Summary
The ability to follow enzyme activity in a cellular context represents a challenging technological frontier that impacts fields ranging from disease pathogenesis to epigenetics. Activity-based probes (ABPs) label the active form of an enzyme via covalent modification of catalytic residues. Here we present an analysis of parameters influencing potency of peptide phosphonate ABPs for trypsin-fold S1A proteases, an abundant and important class of enzymes with similar substrate specificities. We find that peptide length and stability influence potency more than sequence composition and present structural evidence that steric interactions at the prime-side of the substrate-binding cleft affect potency in a protease-dependent manner. We introduce guidelines for the design of peptide phosphonate ABPs and demonstrate their utility in a live-cell labeling application that specifically targets active S1A proteases at the cell surface of cancer cells.
Introduction
The creation and implementation of high-throughput nucleic acid analysis techniques have revolutionized medicine and biology. The developments of affinity capture and protein binding arrays have allowed similar analyses of protein levels and interactions. However, such gene and protein profiling data does not always reflect the dynamic milieu that one finds at the cellular level in vivo. Furthermore, enzymatic activity adds an additional layer of complexity in functionality that cannot currently be addressed using available techniques.
The need to examine enzyme activity is particularly relevant to the study of proteolytic enzymes in biological systems. While synthetic substrates can be used to study individual proteases in vitro, their efficacy in studying multiple proteases in a complex mixture is limited. For example, the caspase family of cysteine proteases have highly overlapping substrate specificities, thus multiple caspases can often cleave the same synthetic substrate (McStay et al., 2008). This complicates functional analyses of individual caspases during apoptosis. However, covalent labeling of active proteases allows one to monitor individual proteases in the context of the whole cell. Technologies based on covalent inhibitors are emerging for activity-based proteomics. Phosphonate inhibitors of serine proteases form the basis of one such technology (Sienczyk and Oleksyszyn, 2009).
Diisopropylfluorophosphonate (DFP) is one of the earliest irreversible inhibitors described for serine proteases, and radiolabeled DFP was one of the first activity-based probes (ABPs) described (Powers et al., 2002). More recent studies have modified DFP to include a detection tag for purification and/or visualization purposes (Liu et al., 1999). Biotin and fluorophores are the most common tags used, and active enzyme profiles of several types of human tissues and tumor types have been created using such molecules (Jessani et al., 2005). While these efforts are valuable to the field, these studies identify a preponderance of serine hydrolases that are not proteases, and ABPs specific for proteases have been difficult to achieve.
The specificity of DFP can be improved in two ways. Replacing the highly electronegative fluorine atoms with two phenoxy groups reduces reactivity and increases stability (Powers et al., 2002). Further modifying these functional groups modulates the electrophilicity and shape of the phosphonate reactive center, which can add selectivity to the compound. For example, incorporating different electron withdrawing or donating groups onto the phenyl rings of diphenyl phosphonates (DPPs) results in differential specificity between the urokinase-type plasminogen activator (uPA) and trypsin (Sienczyk and Oleksyszyn, 2006). A second way to reduce DFP promiscuity is to include a short peptide, which helps the inhibitor bind to the active site of the targeted protease. By utilizing substrate cleavage data and combinatorial peptide libraries, the peptide sequence can direct the inhibitor towards a target protease or group of proteases (Lim and Craik, 2009). This methodology was used to design specific ABPs for Granzymes A and B (Mahrus and Craik, 2005).
Despite the aforementioned tunable properties, the emergence of peptide phosphonates as the premier ABPs for specific serine proteases has yet to occur. One can imagine a platform where peptide phosphonates are incorporated into existing microarray technology to create high-throughput assays to monitor active proteases at the bench or in the clinic. However, such a technology has yet to be developed, and the utility of peptide phosphonates has come into question.
Here we describe the synthesis, evaluation, and application of peptide phosphonate inhibitors designed to target S1A family proteases. This subfamily of Clan PA proteases contains the trypsin-like enzymes, and it is the largest family of proteases in higher eukaryotes (Rawlings et al., 2010). These proteases play many important roles in biology and disease progression, and thus are of interest to many fields. This work examines the effects of peptide sequence and length on inhibition of two similar S1A proteases, thrombin and MT-SP1/matriptase, via enzymatic, structural, and imaging methods. We find that peptide length and leaving group sterics are large determinants of potency, while sequence composition contributes to a lesser degree. Our findings suggest general guidelines for the design of phosphonate ABPs that are optimized for S1A proteases. By applying these guidelines, we demonstrate that peptide phosphonates can quantitatively label and follow proteases on the surface of cancer cells, a novel use of ABPs that can be applied to many systems.
Results
2.1 Improvement to the synthesis of peptide phosphonates
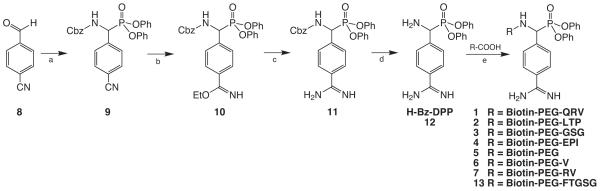
Several modifications to published protocols have improved the synthesis of the diphenyl phosphonate ester of 4-amidinophenyl-glycine (H-(AmPhg)P(OPh)2, for brevity referred to herein as H-Bz-DPP, 12) and the final biotinylated peptide phosphonate probes (Oleksyszyn et al., 1994). The synthesis of 12 (Figure 1) has been reported previously, though isolation of pure compound has been difficult. Briefly, compounds 9-11 were synthesized as described in (Mahrus and Craik, 2005; Oleksyszyn et al., 1994). Published methods describe isolation of 11 by diethyl ether precipitation, however, under the basic reaction conditions to produce 11, the phosphonate center is not stable. Hydrolysis of one of the phenyl ester groups can occur and be replaced with either an ethyl or methyl ester adduct. Reducing reaction c from 5 to 2 days prevented accumulation of the side product as monitored by LC-MS. Longer incubations increased levels of the undesired product. Direct hydrogenation of 11 followed by HPLC produced pure 12. Alternatively, 11 and 12 were separable by silica gel flash chromatography (10:1 CH2Cl2:MeOH and 5:1 CH2Cl2:MeOH with ninhydrin staining, respectively) at Rf ≈ 0.3.
Figure 1.
General synthesis of 4-amidinophenyl-glycine that incorporates benzamidine at the P1 position. Reagents and conditions: (a) P(OPh)3, benzyl carbamate, HOAc, 1 h at rt, 1 h at 85 C; (b) dry EtOH, CHCl3, HCl in dioxane, 5 d at 4 C; (c) NH3 in dioxane, dry EtOH:dioxane (1:1), 2 d at rt; (d) H2, cat. Pd(10% on C), HCl, EtOH, 6 h at rt, (e) EDAC, HOBt, TFE:DCM, 6 h at rt. See also Table S1.
Improvements to the synthesis of the final biotinylated peptide diphenyl phosphonate probes include (1) separate construction of the biotinylated peptide moiety and (2) optimization of the coupling reaction of the peptide to 12 to increase yield and reduce reaction time. Instead of building the probe by extending 12 one amino acid at a time as described in (Boduszek et al., 1994) and (Jackson et al., 1998), biotinylated peptides were synthesized on 2-chlorotrityl chloride resin by standard Fmoc chemistry, cleaved under mild acidic conditions to retain the protecting groups, and coupled to the phosphonate. Previous reports indicate this phosphonate coupling reaction to be time consuming (12-48 hours) and low yielding (5-30%)(Oleksyszyn et al., 1994). Optimization of this reaction proceeded with a TFE:CH2Cl2 (2:5) solvent mixture to reduce the reaction time from overnight to 6 hr, where DMF had been used previously. Treatment with the coupling agent EDAC resulted in the greatest yield of product (Table 1) as compared to PyBOP or DCC (~20%). EDAC protects the base-sensitive phosphonate from hydrolysis since it is typically utilized in the pH range of 4-6. Following the coupling reaction, deprotection of the amino acid side chains followed. HPLC purification was applied to both the protected and deprotected biotinylated peptide phosphonates. As a result, a combination of TFE:CH2Cl2 and EDAC resulted in a fast, efficient coupling that protected the integrity of the phosphonate center and improved product yield (Table 1).
