Abstract
5-fluorouracil (5-FU), 5-fluorodeoxyuridine (5-dUrd) and raltitrixed (RTX), are anticancer agents that target thymidylate synthase (TS) thereby blocking the conversion of dUMP into dTMP. In budding yeast, 5-FU promotes a large increase in the dUMP/dTMP ratio leading to massive polymerase-catalyzed incorporation of uracil (U) into genomic DNA, and to a lesser extent 5-FU, which are both excised by yeast uracil DNA glycosylase (UNG) leading to DNA fragmentation and cell death. In contrast, the toxicity of 5-FU and RTX in human and mouse cell lines does not involve UNG, but instead, other DNA glycosylases that can excise uracil derivatives. To elucidate the basis for these divergent findings in yeast and human cells, we have investigated the how these drugs perturb cellular dUTP and TTP pool levels and the relative abilities of three human DNA glycosylases (hUNG2, hSMUG1 and hTDG) to excise various TS drug-induced lesions in DNA. We found that 5-dUrd only modestly increases the dUTP/dTTP pool levels in asynchronous MEF, HeLa, and HT-29 human cell lines when growth is in standard culture media. In contrast, treatment of chicken DT40 B cells with 5-dUrd or RTX resulted in large increases in the dUTP/TTP ratio. Surprisingly, even though UNG is the only DNA glycosylase in DT40 cells that can act on U/A base pairs derived from dUTP incorporation, an isogenic ung−/− DT40 cell line showed little change its sensitivity to RTX as compared to control cells. In vitro kinetic analyses of the purified human enzymes show that hUNG2 is the most powerful catalyst for excision of 5-FU and U regardless of whether it is found in base pairs with A, G or present in ssDNA. Fully consistent with the in vitro activity assays, nuclear extracts isolated from human and chicken cell cultures show that hUNG2 is the overwhelming activity for removal of both U and 5-FU, despite its bystander status with respect to drug toxicity in these cell lines. The diverse outcomes of TS inhibition with respect to nucleotide pool levels, nature of the resulting DNA lesion, and the DNA repair response are discussed.
The antimetabolites 5-fluorouracil (5-FU)1, 5-flurodeoxyuridine (5-dUrd) and raltitrexed (RTX) are widely used for the treatment of colorectal, breast, and head and neck cancers (1, 2, 3). Fluoropyrimidines are metabolized much like uracil and deoxyuridine and can be enzymatically converted to the active metabolite 5-FdUMP (Fig. 1). A binary complex between 5-FdUMP and 5,10-methylenetetrahydrofolate irreversibly inhibits thymidylate synthase (TS), blocking de novo production of dTMP and also resulting in accumulation of dUMP. The resulting thymine nucleotide pool deficiency brought about by fluoropyrimidine drugs was originally thought to induce the therapeutic effect by a process called “thymineless death”. However, an additional hallmark of fluoropyrimidine treatment is the polymerase catalyzed incorporation of dUMP and 5-F-dUMP into DNA resulting in U/A, 5-FU/A and 5-FU/G base pairs which are substrates for various uracil DNA repair glycosylases (1, 4, 5). Similarly, RTX is a TS-specific folate mimic that also prevents TMP synthesis, but unlike fluoropyrimidines, RTX only results in the accumulation of dUMP and U/A base pairs in DNA. Thus, the toxicity mechanisms of these drugs will depend on the dUTP, 5-F-dUTP and TTP pool levels, as well as the relative specificities of the cellular uracil DNA glycosylase activities towards these uracil containing base pairs.
Figure 1.
Pathways of 5-FU metabolism and DNA toxicity. Metabolites are shown in bold and enzymes in italics: 5-FU, 5-fluorouracil; 5-FdUrd, 5-fluorodeoxyuridine; 5-FdUMP, 5-fluorodeoxyuridine monophosphate; 5-F-dUTP, 5-fluorodeoxyuridine triphosphate; dUTP, deoxyuridine triphosphate; dUMP, deoxyuridine monophosphate; dTMP, deoxythymidine monophosphate; dTTP, deoxythymidine triphosphate; TP, thymidine phosphorylase; NMK, nucleotide monophosphate kinase; NDK, nucleotide diphosphate kinase; TK, thymidine kinase; dUTPase, dUTP nucleotidohydrolase; TS, thymidylate synthase; DPDH, dihydropyrimidine dehydrogenase.
Several previous studies have investigated which DNA glycosylase enzymes are responsible for excising uracil and 5-FU bases in the context of U/A, 5-FU/A and 5-FU/G base pairs, and which are responsible for the toxic effects of the drug (6, 7, 8, 9, 10, 11). In a study performed in budding yeast, Seiple and coworkers showed that deletion of the base excision repair enzyme uracil DNA glycosylase (UNG) resulted in a huge accumulation of U in the yeast genome during treatment with 5-FU (~ 4% of genomic thymidine levels), and that ung- yeast were protected against the cytotoxic effects of 5-FU (8). This study thus established two mechanistic aspects of 5-FU toxicity in the yeast system: (i) 5-FU treatment results in elevated dUTP and accumulation of U in genomic DNA, and (ii) 5-FU toxicity is dependent on excision of U by yeast UNG (UNG is the only enzyme in yeast that removes uracil from DNA).
In contrast to the yeast findings, 5-FU toxicity studies using ung+/ung+ and ung−/ung− mouse embryonic fibroblasts (MEFs) indicated that UNG was not involved in the toxicity mechanism (7, 9). Since there are four different DNA glycosylases in both mice and humans capable of excising U and 5-FU from DNA (UNG, SMUG1, TDG, MBD42/MED1)(12), it is perhaps not surprising that the role of yeast UNG could be supplanted by the combined activities of SMUG1 and TDG in MEF cells (9, 10). From these combined studies, distinct roles for UNG, TDG and SMUG were suggested. UNG was indicated to have no role in preventing or precipitating the toxic effects of 5-FU in MEFs (7, 9), a conclusion also obtained in another study using RTX and HEK293 cells (13). In the MEF cell studies, the glycosylase activity of TDG precipitated 5-FU toxicity in a manner analogous to UNG in yeast, while 5-FU and U excision by SMUG1 was indicated to be protective against 5-FU toxicity.
To further understand these complex findings, we now examine the extent to which fluoropyrimidines and RTX alter the dUTP and TTP nucleotide pools in human, mouse and DT40 chicken cells, and we measure the specific activities of the DNA glycosylases hUNG2, hSMUG1 and hTDG against the diverse lesions that are generated from these drugs. In human and mouse cells, we find only small increases in the dUTP/TTP ratio after drug treatment in standard media, but much larger increases using dialyzed media that is depleted in folate and thymidine. Thus previous studies using standard media may not have induced high dUTP levels, and consequently, genomic uracil incorporation during S phase would have been limited. In contrast, 5-FdUrd and RTX induce a large increase in the dUTP/TTP ratio DT40 chicken cells. Despite UNG being the only U/A glycosylase activity in DT40 cells, the chicken UNG was found to provide no protective effect against the toxic effects of RTX, similar to results with human cells. Although UNG action is curiously irrelevant to drug toxicity in both human and chicken cells (vide supra), in vitro kinetic analyses of purified hUNG2, hSMUG1 and hTDG, as well as activity measurements using nuclear extracts, established that hUNG2 is paradoxically the primary cellular activity that removes U and 5-FU from DNA. We suggest that human and chicken cells are tolerant to U/A base pairs at the density they are introduced during drug treatment, and that the repair activity of UNG during S phase is masked by this tolerance. The UNG independent toxicity of the TS drugs with DT40 cells indicates that the killing mechanism is independent of both uracil base excision repair and mismatch repair (which acts on 5-FU/G pairs).
Experimental Procedures
Cloning, Expression, and Purification of hUNG2, hTDG and hSMUG1
The gene for full-length hUNG2 (939-bp) was amplified from cDNA from the Invitrogen Ultimate ORF collection (Clone ID: IOH43486). The gene for hSMUG1 was obtained from GenScript (813-bp). Both genes were ligated into a pET-19b vector (Novagen) and sequences were confirmed by sequencing both DNA strands. Both proteins were expressed as a N-terminal His10-fusion of which the expression and purification protocols were identical. E. coli C41 cells (Lucigen) were transformed with the pET-19b plasmid dsDNA and grown in LB media at 37°C. Upon reaching an OD600nm of 0.6, the temperature was lowered to 25°C and expression was induced via addition of 1 mM IPTG. After 5 hours of expression at 25°C, cells were harvested via centrifugation (4000 × g) and resuspended in lysis buffer containing 50 mM NaH2PO4 (pH 7.5), 500 mM NaCl, 0.1% Triton X-100, and 20 mM imidazole. Cells were lysed with a microfluidizer reaching ~ 20,000 psi. The resulting cell lysate was clarified via centrifugation (40,000 × g) and the supernatant loaded onto Ni-NTA resin (Qiagen) at 4°C. The unbound protein was washed away with lysis buffer and bound protein was eluted with lysis buffer containing 500 mM imidazole. Eluted protein was dialyzed into 20 mM Hepes-OH pH 8.0, 200 mM NaCl, 0.1 mM EDTA, 2 mM DTT and the His10-tag was removed via protease. Crude protein was then dialyzed against Buffer A (20 mM Hepes-OH pH 8.0, 50 mM NaCl, 1 mM DTT, 0.1 mM EDTA, and 5% glycerol) and loaded onto a SP-Sepharose FF cation-exchange column (GE Healthcare). Bound protein was eluted with a 0–100% linear gradient of Buffer A + 1 M NaCl. Fractions were analyzed via SDS-PAGE (coomassie-blue staining) and judged to be ~99% pure for both hUNG2 and hSMUG1. Protein mass was verified using MALDI-TOF mass spectrometry. Pure protein was dialyzed into storage buffer (20 mM Hepes-OH (pH 7.5), 100 mM NaCl, 0.2 mM EDTA, 1 mM DTT, and 10% glycerol), flash frozen, and stored at −80°C. Purified recombinant human TDG was a gift of Dr. Alex Drohat (14, 15).
