Abstract
Purpose
Previous studies in our laboratory have shown the progressive methylation and suppression of the gene encoding protein tyrosine phosphatase, PTPRO, in the livers of rats fed a methyl-deficient diet that induces hepatocarcinogenesis. Subsequently, we observed the methylation of PTPRO in primary human lung tumors and also showed its potential tumor suppressor characteristics. The present study was undertaken to investigate whether the truncated form of PTPRO (PTPROt), specifically expressed in naïve B lymphocytes, was also methylated and suppressed in chronic lymphocytic leukemia (CLL), a disease generally affecting B lymphocytes.
Experimental Design and Results
Initial screening showed that 60% of the 52 CLL samples analyzed using methylation-specific PCR assay were methylated compared with B lymphocytes from normal individuals, which were not methylated. The expression of PTPROt, as measured by semiquantitative reverse transcription-PCR, inversely correlated with methylation in the few samples tested. Analysis of additional samples (n = 50) by combined bisulfite restriction analysis showed that the PTPRO CpG island was methylated in 82% of patients with CLL compared with B lymphocytes from normal individuals. Furthermore, overall expression of PTPRO was reduced in CLL relative to normal lymphocytes. The PTPRO gene was also suppressed by methylation in the CLL cell lineWaC3CD5, where it could be reactivated upon treatment with the DNA hypome-thylating agent 5-AzaC. Ectopic expression of PTPROt in a nonexpressing cell line increased growth inhibition with fludarabine treatment, a therapy commonly used for CLL.
Conclusion
This study reveals the potential role of PTPRO methylation and silencing in CLL tumorigenesis and also provides a novel molecular target in the epigenetic therapy.
Chronic lymphocytic leukemia (CLL) is the most common form of adult leukemia in the Western hemisphere, with an incidence of 17,000 new cases per year. The median age of patients with CLL is 68 years, although 10% of patients were under the age of 50 at diagnosis. CLL is characterized by the expansion of a single monoclonal B cell population expressing surface membrane CD19, CD20, CD23, dim surface immuno-globulin, and CD5. CLL can be divided into two subsets based on IgVH mutational status, which clinically predicts indolent or more aggressive disease. Until recently, B-CLL was thought to be a static disease characterized by clonal excess of B cells resulting from decreased apoptosis rather than increased proliferation (1, 2). With the recognition of the importance of IgVH mutational status, it has also become apparent that the highly aggressive unmutated subset has a more dynamic disorder. Patients with IgVH unmutated disease are characterized by having shortened telomere lengths (3, 4), repetitive B cell receptor signaling, and a high risk of chromosomal abnormalities (reviewed in ref. 5). The subset of patients with IgVH unmutated CLL showed faster than anticipated turnover rates resulting in fastidious and aggressive clones of disease. In vivo evidence of such rapid CLL turnover associated with rapid disease progression was recently shown in studies using heavy water (6).
Although several studies have shown a variety of genetic abnormalities in CLL including deletions of 11q, 13q, 17p, and trisomy 12, the epigenetic mechanism of gene regulation has only recently been appreciated. Epigenetic silencing of genes is a common mechanism by which tumor suppressor genes are silenced in cancer. In a genome-wide scan for methylation, we recently identified several genes that are differentially and tumor-specifically methylated in CLL (7). As part of a follow-up to this work, both our group and others showed differential methylation of TWIST2, a gene that potentially silences p53 and ZAP-70, between IgVH-mutated and IgVH-unmutated CLL (8). These studies, along with others characterizing the hypomethylation of tcl-1 (9) and bcl-2 (10, 11), suggest that altered expression of genes by methylation probably plays a pivotal role in the pathogenesis of CLL.
Given the critical role of tumor suppressor genes, including a subset of phosphatases, our laboratory has actively pursued the silencing of such genes in several tumor types. Indeed, we have shown that protein tyrosine phosphatase receptor type O (PTPRO) is suppressed by methylation in rat hepatocellular carcinoma (12) and human primary lung cancer (13). In these solid tumors, in vitro and in vivo data suggest that PTPRO acts as a tumor suppressor gene to inhibit the growth of transformed cells. A truncated isoform of PTPRO (PTPROt) that is specifically expressed in B lymphocytes was recently implicated in large cell lymphoma and exhibited a similar growth inhibition behavior (14). Previous studies have shown that several members of the protein tyrosine phosphatase family, including PTPRZ1 and PTPRN2, were methylated in CLL (7). Additionally, signal transduction through a variety of tyrosine kinases contributes significantly to B cell and CLL cell survival. Therefore, we hypothesized that PTPROt may be silenced in CLL through methylation, and that reexpression of this gene may enhance apoptosis and B cell signaling. Here, we show that PTPROt is indeed methylated in a variety of patients with CLL and its expression may promote growth inhibition.
