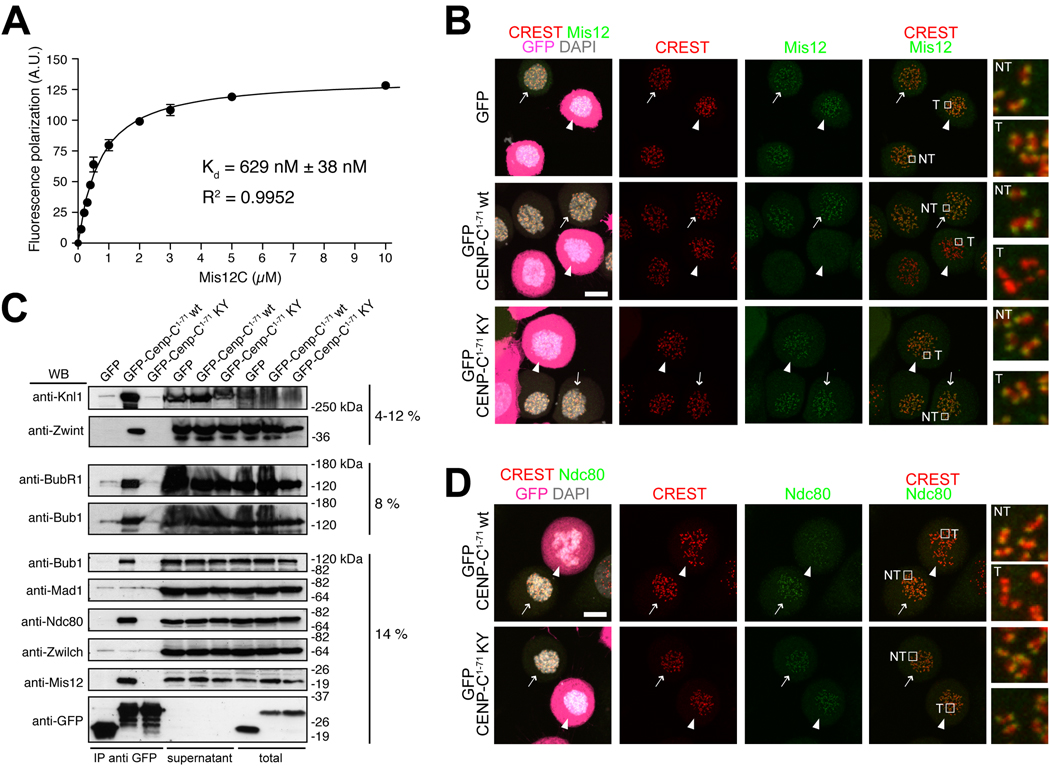
To gain an indicative estimate of the binding affinity of the interaction, we carried out a fluorescence polarization assay on the interaction of Mis12C with a synthetic fluorescein-conjugated peptide corresponding to Cenp-C1-21. The dissociation constant (Kd) of the interaction was 629 ± 38 nM (Figure 4A). Thus, Cenp-C1–71 binds directly to Mis12C in vitro with high affinity. Based on these findings, we reasoned that over-expression of the N-terminal segment of Cenp-C in cells might prevent Mis12C recruitment to kinetochores through competition with endogenous Cenp-C. GFP-Cenp-C (full length) stained kinetochores when expressed in HeLa cells after transient transfection, indicating that GFP fusions at the N-terminus of Cenp-C do not interfere with Cenp-C kinetochore recruitment (Figure S4). GFP-Cenp-C1–71, on the other hand, was not recruited to kinetochores and showed diffuse cytosolic staining in mitotic HeLa cells (Figure 4B). This is in agreement with previous studies showing that the N-terminal region of Cenp-C is dispensable for kinetochore targeting [3, 10, 18].
Figure 4. Cenp-C1–71 disrupts outer kinetochore assembly.