Table 1.
Yields and masses of each phosphonate probe synthesized
| compound (biotin-PEG-R) |
percent yield | m/z calculated (found) |
|---|---|---|
| R=QRVBz-DPP (1) | 72 | 1308.61 (656.34) |
| R=LTPBz-DPP (2) | 56 | 1236.57 (1237.12) |
| R=GSGBz-DPP (3) | 65 | 1126.46 (1126.92) |
| R=EPIBz-DPP (4) | 57 | 1264.56 (1266.31) |
| R=Bz-DPP (5) | 25 | 925.38 (926.7) |
| R=VBz-DPP (6) | 20 | 1024.45 (1025.82) |
| R=RVBz-DPP (7) | 77 | 1180.55 (1181.64) |
| R=FTGSGBz-DPP (13) | 40 | 1374.57 (1376.27) |
2.2 Increasing peptide length improves phosphonate inhibition of serine proteases
Having optimized synthesis of the phosphonate inhibitors, the inhibitors were next characterized in vitro. Previous work using positional scanning, synthetic combinatorial library (PS-SCL) profiling had identified RKSR as the preferred P4-P1 tetrapeptide sequence for MT-SP1 and LTPR as the preferred sequence for thrombin (Bhatt et al., 2007; Harris et al., 2000). Combining this data with other validated substrates and structural information led to the creation of a series of peptide phosphonates that were designed to target MT-SP1 and thrombin (Table 1). Two sequences, QRVBz (1) and LTPBz (2) were rationally designed to maximize specificity for the two S1A proteases. A peptide element designed to be moderately effective against both proteases (GSGBz 3) was also synthesized, as was a fourth sequence (EPIBz 4) containing sub-optimal amino acids for both proteases at each P2-P4 position. Thus, a series of peptides were incorporated to create ABPs that were predicted to cover a range of activities against both MT-SP1 and thrombin.
IC50 values were calculated for each inhibitor against both MT-SP1 and thrombin, and kinact/KI values were calculated for inhibitors of a representative subset against MT-SP1 (Table 2). For MT-SP1, both the IC50 and kinact/KI values for the tetrapeptide inhibitors trended as expected, with the optimal sequence QRVBz-DPP (1) inhibiting best at 0.37uM, and the non-optimal sequence EPIBz-DPP (4) inhibiting worst at 76uM (Table 2a). These values were in agreement with kcat values measured for synthetic fluorogenic substrates with sequences corresponding to each inhibitor (Supplementary Table 1). This correlation between kcat and inhibitory potency has been previously observed (Drag et al., 2010).
Table 2.
Inhibition kinetics of phosphonate probes against MT-SP1 and thrombin. MT-SP1 and thrombin were incubated with varying concentrations of inhibitor and activity was monitored upon addition of substrate. See also Table S2 and Figure S1
| Enzyme | Compound | Inhibitor | IC50 (μM) | |
|---|---|---|---|---|
| A | MTSP1 | 5 | Bz | 3.5 ±0.6 |
| 6 | VBz | 8 ±0.1 | ||
| 7 | RVBz | 0.56 ±0.09 | ||
| 1 | QRVBz | 0.37 ±0.2 | ||
| 3 | GSGBz | 14 ±1 | ||
| 2 | LTPBz | 1.1 ±0.02 | ||
| 4 | EPIBz | 76 ±1 | ||
| 13 | FTGSGBz | 3.5 ±0.5 | ||
|
| ||||
| B | MTSP1 F99A | 5 | Bz | 1.5 ±0.04 |
| 6 | VBz | 1.3 ±0.06 | ||
| 7 | RVBz | 0.23 ±0.01 | ||
| 1 | QRVBz | 0.22 ±0.01 | ||
| 3 | GSGBz | 9.2 ±0.4 | ||
|
| ||||
| C | thrombin | 5 | Bz | 0.097 ±0.01 |
| 6 | VBz | 0.28 ±0.05 | ||
| 7 | RVBz | 0.068 ±0.04 | ||
| 1 | QRVBz | 0.13 ±.002 | ||
| 3 | GSGBz | 0.027 ±0.01 | ||
| 2 | LTPBz | 1.8 ±0.5 | ||
| 4 | EPIBz | 0.76 ±0.2 | ||
| Compound | Inhibitor | kinact/KI (M−1s−1) | |
|---|---|---|---|
| D | 5 | Bz | 50 ±2 |
| 6 | VBz | 60 ±10 | |
| 7 | RVBz | 490 ±40 | |
| 1 | QRVBz | 200 ±60 | |
| 3 | GSGBz | 26 ±0.5 | |
| 4 | EPIBz | 11.7 ±3 |
Each of the peptide phosphonates tested proved to be slow inhibitors of MT-SP1. The fastest inhibitor, QRVBz-DPP (1), was only 200M−1s−1. Previous studies have reported values approaching 37,000M−1s−1 for phosphonate inhibition of serine proteases (Powers et al., 2002). The data shown here indicates that diphenyl phosphonate ABPs did not inhibit MT-SP1 rapidly, and either long incubation times or high phosphonate concentrations were needed to reach complete inhibition. While optimizing the amino acid composition of the peptide improved inhibition, we were unable to gain a large degree of selectivity. The best inhibitor for MT-SP1, 1, was better than optimal inhibitors designed against thrombin or uPA, another S1A protease (data not shown), further evidence suggesting that sequence composition plays a minor role in inhibitory potency for certain proteases.
The minor contribution due to sequence is reiterated by the ability of the peptide phosphonates to label proteases in vitro. The inhibitors were incubated with MT-SP1 and thrombin at increasing concentrations for 16 hours, and biotin-labeled proteases were detected via a Western blot using avidin-HRP. Figure S1 shows that the ability of the ABPs to label both proteases was also not strongly dependent on sequence composition. GSGBz-DPP (3), designed to confer intermediate potency for MT-SP1 and thrombin, labeled more efficiently than the ABPs with optimal sequences for each protease. This observation is partly explained by the greater stability of 3 over time in aqueous solution (Table S1), indicating that sequence stability was an additional parameter for labeling efficiency. Additionally, QRVBz, the best kinetic inhibitor of MT-SP1, contains a second cleavage site after the P3 Arg. This cleavage removed the biotin from the ABP, rendering it undetectable by avidin-HRP and complicating analysis. This secondary cleavage event is evidenced by the appearance of an 846Da fragment upon post-incubation LC-MS analysis.
In addition to the ABPs described above that were designed to test the effects of amino acid sequence on inhibition, three additional ABPs were synthesized to test the effects of peptide length on potency. Previous reports have noted a dependence of peptide length on the mechanistic rate constant k2, which suggest a possible effect of length on peptidyl DPP potency (Case and Stein, 2003). A mono-, di-, and tripeptide phosphonate was synthesized for the QRVBz-DPP probe (1), as this sequence proved to inhibit both proteases well (Table 2). A clear correlation was observed between increasing peptide length and improvement in inhibition, with a 10 fold increase between the IC50 of the longest inhibitor (QRVBz-DPP, 1) and the shortest inhibitor (Bz-DPP, 5), from 0.37uM to 3.5uM. A similar trend was seen in the kinact/KI data across the same parameters.