Materials and Methods
Patient sample processing and cell culture
All the patients enrolled in this study had immunophenotypically defined B-CLL as outlined by the modified 1996 National Cancer Institute criteria. Two sets of samples were used for this study: the first set of 52 samples lacked detailed molecular characterization and were used as a pilot set to test the occurrence of PTPRO methylation in CLL. The second set of 50 samples were obtained from patients enrolled on the intergroup phase III trial, E2997. These samples were obtained at study entry and was restricted to patients that had CLL which met the 1996 National Cancer Institute criteria for initiating treatment, and who had not previously received chemotherapy for CLL. Blood was obtained from patients following informed consent under a protocol approved by the Ohio State University Cancer Institutional Review Board. B-CLL cells were isolated immediately following donation using Ficoll density gradient centrifugation (Ficoll-Paque Plus, Amersham Biosciences). Isolated mononuclear cells were incubated in RPMI 1640 supplemented with 10% heat-inactivated fetal bovine serum (FBS, Hyclone Laboratories), 2 mmol/L of L-glutamine (Invitrogen), and penicillin (100 units/mL)/streptomycin (100 μg/mL; Sigma-Aldrich) at 37°C in an atmosphere of 5% CO2. Samples used were >90% B cells as determined by CD19 surface staining and fluorescence-activated cell sorting analyses. For those samples with <90% B cells, negative selection was applied to deplete non–B cells using B cell Isolation Kit II (Miltenyi Biotec, Inc.) according to the manufacturer-suggested protocol. Briefly, isolated mononuclear cells were counted, and centrifuged to get the pellet. Cells (108) were resuspended in 400 μL of cold MACS buffer (pH 7.2; PBS, supplemented with 0.5% bovine serum albumin and 2 mmol/L EDTA). Biotin-antibody cocktail (100 μL) was added and mixed with the cell suspension before incubating at 4°C for 10 min. Buffer (300 μL) and antibiotin microbeads (200 μL) were then added and the cell suspension was mixed well and incubated for an additional 15 min at 4°C. Cells were then washed with 10× volume of MACS buffer and pelleted off at 300 × g for 10 min. Cells were resuspended in 500 μL of MACS buffer and separated by MS magnetic column.
Normal B lymphocytes were separated using positive selection. Briefly, leukocyte filters were obtained from the Red Cross and back-flushed with sterile PBS to obtain the cells. The cells were then separated by density gradient centrifugation using Ficoll. The leukocyte fraction was washed twice with PBS and resuspended. B cells were depleted with CD19 antibody–labeled beads (Miltenyi Biotec), which uses a direct magnetic labeling system for the isolation of B cells from human peripheral blood mononuclear cells. Because these cells were derived from the peripheral blood of normal volunteers, they were predominantly CD19+ memory B cells. The T cells used in this study were obtained by incubating mononuclear cell isolates from healthy volunteers with CD3 microbeads (Miltenyi Biotec). Magnetically labeled cells were isolated by passing the cell suspension over an LS column (Miltenyi Biotec) with a purity of >80% in pilot experiments done by our laboratory.
The previously characterized CLL cell line, WaC3CD5 (15), was maintained in RPMI 1640 (Invitrogen) supplemented with 10% heat-inactivated FBS (Invitrogen), 2 mmol/L of L-glutamine (Invitrogen), penicillin (100 units/mL), and streptomycin (100 μg/mL) at 37°C in a humidified incubator with 5% CO2.
RNA isolation and reverse transcription-PCR
Total RNA was isolated using the guanidinium isothiocyanate-acid phenol method (16). The RNA was treated with DNase I using standard protocols. DNase-treated RNA (3 μg) was reverse-transcribed using random hexamers and Moloney murine leukemia virus reverse transcriptase according to the instructions in the Gene-Amp RNA PCR kit (Perkin-Elmer). An aliquot of the cDNA was used for reverse transcription-PCR (RT-PCR) with primers specific for PTPRO (PTPROt-F: GGGGATGCTTCACCTGCTTA and PTPRO-R: ACCATTGTTGAGACGGCTATGAACG) or the internal control 18S rRNA (18S-F: TCAAGAACGAAAGTCGGAGG and 18S-R: GGACATCTAAGGGCATCACA). The qualitative PCR (cell lines) was done by using cDNA equivalent to 150 ng (for 18S rRNA) and 300 ng (for PTPRO) of RNA. Each 25 μL reaction mix consisted of 1× ThermoPol buffer (New England Biolabs), 0.2 mmol/L of deoxynucleo-tide triphosphate, 10 pmol of each primer, and 1 unit of Taq DNA polymerase (New England Biolabs). The cycling conditions were as follows: denaturation at 94°C, annealing at 54.5°C (for PTPRO) or 65°C (for 18S rRNA) and extension at 72°C with a total of 32 cycles (for PTPRO) and 25 cycles (for 18S rRNA). The PCR products were separated on 1.5% agarose stained with ethidium bromide and imaged under UV light using Kodak Digital Science 1D software. The SYBR Green real-time PCR was done using cDNA equivalent to 12 ng (for 18S rRNA) and 150 ng (for PTPRO) of RNA. The 10 μL reaction done using SYBR Green Master mix (Applied Biosystems) was run on Mx3000P (Stratagene). The data, represented as copies of PTPRO normalized to copies of 18S rRNA, were calculated using standard curves generated by serial dilutions of cloned PTPRO or 18S rRNA cDNA.