(A) Fluorescence anisotropy measurements of the binding affinity of Mis12Mini for a synthetic fluorescein-conjugated peptide encompassing residues 1-21 of Cenp-C. (B) Direct fluorescence (GFP, DAPI) or indirect immuno-fluorescence from the indicated species in HeLa cells. In this and subsequent panels, arrows indicate non-transfected GFP-negative cells whereas arrowheads indicate transfected GFP-positive cells. Localization of Mis12 in GFP transfected cells is normal. Mis12 is displaced from kinetochores in cells expressing the wild type sequence of GFP-Cenp-C1–71. The construct localizes in the cytosol but it is not enriched at kinetochores. Mis12 is normally localized in cells expressing the double point mutant GFP-Cenp-C1–71K10A-Y13A mutant (abbreviated as KY). Scale bar = 10 µM. (C) Lysates from HeLa cells expressing the indicated GFP species were used for immuno-precipitations using an anti-GFP antibody. Several outer kinetochore proteins interact with wild type GFP-Cenp-C1–71 but not with its mutated form. Content of IPs from 1.5 mg of cell lysates, 45 µg of supernatants and 45 µg of total cell lysates were loaded on gel. (D) As in B, but detecting Ndc80.
In agreement with our in vitro analysis (Figures 1–3), immuno-precipitates revealed Mis12, Ndc80, Knl1, and the Knl1-associated subunit Zwint, as binding partners of GFP-Cenp-C1–71 (Figure 4C and Figure S5F) Because GFP-Cenp-C1–71 does not localize to kinetochores, its over-expression is expected to prevent kinetochore localization of its binding partners during mitosis. Indeed, kinetochore localization of Mis12, Knl1 and Zwint was abrogated upon expression of GFP-Cenp-C1–71 (Figure 4B and Figure S5). Kinetochore localization of Ndc80 was very significantly reduced, although small residual amounts of this protein remained visible (Figure 4D). On the other hand, mislocalization of outer kinetochore subunits was never observed upon expression of the GFP-Cenp-C1–71-K10A/Y13A double mutant (Figures 4B, 4D and S5). These results demonstrate that the over-expression of GFP-Cenp-C1–71 prevents the recruitment of the KMN network to the outer kinetochore.
Impaired chromosome segregation in cells expressing Cenp-C1–71
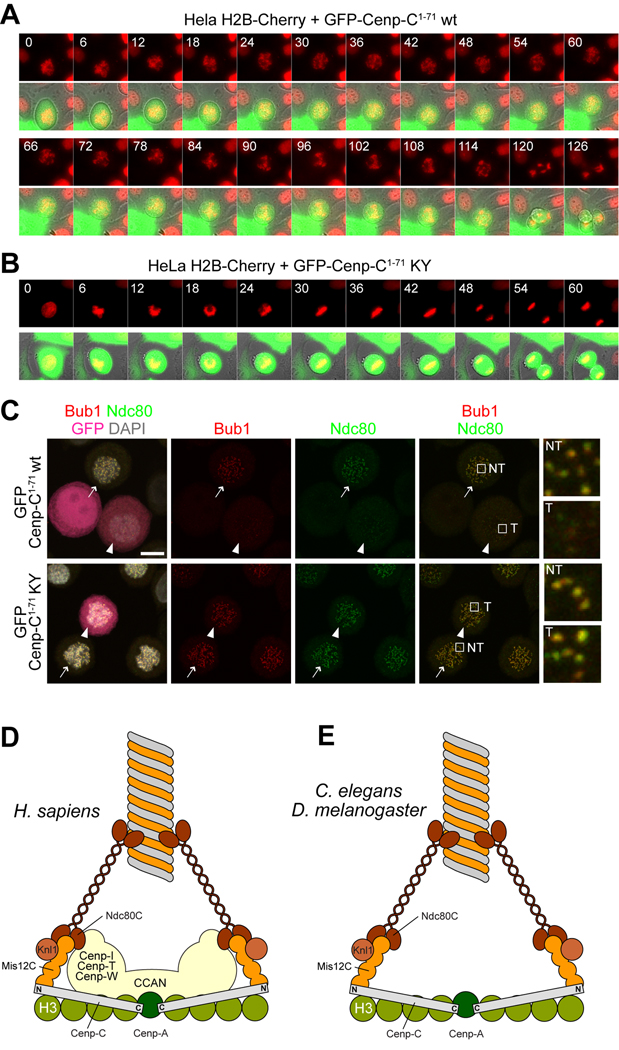
Next, we monitored the progression of cells expressing Cenp-C1–71 or Cenp-C1–71-K10A/Y13A through mitosis. We performed time-lapse video-microscopy on HeLa cells stably expressing histone H2B-Cherry to mark chromosomes and transfected with GFP-Cenp-C1–71 or GFP-Cenp-C1–71-K10A/Y13A (Figure 5A–B). In 6 out of 6 cells transfected with the GFP-Cenp-C1–71 construct, we observed a dramatic chromosome segregation phenotype. Chromosomes in these cells were unable to form a clear metaphase plate. All cells exited mitosis prematurely in the absence of a metaphase plate. None of these effects were observed in cells expressing the GFP-Cenp-C1–71-K10A/Y13A construct, which instead divided normally (16 cells observed).