It was clear that not all positions affected MT-SP1 inhibition equally. The addition of a P3 residue increased potency by an order of magnitude (from 8uM to 0.56uM), while only modest changes were observed by the addition of P2 or P4. This was consistent with previously observed substrate:enzyme binding interactions with serine proteases, in which much of the binding energy is contributed by the P1 amino acid interacting with the S1 pocket of the protease. This also correlated with known MT-SP1 substrate data and with crystallographic observations about the S4 and S3 pockets (Bhatt et al., 2007; Friedrich et al., 2002). Friedrich et al. observed that the deep S3 pocket of MT-SP1 could accommodate a Lys or Arg residue from either P3 or P4 of a substrate while the other amino acid is solvent exposed. This phenomenon could explain the modest increase in inhibition seen when increasing from a tripeptide- to a tetrapeptide phosphonate (Friedrich et al., 2002).
A similar trend was seen in IC-50 values measured against thrombin (Table 2c) and uPA (data not shown). Again, it is notable that QRVBz-DPP (1), the ideal MT-SP1 inhibitor, was more potent than the ideal thrombin inhibitor LTPBz-DPP (2) by an order of magnitude (0.125uM for 1 compared to 1.1uM for 2). Sequence data alone does not appear to be sufficient to design ABPs that can distinguish between these two proteases, though other proteases with much more stringent substrate binding pockets have shown to be amenable to sequence-derived specificity (Mahrus and Craik, 2005).
To determine if potency continued to improve beyond P4, a hexapeptide ABP was synthesized and tested against MT-SP1. The preferred P5 and P6 amino acids for MT-SP1 were added to the most stable inhibitor (GSGBz 3) to create 13 (FTGSGBz). 13 is 25% more potent than 3 against MT-SP1, verifying that further increasing length improved potency.
Interestingly, each of the ABPs synthesized for this study inhibited thrombin at much lower concentrations than MT-SP1. We next sought to obtain a structural basis for the improvement in inhibition observed against thrombin.
2.3 Structure of MT-SP1 bound to Bz-DPP
The structure of MT-SP1 bound to Bz-DPP phosphonate was solved to 1.19Å resolution (Figure 2, Table 3). The protein was incubated with saturating ABP concentrations overnight at room temperature and purified via size exclusion chromatography. Attempts to crystallize MT-SP1 with Ac-QRVBz-DPP only produced crystals containing Bz-DPP. A possible explanation is that nonspecific hydrolysis of the peptide bond occurred during the several rounds of seeding and crystal growth that were necessary to obtain high-quality crystals, all of which took place in an aqueous environment at room temperature over several months. The truncated inhibitor may have also resulted from incomplete purification of the Bz-DPP (12) starting material from the full-length ABP.
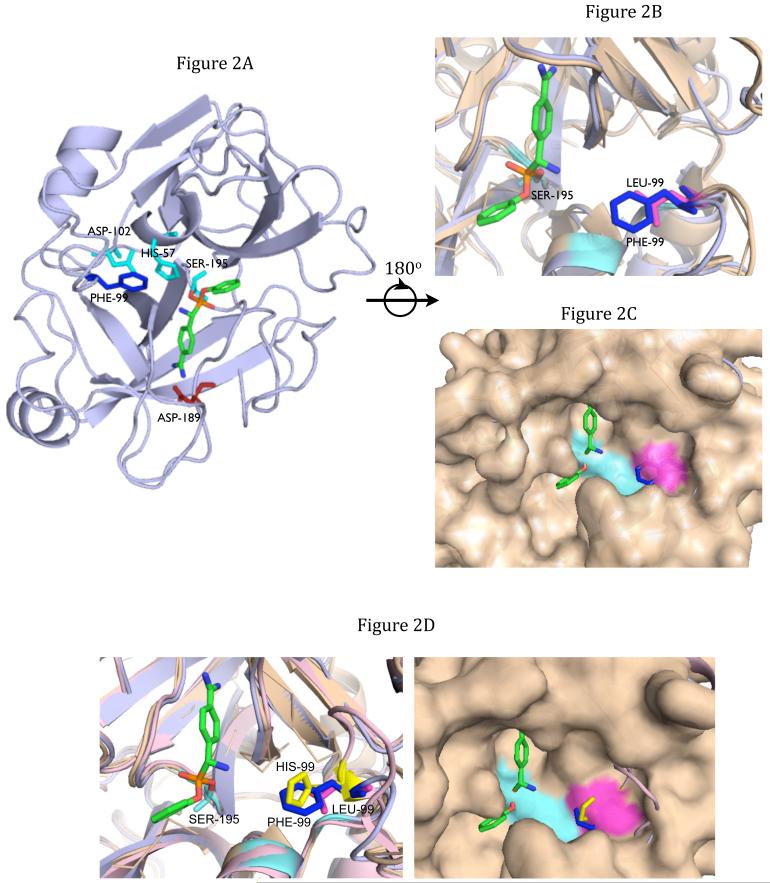
Figure 2.
a: Structure of the catalytic domain of MT-SP1 bound to Benzamidine phosphonate. Numbered amino acid residues correspond to chymotrypsin protease numbering.
b: Close-up ribbon of thrombin (tan) overlaid onto MT-SP1 (light blue). Thrombin L99 purple, MT-SP1 F99 blue, catalytic triad cyan.
c: Thrombin (tan) surface overlaid onto MT-SP1 (light blue). Thrombin L99 purple, MT-SP1 F99 blue, catalytic triad cyan.
d: Thrombin surface (tan) overlaid onto MT-SP1 (light blue) and uPA (pink). uPA His99 (yellow) occupies prime-side pocket similarly as MT-SP1 Phe99 (blue). Thrombin Leu99 purple, catalytic triad cyan. See also Figure S2.
Table 3.
Data collection and refinement statistics
| Structure | MSTP1-covalent adduct |
|---|---|
| PDB ID | 3NCL |
| Data set | ALS290709 |
| Data statistics | |
| wavelength | 1.11587Å |
| space group | C2 |
| cell dimensions | a=79.9Å b=80.2Å c=40.5Å β=95.8° |
| resolution (last shell) | 54.6-1.19Å (1.25-1.19Å) |
| unique reflections | 58,355 (1,597) |
| redundancy | 6.7 (3.8) |
| completeness | 76.4% (14.5%) |
| mean I/s(I) | 21.0 (5.0) |
| Rsym | 6.5% (15.0%) |
| Refinement statistics | |
| resolution range | 54.6-1.19Å |
| reflections used work (test) | 56,653 (1,530) |
| Rfree/Rfac | 20.3% / 19.4% |
| overall figure of merit | 0.915 |
| overall Bwilson | 12.7Å2 |
| protein atoms | 1900, 21.8Å2 |
| ligand atoms | 21, 34.8Å2 |
| solvent atoms | 221, 38.9Å2 |
| r.m.s.d. bonds | 0.016Å |
| r.m.s.d. angles | 1.010° |
| Ramachandran analysis | |
| residues in preferred regions | 96.4% |
| residues in allowed regions | 3.6% |
| outliers | 0% |
This is the highest resolution MT-SP1 structure to date, which allows for visualization of all bonds. The phosphonate bound in the solvent exposed binding pocket in the expected conformation, with the phosphorous atom bound to the active site Ser195 and Bz in the S1 pocket (Figure 2a). The positively charged guanidine group of Bz formed two hydrogen bonds with the negatively charged Asp202. The N-terminal end of the inhibitor pointed down the substrate binding pocket, and the phenyl ring is solvent exposed.
A structure of thrombin (PDB: 1QUR) (Steinmetzer et al., 1999) was overlaid onto the MT-SP1-Bz-DPP structure, and the probe fit similarly into the binding pocket (Figure 2b). Inspection of the placement of the phenyl rings of the phosphonate revealed one important difference in the active site architecture. In MT-SP1 residue Phe99 lay in the region where the leaving group phenyl ring binds during catalysis. In thrombin, a smaller Leu99 occupied this location, forming a larger pocket than seen in MT-SP1. The Phe was only 2Å away from the phosphorous, thus we hypothesized that Phe99 caused steric interference with the leaving phenyl ring, slowing orientation of the inhibitor in the active site and slowing inhibition of MT-SP1 relative to that of thrombin.