Bisulfite conversion and methylation-specific PCR
Isolation of genomic DNA and treatment with sodium bisulfite were done according to protocols optimized in our laboratory (17–19). The methylation-specific PCR (MSP) was done as described earlier (13). Briefly, bisulfite-converted DNA was used for PCR amplification with nonmethylation-specific primers (hPTP-UF: 5′-ATGTTTTTGGAG-GATTTTGGGT-3′and hPTP-UR: 5′-ATACCCCATCACTACACAAACA-3′) and methylation-specific primers (hPTP-MF: 5′-CGTTTTTGGAG-GATTTCGGGC and hPTP-MR: 5′-AAAACACGACTACGCTAACG-3′). An aliquot of the PCR product was separated on 2.5% agarose. The ethidium bromide–stained DNA was visualized under UV light and captured using a Kodak 1D imaging system.
Combined bisulfite restriction analysis and bisulfite sequencing
Nested PCR (as described earlier; ref. 13) was done on bisulfite-converted DNA to amplify the PTPRO CpG island from −208 to +236 bp with respect to the transcription start site. The PCR product was purified using a gel extraction kit (Qiagen). The purified PCR product was used either for combined bisulfite restriction analysis (COBRA) or bisulfite sequencing. For COBRA, an aliquot of the PCR product was incubated at 65°C in the presence or absence of TaqI for 1 h in a reaction containing 1× React 2 (Invitrogen). The entire digestion mix was loaded onto 2.5% agarose and the ethidium bromide–stained DNA visualized under UV light was captured by using a Kodak 1D imaging system. For semiquantitative COBRA, bisulfite-converted DNA from a completely methylated sample (CpGenome; Chemicon, Int.) and another from an unmethylated sample (a cell line in which the PTPRO CpG island was not methylated) were used. These DNA were mixed in different proportions so as to obtain a series of samples with a known percentage of methylation at the PTPRO CpG island (used as standards). COBRA was done on these standards as well as the primary cells using TaqI as the restriction enzyme (as described above). A plot of percentage methylation (known) versus the fraction of PCR product digested by TaqI (M/U + M) was then used to compute the percentage of methylation in the normal/CLL sample. For bisulfite sequencing, the purified PCR product was cloned into pDrive vector according to the PCR cloning kit (Qiagen). A few randomly selected clones were subjected to automated sequencing. Direct sequencing was done using the Thermo Sequenase Radiolabeled terminator cycle sequencing kit with the primer hGlepp1-BS-F3 (5′-TAGGGGGATTGGAAAGGTAG-3′) following the protocol of the manufacturer.
Construction of Flag-tagged PTPROt expression vector
The wild-type (WT) and catalytic site (CS) mutant of the PTPU2L gene was used as a template to PCR-amplify PTPROt using the primers PTPt-EcoRI (5′-ATGAATTCCAATGGTTACAGAGATGA-3′) and PTP-R-Bam (5′-CTGGATCCCTTGCTAACATTCTCG-3′). The PCR product digested with EcoRI and BamHI was cloned into the same site of p3XFLAG-CMV-14 expression vector (Sigma). The PTPROt gene along with the Flag epitope at the 3′-end, excised from the recombinant plasmid by digestion with NotI and SphI, was subcloned into the NotI and BamHI sites of pRetro-On plasmid.
Generation of WaC3CD5 cells stably expressing PTPROt
Exponen-entially growing Phoenix amphotropic cells were transfected with either pRetro-On (vector control) or PTPRO (WT and CS) cloned in pRetro-On using calcium phosphate coprecipitation (20, 21). Forty-eight hours after transfection, 1.0 mL of the viral supernatant was mixed with 1.0 mL of WaC3CD5 cells (3 × 105 cells/mL) and 16 μg of polybrene in each well of a six-well plate. The plate was centrifuged at 2,300 rpm for 90 min at room temperature in a Beckman GPH rotor 3.7. Following centrifugation, 2.0 mL of growth media (RPMI 1640, 2 mmol/L glutamine, 10% FBS, and penicillin-streptomycin) was added to each well and cells were incubated for 24 h. The viral particles were then washed and cells were allowed to grow in growth media for 48 h followed by selection in media containing 2 μg/mL of puromycin. During the selection process, viable cells were separated from cell debris using Histopaque-1077 (Sigma) according to the protocol of the manufacturer. The selected cells were subjected to doxycycline induction for 24 and 48 h. Expression of the flag-tagged protein was monitored by Western blot analysis using anti-Flag M2 antibody (Sigma). There was, however, no difference between basal and induced expression (data not shown). Individual clones were then selected by limited dilution and used in the absence of doxycycline.
3-(4,5-Dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay to assess the effect of fludarabine on the growth of WaC3CD5 cells
Wac3CD5 cells (vector control and PTPROt expressing) were seeded in 96-well plate at 3 × 105 cells/100 μL in each well. The cells were either left untreated or were treated with 1, 2, and 5 μmol/L of F-Ara-A (obtained commercially from Berlex Oncology) for 48 h. The 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay–based Cell Proliferation kit II (Roche) was used according to the manufacturer’s protocol to measure the metabolic activity of the cells as an indicator of cell growth. The difference in growth of F-Ara-A–treated cells (as measured by the change in absorbance of the solubilized tetrazolium salt) is represented in arbitrary units relative to the respective untreated cells.