Figure 5. Cenp-C1–71 disrupts the spindle checkpoint.
(A) HeLa cells co-expressing histone H2B-Cherry and wild type GFP-Cenp-C1–71 were filmed as they transited through mitosis. Cells attempted congression but failed to align and exited aberrantly, indicative of spindle checkpoint failure. Scale bar = 10 µM. (B) HeLa cells co-expressing histone H2B-Cherry and mutant GFP-Cenp-C1–71-K10A-Y13A divided normally. (C) Direct fluorescence (GFP, DAPI) or indirect immuno-fluorescence from the indicated species in HeLa cells. In this and subsequent panels, arrows indicate non-transfected GFP-negative cells whereas arrowheads indicate transfected GFP-positive cells. Localization of Bub1 and Ndc80 is severely impaired in cells expressing GFP-Cenp-C1–71, whereas it is normal in cells expressing the double point mutant GFP-Cenp-C1–71K10A-Y13A mutant (abbreviated as KY). Scale bar = 10 µM. (D) A model for the interaction of inner and outer kinetochore in mammalian cells. A “two-hand” model predicts that Ndc80 is recruited in part via Mis12C, but also through a pathway that depends on the CCAN. Only the globular C-terminal domain of Knl1 is shown. (E) The CCAN is not present in D. melanogaster or C. elegans, suggesting that the interaction of the Mis12C with Cenp-C is the only contact between the inner and outer kinetochore.>
Cells expressing GFP-Cenp-C1–71 appear to be unable to mount a strong checkpoint response, as they exit mitosis prematurely with many unattached or incorrectly attached chromosomes (Figure 5A,B). The Mis12C has been shown to be important for (direct or indirect) kinetochore recruitment of several checkpoint components [19, 27]. Correspondingly, we found that besides KMN subunits, the GFP-Cenp-C1–71 precipitates also contain Bub1 and BubR1 (Figure 4B). Indeed, both Bub1 and BubR1 were displaced from kinetochores in HeLa cells expressing GFP-Cenp-C1–71 (Figure 5C and S5B). Although Mad1 and Zwilch, two additional checkpoint components, were not identified in the GFP-Cenp-C1–71 immuno-precipitates, they too were variably mislocalized upon expression of GFP-Cenp-C1–71 (Figure S5C–E), in agreement with a previously established requirement for the Mis12C on their kinetochore recruitment [19, 27]. Because Ndc80 is required for kinetochore localization of Zwilch and Mad1, we suspect that its residual kinetochore localization in cells expressing GFP-Cenp-C1–71 is responsible for residual Mad1 and Zwilch recruitment (Figure S5C–E). These residual amounts of checkpoint proteins, in turn, might explain the residual checkpoint response observed in Figure 5A,B.