DNA Substrates
All 5’-FAM containing oligonucleotides were synthesized on an Applied Biosystems 394 DNA synthesizer using standard phosphoramidites and supports obtained from Glen Research as previously described(16). Unlabeled oligonucleotides were obtained from Integrated DNA Technologies.
Steady-state kinetics
hUNG2 and hSMUG1 kinetics experiments were conducted at room temperature (~ 22 °C) in HEMN buffer (20 mM Hepes-OH, pH 7.5, 100 mM NaCl, 2.5 mM MgCl2, 0.2 mM EDTA, 1 mM DTT, and 0.1 mg/mL BSA) under conditions of [E] << Km with titrated [S]. Reactions were ran for 5 – 30 minutes and were quenched by adding an equal volume of 25:24:1 phenol:chloroform:isoamyl alcohol (PCA) and vortexing. To process the reactions, an aliquot of the aqueous layer (containing the DNA) was then extracted and the DNA backbone cleaved at abasic sites via addition of 100 mM NaOH followed by heating at 95 °C for 10 minutes. Formamide was added to >95% and the samples were ran on 7 M urea 19% denaturing PAGE to separate full-length substrate (19-mer) from cleaved product (9-mer). The 5’-FAM label of substrate and product was imaged in gel using a Typhoon (GE Healthcare) and quantitated using Quantity One (BioRad). Reaction rate (kobs) was plotted against DNA substrate concentration and kinetic parameters determined by fitting the data to eq 5, where [S] is the substrate concentration, kcat is the maximum reaction rate, and Km is the substrate concentration at ½ kcat.
| (5) |
Single-turnover kinetics
Single-turnover kinetic experiments on hSMUG1 were performed manually at room temperature in HEMN buffer under conditions of [E] >> [S]. For slower reactions, experiments were performed by rapidly mixing equal volumes of enzyme and substrate using a hand held pipet, and quenched by adding an equal volume of 0.5 M HCl from a second pipet. For more rapid time courses, a rapid chemical quench-flow instrument was used (KinTek). An equal volume of 25:24:1 PCA was added to extract all protein and an aliquot of the aqueous layer (containing DNA) was removed for further processing. The DNA backbone was cleaved at abasic sites via addition of NaOH to obtain pH > 10 followed by heating at 95 °C for 10 minutes. Substrate turnover was analyzed exactly as described for the steady-state measurements. The maximum single-turnover cleavage rate (kmax) was determined by plotting kobs versus enzyme concentration ([E]) and fitting the data to eq 6, where KD is [E] at one-half of kmax.
| (6) |
Preparation of Nuclear and Total Cell Extracts
Mouse embryonic fibroblasts (MEF), HeLa, and HT-29 colon cancer cells were grown adherently at 37 °C and 5% CO2 in either DMEM (MEF, HeLa) or RPMI (HT-29) media supplemented with 10% fetal bovine serum and 1% streptomycin/penicillin. Cells were plated at ~ 20% confluence and allowed to grow for 48 hrs in the absence and presence of 1 µM 5-FU. Cells were then harvested by trypsinization and counted using a hemocytometer. Nuclear extracts were prepared from 5-FU treated and untreated cells as previously described with slight modifications (17). Trypsinized cells were harvested via centrifugation at 200g @ 4 °C and washed 2× with 50 cell-volumes of ice-cold PBS. All subsequent steps were performed at 4 °C. Pelleted cells were resuspended in one packed cell volume of buffer consisting of 10 mM Hepes-OH pH 7.5, 10 mM NaCl, 1.5 mM MgCl2, 0.5 mM PMSF, and 1 mM DTT and cells were allowed to swell on ice for 15 minutes. Swollen cells were lysed by rapidly forcing them through a 25 5/8-gauge needle. Five passes were sufficient to obtain > 95% cell lysis as judged via trypan blue staining under a light microscope. Crude nuclei were pelleted from the cell lysate via centrifugation at 800g for 5 minutes. This crude nuclei pellet was resuspended in 2/3 of one packed-cell-volume (previously determined from whole-cell pellet) of buffer consisting of 20 mM Hepes-OH, 420 mM NaCl, 25% glycerol, 1.5 mM MgCl2, 0.2 mM EDTA, 0.5 mM PMSF, and 1 mM DTT followed by incubation on ice for 30 minutes with gentle agitation. Insoluble debris was pelleted via centrifugation at 16,000g for 5 minutes. The nuclear extract (supernatant) was dialyzed against 1000-volumes of Buffer Z (20 mM Hepes-OH pH 7.5, 100 mM NaCl, 2 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 5% glycerol) for 4 hrs at 4 °C using a 3,500 Da MWCO membrane (Pierce). Dialyzed extracts were flash frozen and stored at −80 °C until use. Protein concentration was determined using the Bradford assay (BioRad) with BSA as the standard.
Cell extracts of DT40 AID−/− and UNG−/−AID −/− chicken cells (a gift from Patricia J. Gearhart, National Institute of Aging) were prepared from cells grown in suspension at 37°C and 5% CO2 in chicken cell media (RPMI 1640, 10% fetal bovine serum (FBS), 1% chicken serum, 50 µM β-mercaptoethanol, 1% penicillin/streptomycin). Cells were plated at ~25% confluence and allowed to grow for 24 hours in the absence and presence of either 7.5 µM 5-fluorodeoxyuridine (5-FdUrd) or 0.5 µM raltitrexed (RTX) in the presence of either chicken cell media or folate-free chicken cell media (folate-free RPMI 1640, 10% dialyzed FBS, 1% dialyzed chicken serum, 50 µM β-mercaptoethanol, 80 nM 5-methyl tetrahydrofolate, and 1% penicillin/streptomycin). After incubation, cells were transferred to 15 mL Falcon tubes and spun down for 5 minutes at 1500 rpm at 4°C. The supernatant was discarded and the cells were washed with 1X PBS. Again, the supernatant was discarded and the cells were resuspended in 125 µL CelLytic M (Sigma). Cells were incubated, while rocking, for 15 minutes at room temperature. Immediately following incubation, the cells were centrifuged for 15 minutes at 14,000 rpm at 4°C. The supernatant containing the soluble protein fraction was transferred to a pre-chilled Eppendorf. The concentration of each extract was determined by a Bradford Assay (BioRad) using BSA as a standard. Extracts were frozen and stored at −80°C.
Base Excision Activity of Cell Extracts
For nuclear extracts prepared from human or mouse cells, activity assays were performed at 37 °C in nuclear extract buffer containing 20 mM Hepes-OH, pH 7.5, 100 mM NaCl, 2 mM EDTA, 1 mM DTT, and 0.1 mg/mL BSA. Nuclear extracts from untreated or 5-FU treated cells (0.2 µg/µL final concentration in 20 µL) were pre-incubated at room temperature in the presence or absence of UGI (1 µM final concentration) for 20 minutes before initiating the reaction by addition of DNA substrate (1 µM final concentration). Aliquots of the reactions were quenched at 3 hours by adding an equal volume of 25:24:1 PCA and vortexing. Samples were processed and analyzed exactly as described for the steady-state experiments. For DT40 cell extracts, an extract volume containing 5 µg of total protein was added to TEN-X Buffer (10 mM Tris-HCl, pH 7.5; 100 mM NaCl; 1 mM EDTA; 0.2% Triton-X 100), in the absence or presence of 1 µM UGI, and the reaction was initiated by the addition of 15 µL of fluorescent hairpin DNA substrate (300 nM, 150 µL final volume):
5'- FAM-GsCsAUUAAGAAG-(PEG)6-CUUCUUAATsGsCs-DAB- 3'
This sequence contains a polyethylene glycol linker, phosphorothioate linkages (s) to block exonuclease activities present in cell extracts, and a fluorescein (FAM) and dabcyl (DAB) fluorophore-quench pair. The removal of multiple uracils from U/A base pairs by UNG results in separation of the DNA strands and an increase in fluorescence due to the separation of the fluorophore-quench pair (16). After mixing, the reaction solution was immediately placed in fluorescence cuvette, and time-based acquisition of the fluorescence intensity at 520 nm was performed for 600 seconds with readings taken every 10 seconds (FluoroMax 3 fluorimeter) using an excitation wavelength of 494 nm (2.5 second integration time, excitation slit = 1 nm, emission slit = 4 nm). The time courses were fitted using Prism and the velocities (FU/s) were taken from the linear slopes after the initial lag phase that results from distributive cleavage of the uracil sites.