Statistical methods
Descriptive statistics are presented for the proportions of methylated samples, with exact binomial confidence intervals.
Results
PTPROt is methylated in CLL cell lines
We have previously reported that PTPRO is methylated and suppressed in primary rat hepatomas (12) and primary human hepatocellular carcinoma.5 Furthermore, we showed methylation and silencing of PTPRO in a large number of human lung cancer cell lines and in primary human lung tumors (13). Functional studies in a non–small cell lung cancer cell line also suggested the potential of this tyrosine phosphatase as a growth suppressor. B lymphocytes express PTPROt, a tissue-specific isoform of PTPRO, which is involved in cell cycle arrest (14). We studied the expression and methylation of PTPRO in the three previously published CLL cell lines WaC3CD5, Mec1, and Mec2 by subjecting total RNA to RT-PCR using primers specific for the full-length and truncated isoforms of PTPRO. PTPROt was only expressed in Mec2 but not in WaC3CD5 and Mec1 (Fig. 1A, left). As expected, the full-length isoform was not expressed in either of these cell lines (data not shown). We next did bisulfite genomic sequencing to determine whether the CpG island located in the promoter of PTPRO was differentially methylated in these cells. For this purpose, primers with no CpG bias were used to amplify the PTPRO CpG island spanning −208 to +236 from bisulfite-treated DNA in which all unmethylated cytosines were converted to thymines. Cloning of the PCR products and sequencing of a few randomly selected clones showed dense methylation of CpG dinucleo-tides between +105 and +168 in all three cell lines, with pockets of methylation in the upstream region spanning the promoter only in the nonexpressing Mec1 and WaC3CD5 cells (Fig. 1B). There was also distinct hypomethylation of the three CpG dinucleotides between +140 and +154 in the expressing Mec2 cells. The nonexpressing Mec1 and WaC3CD5 cells were then treated with the hypomethylating agent 5-Aza-deoxycytidine (decitabine), which induces degradation of the maintenance DNA methyltransferase Dnmt1, resulting in the loss of its activity (22). Analysis of PTPROt expression by RT-PCR showed reexpression of the gene upon treatment of WaC3CD5 cells with 5-AzaC, whereas expression of the phosphatase gene was not affected by this treatment in Mec1 cells (Fig. 1A, right; and data not shown). It is conceivable that one or more of the key transcription factors involved in PTPROt expression are lacking in Mec1 cells. To determine whether the reexpression of PTPROt was associated with demethylation of the PTPRO CpG island, and was not simply an effect secondary to the activation of a transcriptional regulator, we analyzed methylation status of the PTPRO CpG island in untreated cells as well as in cells treated with 5-AzaC by COBRA. The 440 bp region of the PTPRO CpG island amplified from bisulfite-converted DNA contains one TaqI site that would be retained only if the original DNA was methylated. Amplification of the CpG island in cells exhibiting CpG methylation followed by its digestion with TaqI will result in two smaller fragments that can be visualized by agarose gel electrophoresis. This assay showed the dose-dependent reduction of methylation at the TaqI site in the PTPRO CpG island in WaC3CD5 cells upon AzaC treatment (Fig. 1C), further reinforcing the role of methylation in regulating the expression of PTPROt in these cells. Direct sequencing done on the pooled PCR product also showed demethylation at other CpG dinucleotides within the CpG island (Fig. 1D).
Fig. 1.
Expression and methylation of PTPROt in leukemia cell lines. A, total RNA from three CLL cell lines was used for RT-PCR with primers specific for PTPROt.18S rRNA was used as a loading control. Total RNA from brain was used as a positive control. B, the PTPRO CpG island was amplified from bisulfite-converted DNA. The PCR product was cloned in aTA cloning vector and 9 to 10 clones were subjected to automated sequencing. The nucleotide positions of each cytosine (with respect to the transcription start site) in a CpG dinucleotide context within the region analyzed are denoted. Filled boxes, methylated cytosines. C, the PTPRO CpGisland was amplified from bisulfite-treated DNA ofWaC3CD5 cells (control or treated with different doses of 5-AzaC for 10 d) using primers devoid of CpG bias. The PCR product was digested withTaqI (to test for methylation) or withTsp509 I (to test for complete bisulfite conversion), separated on agarose gel and stained with ethidium bromide. D, the bisulfite PCR product ofWaC3CD5 cells (control and treated with 5.0 μmol/L 5-AzaC for 10 d) was subjected to sequencing using theThermo Sequenase RadiolabeledTerminator Sequencing kit. The presence of thymine and cytosine at the same position (arrows) in the 5-AzaC – treated sample denotes partial demethylation of the cytosine.