To test this hypothesis, MT-SP1 Phe99 was mutated to Ala to create MT-SP1 F99A, and IC50 measurements with the same series of inhibitors were repeated (Table 2b). The IC-50 values for MT-SP1 F99A fell between that of MT-SP1 and thrombin in each case, indicating the size of the prime side pocket in MT-SP1 influenced phosphonate inhibition.
When the structure of uPA (PDB: 3KGP) (Zhang et al., 2010), an S1A protease with slow kinetics of phosphonate inhibition similar to those of MT-SP1 was overlaid with thrombin, a similar steric hindrance was observed (Fig 2d). In the prime-side binding pocket for uPA, His99 protruded exactly where Phe99 did in MT-SP1. Thrombin had a larger prime-side binding pocket than either MT-SP1 or uPA, due to the Leu found at residue 99. The smaller pocket in uPA, as with MT-SP1, may explain the slow inhibition of uPA by DPPs. This reinforces the idea that the steric fit of the leaving group into the prime side pocket has an important effect on potency.
2.4 Labeling of cell surface proteases by ABPs
Having determined the kinetic and labeling properties of the ABPs in vitro, their potential for labeling active proteases on the surface of live immortalized cancer cells was examined. The most stable inhibitor (13) and the most potent inhibitor (1) were used to label the cell surface proteases of two different cell lines grown in culture. These cell lines represented epithelial cancer types from two different species: PC3 cells, derived from human prostate cancer, and PDAC2.1 cells, derived from a transgenic mouse model of pancreatic ductal adenocarcinoma (Nolan-Stevaux et al., 2009). Live cells were incubated with phosphonate overnight in serum free media at 37°C. Streptavidin conjugated with AlexaFluor488 dye was used to fluorescently tag the ABPs. Cells were examined for the presence of labeled proteases either by fluorescent microscopy or by flow cytometry.
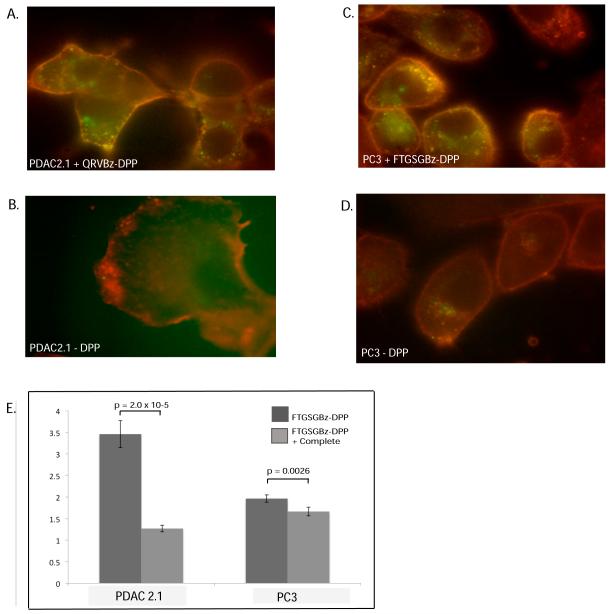
As seen in Figure 3, green fluorescent labeling was observed in PC3 and PDAC cells that had been exposed to ABPs, indicating the presence of active S1A proteases on the cell surface. PDAC2.1 (Figure 3a) cells labeled discrete foci on the cell surface. In the case of PC3 cells (Figure 3b), these labeled proteases were localized to one end of each cell. In addition to cell surface labeling, punctate labeling was also observed just internal to the plasma membrane. These puncta co-localized with membrane staining dye, indicative of internalization of labeled surface proteases. Internalization was also occasionally observed in PDAC2.1 cells and more frequently observed in MCF7 cells, a human breast cancer cell line (Figure S2) This result indicates that ABPs can visualize and follow active serine proteases on the surface of live cells, a novel use for peptide phosphonates.
Figure 3.
Cell surface labeling of active serine proteases.
PDAC2.1 and PC3 cells were imaged in the presence (3A, 3C) or absence (3B, 3D) of ABP. Cell membranes were stained with tetramethylrhodamine-conjugated wheat germ agglutinin (red), and labeled proteases were stained with streptavidin-AlexaFluor 488 (green). Colocalization of labeled proteases with membrane is seen at the cell surface and in discrete puncta, but only in the presence of ABP. Flow cytometry was used to quantify labeled streptavidin on the surface of cells (panel E). Averaged mean fluorescence intensity values are plotted normalized to background staining of labeled streptavidin in the absence of ABP. P-values correspond to a two-sample T-test, and error bars correspond to the addition of the Relative Standard Error values for the two averages multiplied by the normalized mean of each sample.
After demonstrating the ability to label cell surface proteases, the ability to quantify the active form of these proteases was tested. Cells were grown, exposed to phosphonate overnight at 37°C, and fluorescently tagged with labeled streptavidin. Fluorescence was quantified using flow cytometry on the live cells. Fluorescence increased relative to background when cells had been incubated with an ABP, and this labeling was reduced in the presence of protease inhibitors (Figure 3e). These results demonstrate that ABPs can be used to quantify active proteases on the surface of live cells, and that labeling can be directly attributed to proteolysis.
Collectively, the cell-based experiments show that peptide phosphonates can be used to quantitatively label active cell surface proteases on different cell types. Live cell imaging can also be used in conjunction with ABP labeling to determine the localization of active proteases in the context of a cell or group of cells.
Discussion
This study presents additional guidelines for the synthesis, design, and application of peptide phosphonate ABPs to those described previously (Oleksyszyn and Powers, 1994). Our results show that by focusing on peptide length, stability, and prime side sterics, the lifetime and utility of these ABPs can be improved dramatically. By following these guidelines, a novel protease imaging application using ABPs was developed. These experiments demonstrate that ABPs can be used to quantify, image, and follow proteases on cell surfaces.
We have improved the utility of phosphonate ABPs by combining synthetic, kinetic, and structural data. Obtaining reagent quantities of material has been a significant barrier to using these molecules in numerous applications. The improved synthesis methodology presented here both increases reproducibility of ABP production and increases yields up to seven fold compared to previous methods. The robust generation of ABPs greatly improves their utility, and synthesizing greater quantities of these probes enabled a systematic study of their potency.
Three major factors were found to contribute to peptide phosphonate potency: peptide stability, length, and prime side sterics. Peptide sequence is often viewed as a primary determinant of potency, and reports of sequence-based selectivity exist for Granzymes A and M, two S1A proteases with highly selective and differing substrate specificities at P2-P4 (Mahrus and Craik, 2005). However, for the majority of trypsin-fold proteases, much of the substrate binding energy of is contributed via binding of a basic P1 amino acid in the deep S1 pocket of the active site. Because of the strong P1 contribution, the P2-P4 sequence often plays a minor role. Peptide stability, however, was found to contribute significantly to ABP functionality. Inhibitors containing the amino acid Pro were especially unstable and problematic for both synthesis and purification, while long, charged side chains like Arg were found to react with the electrophilic phosphonate, subsequently inactivating the inhibitor. Additionally, the use of basic residues like Arg upstream of P1 can result in a secondary cleavage site for trypsin-like proteases, which removes the probe from the reactive diphenylphosphonate moiety. Poorly fitting residues can have a detrimental effect on inhibition, as seen by the slow kinetics of the EPIBz (4) probe against MT-SP1, thrombin, and uPA (Table 2 and data not shown). Therefore, the optimal peptide element should be composed of the most stable residues tolerated by the protease.