Single Nucleotide Extension (SNE) Experiment for Measurement of Intracellular dUTP, 5-F-dUTP and TTP
MEF, HeLa, HT-29 colon cancer cells, DT40 AID−/−, and DT40 AID−/−UNG−/− colon cancer cells were grown as described above. dNTPs were prepared as previously described with some modifications (18). Prior to drug treatment, ~105 cells were plated to a final volume of 3 mL. After 72 hours (HT-29 and HeLa cells) or 42 hours (MEF and DT40 cells), the media was changed. In half of the samples, the media remained the same; in the other half, the media was changed to folate-free DMEM (MEF, HeLa) or RPMI (HT-29) with 10% dialyzed FBS and 1% P/S and supplemented with 80 nM 5-methyl tetrahydrofolate or folate-free chicken media (DT40). Cells in each type of media were left untreated or treated with 7.5 µM 5-FdUrd or 0.5 µM RTX. After 24-hour incubation, drug-containing media was removed and cells were washed with their corresponding, drug-free media and then allowed to incubate for two hours longer in the same incubation conditions. Following the second incubation, the media was removed and 400 µL cold 60% methanol was incubated in the wells for 1 hour at 4 °C in order to extract the dNTPs from adherent cell lines. Suspension cell lines were first washed with 1X PBS, counted using a Scepter Automated Cell Counter (Millipore), centrifuged at 1500 rpm for 5 minutes, and then resuspended in 400 µL cold methanol in order to extract dNTPs. Following incubation, the liquid in the wells was removed and applied to a Microcon microcentrifuge filter with a molecular weight cutoff of 10 kDa and allowed to centrifuge at 14,000 rpm for 15 minutes. dNTPs were stored at −20 °C and were stable for no more than one week. Two 75 µL aliquots of each dNTP extraction were evaporated to dryness. To one aliquot, 40 µL of dUTPase buffer (34 mM Tris-HCl, pH 8.0; 10 mM MgCl2; 0.5 mM EDTA; 0.25 mg/mL BSA) containing 20 ng dUTPase was added. To the second aliquot was added 40 µL of dUTPase buffer without dUTPase. The samples were allowed to incubate at 37 °C for 20 minutes. Then, 60 µL of 100% methanol was added and the samples were again evaporated to dyness under vacuum. To the dried down dNTPs, 100 µL of SNE buffer (34 mM Tris-HCl, pH 8.0; 10 mM MgCl2; 0.2 mM EDTA; 0.25 mg/mL BSA) containing 32 nM FAM-labeled primer/template and 50 U Moloney murine leukemia virus reverse transcriptase (NEB) was added and allowed to incubate for 45 minutes at 42 °C. To specifically assess dUTP and TTP levels, a DNA primer/template sequence with a single adenine nucleotide overhang was used:
-
5′FAM-TGTTCTATGTTCATACACCACA-3′
3′-ACAAGATACAAGTATGTGGTGTA-5’
Following incubation with the polymerase, 50 µL of formamide containing 0.25% xylene cyanol and bromophenol blue was added to each sample followed by incubation at 95 °C for 20 minutes. 12 µL of each sample was then applied to a 20% denaturing polyacrylamide gel containing 7 M urea at 15W/gel and 45 °C for about 2 hours to separate the unextended primer from the primer that was extended by a single thymidine or deoxyuridine nucleotide. Gels were imaged on a Typhoon imager (GE Healthcare). Control experiments were perform with dGTP, dCTP and dATP to confirm that this template primer is specifically extended by TTP, dUTP and 5-FdUTP
SNE Data Analysis
Single nucleotide extension was quantified using Quantity One software (BioRad). Standard curves (generated in triplicate, see Supplemental Figure S1) were obtained by measuring the percent extension of increasing concentrations of dUTP and TTP standards (Roche Diagnostics). After calculation of the mean and standard deviation of the three replicate measurements, a standard curve was generated using a second-order polynomial fit (GraphPad Prism). The average percent extension of the dNTPs isolated from the cell extracts was determined in triplicate and compared to these standard curves to determine the concentration of TTP or [dUTP + 5-F-dUTP] in each sample. For each sample, the amount of TTP was calculated as the amount of extension observed after dUTPase pretreatment (both dUTP and 5-F-dUTP are substrates for dUTPase and the polymerase after treatment with 5-FdUrd but only dUTP is a substrate after treatment with RTX). The [dUTP + 5-F-dUTP] was calculated by difference, i.e [dUTP + 5-F-dUTP] extension = [extension before dUTPase treatment] - [extension after dUTPase pretreatment]. Concentrations were converted to pmol of dNTP per million cells.
Toxicity of 5-FdUrd and RTX to DT40 Cells
DT40 AID−/− and AID−/−UNG−/− were grown as described above. In a 96-well plate, cells were plated at a concentration of 50,000 cells/well and brought up to a final volume of 90 µL in either chicken media or folate-free chicken media. 10 µL of 10X drug stocks (made up in folate-free RPMI 1640) were added to each well in triplicate. Final drug concentrations were in the range 10 pM to 10 µM. Cells were allowed to incubate for 72 hours at 37°C and 5% CO2. Following incubation, 10 µL of 5 mg/mL MTT (3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide in 1X PBS) was added to the cells. Cells were allowed to incubate for 4 more hours in the same conditions. During this time, formazan crystals formed and precipitated. After incubation, crystals were resuspended in 100 µL resuspension buffer (isopropanol containing 10% Triton X-100 and 0.1 M HCl) and the absorbance was read on a plate reader at 570–650 nm. Triplicate measurements were averaged and the standard error was determined. IC50 values were calculated from curve fitting to eq 7.
| (7) |
Results
5-FdUrd Does Not Significantly Increase [dUTP] in Human and Mouse Cell Lines in Standard Culture Media
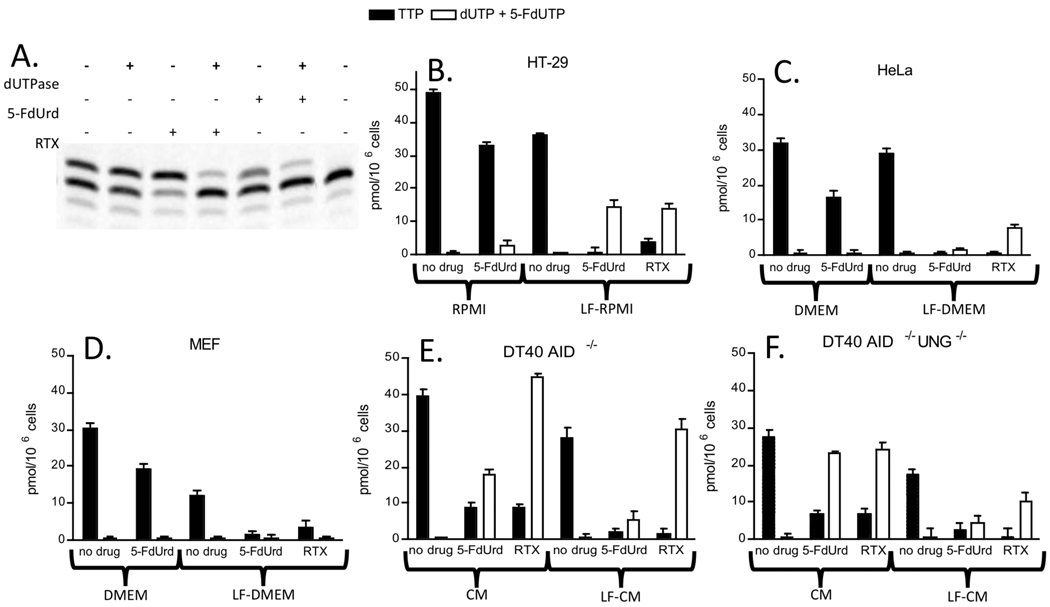
We were curious why previous studies in mammalian cell lines found no role for UNG in the toxicity of 5-FdUrd in human cell culture, even though UNG was shown to play a significant role in the toxicity of 5-FU in yeast (8). An important prerequisite event that influences the toxicity mechanism is the magnitude by which the ratio [dUTP]/[TTP] is increased during fluoropyrimidine treatment. If the post-treatment ratio is large, then massive amounts of dUTP can be incorporated into cellular DNA during S phase, as observed in yeast(8). Given the mechanistic importance of this ratio, we used a single nucleotide extension (SNE) assay to measure the levels of [dUTP + 5-F-dUTP] and TTP present in MEF, HeLa, and HT-29 and DT40 cell extracts before and after treatment with 5-FdUrd and RTX (19). The SNE assay measures the amount of polymerase-catalyzed extension of a DNA primer/template containing a single deoxyadenosine overhang on the template strand. Thus, total TTP and [dUTP + 5-F-dUTP] levels in a cell extract are measured in an extension reaction using this primer/template. The specific levels of TTP are obtained by pretreatment of the extracts with dUTPase, specifically converting dUTP and 5-F-dUTP to dUMP and 5-F-dUMP, respectively. The combined levels of dUTP and 5-F-dUTP are calculated by difference [dUTP + 5-F-dUTP] = [TTP + dUTP + 5-F-dUTP] – [TTP])(19) (Fig. 2A).