Methylation and expression of PTPRO in human primary CLL
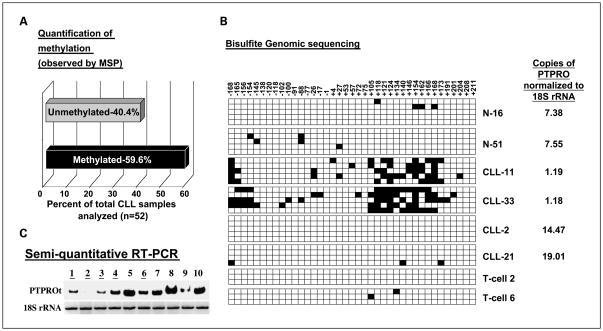
Several groups have noted relatively high levels of methylation in cell lines as compared with primary tumor cells. Therefore, we determined if methylation of PTPROt occurred in primary CLL cells. We initially analyzed 52 CLL samples for PTPRO methylation by MSP. Genomic DNA from these samples was treated with a bisulfite reagent to convert all unmethylated cytosines to thymines. Primers specific for unmethylated and methylated DNA were then used to amplify the respective DNA populations. The results showed that 59.6% of the CLL samples (n = 52) were methylated whereas the normal B lymphocytes (n = 9) were essentially methylation-free (Fig. 2A). To establish the methylation status of PTPRO gene, we did bisulfite genomic sequencing on two samples from each group (normal B lymphocytes, unmethylated CLL, and methylated CLL). Patient samples (nos. 11 and 33) that were methylated by MSP showed high level of methylation between CpG sites +105 and +172, the region that was also densely methylated in WaC3CD5 cells (see Fig. 2B). As anticipated, the normal B lymphocytes (N-16 and N-51) and CLL samples that were unmethylated as shown by MSP (CLL-2 and CLL-21) showed no or very little methylation throughout the region analyzed (Fig. 2B). To rule out the possibility that methylation observed in the peripheral blood lymphocytes (PBL) of patients with CLL was contributed exclusively by the T cell population, we did bisulfite genomic sequencing of T cells (T cell 2 and T cell 6). The results (Fig. 2B) showed that the PTPRO CpG island is methylation-free in these cells, confirming that it is indeed methylated in the B cell population of a subset of patients with CLL.
Fig. 2.
Methylation and expression of PTPRO in primary human CLL. A, quantification of PTPROt methylation in primary human CLL samples as studied by MSP. B, bisulfite-treated DNA from PBLs of normal volunteers (N), PBLs of patients with primary CLL (CLL), andTcells of normal volunteers (T) were used for bisulfite genomic sequencing as described in Fig. 1B. The nucleotide positions of each cytosine (with respect to the transcription start site) in a CpG dinucleotide context within the region analyzed are denoted. Filled boxes, methylated cytosines. The expression of PTPROt in copies normalized to 18S rRNA is indicated next to each sample. C, total RNA from CD19+ selected B lymphocytes from five methylated (lanes1, 2, 3, 4, and 6) and five unmethylated (lanes 5, 7, 8, 9, and10) samples were used for radioactive RT-PCR with primers specific for PTPROt.18S rRNA was used as a loading control.
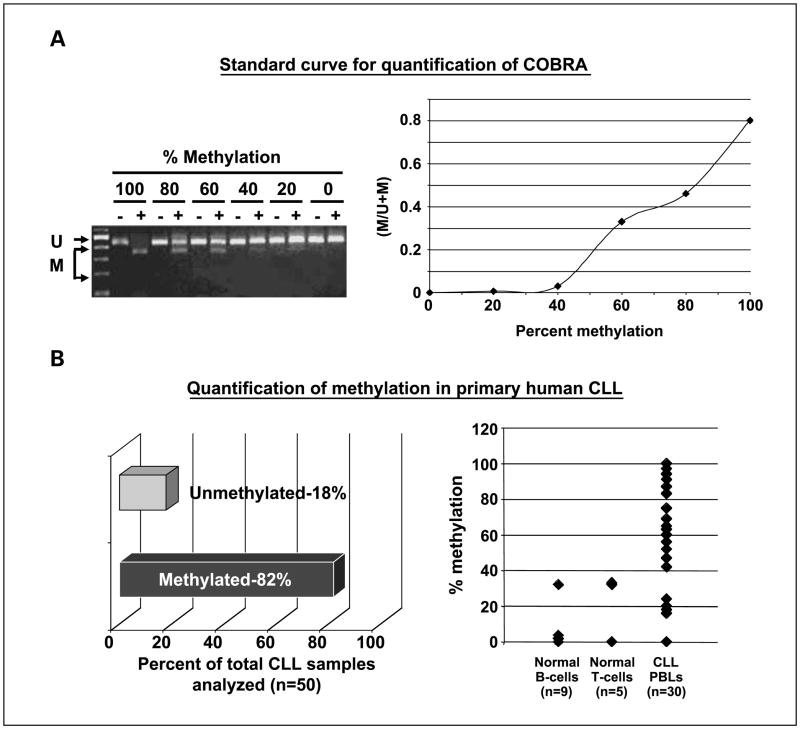
To determine if the methylation observed in CLL samples inversely correlates with the expression of PTPROt as observed in the WaC3CD5 CLL cell line, we studied its expression by semiquantitative radioactive RT-PCR in CD19-selected B lymphocytes from five methylated and five unmethylated samples. The results showed differential expression among these samples (Fig. 2C). The expression in the methylated samples was lower than that in the unmethylated samples. The aberrant PTPRO methylation in CLL, which inversely correlated with its expression, prompted more detailed examination of 50 additional CLL samples. Characterization of these samples from patients enrolled on the intergroup phase III trial (E2997) is detailed in Table 1. Here, we used quantitative COBRA to study the methylation