Conclusions
We report that the N-terminal region of Cenp-C contains a crucial link between the inner and outer kinetochore (Figure 5D). The identification of Mis12C as a direct binding partner of Cenp-C is consistent with recent super-resolution analyses of the kinetochore [28–30]. These have indicated that the N-terminal region of Cenp-C is positioned on a plane, perpendicular to the inter-kinetochore axis, which also hosts Mis12C subunits.
The evolutionary conservation of the Mis12-binding motif suggests that its role may be universal. In certain organisms, such as C. elegans and D. melanogaster, no homologs of the CCAN subunits, with the exception of Cenp-C, have been so far identified. In these organisms, the interaction between Cenp-C and Mis12C might provide the only linkage between the inner and outer kinetochore (Figure 5E), as strongly suggested by an accompanying paper reporting an analysis of the interaction of Cenp-C and Mis12 complex subunits in D. melanogaster [31].
In most other organisms, including humans, additional contacts between the CCAN and the KMN network likely exist. Kinetochore recruitment of the Ndc80C in the absence of Knl1 is unaltered, and it is reduced, but not eliminated, upon depletion of Mis12C subunits [19, 27, 32–34]. We confirm these previous observations, because we consistently observed residual kinetochore levels of Ndc80 in cells expressing a dominant-negative construct of Cenp-C. These observations justify a two-hand model of outer kinetochore assembly [25, 33] in which Ndc80C is recruited via the Mis12C as well as via another point of contact with the CCAN, probably involving proteins such as Cenp-I, Cenp-T and Cenp-W [19, 35].
How does Cenp-C act as a linker between the inner and outer kinetochore? There are DNA-binding and dimerization domains in the central and C-terminal region of Cenp-C [2–10]. One such domain binds directly to a Cenp-A-containing specialized centromeric nucleosome [5] (Figure 1A). A crucial question for future studies is whether the interaction of Cenp-C with Mis12 occurs on the same specialized Cenp-A-containing centromeric nucleosome or rather on neighbouring H3-containing nucleosomes. The reported association of Cenp-C and additional centromeric proteins with H3-containing nucleosomes supports the latter hypothesis [36]. In this scheme, Cenp-C might act as a ruler in the inner kinetochore plate (Figure 5D–E). We speculate that Cenp-C contributes to docking the outer kinetochore KMN network at the required distance from a “centre” marked by the Cenp-A nucleosome(s).
The significance of Cenp-C’s rigidifying the Mis12 complex is unclear and is also an important subject for future study. Structural transitions in the Mis12 complex have been observed in super-resolution studies in response to kinetochore-microtubule attachment state [30]. Our results suggest that interaction with inner kinetochore subunits also modulates the conformation of this complex. It is tempting to speculate that Cenp-C binding biases Mis12 towards a conformation suitable for interaction with other cellular factors specifically when it is localized to kinetochores.
Remarkably, the Aurora-B containing chromosome-passenger complex, which regulates crucial aspects of spindle checkpoint signalling and outer kinetochore function, also associates with H3-containing nucleosome through a modification of the H3 N-terminal tail (reviewed in reference [37]). Aurora B localization is perturbed when Cenp-C is depleted, or when the interaction of the Mis12 complex with HP1 (heterochromatin protein 1, a histone-tail binding protein) is affected [22, 38]. All these clues point to the existence of a specialized structure in the inner kinetochore holding H3-containing and Cenp-A-containing nucleosomes in close proximity and possibly interacting through Cenp-C and other subunits.