Figure 2.
Measurement of [dUTP + 5-F-dUTP] and TTP levels in HT29, MEF, HeLa and DT40 cell extracts in the presence and absence of 5-FdUrd (7.5 µM) and RTX (0.5 µM). (A) Denaturing polyacrylamide gel analysis of template extension reaction using NTP extracts obtained from HT29 cells cultured in the absence and presence of 5-FdUrd using low folate RPMI media with dialyzed FBS. T, DNA template primer. The minor n - 1 band in each lane arises from pyrophosphorolysis of the template. (B), (C), (D) TTP and [dUTP + 5-F-dUTP] levels (pmol/106 cells) measured in methanol extracts obtained from HT29, HeLa or MEF cells grown in the absence or presence of 5-FdUrd or RTX using standard or low folate (LF) RPMI or DMEM media. (E, F) TTP and [dUTP + 5-F-dUTP] levels (pmol/106 cells) measured in methanol extracts obtained from DT40 cells grown in the absence or presence of 5-FdUrd or RTX using standard or low folate (LF) chicken media (CM).
When MEF, HeLa, or HT-29 cells were treated with 7.5 µM 5-FdUrd in standard culture media (either DMEM or RPMI containing 10% FBS and 1% P/S), we found no significant change in the combined dUTP and 5-F-dUTP levels, and only a modest ~2-fold decrease in the TTP levels in the extracts (Figure 2B, 2C, 2D). This concentration of 5-FdUrd used in these experiments is at least 500 fold greater than its typical IC50 in cell culture (7), and these cell lines and media match those used in previous studies of glycosylase-mediated fluoropyrimidine toxicity (7, 9, 10).
5-F-dUrd and RTX Significantly Increase the dUTP/TTP when Human and Mouse Cells are Grown in Media Containing Dialyzed FBS
Normal culture media contains high concentrations of folate (2.5 µM) and thymidine, which could antagonize the effects of the TS drugs. Consistent with this expectation, significantly higher dUTP/TTP ratios were obtained using media containing dialyzed FBS and physiological levels of 5-methyl tetrahydrofolate (80 nM) (Fig. 2B, C, D). Under these conditions, marked decreases in TTP levels are observed in all cell lines, and in the case of HT-29 cells, a large increase in the dUTP level occurs as well ([dUTP]/[TTP] ~ 15). The equal or greater levels of dUTP detected with RTX as compared to 5-F-dUrd strongly suggests that the majority of the uridine triphosphate pool in the presence of 5-dUrd is comprised of dUTP and not 5-F-dUTP.
Cytotoxic Effects of of 5-dUrd and RTX in DT40 Chicken Cells is not Dependent on UNG Activity
Given the redundancy of glycosylase activities in human cells, and their complex roles in 5-FU and RTX toxicity (7, 9, 10), we turned to a simpler cell system to further elucidate the roles of dUTP pool levels and UNG in the toxicity of RTX. Chicken DT40 B cells offer several experimental advantages over the human cell lines. First, and much like yeast, DT40 cells do not have any other glycosylase activity other than UNG that can remove uracil from U/A base pairs (see Supplemental Figure S2) (20). Second, these rapidly dividing cells spend a much greater time in S phase of the cell cycle, increasing the likelihood that an S phase specific enzyme such as UNG would play a role in drug mechanism. Third, an ung−/− DT40 strain is available for directly testing the role of UNG in the absence of any other glycosylase activity. In addition, since RTX only introduces U/A base pairs into DNA, the role of mismatch repair pathways in the toxicity mechanism is expected to be absent. Thus, these cells provide a controlled system for addressing the role of UNG in repairing or precipitating the toxicity of U/A base pairs.
For comparisons using isogenic cell lines, we use the control DT40 cell line that has a knock-out in the cytidine deaminase enzyme AID (aid−/−), and the corresponding UNG-deficient double knock-out (aid−/− ung−/−). The use of this strain does not impact the experimental design becasue the wild-type DT40 strain showed no marked difference in its response to RTX as compared to the aid−/− strain (data not shown). In general, we found that both DT40 cell lines showed a much larger increase in the dUTP/TTP ratio upon treatment with 5-dUrd and RTX as compared to the human cell lines, both using normal and dialyzed chicken media (Fig. 2E, 2F). The one exception was the comparison between HeLa cells and DT40 aid−/− ung−/− cells using dialyzed media, where the two cell lines responded similarly. Also, treatment with RTX resulted in greater dUTP/TTP ratios as compared to 5-dUrd.
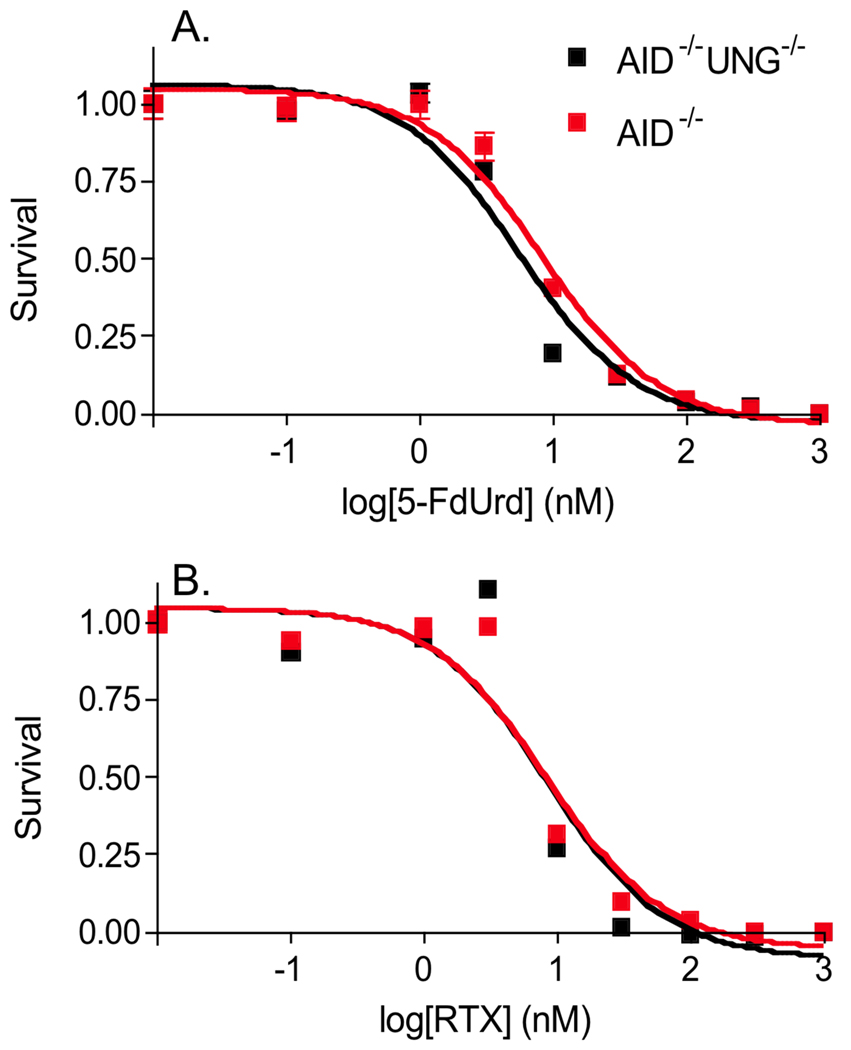
We measured the IC50 values for these drugs in normal and dialyzed chicken media using the control and UNG deletion cell lines (Fig. 3). The drugs are highly active in the control strain, with an IC50 value of 8 ± 1 nM in normal media. The UNG deficient strain shows a similar IC50 as the control strain for both 5-dUrd and RTX (6 and 9 nM, respectively), indicating that UNG does not play a discernable protective or toxic role in the drug mechanism. Thus, even with dUTP/TTP levels as high as 30/1, and the absence of other glycosylases, a role for in UNG in generating toxic abasic site lesions from 5-FU/A or U/A base pairs is not evident (see Discussion). Similar results were obtained using dialyzed chicken media.
Figure 3.