of each sample to facilitate the quantification of methylation using a standard curve as described in Materials and Methods. As observed earlier, a large number of patients with CLL (41 of 50, 82%; 90% confidence interval, 71–90%) showed >40% methylation (cutoff was selected based on the linear range of the standard curve above the sensitive detection level of the assay; Fig. 3A and B). The discrepancy in the incidence of methylation between the two sets of samples (59.6% versus 82%) could be due to differences in the sensitivities of the assays or clinical features of the samples analyzed (see Discussion). We next determined whether PTPROt was also down-regulated in a majority of the CLL samples. For this purpose, we used SYBR Green real-time RT-PCR to measure the expression of PTPROt (normalized to 18S rRNA) in primary CLL samples (n = 37), which gave high quality RNA for the assay. CD19+ selected B lymphocytes (n = 3) and PBLs from normal individuals (n = 7) were used as control samples. The data (Fig. 4) showed an overall down-regulation of PTPROt. Twenty-nine methylated CLL samples exhibited drastically reduced expression of PTPROt. It is not known whether the two PTPRO-expressing samples exhibit distinct pathologic characteristics. PTPROt was also suppressed in six unmethylated samples. The latter samples could represent early stages of CLL in which PTPROt may be suppressed due to transcriptional repression prior to promoter methylation. It is interesting that five of these samples were indeed early stage disease, i.e., either Rai stage 1 or 2. These observations are consistent with the general down-regulation of expression of other tumor suppressor genes in tumors relative to matching normal cells/tissues, and suggest the functional significance of PTPROt methylation and suppression in CLL.
Table 1.
Biological characterization of the B-CLL panel (n = 50) derived from E2997
| Rai stage | |
| 0 | 1 |
| 1 | 10 |
| 2 | 17 |
| 3 | 9 |
| 4 | 12 |
| Mutated (<98% homology with germ line) | 9 |
| Unmutated (≥98% homology with germ line) | 37 |
| del(17p) | 4 |
| del(11q) | 11 |
| del(6q) | 3 |
| t(12) | 10 |
| Normal | 9 |
| del(13q) | 13 |
| CD38 < 30% | 21 |
| CD38 ≥ 30% | 28 |
| Median CD38 (range) | 36% (0–99%) |
| Median immunoblot ZAP-70 (range) | 0.291 (0.000–2.215) |
| Median flow ZAP-70 (range) | 0.31 (0.09–0.80) |
Fig. 3.
Quantification of PTPRO methylation in primary CLL cells by semiquantitative COBRA. A, COBRA was done withTaqI restriction enzyme on unmethylated and methylated DNA mixed in known proportions (left). The standard curve derived from this experiment was used to estimate the methylation level in each CLL sample (right). B, genomic DNA from B cells, Tcells, and PBLs from patients with CLL was used for COBRA. DNA was treated with bisulfite reagent followed by PCR amplification of the PTPRO CpG island using unbiased primers. The PCR product was then digested withTaqI. Any digested product would indicate that the original DNA was methylated. Quantification of the percentage of CLL samples that were qualitatively unmethylated or methylated (left); quantification of the percentage methylation at the PTPRO CpG island within each sample, as determined by semiquantitative COBRA (right).
Fig. 4.
Real-time RT-PCR for the expression of PTPROt. Total RNA isolated from normal B cells and PBLs from normal individuals and patients with CLL were used for real-time RT-PCR with SYBR Green chemistry.18S rRNAwas used as an internal control.