Methods
See Experimental Procedures in Supplemental Material
Supplementary Material
Acknowledgments
We thank Sebastiano Pasqualato, Silvia Monzani, and Lucia Massimiliano for providing reagents and the members of the Musacchio laboratory for many helpful discussions. Work in the Musacchio laboratory is generously funded by the Association for International Cancer Resarch (AICR), the FP7 European Research Council contract KINCON and the Integrated Project MitoSys (grant agreement n° 241548), the Italian Association for Cancer Research (AIRC), the Cariplo Foundation, and the Human Frontier Science Program. Work in the Nogales laboratory is funded by NIGMS. G.M.A. is partially funded by an NIH training grant. E.S. is supported by a fellowship of the Italian Foundation for Cancer Research (FIRC). E.N. is a Howard Hughes Medical Institute Investigator.
Footnotes
This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Santaguida S, Musacchio A. The life and miracles of kinetochores. EMBO J. 2009;28:2511–2531. doi: 10.1038/emboj.2009.173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Trazzi S, Bernardoni R, Diolaiti D, Politi V, Earnshaw WC, Perini G, Della Valle G. In vivo functional dissection of human inner kinetochore protein CENP-C. J Struct Biol. 2002;140:39–48. doi: 10.1016/s1047-8477(02)00506-3. [DOI] [PubMed] [Google Scholar]
- 3.Trazzi S, Perini G, Bernardoni R, Zoli M, Reese JC, Musacchio A, Della Valle G. The C-terminal domain of CENP-C displays multiple and critical functions for mammalian centromere formation. PLoS One. 2009;4:e5832. doi: 10.1371/journal.pone.0005832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen RL, Espelin CW, De Wulf P, Sorger PK, Harrison SC, Simons KT. Structural and functional dissection of Mif2p, a conserved DNA-binding kinetochore protein. Mol Biol Cell. 2008;19:4480–4491. doi: 10.1091/mbc.E08-03-0297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carroll CW, Milks KJ, Straight AF. Dual recognition of CENP-A nucleosomes is required for centromere assembly. J Cell Biol. 2010;189:1143–1155. doi: 10.1083/jcb.201001013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Song K, Gronemeyer B, Lu W, Eugster E, Tomkiel JE. Mutational analysis of the central centromere targeting domain of human centromere protein C, (CENP-C) Exp Cell Res. 2002;275:81–91. doi: 10.1006/excr.2002.5495. [DOI] [PubMed] [Google Scholar]
- 7.Meluh PB, Koshland D. Budding yeast centromere composition and assembly as revealed by in vivo cross-linking. Genes Dev. 1997;11:3401–3412. doi: 10.1101/gad.11.24.3401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sugimoto K, Yata H, Muro Y, Himeno M. Human centromere protein C (CENP-C) is a DNA-binding protein which possesses a novel DNA-binding motif. J Biochem. 1994;116:877–881. doi: 10.1093/oxfordjournals.jbchem.a124610. [DOI] [PubMed] [Google Scholar]
- 9.Yang CH, Tomkiel J, Saitoh H, Johnson DH, Earnshaw WC. Identification of overlapping DNA-binding and centromere-targeting domains in the human kinetochore protein CENP-C. Mol Cell Biol. 1996;16:3576–3586. doi: 10.1128/mcb.16.7.3576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lanini L, McKeon F. Domains required for CENP-C assembly at the kinetochore. Mol Biol Cell. 1995;6:1049–1059. doi: 10.1091/mbc.6.8.1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carroll CW, Silva MC, Godek KM, Jansen LE, Straight AF. Centromere assembly requires the direct recognition of CENP-A nucleosomes by CENP-N. Nat Cell Biol. 2009;11:896–902. doi: 10.1038/ncb1899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Saitoh H, Tomkiel J, Cooke CA, Ratrie H, 3rd, Maurer M, Rothfield NF, Earnshaw WC. CENP-C, an autoantigen in scleroderma, is a component of the human inner kinetochore plate. Cell. 1992;70:115–125. doi: 10.1016/0092-8674(92)90538-n. [DOI] [PubMed] [Google Scholar]