Peptide length, rather than sequence, had a more noticeable contribution to improving inhibitory capabilities. Increasing length increased potency, most notably at the P3 position. This agrees with earlier findings (Oleksyszyn and Powers, 1991). The data suggests that when designing peptide phosphonate inhibitors, including a longer peptide will improve inhibition more reliably than modifying the sequence. The sequence composition data and stability observations indicate that phosphonate inhibitors of trypsin-fold proteases should contain at least a tetrapeptide composed solely of stable amino acids.
The third factor contributing to potency is the steric fit of the phosphonate leaving group in the prime side pocket of the protease. Atomic-resolution structural data showed the prime-side pocket is smaller in MT-SP1 than in thrombin. This pocket is adjacent to the catalytic S195, exactly where the leaving group phenyl ring would reside. This smaller pocket in MT-SP1 is due to the presence of a bulky Phe at residue 99. Mutagenesis experiments confirmed that the prime side pocket is an important binding determinant for DPPs and MT-SP1. The data indicates that the leaving group of the phosphonate can have a large effect on inhibition. When designing a phosphonate ABP specific for a protease, multiple leaving groups should be tested on a stable peptide scaffold to find the best inhibitor.
The data shown here provide important information for the design of peptide phosphonate ABPs with both broad and narrow specificity. We show that the peptide element (in stability and length) and the leaving group (in reactivity and sterics) contribute to inhibitory potency of phosphonate ABPs. When designing a broad-specificity phosphonate ABP, one should start with a scaffold containing at least four stable amino acids and then consider varying the leaving group if improved potency is desired. When designing an inhibitor for a specific S1A protease, substrate cleavage data should be viewed as a starting point. Varying the leaving group may result in larger differences in potency, even for closely related proteases. Interestingly, kinetic data indicates that varying the reaction time may label different sets of proteases, with longer incubations resulting in larger sets of labeled proteases. Therefore, reaction time may also influence selectivity. However, these experiments suggest that only in rare instances will true specificity be engineered by varying peptide composition alone.
By following the guidelines presented here, several broad-spectrum ABPs for S1A proteases were produced in high yields. We used the most potent (1) and stable (13) ABPs to develop a novel method of imaging proteases on the surface of cancer cells. ABP-labeled proteases can be visualized and quantified with fluorescently conjugated streptavidin. This approach found that levels of protease activity vary by cell type, and that the location of labeled proteases can be tracked through the cell (Figure 3). Protease activity and localization information can be leveraged to obtain new information about protease function.
The ability to study the localization of active proteases is a novel use of phosphonate activity-based probes, and offers a promising way to examine the biology of cell surface protease activity. Previous studies with cysteine protease ABPs have been successful at labeling and visualizing lysosomal cathepsins and extracellular cathepsins in tumors (Blum et al., 2005; Joyce et al., 2004). The current study extends the use of ABPs to serine proteases on the surface of live cells. Differences in labeling and localization varied between the cell types, a conclusion previously observed with MT-SP1 using specific antibody-based probes (Darragh et al., 2010). These broad-spectrum ABPs may be used to highlight cell-specific differences in global S1A protease function.
For example, cell surface proteases have been implicated in metastasis, and it is interesting to note that active protease localization was polarized. Notably, high concentrations of enzymes were observed at one edge of the cells distal to cell-cell junctions. In addition, correlating the degree of labeling with a cellular phenotype such as metastatic potential may yield new information about cancer cell biology.
Though the pattern varied, internalization of proteases was observed in all cell types, as evidenced by the co-localization of membrane and ABP in puncta just inside the cell membrane. We hypothesize two explanations for this observation. First, many proteins undergo natural trafficking to and from the cell surface, modulating cellular interactions with the outside environment. Trafficking has been implicated previously in the regulation of proteases in the cell (Ghosh et al., 2003). ABPs could thus be used to track the movement and function of active proteases in many complex biological systems, including cancer biology, where protease activity is frequently dys-regulated. Alternatively, the ABPs themselves may cause proteases to become internalized. Ligand-dependent endocytosis has been observed with other cell surface receptors (Behrendt, 2004; Ghosh et al., 2003). The molecular mechanism of cell surface protease internalization observed via ABPs invites further investigation.
In summary, through synthetic, kinetic, and structural insights, we have developed additional guidelines that define and expand the use of peptide phosphonate ABPs. By following these guidelines, we have created a potent pan-S1A protease probe and developed a general methodology for the study of active proteases at the surface of cells. This technology could allow for future insights into the role of proteases in cancer, and has potential applications to other fields in biology.
Materials and Methods
Materials
MT-SP1 was expressed and purified as described previously and stored at −20°C in 50mM Tris pH 8.0, 50mM NaCl, 10% glycerol (Takeuchi et al., 1999). Mutants for crystallography and kinetic studies were expressed, purified, and stored in the same manner. Thrombin was purchased from Sigma and stored at −20°C in 50mM Tris pH 8.0, 50mM NaCl, 0.1 mg/mL BSA. The MT-SP1 substrates spectrazyme-tPA and spectrafluor-tPA were purchased from American Diagnostica and stored at −20 at 10 mM in H20.
The materials for peptide synthesis including PyBOP, EDAC, HOBt, Fmoc-PEG20atom-OH, and 2-chlorotrityl chloride resin were purchased from NovaBiochem. All other chemicals were purchased from Sigma unless otherwise noted. Solvents including anhydrous ethanol (EtOH), choloroform (CHCl3), 1,4-dioxane, diethyl ether (Et2O), dichloromethane (DCM), trifluroethanol (TFE), trifluoroacetic acid (TFA) and N,N-dimethylformamide (DMF) were used as received. The peptides were synthesized following standard Fmoc-SPPS procedure on 2-chlorotrityl chloride resin.
Reactions were analyzed by LC-MS performed on a Waters Alliance liquid chromatography system with a Waters Micromass ZQ single-quadrupole mass spectrometer. HPLC purifications were carried out using an Agilent 1200 series system with C18 reversed-phase columns (Waters). Mobile phase consisted of 99.9:0.1% water/trifluoroacetic acid (solvent A) and 95:4.9:0.1% acetonitrile/water/trifluoroacetic acid (solvent B). All final compounds were characterized by Matrix-assisted laser desorption ionization time-of-flight (MALDI-TOF) mass spectrometry using an ABI 4700 MALDI-TOF-TOF mass spectrometer.
Compound numbers in bold refer to the structures shown in Figure 1. Diphenyl [N-(benzyloxycarbonyl)amino](4-cyanophenyl)methanephosphonate (9) and NHS-biotin were synthesized according to literature procedure.(ref) No attempts were made to resolve the D,L-(4-AmPhGly)P(OPh)2 diastereomers.
Diphenyl-[N-(benzyloxycarbonyl)amino](4-ethylesterphenyl)methane phosphonate hydrochloride (10)
A batch of 9 (3.0g, 6.0mmol) was dissolved in 100 mL CHCl3 and placed in a 0° C bath. Next Ar was passed over the solution and under the flow of Ar, 3 mL of anhydrous EtOH (60mmol) and 100 mL of 4 M HCl in dioxane were added. The reaction was stirred under argon at 4°C for 5 days, after which 10 formed as a fine white precipitate formed. The precipitate was filtered, washed with Et2O, and dried under reduced pressure (2.32g, 72%). Mass calcd for C30H29N2O6P: 544.18; found 545.45 (M+H)+.