Toxicity of 5-FdUrd and RTX to DT40 cell lines that express or are deficient in UNG activity. The IC50 values for 5-FdUrd are 6 ± 1 nM (aid−/− ung−/−) and 8 ± 1 nM (aid−/−) and for RTX 9 ± 2 nM (aid−/− ung−/−) and 8 ± 2 nM (aid−/−). The error bars are standard errors from three replicate determinations.
hUNG2 is the Primary Activity for Removal of 5-FU and U from DNA
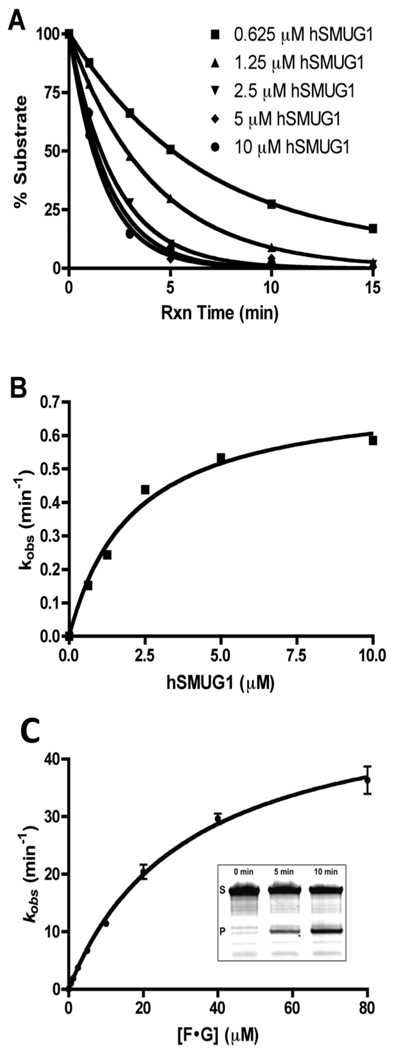
One possible mechanism for the lack of involvement of UNG in the toxicity of 5-F-dUrd and RTX is that its activity level is low compared to other glycosidases, and/or it is poorly effective at the removal of 5-FU lesions. To investigate this question, steady-state and single-turnover kinetic experiments were performed on hUNG2, hSMUG1, and hTDG using substrates in which 5-FU and U were paired with A or G, or were presented in the context of ssDNA (Table 1), Representative steady-state and single turnover kinetic data for hUNG2 and hSMUG1 reacting with 5-FU/A and 5-FU/G are shown in Figure 4.
Table 1.
Sequences and abbreviations of substrates used in this worka
| Substrate Abbreviation | Sequence |
|---|---|
| ssU | 5′ FAM-CACTGCTCAUGTACAGAGC 3′ |
| ssF | 5′ FAM-CACTGCTCAFGTACAGAGC 3′ |
| U/G | 5′ FAM-CACTGCTCAUGTACAGAGC 3′ |
| 3′ GTGACGAGTGCATGTCTCG 5′ | |
| U/A | 5′ FAM-CACTGCTCAUGTACAGAGC 3′ |
| 3′ GTGACGAGTACATGTCTCG 5′ | |
| F/G | 5′ FAM-CACTGCTCAFGTACAGAGC 3′ |
| 3′ GTGACGAGTGCATGTCTCG 5′ | |
| F/A | 5′ FAM-CACTGCTCAFGTACAGAGC 3′ |
| 3′ GTGACGAGTACATGTCTCG 5′ | |
| T/G | 5′ FAM-CACTGCTCATGTACAGAGC 3′ |
| 3′ GTGACGAGTGCATGTCTCG 5′ | |
| C/G | 5′ FAM-CACTGCTCACGTACAGAGC 3′ |
| 3′ GTGACGAGTGCATGTCTCG 5′ |
Abbreviations: U, deoxyuridine; F, 5-fluorodeoxyuridine; ss, single-stranded; FAM, 6-carboxyfluorescein.
Figure 4.
Representative single-turnover and steady-state measurements of glycosylase activity. (A) Single-turnover kinetic time courses for hSMUG1 catalyzed excision of 5-Fu from the F/A duplex as a function of enzyme concentration. (B) hSMUG1 concentration dependence of the observed rate constants. The kinetic parameters are reported in Table 2. (C) Steady-state kinetics for UNG2 excision of 5-FU from the F/G duplex (Table 1). Upper and lower bands in gel correspond to substrate and product bands, respectively.
Comparison of the steady-state kcat values for hUNG2, hSMUG1 and hTDG3 (Figure 5A and Table 2), shows that hUNG2 is able to process both 5-FU and U lesions at a rate at least 100-fold greater than either hSMUG1 or hTDG independent of the context of the lesion. Comparing activities under kcat/Km conditions, we again find that hUNG2 is superior in the excision of all substrates except for the F/G mispair, where the kcat/Km of hUNG2 is comparable to hSMUG1, and ten times slower than hTDG (Fig. 5B)(Table 2).
Figure 5.
In vitro kinetic excision parameters of purified hSMUG1 and hTDG relative to hUNG2 for an array of DNA substrates. (A) kcat of hSMUG1 and hTDG relative to hUNG2. (B) kcat/Km of hSMUG1 and hTDG relative to hUNG2. Kinetic parameters for hTDG were previously reported (14, 15) under identical buffer and substrate conditions.
Table 2.
Kinetic parameters of hUNG2, hSMUG1, and hTDG on 5-FU (F) and U containing DNA substrates.a
| hUNG2 | hSMUG1 | hTDGb | ||||||
|---|---|---|---|---|---|---|---|---|
| Substrate | kcat (min−1) | Km (µM) | kcat (min−1) | Km (µM) | kmax (min−1) | kcat (min−1) | Km(µM) | kmax(min−1) |
| ssU | 2193 ± 39 | 15 ± 1 | 5.6 ± 0.1 | 12 ± 1 | ≥ 5.6 | NA | NA | NA |
| U•G | 830 ± 30 | 3.3 ± 0.3 | 7.0 ± 0.3 | 1.8 ± 0.3 | 209 ± 16 | 0.005 ± 0.001 | ~ 0.01 | 2.2 ± 0.3 |
| U•A | 171 ± 5 | 3.3 ± 0.2 | 0.39 ± 0.01 | 0.40 ± 0.02 | 14 ± 1 | ND | ND | 0.0033 ± 0.0001 |
| ssF | 90 ± 4 | 64 ± 5 | < 0.1 | ~ 10 – 20 | ND | ND | ND | ~ 1c |
| F•G | 51 ± 2 | 32 ± 3 | 0.48 ± 0.02 | 0.7 ± 0.1 | 40 ± 6 | 0.63 ± 0.17 | 0.039 ± 0.008 | 278 ± 35 |
| F•A | 39 ± 2 | 39 ± 5 | < 0.1 | ~ 1 | 0.7 ± 0.1 | ND | ND | 1.08 ± 0.08 |
Because previous data for hTDG and hSMUG1 have shown that their steady-state turnover is greatly limited by slow release of the abasic reaction product, we wanted to check their activity with hUNG2 when these enzymes were provided in excess over the DNA substrate (14, 15, 21, 22). Thus, single-turnover measurements were performed in the presence of excess, rate-saturating amounts of hSMUG1 and hTDG. Even under very nonrealistic conditions where hSMUG1 was present at a ~ 200-fold greater concentration than its substrate site, its most favored substrates (U/G and F/G) were cleaved more slowly than catalytic amounts of hUNG2 (compare kcat and kmax values in Table 2). With hTDG, single-turnover measurements show that it can excise 5-FU from a 5-FU/G base pair about 5-times faster than the kcat value for hUNG (Table 2) (14, 15, 22). However, these conditions of [hTDG]/[S] >> 1 are not realistic for hTDG within the cell nucleus (see cell lysate results and Discussion). We note in this context that the kmax of hUNG2 for a 5-FU/G base pair is comparable to its kcat because product release is not rate-limiting for the 5-FU/G substrate. Previous work has shown that the catalytic activity of hTDG correlates well with the pyrimidine leaving group pKa (14). In contrast, the active sites of hSMUG1 and hUNG2 sterically preclude most C5 substituted uracil analogues (23, 24). The ability of 5-FU to enter the active site of hUNG2 and hSMUG1 likely reflects the relatively small increase in the van der Waals radius of the C5 fluorine substituent of 5-FU (1.35 Å) as compared to the C5 proton substituent of U (1.20 Å).
5-FU Excision Activity of Human and Chicken Cell Extracts
Knowledge of the in vitro activity profiles of hUNG2, hSMUG1, and hTDG on 5-FU and U lesions, combined with the use of the potent and specific protein inhibitor of UNG2 (UGI), provides the experimental tools necessary to determine the relative abundance of hUNG2, hSMUG1, and hTDG in human nuclear extracts prepared in the presence and absence of drug. In addition, similar experiments can be performed to investigate UNG activity levels in extracts prepared from aid−/− and aid−/− ung−/− DT40 cells in the presence and absence of drug.