- 13.Cheeseman IM, Niessen S, Anderson S, Hyndman F, Yates JR, Oegema K, Desai A. A conserved protein network controls assembly of the outer kinetochore and its ability to sustain tension. Genes Dev. 2004;18:2255–2268. doi: 10.1101/gad.1234104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Westermann S, Cheeseman IM, Anderson S, Yates JR, 3rd, Drubin DG, Barnes G. Architecture of the budding yeast kinetochore reveals a conserved molecular core. J Cell Biol. 2003;163:215–222. doi: 10.1083/jcb.200305100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.De Wulf P, McAinsh AD, Sorger PK. Hierarchical assembly of the budding yeast kinetochore from multiple subcomplexes. Genes Dev. 2003;21:21. doi: 10.1101/gad.1144403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pinsky BA, Tatsutani SY, Collins KA, Biggins S. An Mtw1 complex promotes kinetochore biorientation that is monitored by the Ipl1/Aurora protein kinase. Dev Cell. 2003;5:735–745. doi: 10.1016/s1534-5807(03)00322-8. [DOI] [PubMed] [Google Scholar]
- 17.Desai A, Rybina S, Muller-Reichert T, Shevchenko A, Shevchenko A, Hyman A, Oegema K. KNL-1 directs assembly of the microtubule-binding interface of the kinetochore in C. elegans. Genes Dev. 2003;17:2421–2435. doi: 10.1101/gad.1126303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Milks KJ, Moree B, Straight AF. Dissection of CENP-C-directed centromere and kinetochore assembly. Mol Biol Cell. 2009;20:4246–4255. doi: 10.1091/mbc.E09-05-0378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Liu ST, Rattner JB, Jablonski SA, Yen TJ. Mapping the assembly pathways that specify formation of the trilaminar kinetochore plates in human cells. J Cell Biol. 2006;175:41–53. doi: 10.1083/jcb.200606020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kwon MS, Hori T, Okada M, Fukagawa T. CENP-C is involved in chromosome segregation, mitotic checkpoint function, and kinetochore assembly. Mol Biol Cell. 2007;18:2155–2168. doi: 10.1091/mbc.E07-01-0045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Przewloka MR, Zhang W, Costa P, Archambault V, D'Avino PP, Lilley KS, Laue ED, McAinsh AD, Glover DM. Molecular analysis of core kinetochore composition and assembly in Drosophila melanogaster. PLoS ONE. 2007;2:e478. doi: 10.1371/journal.pone.0000478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Orr B, Sunkel CE. Drosophila CENP-C is essential for centromere identity. Chromosoma. 2010 doi: 10.1007/s00412-010-0293-6. [DOI] [PubMed] [Google Scholar]
- 23.Tanaka K, Chang HL, Kagami A, Watanabe Y. CENP-C functions as a scaffold for effectors with essential kinetochore functions in mitosis and meiosis. Dev Cell. 2009;17:334–343. doi: 10.1016/j.devcel.2009.08.004. [DOI] [PubMed] [Google Scholar]
- 24.Ciferri C, Pasqualato S, Screpanti E, Varetti G, Santaguida S, Dos Reis G, Maiolica A, Polka J, Deluca JG, De Wulf P, et al. Implications for kinetochore-microtubule attachment from the structure of an engineered Ndc80 complex. Cell. 2008;133:427–439. doi: 10.1016/j.cell.2008.03.