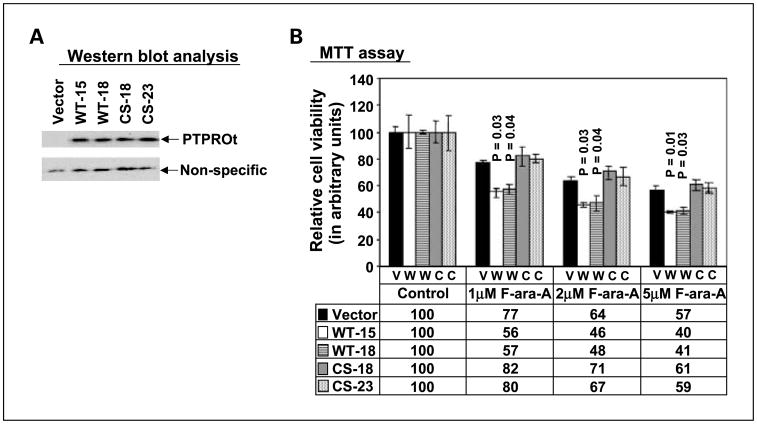
Ectopic expression of PTPROt in a CLL cell line increases its susceptibility to fludarabine
The function of PTPROt in CLL and other B cell diseases has not been characterized. Mechanistic interrogation of PTPROt in the WaC3CD5 cell line was pursued by ectopic expression of flag-tagged PTPROt. We also expressed the catalytically inactive C → S mutant (CS mutant) to determine whether phosphatase activity was necessary for the function of PTPROt. It is noteworthy that the expression of WT protein was low in all clones compared with the CS clones, some of which showed high level of expression (data not shown). Only clones with a comparable expression of WT and CS mutant PTPROt were used for further studies (Fig. 5A). Comparable growth kinetics of the parent cell line, wild-type and mutant PTPROt–transfected cell lines suggested limited influence of PTPROt (when expressed at low levels) on the proliferation rate of CLL cells. We hypothesized that the presence of PTPRO may positively influence drug-induced growth inhibition. To test this possibility, the vector-transfected and PTPROt-expressing cells were treated with different doses of fludarabine, a purine analogue commonly used in CLL therapy. The proliferation of cells was assessed by a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazo-lium bromide assay, which was used as an index of active cellular metabolism. The data (Fig. 5B) shows the growth inhibition of the PTPROt WT–expressing cells (relative to the vector transfected controls) with increasing fludarabine dose. Furthermore, the mutant-expressing cells were not significantly affected by the treatment. This study shows that expression of PTPROt (as observed in normal B lymphocytes) may enhance growth inhibition promoted by fludarabine.
Fig. 5.
Effect of ectopic expression of PTPROt on fludarabine sensitivity of the CLL cell lineWaC3CD5. A, ectopic expression of PTPROt in the CLL cell line WaC3CD5 (two clones each ofWTand CS) was monitored by subjecting whole cell extracts from vector-transfected and PTPROt-expressing cells toWestern blot analysis with anti-Flag antibody. Equal amounts of a nonspecific band detected by the same antibody indicates equal protein loading. B, WaC3CD5 cells (vector, WT, and CS) were treated with different concentrations of F-Ara-A for 48 h. Cell viability was assessed by 3-(4,5-dime-thylthiazol-2-yl)-2,5-diphenyltetrazolium bromide assay and is expressed in arbitrary units relative to control (untreated) cells.
Discussion
Although protein tyrosine kinases have long been recognized as key players in oncogenesis, the potential role of protein tyrosine phosphatases in the initiation and progression of cancer is gaining increasing recognition (see refs. 23–25 for recent reviews). We have shown, for the first time, the methylation-mediated suppression of PTPROt in CLL. This is the first detailed report analyzing the methylation and suppression of a protein tyrosine phosphatase in a large cohort of patients. Using MSP, COBRA, and bisulfite sequencing we have shown that this gene is methylated in a large subset of patients with CLL but not in normal B or T lymphocytes. It is worth mentioning that the clinically unselected series of patients (first set of 52 samples) showed a lower frequency of methylation compared with the clinically selected series of patients (second set of 50 patients) meeting the National Cancer Institute criteria to initiate therapy. Therefore, it is possible that PTPRO methylation is associated with clinically aggressive disease. A detailed analysis of samples from consecutive and clinically unselected CLL series would be required to address this issue, which is beyond the scope of this article. It is noteworthy that PTPROt expression is also suppressed in a large population of CLL samples. Expression of PTPROt, which is predominantly expressed in B lymphocytes, is known to be developmentally regulated with the highest expression observed in naïve B cells (14). It is interesting that leukemia clones from about half of the patients represent an expansion of postgerminal center memory B cells (26, 27), in which expression of PTPROt is reduced (14). These observations suggest the importance of PTPROt silencing in the pathogenesis of CLL.
The PTPRO gene also encodes a larger isoform (PTP-U2/Glepp1) that is primarily expressed in the brain and kidney in which it plays a role in neuronal axon growth/guidance and glomerular filtration, respectively (28–30). An earlier report from our laboratory has shown the growth-suppressive and proapoptotic potential of this isoform in lung cancer in which it was also methylated and suppressed (13). It is noteworthy that ectopic expression of PTPROt in a nonexpressing CLL cell line leads to increased growth inhibition when cells are exposed to fludarabine. This observation probably accounts for the reduced sensitivity of proliferating clonal CLL cell lines to fludarabine-based therapy in CLL. Examination of PTPROt expression in a large subset of uniformly treated patients with CLL will, however, be required to confirm the importance of this gene in predicting the response to therapy.
In addition to its significance as a diagnostic and prognostic marker, PTPROt may be used as a therapeutic target to increase the sensitivity of cytotoxic drugs in clinical use. Because the gene is suppressed by methylation in a majority of patients, use of epigenetic drugs that are rapidly gaining importance and application in the clinic for a variety of cancers can be used to induce its expression. The present study has shown that the gene can be reactivated by demethylation in the CLL cell line WaC3CD5. Furthermore, the effect of PTPROt expression on the action of fludarabine also opens up a new avenue to explore combination therapy with epigenetic drugs, especially in resilient cases in which PTPROt is suppressed. It is interesting that most classical protein tyrosine phosphatases harbor a CpG island (31). Although methylation of only a few protein tyrosine phosphatases has been reported in human cancer (see refs. 24, 31 for reviews), the presence of a CpG island in the regulatory regions of other protein tyrosine phosphatases suggests that the expression of at least some of these genes may also be regulated by an epigenetic mechanism and could be potential targets for cancer therapy.