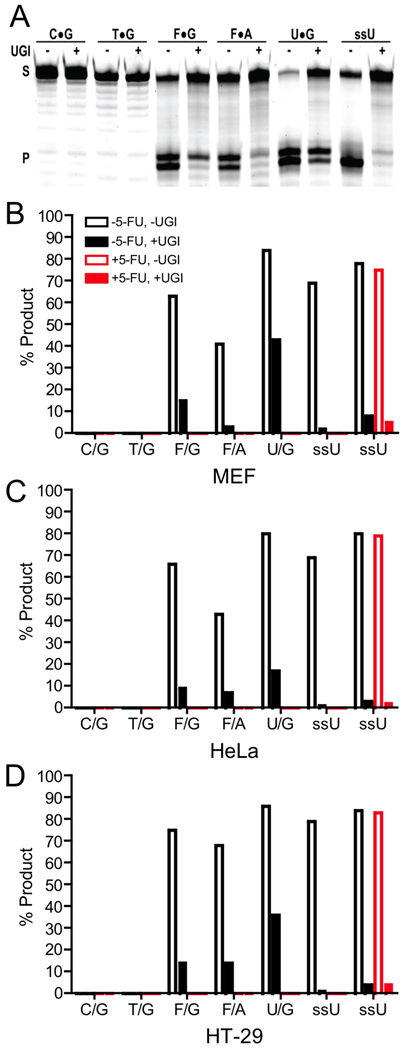
Purified nuclei from MEF, HT-29, and HeLa cells were prepared and analyzed for their ability to excise lesions from the array of DNA substrates in Table 1. Deconvolution of the relative activity contributions of each enzyme was performed based on the following rationale. First, the contribution of UNG2 to substrate cleavage can be easily assessed by performing reactions in the absence and presence of UGI (25). [Contrary to a previous report (21), we find that UGI does not inhibit either SMUG1 or TDG (Supplemental Figure S3)]. Second, the specific SMUG1 activity in the extracts may be assessed from the residual ssU activity remaining in the presence of UGI, because SMUG1 is the only enzyme capable of processing ssU when UNG is inhibited (Table 2)(Supplemental Figure S4). TDG activity may be estimated from the F/A activity remaining after subtracting the contributions of both UNG2 and SMUG1 because it is the only remaining glycosylase capable of processing this lesion (Table 2) (26, 27, 28). Finally, the fractional contribution of TDG to F/G excision may be estimated using its fractional contribution to F/A excision (Supplemental Figure S4). The small residual F/G excision activity (< 5%), after subtraction of UNG2, SMUG1, and TDG activities, may arise from MBD4.
As seen in Figure 6A, B, C, and D, UNG2 is the major glycosylase activity present in nuclear extracts of MEF, HeLa, and HT-29 cell lines using substrates with F/G, F/A and ssU lesions, regardless of whether the cell extracts were prepared from cell cultures grown in the absence or presence of 5-FU. We are able to conclude from the low levels ssU excision in the presence of UGI that SMUG1 contributes less than 6% to the excision of 5-FU in any of the cell lines tested (Supplemental Fig. S4). This finding is consistent with a previous report classifying SMUG1 as a low-abundance protein (29). The low SMUG1 contribution suggests that hTDG is the major processor of F/A lesions when UNG2 is absent, also agreeing with a previous study using MEF cell extracts (10). A lack of measurable T/G excision by hTDG in these extracts is not surprising because hTDG excises F/G lesions ~2000-fold more readily than T/G lesions (15). Furthermore, based on the relative excision activity of hTDG with F/A and F/G lesions (Supplemental Fig. S4), we conclude that hTDG provides the majority of the F/G excision activity when UNG2 is absent.4 These interpretations are not biased by the fact that steady-state catalysis by hTDG is limited by slow product release*, because the addition of recombinant human apyrimidinic endonuclease to remove the inhibitory abasic product from the reactions did not alter the outcome that UNG was the predominant activity present in the extracts (Supplemental Figure S6).5
Figure 6.
Excision of various DNA lesions by nuclear extracts obtained from cells grown in the absence (black bars) and presence (red bars) of 1 µM 5-FU for 48 h. (A) Representative gel of various DNA substrates incubated with MEF nuclear extracts in the presence and absence of the specific inhibitor of UNG (UGI). Top and bottom bands correspond to substrate (S) and product (P) bands, respectively. Identity of the central base pair of the substrate is listed above each lane (−/+ indicates the absence or presence of UGI). The two product bands are due to fractional removal of the terminal deoxyribose sugar during hot alkali processing of abasic sites. (B, C, D) Percent cleavage of the indicated DNA constructs by MEF, HeLa, and HT-29 by nuclear extracts obtained from cells grown in the absence and presence of 5-FU. Activity measurements were performed in the presence and absence of UGI to determine the contribution of UNG to the observed cleavage activity. The identical ssU activities of extracts prepared from cultures grown in the absence and presence of 5-FU indicates that UNG is not down regulated in the presence of the drug.
Given a previous report that nuclear UNG protein levels were down regulated by 5-dUrd (30), we compared the contribution of UNG activity in nuclear extracts prepared from cultures that had been grown with and without 1 µM 5-FU (Fig. 6, compare black and red bars). However, we did not find any measureable effect of 5-FU on the activity levels of nuclear UNG2 in these cell lines. We also measured the UNG activity in chicken cell extracts prepared in the absence and presence of 0.5 µM RTX for 24 h (Figure S2). Similar to the results with 5-FU and human cells, no decrease in UNG activity was observed.
Discussion
Reconciling the Role of DNA Glycosylases in 5-FU Toxicity
A key initial event in determining the fate of a cell during fluoropyrimidine treatment is the magnitude of the perturbations in the ratios dUTP/TTP and 5-F-dUTP/TTP (Figure 1). Mechanistically, a large increase in these ratios requires (i) efficient inhibition of thymidylate synthase, (ii) inefficient action of dUTPase and dihydropyrimidine dehydrogenase, and (iii) active nucleoside kinases. Although pyrimidine metabolism involves far more complexity than these three components, if any of these mechanistic requirements are not met, the nucleotide ratios may not be altered significantly, and the toxicity mechanism may affected.
The above mechanistic considerations may provide an explanation for the different requirements for DNA glycosylases that have been reported in budding yeast and mammalian cell culture systems. In an UNG deletion yeast strain, genomic levels of U and 5-FU were found to be 1 per 100 and 1 per 10,000 nucleotides, respectively, after treatment with 5-FU, whereas undetectable levels of uracil were found in the wild-type strain (8). These findings, combined with the observation that the wild-type yeast strain was 10-fold more sensitive to 5-FU than the ung deletion strain, require a large fluoropyrimidine-induced increase in the dUTP/TTP pool in yeast, and a significant role of UNG catalyzed uracil excision in the toxicity mechanism. In contrast, the present study finds only small changes in the [dUTP + 5-F-dUTP]/TTP ratio upon treating three mammalian cell lines with 5-dUrd using standard cell culture media, but much more significant increases in this ratio when using dialyzed FBS media containing low folate levels (Fig. 2). We suspect that the high folate and thymidine levels in normal media may be strongly antagonistic to action of fluoropyrimidines and RTX via the pyrimidine salvage pathway and direct competitive inhibition. Indeed, the consistently low ratio dUTP/TTP measured in cells grown in standard media is fully consistent with the previously determined low levels of U and 5-FU in genomic DNA isolated from mammalian cells treated with 5-fluorodeoxuridine. Although these genomic levels vary widely in different studies (0.01 to 1 uracil per 104 genomic nucleotides) (9, 10, 13, 31), the levels are at least 100-fold lower than found in yeast. Moreover, in mammalian cells the genomic levels of 5-FU are typically detected at nearly an order of magnitude greater density than uracil, whereas in yeast the uracil levels greatly exceed that of 5-FU (8). Thus, the glycosylase-mediated toxicity of fluoropyrimidines in yeast is dominated by a large rise in dUTP levels, dense incorporation of dUTP (but not 5-F-dUTP) into DNA, UNG excision, and ultimately, whole scale fragmentation of the yeast genome(8). In contrast, mammalian cells differ in two key respects. First, the large perturbation in the nucleotide pool levels never occurs in standard culture media, and second, the level of genomic 5-FU is an order of magnitude higher than in yeast, and even exceeds the level of genomic U. Accordingly, 5-FU cytotoxicity in MEF cells correlates more closely with the processing of 5-FU rather than U in genomic DNA (9, 10).
Variable Roles of Uracil DNA Glycosylases in Drug Mechanism
The redundancy of DNA glycosylases that act on U and 5-FU in mammalian cells leads to the possibility that the fate of these lesions may depend on the initiating glycosylase. In the case of UNG2, which we have shown is the most globally active uracil glycosylase in nuclear extracts, encounter with a sparse uracil or 5-FU residue during S phase DNA replication is easily repaired given the abundance and activity of the enzyme. Unlike the yeast system, these UNG initiated repair events may not be dense enough to elicit fork arrest or double strand breaks, and accordingly, UNG activity does not precipitate drug toxicity. The low levels of U and 5-FU that escape detection of UNG during S phase are then intercepted during G1 or G2 by the redundant activity of SMUG1, accounting for the fluoropyrimidine hypersensitivity of SMUG1 deficient cells(9). Finally, encounter of a 5-FU/G lesion by TDG appears to be cytotoxic, based on the decrease in fluoropyrimidine sensitivity of cells where TDG activity has been knocked down using RNA silencing. These results suggest that lesion excision by TDG during G1 or G2 results in faulty repair, DNA strand breaks or the initiation of an apoptotic signaling pathway.