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Petrovic A, Pasqualato S, Dube P, Krenn V, Santaguida S, Cittaro D, Monzani S, Massimiliano L, Keller J, Tarricone A, et al. The MIS12 complex is a protein interaction hub for outer kinetochore assembly. J Cell Biol. 2010;190:835–852. doi: 10.1083/jcb.201002070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Johnston K, Joglekar A, Hori T, Suzuki A, Fukagawa T, Salmon ED. Vertebrate kinetochore protein architecture: protein copy number. J Cell Biol. 2010;189:937–943. doi: 10.1083/jcb.200912022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.McAinsh AD, Meraldi P, Draviam VM, Toso A, Sorger PK. The human kinetochore proteins Nnf1R and Mcm21R are required for accurate chromosome segregation. Embo J. 2006;25:4033–4049. doi: 10.1038/sj.emboj.7601293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Joglekar AP, Bloom K, Salmon ED. In Vivo Protein Architecture of the Eukaryotic Kinetochore with Nanometer Scale Accuracy. Current Biology. 2009:1–6. doi: 10.1016/j.cub.2009.02.056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Schittenhelm RB, Heeger S, Althoff F, Walter A, Heidmann S, Mechtler K, Lehner CF. Spatial organization of a ubiquitous eukaryotic kinetochore protein network in Drosophila chromosomes. Chromosoma. 2007;116:385–402. doi: 10.1007/s00412-007-0103-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wan X, O'Quinn RP, Pierce HL, Joglekar AP, Gall WE, DeLuca JG, Carroll CW, Liu ST, Yen TJ, McEwen BF, et al. Protein architecture of the human kinetochore microtubule attachment site. Cell. 2009;137:672–684. doi: 10.1016/j.cell.2009.03.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Przewloka MR, Venkei Z, Bolanos-Garcia V, Debski J, Dadlez M, Glover DM. Current Biology This issue. 2011 doi: 10.1016/j.cub.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Kline S. The human Mis12 complex is required for kinetochore assembly and proper chromosome segregation. The Journal of Cell Biology. 2006;173:9–17. doi: 10.1083/jcb.200509158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Cheeseman IM, Hori T, Fukagawa T, Desai A. KNL1 and the CENP-H/I/K Complex Coordinately Direct Kinetochore Assembly in Vertebrates. Mol Biol Cell. 2008;19:587–594. doi: 10.1091/mbc.E07-10-1051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kiyomitsu T, Obuse C, Yanagida M. Human Blinkin/AF15q14 is required for chromosome alignment and the mitotic checkpoint through direct interaction with Bub1 and BubR1. Dev Cell. 2007;13:663–676. doi: 10.1016/j.devcel.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 35.Hori T, Okada M, Maenaka K, Fukagawa T. CENP-O class proteins form a stable complex and are required for proper kinetochore function. Mol Biol Cell. 2008;19:843–854. doi: 10.1091/mbc.E07-06-0556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hori T, Amano M, Suzuki A, Backer CB, Welburn JP, Dong Y, McEwen BF, Shang WH, Suzuki E, Okawa K, et al. CCAN makes multiple contacts with centromeric DNA to provide distinct pathways to the outer kinetochore. Cell. 2008;135:1039–1052. doi: 10.1016/j.cell.2008.10.019. [DOI] [PubMed] [Google Scholar]
- 37.Vader G, Lens SM. Chromosome segregation: taking the passenger seat. Curr Biol. 2010;20:R879–R881. doi: 10.1016/j.cub.2010.09.037. [DOI] [PubMed] [Google Scholar]
- 38.Kiyomitsu T, Iwasaki O, Obuse C, Yanagida M. Inner centromere formation requires hMis14, a trident kinetochore protein that specifically recruits HP1 to human chromosomes. J Cell Biol. 2010;188:791–807. doi: 10.1083/jcb.200908096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Maskell DP, Hu XW, Singleton MR. Molecular architecture and assembly of the yeast kinetochore MIND complex. J Cell Biol. 2010;190:823–834. doi: 10.1083/jcb.201002059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hornung P, Maier M, Alushin GM, Lander GC, Nogales E, Westermann S. Molecular Architecture and Connectivity of the Budding Yeast Mtw1 Kinetochore Complex. J Mol Biol. 2010 Nov 12; doi: 10.1016/j.jmb.2010.11.012. [Epub ahead of print] [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.