Diphenyl amino(4-amidinophenyl)methanephosphonate, trifluoroacetic acid salt (12)
A batch of 10 (1.0g, 1.8mmol) was dissolved in 25 mL 1,4-dioxane and 25 mL anhydrous EtOH. Next the reaction was purged with Ar and 10mL of 0.5M NH3 in dioxane (5 mmol) was added dropwise. The reaction was stirred under argon at 23° C for two days, and the solvent was removed completely under vacuum to afford the gummy white crude intermediate, diphenyl [N-(benzyloxycarbonyl)amino](4-amidinophenyl) methanephosphonate 11. This intermediate was dissolved in anhydrous EtOH (80mL) and concentrated HCl (305μL, 3.7mmol) and hydrogenated over 10% palladium on activated carbon for 5 h at room temperature. The catalyst was separated by filtration, and the solvent was removed under reduced pressure. The white powder thus isolated was dissolved in HPLC solvent (water with 0.1% TFA) with the aid of DMF and purified by reverse-phase HPLC. Lyophilization of fractions containing product afforded 0.49 g (27%) of 12 as a white powder. Mass calcd for C20H20N3O3P: 381.12, m/z found: 382.01 (M+H)+.
Coupling of the biotinylated peptide to 12. General procedure
To a DCM/TFE(5:1) solution containing the biotin-PEG-peptide (0.03 mmol), EDAC (0.03 mmol) and HOBt (0.03 mmol), was added H-Bz-DPP 12 (0.03 mmol). After 2 h, an additional eq of EDAC and HOBt was added and the mixture stirred for 8 hr. Next the solvent was removed, the isolated oil was dissolved in MeCN/water (1:3), and purified by reverse-phase HPLC. The fractions with product were lyophilized down to yield the desired product as a white powder. The final probes were dissolved in DMSO and stored at −20° C. The concentration of the solution was determined by HABA biotin quantification kit (Pierce).
Biotin-PEG-Gln-Arg-Val-Bz-DPP·TFA (1)
Reaction of Biotin-PEG-Gln(Trt)-Arg(Pbf)-Val-OH (0.10 g, 0.11 mmol), with 12 yielded 39 mg (72% yield) of Biotin-PEG-Gln(Trt)-Arg(Pbf)-Val-DPP as a white powder. Mass calcd for C92H119N14O18PS2: 1802.80, m/z found: 1802.10 (M+H)+. Next the powder was resuspended in 95% TFA, 2.5% TIS, 2.5% water and agitated for 2 hours, to obtain the desired probe 1. The product was precipitated into cold ether, pelleted by centrifugation, and purified by reversed-phase HPLC. Lyophilization of fractions containing product afforded 22 mg (79%) afforded 1 as a white powder. Mass calcd for C60H89N14O15PS: 1308.61, m/z found: 656.34 (M+2H)2+.
Biotin-PEG-Leu-Thr-Pro-Bz-DPP (2)
The protected peptide Biotin-PEG-Leu-Thr(tBu)-Pro-OH was reacted with 12 and resulted in Biotin-PEG-Leu-Thr(tBu)-Pro-Bz-DPP as a white powder (22 mg, 56% yield). Mass calcd for C63H93N10O15PS: 1292.63, m/z found: 1293.29 (M+H)+. Next the powder was resuspended in 95% TFA, 2.5% TIS, 2.5% water and agitated for 2 hours, to obtain the desired probe 2. The product was precipitated into cold ether, pelleted by centrifugation, and purified by reversed-phase HPLC. Lyophilization of fractions containing product afforded 8 mg (36% yield). Mass calcd for C59H85N10O15PS: 1236.57, m/z found: 1237.12 (M+H)+.
Biotin-PEG-Gly-Ser-Gly-Bz-DPP (3)
Reaction of Biotin-PEG-Gly-Ser-Gly-OH with 12 resulted in 3 as a white powder (22 mg, 65%). Mass calcd for C51H71N10O15PS: 1126.46, m/z found: 1126.92(M+H)+.
Biotin-PEG-Glu-Pro-Ile-Bz-DPP (4)
Biotin-PEG-Glu(OtBu)-Pro-Ile-OH was reacted with 12 to afford 23 mg (57% yield) of Biotin-PEG-Glu(OtBu)-Pro-Ile-Bz-DPP. Mass calcd for C64H93N10O16PS: 1320.62, m/z found: 1321.43 (M+H)+. Then the powder was resuspended in 95% TFA, 2.5% TIS, 2.5% water and agitated for 2 hours to obtain the desired probe. The product was precipitated into cold ether, pelleted by centrifugation, and purified by reversed-phase HPLC. Lyophilization of fractions containing product afforded 20 mg (90% yield) of 4. Mass calcd for C60H85N10O16PS: 1264.56, m/z found: 1266.31 (M+H)+.
Biotin-PEG-Bz-DPP (5)
Reaction of Biotin-PEG-OH with 12 yielded 5, which was isolated as white powder. (7mg, 25%). Mass calcd for C44H60N7O11PS: 925.38, m/z found: 926.17 (M+H)+.
Biotin-PEG-Val-Bz-DPP (6)
A batch of Biotin-PEG-Val-OH was reacted with 12 to afford 6 as a powder after HPLC purification (7mg, 20%). Mass calcd for C49H69N8O13PS: 1024.45, m/z found: 1025.82 (M+H)+.
Biotin-PEG-Arg-Val-Bz-DPP (7)
Reaction of Biotin-PEG-Arg(Pbf)-Val-OH with 12 afforded Biotin-PEG-Arg(Pbf)-Val-OH as a white powder (34 mg, 77%). Mass calcd for C68H99N12O17PS2,: 1450.64, m/z found: 1451.24 (M+H)+. Then the powder was dissolved in 95% TFA, 2.5% TIS, 2.5% water and agitated for 2 hours to obtain the desired probe. The product was precipitated into cold ether, pelleted by centrifugation, and purified by reversed-phase HPLC. Lyophilization of fractions containing product afforded 24 mg (90% yield) of 7. Mass calcd for C55H81N12O14PS: 1180.55, m/z found: 1181.64 (M+H)+.
Biotin-PEG-Phe-Thr-Gly-Ser-Gly-Bz-DPP (13)
Reaction of Biotin-PEG-Phe-Thr-Gly-Ser-Gly-OH with 12 afforded 13 as a white powder (45 mg, 40%). Mass calcd for C64H87N12O18PS: 1374.57, m/z found: 1376.27 (M+H)+.
Inhibition assays
All kinetic fluorescence measurements were taken in duplicate using a SpectraMax Gemini fluorescence spectrometer (Molecular Devices) with an excitation wavelength of 380nm, an emission wavelength of 460 nm, and a 435 nm cutoff filter. A solution of inhibitor was serially diluted over an appropriate concentration range and incubated with enzyme. Substrate was added at the end of 4 hours to initiate the reaction, and IC-50s were calculated. MT-SP1 was used at 0.2nM in a buffer containing 50mM Tris pH 8.0, 50mM NaCl, 0.01% Tween-20, with 200uM spectrafluor-tPA as the substrate. Thrombin was used at 0.5nM in a buffer containing 50mM Tris pH 8.0, 50mM NaCl, 0.01% Tween-20, 0.1 mg/mL BSA, with 200uM Boc-b-benzyl-Asp-Pro-Arg-AMC as the substrate. All reactions were run in duplicate.
Steady-state kinetics were used to determine the observed rate constants for the inhibition reaction. The inhibitors were serially diluted in a 96 well plate at an appropriate range of concentrations. Enzyme was added in hour intervals over 8-10 hours. The reaction was initiated by the addition of 200uM substrate, and the Vmax recorded on a SpectraMax fluorescence spectrometer. Kobs was determined at each inhibitor concentration by plotting Vmax vs time, and KIapp was determined by plotting Kobs vs [1]. Kinact/KI was determined using the equation
Crystallization
Crystals were grown at room temperature by vapour diffusion in hanging drops. A combination of micro and macro seeding was used to grow large single crystals in 4.0 M Na Formate at pH=7.0 and 25 mM FeCl3 as additive. Crystals belong to monoclinic space group C2 with one protease-inhibitor complex in the asymmetric unit corresponding to a solvent content of 50% and diffracted to better than 1.2Å resolution.