Consistent with the idea that the identity of the initiating repair glycosylase can determine a lesion’s fate, it is not surprising that mismatch repair (MMR) is strongly implicated in exerting 5-FU cytotoxicity (6, 32, 33). Unlike uracil BER, that excises and replaces only a single damaged nucleotide, MMR recognition of a 5-FU/G or U/G mismatch directs a DNA nick ~ 250 – 1000 bp from the mismatch by a complex of MMR proteins(11). The entire length of the DNA strand from the nick to the mismatch is then degraded and synthesized anew, increasing the chances of incorporating a new lesion during the repair process. Independent of this futile repair cycle, MMR has also been shown to exert 5-FU cytotoxicity via the signaling of cell-cycle arrest and apoptosis during lesion detection (28).
The report that hUNG is down regulated in many, but not all, human cell lines during treatment with 5-dUrd adds an additional layer of complexity to the understanding of drug mechanism (30). This down regulation could represent a survival strategy in some transformed cell lines that allows evasion of what would be an otherwise toxic effect of uracil residues introduced during S phase DNA synthesis. Consistent with this idea, HeLa cells do not show this same hUNG2 down regulation, and are protected against the toxicity of 5-dUrd when hUNG2 is silenced using siRNA methods (30). This effect with 5-dUrd is not recapitulated in our studies using 5-FU or RTX in the same cell lines, as well as in chicken cells. These apparently conflicting results may reflect significant differences in the cellular response to 5-FU and RTX as opposed to 5-dUrd.
Toxicity of 5-dUrd and RTX to DT40 Cells is Independent of Uracil BER and MMR
The DT40 chicken cell line provides a useful system to explore the relative role of uracil excision repair and mismatch repair in the toxicity mechanisms of 5-dUrd and RTX. The similar IC50 values of these drugs, the lack of involvement of UNG, and the similar perturbations in dUTP/TTP with both drugs, strongly suggests a common cell killing mechanism that may be related to the perturbation in the nucleotide pool levels. Moreover, a significant involvement of MMR in the toxicity mechanism is not indicated because 5-FdUrd should activate this pathway through the incorporation of 5-F-dUTP*, while RTX (which only produces dUTP), should not. These results in DT40 cells are most consistent with the original thymineless death mechanism for fluoropyrimidine action.
Implications for Cancer Therapy
The different cytotoxic mechanisms of fluoropyrimidines in yeast, mammalian and chicken cells suggests that the pathway for cell killing might be pharmacologically selected with appropriate compounds. The variable results with different TS inhibitors and different cell types suggests that it will be difficult to predict the clinical outcome for a given tumor and drug without knowledge of the pathways that are involved. Selection of a different killing pathway would be beneficial if a tumor is defective in a DNA repair pathway that is otherwise required for the cytotoxic effect. One established example is MMR deficient colon cancers that are resistant to 5-FU (33). If the killing mechanism could be switched to a UNG-dependent killing pathway (as observed in HeLa cells), the efficacy of the drug might be restored. One possible approach to this end would be to inhibit the enzyme dUTPase, which can lead to large increases in dUTP levels during fluoropyrimdine treatment(34). Such perturbations could shift the toxicity mechanism to one that involves UNG, as already observed in yeast.
Supplementary Material
Acknowledgement
We thank Dr. Alexander Drohat for the TDG enzyme used in these studies.
Footnotes
This work was supported by grant GM056834 from the National Institutes of Health to J.T.S. and a Ruth L. Kirschstein National Research Service Award (F31 GM083623) to J. B. P.
Abbreviations: DMEM, Dulbecco’s Modified Eagle Medium; RPMI, Roswell Park Memorial Institute medium; CM, chicken media; LF-CM, low folate chicken media; SMUG1, single-strand selective monofunctional uracil DNA glycosylase; UNG2, nuclear uracil DNA glycosylase; TDG, thymine DNA glycosylase; TS, thymidylate synthase; dUTP, deoxyuridine triphosphate; 5-F-dUTP, 5-fluorodeoxyuridine triphosphate; dTTP, deoxythymidine triphosphate; 5-FU, 5-fluorouracil base; U, uracil; MEF, mouse embryonic fibroblast; MBD4, methyl-CpG-binding domain protein 4; FAM, 6-carboxyfluorescein; PBS, phosphate buffered saline; PMSF, phenylmethylsulfonyl fluoride; DTT, dithiothreitol; PCA, phenol:chloroform:isoamyl alcohol; BSA, bovine serum albumin; UGI, uracil DNA glycosylase inhibitor protein;
While the remaining 5-FU DNA glycosylase, MBD4, has been shown to excise 5-FU opposite G in vitro (35, 36), it has yet to be implicated in 5-FU excision in vivo in either mouse or human cell lines (14, 27).
The kinetic properties of hTDG under identical solution conditions to those used here have been previously reported (10, 26, 37).
The higher ratio of U/G to F/G excision as compared to F/A to ssU excision in the MEF cells, but not HeLa and HT-29 extracts, suggests the presence of an increased mismatch activity. One might attribute this additional MEF mismatch activity to the mismatch specific MBD4 glycosylase (35). However, because mSMUG1 has an increased mismatch activity compared to hSMUG1 (36), and the activity of MBD4 is greatly impaired at 100 mM salt (27), we assign this activity to mSMUG1 (see Discussion).
These extract experiments overestimate the contribution of TDG activity in vivo for two reasons. First, the oligonucleotides contained 5-FU lesions in a CpG or CpA sequence context, which are preferred by TDG by 2 to 150-fold as compared to other sequence contexts (14). Secondly, because nuclear extracts were prepared from asynchronous cell cultures, and the expression of the enzymes is cell cycle dependent, the average amount of each enzyme present will be weighted by the fraction of the cells in each stage of the cell cycle. Since TDG is present during the G2/G1 phases (> 70% of the cell-cycle time), and UNG2 is present only in S-phase (< 30% of the average cell-cycle time)(10, 26, 37), then the extract preparations are weighted towards TDG.
Supporting Information Available
Six supporting figures are included. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Longley DB, Harkin DP, Johnston PG. 5-Fluorouracil: Mechanisms of Action and Clinical Strategies. Nat Rev Cancer. 2003;3:330–338. doi: 10.1038/nrc1074. [DOI] [PubMed] [Google Scholar]
- 2.Wyatt MD, Wilson DM. Participation of DNA Repair in the Response to 5-Fluorouracil. Cell Mol. Life Sci. 2009;66:788–799. doi: 10.1007/s00018-008-8557-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Van Triest B, Pinedo HM, Giaccone G, Peters GJ. Downstream Molecular Determinants of Response to 5-Fluorouracil and Antifolate Thymidylate Synthase Inhibitors. Ann. Oncol. 2000;11:385–391. doi: 10.1023/a:1008351221345. [DOI] [PubMed] [Google Scholar]
- 4.Cohen SS. On the Nature of Thymineless Death. Ann. N. Y. Acad. Sci. 1971;186:292–301. doi: 10.1111/j.1749-6632.1971.tb31155.x. [DOI] [PubMed] [Google Scholar]
- 5.Houghton JA, Tillman DM, Harwood FG. Ratio of 2'-Deoxyadenosine-5'-triphosphate/thymidine-5'-Triphosphate Influences the Commitment of Human Colon Carcinoma Cells to Thymineless Death. Clin. Cancer Res. 1995;1:723–730. [PubMed] [Google Scholar]
- 6.Carethers JM, Chauhan DP, Fink D, Nebel S, Bresalier RS, Howell SB, Boland CR. Mismatch Repair Proficiency and in Vitro Response to 5-Fluorouracil. Gastroenterology. 1999;117:123–131. doi: 10.1016/s0016-5085(99)70558-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Andersen S, Heine T, Sneve R, König I, Krokan HE, Epe B, Nilsen H. Incorporation of dUMP into DNA is a Major Source of Spontaneous DNA Damage, while Excision of Uracil is Not Required for Cytotoxicity of Fluoropyrimidines in Mouse Embryonic Fibroblasts. Carcinogenesis. 2005;26:547–555. doi: 10.1093/carcin/bgh347. [DOI] [PubMed] [Google Scholar]