Structure resolution and refinement
Data were collected at beamline 8.3.1 at the Advanced Light Source in Berkeley on a single crystal cryoprotected in mother liquor supplemented with 20% glycerol. The data were indexed, scaled and reduced using Mosflm and Scala in Elves (Holton and Alber, 2004). The structure was solved by molecular replacement using Phaser (McCoy et al., 2007) with the previously solved protease structure (PDB 3BN9) as search probe (Farady et al., 2008). Automatic building and refinement were performed in Phenix (Adams et al., 2002) using Phenix elBow to generate the covalently bound ligand. Manual building was carried out in Coot (Emsley and Cowtan, 2004). The stererochemistry of the final model was validated using MolProbity (Davis et al., 2007).
Western blot labeling analysis
For recombinant protease labeling, enzyme was combined with varying concentrations of phosphonate inhibitor (3mM-30uM) at room temperature overnight. The reaction was stopped by the addition of SDS loading buffer and boiling for 10 minutes. Western blots were developed using the Vectastain ABC elite kit (Vector Labs).
Fluorescent substrate synthesis
Substrates corresponding to each inhibitor were synthesized by solid phase peptide synthesis. ACC-Rink-amide resin was obtained from Kimia Corp. The first amino acid was coupled using 5 equivalents each of amino acid, HATU, and collidine in dry DMF under argon for 16 hours with agitation. The full-length peptide was synthesized using a Symphony Quartet peptide synthesizer (Protein Technologies, Inc.), acetylated with 8 equivalents each of acetic anhydride and DIPEA, and cleaved with 95% TFA/2.5% water/ 2.5% triisopropyl silane. Cleaved peptides were precipitated into cold ether, collected by centrifugation, and purified by HPLC.
Cell culture and propagation
PC3 cells were obtained from the American Type Culture Collection (ATCC) and propagated in F12K Nutrient mixture with Kaighn’s Modification (1X) and L-Glutamine (GIBCO). Media was supplemented with 10% FBS and 1X Penicillin:Streptomycin. MCF7 cells were obtained from the ATCC and propagated in Dulbecco’s Modified Eagle Medium with high-glucose (D-ME H21) without phenol red (GIBCO). Media was supplemented with 10% FBS, 10ug/mL Insulin, and 1X Penicillin:Streptomycin. PDAC2.1 cells were isolated from p48-Cre/+, LSL-KrasG12D/+, Trp53F/+ transgenic mice according to (Nolan-Stevaux et al., 2009). PDAC2.1 cells were propagated in D-ME H21 (GIBCO) supplemented with 10% FBS and 1X Penicillin:Streptomycin.
Flow cytometry
Cells were grown to confluency in 6-well cell culture-treated dishes using complete media appropriate for each specific cell line. Media was then aspirated and replaced with Opti-MEM serum-free medium (GIBCO). All experiments were done in triplicate. FTGSGBz-DPP (13) was added to the appropriate wells at a final concentration of 50mM. To test proteolysis, ABP was added in the presence of 1X Complete Protease Inhibitor Cocktail (Roche) dissolved in Opti-MEM. The cells were then incubated at 37°C for approximately 20hrs. After incubation, ABP-containing media was aspirated and cells were washed 3x with Opti-MEM. Cells were detached from the surface of the wells with Enzyme-free Cell Dissociation Buffer (GIBCO) for 15min. Detached cells were washed 2x with Opti-MEM, resuspended in 200uL, and incubated for 20min with Streptavidin:AlexaFluor 488 conjugate (Invitrogen) at 4°C. Cells were washed 3x with Opti-MEM and resuspended in 500uL Opti-MEM and assayed for fluorescence using a BD FACSCalibur (BD Biosciences). Data was analyzed and population mean fluorescence values were obtained using FlowJo Flow Cytometry Analysis Software (TreeStar, Inc.).
Microscopy
Cells were grown to confluency in 9.5cm2 glass bottom microwell dishes (MatTek) in complete media. Media was then aspirated and replaced with Opti-MEM. Phosphonate was added to each dish to a final concentration of 50mM. FTGSGBz-DPP (13) was incubated for 20hrs at 37°C and QRVBz-DPP (1) was incubated for 3hrs at 37°C, respectively. Cells were washed 3x with Opti-MEM and then incubated for 20min with Streptavidin:AlexaFluor 488 conjugate and tetramethylrhodamine-conjugated wheat germ agglutinin (Invitrogen), simultaneously, at 4°C. Cells were then washed 3x with Opti-MEM and imaged. Fluorescence microscopy was carried out in the wide field using a Nikon Diaphot with a Nikon 60x lens, numerical aperture 1.4, objective and standard interference filter sets (Omega Optical). Images were collected using a 12-bit cooled charge-coupled device camera (Princeton Instruments) interfaced to a computer running Micro-Manager 1.3 software (http://micro-manager.org). Images were processed using Adobe Photoshop to assemble dual color image files. Brightness/Contrast was adjusted, where necessary, to improve image quality and clarity.
Highlights.
-
-
Synthesis of peptide phosphonate ABPs has been optimized to increase yield.
-
-
Peptide length is a key determinant of phosphonate inhibition.
-
-
3-D structure reveals an important steric contribution to phosphonate potency.
-
-
ABPs were shown to label active S1A proteases on the surface of live cancer cells.
Supplementary Material
Acknowledgement
Mass spectrometry was provided by the Bio-Organic Mass Spectrometry Resource at UCSF (A. L. Burlingame, Director) supported by the Biomedical Research Technology Program of the NIH National Center for Research Resources (NIH NCRR P41RR001614 and NRCC RR014606). This work was supported by a NIH/NIGMS award (K12GM081266) to A.E.-R., a NIGMS-IMSD award (R25-GM56847) to C.T., and by the National Institutes of Health (NIH R01CA128765).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Adams PD, Grosse-Kunstleve RW, Hung LW, Ioerger TR, McCoy AJ, Moriarty NW, Read RJ, Sacchettini JC, Sauter NK, Terwilliger TC. PHENIX: building new software for automated crystallographic structure determination. Acta Crystallogr D Biol Crystallogr. 2002;58:1948–1954. doi: 10.1107/s0907444902016657. [DOI] [PubMed] [Google Scholar]
- Behrendt N. The urokinase receptor (uPAR) and the uPAR-associated protein (uPARAP/Endo180): membrane proteins engaged in matrix turnover during tissue remodeling. Biol Chem. 2004;385:103–136. doi: 10.1515/BC.2004.031. [DOI] [PubMed] [Google Scholar]
- Bhatt AS, Welm A, Farady CJ, Vasquez M, Wilson K, Craik CS. Coordinate expression and functional profiling identify an extracellular proteolytic signaling pathway. Proc Natl Acad Sci U S A. 2007;104:5771–5776. doi: 10.1073/pnas.0606514104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blum G, Mullins SR, Keren K, Fonovic M, Jedeszko C, Rice MJ, Sloane BF, Bogyo M. Dynamic imaging of protease activity with fluorescently quenched activity-based probes. Nat Chem Biol. 2005;1:203–209. doi: 10.1038/nchembio728. [DOI] [PubMed] [Google Scholar]
- Boduszek B, Oleksyszyn J, Kam CM, Selzler J, Smith RE, Powers JC. Dipeptide phosphonates as inhibitors of dipeptidyl peptidase IV. J Med Chem. 1994;37:3969–3976. doi: 10.1021/jm00049a016. [DOI] [PubMed] [Google Scholar]