- 8.Seiple L, Jaruga P, Dizdaroglu M, Stivers JT. Linking Uracil Base Excision Repair and 5-Fluorouracil Toxicity in Yeast. Nucleic Acids Res. 2006;34:140–151. doi: 10.1093/nar/gkj430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.An Q, Robins P, Lindahl T, Barnes DE. 5-Fluorouracil Incorporated into DNA is Excised by the Smug1 DNA Glycosylase to Reduce Drug Cytotoxicity. Cancer Res. 2007;67:940–945. doi: 10.1158/0008-5472.CAN-06-2960. [DOI] [PubMed] [Google Scholar]
- 10.Kunz C, Focke F, Saito Y, Schuermann D, Lettieri T, Selfridge J, Schär P. Base Excision by Thymine DNA Glycosylase Mediates DNA-Directed Cytotoxicity of 5-Fluorouracil. PLoS Biol. 2009;7:e91. doi: 10.1371/journal.pbio.1000091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Fischer F, Baerenfaller K, Jiricny J. 5-Fluorouracil is Efficiently Removed from DNA by the Base Excision and Mismatch Repair Systems. Gastroenterology. 2007;133:1858–1868. doi: 10.1053/j.gastro.2007.09.003. [DOI] [PubMed] [Google Scholar]
- 12.Krokan HE, Drabløs F, Slupphaug G. Uracil in DNA--Occurrence, Consequences and Repair. Oncogene. 2002;21:8935–8948. doi: 10.1038/sj.onc.1205996. [DOI] [PubMed] [Google Scholar]
- 13.Luo Y, Walla M, Wyatt MD. Uracil Incorporation into Genomic DNA does Not Predict Toxicity Caused by Chemotherapeutic Inhibition of Thymidylate Synthase. DNA Repair (Amst) 2008;7:162–169. doi: 10.1016/j.dnarep.2007.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morgan MT, Bennett MT, Drohat AC. Excision of 5-Halogenated Uracils by Human Thymine DNA Glycosylase. Robust Activity for DNA Contexts Other than CpG. J. Biol. Chem. 2007;282:27578–27586. doi: 10.1074/jbc.M704253200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Fitzgerald ME, Drohat AC. Coordinating the Initial Steps of Base Excision Repair. Apurinic/apyrimidinic Endonuclease 1 Actively Stimulates Thymine DNA Glycosylase by Disrupting the Product Complex. J. Biol. Chem. 2008;283:32680–32690. doi: 10.1074/jbc.M805504200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jiang YL, Krosky DJ, Seiple L, Stivers JT. Uracil-Directed Ligand Tethering: An Efficient Strategy for Uracil DNA Glycosylase (UNG) Inhibitor Development. J. Am. Chem. Soc. 2005;127:17412–17420. doi: 10.1021/ja055846n. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lee KA, Bindereif A, Green MR. A Small-Scale Procedure for Preparation of Nuclear Extracts that Support Efficient Transcription and Pre-mRNA Splicing. Gene Anal. Tech. 1988;5:22–31. doi: 10.1016/0735-0651(88)90023-4. [DOI] [PubMed] [Google Scholar]
- 18.Horowitz RW, Zhang H, Schwartz EL, Ladner RD, Wadler S. Measurement of Deoxyuridine Triphosphate and Thymidine Triphosphate in the Extracts of Thymidylate Synthase-Inhibited Cells using a Modified DNA Polymerase Assay. Biochem. Pharmacol. 1997;54:635–638. doi: 10.1016/s0006-2952(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 19.Horowitz RW, Zhang H, Schwartz EL, Ladner RD, Wadler S. Measurement of Deoxyuridine Triphosphate and Thymidine Triphosphate in the Extracts of Thymidylate Synthase-Inhibited Cells using a Modified DNA Polymerase Assay. Biochem. Pharmacol. 1997;54:635–638. doi: 10.1016/s0006-2952(97)00259-1. [DOI] [PubMed] [Google Scholar]
- 20.Di Noia JM, Rada C, Neuberger MS. SMUG1 is Able to Excise Uracil from Immunoglobulin Genes: Insight into Mutation Versus Repair. EMBO J. 2006;25:585–595. doi: 10.1038/sj.emboj.7600939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kavli B, Sundheim O, Akbari M, Otterlei M, Nilsen H, Skorpen F, Aas PA, Hagen L, Krokan HE, Slupphaug G. HUNG2 is the Major Repair Enzyme for Removal of Uracil from U:A Matches, U:G Mismatches, and U in Single-Stranded DNA, with hSMUG1 as a Broad Specificity Backup. J. Biol. Chem. 2002;277:39926–39936. doi: 10.1074/jbc.M207107200. [DOI] [PubMed] [Google Scholar]
- 22.Bennett MT, Rodgers MT, Hebert AS, Ruslander LE, Eisele L, Drohat AC. Specificity of Human Thymine DNA Glycosylase Depends on N-Glycosidic Bond Stability. J. Am. Chem. Soc. 2006;128:12510–12519. doi: 10.1021/ja0634829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Kavli B, Slupphaug G, Mol CD, Arvai AS, Peterson SB, Tainer JA, Krokan HE. Excision of Cytosine and Thymine from DNA by Mutants of Human Uracil-DNA Glycosylase. EMBO J. 1996;15:3442–3447. [PMC free article] [PubMed] [Google Scholar]
- 24.Nilsen H, Rosewell I, Robins P, Skjelbred CF, Andersen S, Slupphaug G, Daly G, Krokan HE, Lindahl T, Barnes DE. Uracil-DNA Glycosylase (UNG)-Deficient Mice Reveal a Primary Role of the Enzyme during DNA Replication. Mol. Cell. 2000;5:1059–1065. doi: 10.1016/s1097-2765(00)80271-3. [DOI] [PubMed] [Google Scholar]
- 25.Wang Z, Mosbaugh DW. Uracil-DNA Glycosylase Inhibitor Gene of Bacteriophage PBS2 Encodes a Binding Protein Specific for Uracil-DNA Glycosylase. J. Biol Chem. 1989;264:1163–1171. [PubMed] [Google Scholar]
- 26.Hardeland U, Bentele M, Jiricny J, Schar P. Separating Substrate Recognition from Base Hydrolysis in Human Thymine DNA Glycosylase by Mutational Analysis. J. Biol. Chem. 2000;275:33449–33456. doi: 10.1074/jbc.M005095200. [DOI] [PubMed] [Google Scholar]
- 27.Petronzelli F, Riccio A, Markham GD, Seeholzer SH, Stoerker J, Genuardi M, Yeung AT, Matsumoto Y, Bellacosa A. Biphasic Kinetics of the Human DNA Repair Protein MED1 (MBD4), a Mismatch-Specific DNA N-Glycosylase. J Biol Chem. 2000;275:32422–32429. doi: 10.1074/jbc.M004535200. [DOI] [PubMed] [Google Scholar]
- 28.Meyers M, Wagner MW, Mazurek A, Schmutte C, Fishel R, Boothman DA. DNA Mismatch Repair-Dependent Response to Fluoropyrimidine-Generated Damage. J. Biol. Chem. 2005;280:5516–5526. doi: 10.1074/jbc.M412105200. [DOI] [PubMed] [Google Scholar]
- 29.Nilsen H, Haushalter KA, Robins P, Barnes DE, Verdine GL, Lindahl T. Excision of Deaminated Cytosine from the Vertebrate Genome: Role of the SMUG1 Uracil-DNA Glycosylase. EMBO J. 2001;20:4278–4286. doi: 10.1093/emboj/20.15.4278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Fischer JA, Muller-Weeks S, Caradonna SJ. Fluorodeoxyuridine Modulates Cellular Expression of the DNA Base Excision Repair Enzyme Uracil-DNA Glycosylase. Cancer Res. 2006;66:8829–8837. doi: 10.1158/0008-5472.CAN-06-0540. [DOI] [PubMed] [Google Scholar]
- 31.Canman CE, Lawrence TS, Shewach DS, Tang HY, Maybaum J. Resistance to Fluorodeoxyuridine-Induced DNA Damage and Cytotoxicity Correlates with an Elevation of Deoxyuridine Triphosphatase Activity and Failure to Accumulate Deoxyuridine Triphosphate. Cancer Res. 1993;53:5219–5224. [PubMed] [Google Scholar]
- 32.Meyers M, Wagner MW, Hwang HS, Kinsella TJ, Boothman DA. Role of the hMLH1 DNA Mismatch Repair Protein in Fluoropyrimidine-Mediated Cell Death and Cell Cycle Responses. Cancer Res. 2001;61:5193–5201. [PubMed] [Google Scholar]
- 33.Li LS, Morales JC, Veigl M, Sedwick D, Greer S, Meyers M, Wagner M, Fishel R, Boothman DA. DNA Mismatch Repair (MMR)-Dependent 5-Fluorouracil Cytotoxicity and the Potential for New Therapeutic Targets. Br. J. Pharmacol. 2009;158:679–692. doi: 10.1111/j.1476-5381.2009.00423.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Koehler SE, Ladner RD. Small Interfering RNA-Mediated Suppression of dUTPase Sensitizes Cancer Cell Lines to Thymidylate Synthase Inhibition. Mol. Pharmacol. 2004;66:620–626. doi: 10.1124/mol.66.3.. [DOI] [PubMed] [Google Scholar]
- 35.Bellacosa A. Role of MED1 (MBD4) Gene in DNA Repair and Human Cancer. J. Cell. Physiol. 2001;187:137–144. doi: 10.1002/jcp.1064. [DOI] [PubMed] [Google Scholar]
- 36.Pettersen HS, Sundheim O, Gilljam KM, Slupphaug G, Krokan HE, Kavli B. Uracil-DNA Glycosylases SMUG1 and UNG2 Coordinate the Initial Steps of Base Excision Repair by Distinct Mechanisms. Nucleic Acids Res. 2007;35:3879–3892. doi: 10.1093/nar/gkm372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hengst L, Reed SI. Translational Control of p27Kip1 Accumulation during the Cell Cycle. Science. 1996;271:1861–1864. doi: 10.1126/science.271.5257.1861. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.