- Case A, Stein RL. Mechanistic origins of the substrate selectivity of serine proteases. Biochemistry. 2003;42:3335–3348. doi: 10.1021/bi020668l. [DOI] [PubMed] [Google Scholar]
- Darragh MR, Schneider EL, Lou J, Phojanakong PJ, Farady CJ, Marks JD, Hann BC, Craik CS. Tumor detection by imaging proteolytic activity. Cancer Res. 2010;70:1505–1512. doi: 10.1158/0008-5472.CAN-09-1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davis IW, Leaver-Fay A, Chen VB, Block JN, Kapral GJ, Wang X, Murray LW, Arendall WB, 3rd, Snoeyink J, Richardson JS, et al. MolProbity: all-atom contacts and structure validation for proteins and nucleic acids. Nucleic Acids Res. 2007;35:W375–383. doi: 10.1093/nar/gkm216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drag M, Bogyo M, Ellman JA, Salvesen GS. Aminopeptidase fingerprints, an integrated approach for identification of good substrates and optimal inhibitors. J Biol Chem. 2010;285:3310–3318. doi: 10.1074/jbc.M109.060418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- Farady CJ, Egea PF, Schneider EL, Darragh MR, Craik CS. Structure of an Fab-protease complex reveals a highly specific non-canonical mechanism of inhibition. J Mol Biol. 2008;380:351–360. doi: 10.1016/j.jmb.2008.05.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedrich R, Fuentes-Prior P, Ong E, Coombs G, Hunter M, Oehler R, Pierson D, Gonzalez R, Huber R, Bode W, et al. Catalytic domain structures of MT-SP1/matriptase, a matrix-degrading transmembrane serine proteinase. J Biol Chem. 2002;277:2160–2168. doi: 10.1074/jbc.M109830200. [DOI] [PubMed] [Google Scholar]
- Ghosh P, Dahms NM, Kornfeld S. Mannose 6-phosphate receptors: new twists in the tale. Nat Rev Mol Cell Biol. 2003;4:202–212. doi: 10.1038/nrm1050. [DOI] [PubMed] [Google Scholar]
- Harris JL, Backes BJ, Leonetti F, Mahrus S, Ellman JA, Craik CS. Rapid and general profiling of protease specificity by using combinatorial fluorogenic substrate libraries. Proc Natl Acad Sci U S A. 2000;97:7754–7759. doi: 10.1073/pnas.140132697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holton J, Alber T. Automated protein crystal structure determination using ELVES. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:1537–1542. doi: 10.1073/pnas.0306241101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson DS, Fraser SA, Ni LM, Kam CM, Winkler U, Johnson DA, Froelich CJ, Hudig D, Powers JC. Synthesis and evaluation of diphenyl phosphonate esters as inhibitors of the trypsin-like granzymes A and K and mast cell tryptase. J Med Chem. 1998;41:2289–2301. doi: 10.1021/jm970543s. [DOI] [PubMed] [Google Scholar]
- Jessani N, Niessen S, Wei BQ, Nicolau M, Humphrey M, Ji Y, Han W, Noh DY, Yates JR, 3rd, Jeffrey SS, et al. A streamlined platform for high-content functional proteomics of primary human specimens. Nat Methods. 2005;2:691–697. doi: 10.1038/nmeth778. [DOI] [PubMed] [Google Scholar]
- Joyce JA, Baruch A, Chehade K, Meyer-Morse N, Giraudo E, Tsai FY, Greenbaum DC, Hager JH, Bogyo M, Hanahan D. Cathepsin cysteine proteases are effectors of invasive growth and angiogenesis during multistage tumorigenesis. Cancer Cell. 2004;5:443–453. doi: 10.1016/s1535-6108(04)00111-4. [DOI] [PubMed] [Google Scholar]
- Lim MD, Craik CS. Using specificity to strategically target proteases. Bioorg Med Chem. 2009;17:1094–1100. doi: 10.1016/j.bmc.2008.03.068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y, Patricelli MP, Cravatt BF. Activity-based protein profiling: the serine hydrolases. Proc Natl Acad Sci U S A. 1999;96:14694–14699. doi: 10.1073/pnas.96.26.14694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mahrus S, Craik CS. Selective chemical functional probes of granzymes A and B reveal granzyme B is a major effector of natural killer cell-mediated lysis of target cells. Chem Biol. 2005;12:567–577. doi: 10.1016/j.chembiol.2005.03.006. [DOI] [PubMed] [Google Scholar]
- McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McStay GP, Salvesen GS, Green DR. Overlapping cleavage motif selectivity of caspases: implications for analysis of apoptotic pathways. Cell Death Differ. 2008;15:322–331. doi: 10.1038/sj.cdd.4402260. [DOI] [PubMed] [Google Scholar]
- Nolan-Stevaux O, Lau J, Truitt ML, Chu GC, Hebrok M, Fernandez-Zapico ME, Hanahan D. GLI1 is regulated through Smoothened-independent mechanisms in neoplastic pancreatic ducts and mediates PDAC cell survival and transformation. Genes Dev. 2009;23:24–36. doi: 10.1101/gad.1753809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oleksyszyn J, Boduszek B, Kam CM, Powers JC. Novel amidine-containing peptidyl phosphonates as irreversible inhibitors for blood coagulation and related serine proteases. J Med Chem. 1994;37:226–231. doi: 10.1021/jm00028a004. [DOI] [PubMed] [Google Scholar]
- Oleksyszyn J, Powers JC. Irreversible inhibition of serine proteases by peptide derivatives of (alpha-aminoalkyl)phosphonate diphenyl esters. Biochemistry. 1991;30:485–493. doi: 10.1021/bi00216a026. [DOI] [PubMed] [Google Scholar]
- Oleksyszyn J, Powers JC. Amino acid and peptide phosphonate derivatives as specific inhibitors of serine peptidases. Methods Enzymol. 1994;244:423–441. doi: 10.1016/0076-6879(94)44032-8. [DOI] [PubMed] [Google Scholar]
- Powers JC, Asgian JL, Ekici OD, James KE. Irreversible inhibitors of serine, cysteine, and threonine proteases. Chem Rev. 2002;102:4639–4750. doi: 10.1021/cr010182v. [DOI] [PubMed] [Google Scholar]
- Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sienczyk M, Oleksyszyn J. Inhibition of trypsin and urokinase by Cbz-amino(4-guanidinophenyl)methanephosphonate aromatic ester derivatives: the influence of the ester group on their biological activity. Bioorg Med Chem Lett. 2006;16:2886–2890. doi: 10.1016/j.bmcl.2006.03.002. [DOI] [PubMed] [Google Scholar]
- Sienczyk M, Oleksyszyn J. Irreversible inhibition of serine proteases - design and in vivo activity of diaryl alpha-aminophosphonate derivatives. Curr Med Chem. 2009;16:1673–1687. doi: 10.2174/092986709788186246. [DOI] [PubMed] [Google Scholar]
- Steinmetzer T, Renatus M, Kunzel S, Eichinger A, Bode W, Wikstrom P, Hauptmann J, Sturzebecher J. Design and evaluation of novel bivalent thrombin inhibitors based on amidinophenylalanines. Eur J Biochem. 1999;265:598–605. doi: 10.1046/j.1432-1327.1999.00742.x. [DOI] [PubMed] [Google Scholar]
- Takeuchi T, Shuman MA, Craik CS. Reverse biochemistry: use of macromolecular protease inhibitors to dissect complex biological processes and identify a membrane-type serine protease in epithelial cancer and normal tissue. Proc Natl Acad Sci U S A. 1999;96:11054–11061. doi: 10.1073/pnas.96.20.11054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L, Zhang ZG, Buller B, Jiang J, Jiang Y, Zhao D, Liu X, Morris D, Chopp M. Combination treatment with VELCADE and low-dose tissue plasminogen activator provides potent neuroprotection in aged rats after embolic focal ischemia. Stroke. 2010;41:1001–1007. doi: 10.1161/STROKEAHA.109.577288. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.