The altered methylation profile of PTPRO and its expression in some primary CLL has other important functional implications. Constitutively active signaling pathways and a relationship between low apoptotic index and up-regulated tyrosine phosphorylated proteins have been reported in CLL (26, 32, 33). These studies, therefore, point towards the role of deregulated tyrosine phosphorylation and subsequent signal transduction pathways in the etiology of B-CLL. In this context, there are several reports on aberrant up-regulation of ZAP-70, a T lymphocyte tyrosine kinase (34), in a subset of patients with CLL with unmutated IgVH status (35) as well as the up-regulation and constitutive activation of the Src kinase Lyn (36) in CLL. Such up-regulation and/or activation could be a result of the altered tyrosine phosphorylation of these enzymes. The identification of these or other CLL-specific phosphoproteins that are potential substrates of PTPROt could thus provide additional downstream targets for drug development. Indeed, a recent report has revealed that the protein tyrosine kinase SYK, a mediator of signaling through the B cell receptor, is one such substrate (37). It would be of considerable interest to delineate the role of altered phosphorylation of the PTPROt substrates in CLL tumorigenesis.
Acknowledgments
Grant support: Leukemia and Lymphoma Society of America, R21CA110496 (J.C. Byrd), CA 86978 and CA101956 (S.T. Jacob), and the D. Warren Brown Foundation.
Footnotes
Ghoshal K, Majumder S, DattaJ, Kuttay H, Frankel W, Jacob ST, unpublished data.
References
- 1.Munk Pedersen I, Reed J. Microenvironmental interactions and survival of CLL B-cells. Leuk Lymphoma. 2004;45:2365–72. doi: 10.1080/10428190412331272703. [DOI] [PubMed] [Google Scholar]
- 2.Caligaris-Cappio F, Hamblin TJ. B-cell chronic lymphocytic leukemia: a bird of a different feather. J Clin Oncol. 1999;17:399–408. doi: 10.1200/JCO.1999.17.1.399. [DOI] [PubMed] [Google Scholar]
- 3.Vuillier F, Claisse JF, Vandenvelde C, et al. Evaluation of residual disease in B-cell chronic lymphocytic leukemia patients in clinical and bone-marrow remission using CD5–19 markers and PCR study of gene rearrangements. Leuk Lymphoma. 1992;7:195–204. doi: 10.3109/10428199209053623. [DOI] [PubMed] [Google Scholar]
- 4.Robertson LE, Huh YO, Butler JJ, et al. Response assessment in chronic lymphocytic leukemia after fludarabine plus prednisone: clinical, pathologic, immunophenotypic, and molecular analysis. Blood. 1992;80:29–36. [PubMed] [Google Scholar]
- 5.Keating MJ, Chiorazzi N, Messmer B, et al. Biology and treatment of chroniclymphocyticleukemia. Hematology Am Soc Hematol Educ Program. 2003:153–75. doi: 10.1182/asheducation-2003.1.153. [DOI] [PubMed] [Google Scholar]
- 6.Messmer BT, Messmer D, Allen SL, et al. In vivo measurements document the dynamic cellular kinetics of chronic lymphocytic leukemia B cells. J Clin Invest. 2005;115:755–64. doi: 10.1172/JCI23409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rush LJ, Raval A, Funchain P, et al. Epigenetic profiling in chronic lymphocytic leukemia reveals novel methylation targets. Cancer Res. 2004;64:2424–33. doi: 10.1158/0008-5472.can-03-2870. [DOI] [PubMed] [Google Scholar]
- 8.Raval A, Lucas DM, Matkovic JJ, et al. TWIST2 demonstrates differential methylation in immunoglobulin variable heavy chain mutated and unmutated chronic lymphocytic leukemia. J Clin Oncol. 2005;23:3877–85. doi: 10.1200/JCO.2005.02.196. [DOI] [PubMed] [Google Scholar]
- 9.Yuille MR, Condie A, Stone EM, et al. TCL1 is activated by chromosomal rearrangement or by hypomethylation. Genes Chromosomes Cancer. 2001;30:336–41. doi: 10.1002/gcc.1099. [DOI] [PubMed] [Google Scholar]
- 10.Packham G, Stevenson FK. Bodyguards and assassins: Bcl-2 family proteins and apoptosis control in chronic lymphocytic leukaemia. Immunology. 2005;114:441–9. doi: 10.1111/j.1365-2567.2005.02117.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hanada M, Delia D, Aiello A, Stadtmauer E, Reed JC. bcl-2 gene hypomethylation and high-level expression in B-cell chronic lymphocytic leukemia. Blood. 1993;82:1820–8. [PubMed] [Google Scholar]
- 12.Motiwala T, Ghoshal K, Das A, et al. Suppression of the protein tyrosine phosphatase receptor type O gene (PTPRO) by methylation in hepatocellular carcinomas. Oncogene. 2003;22:6319–31. doi: 10.1038/sj.onc.1206750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Motiwala T, Kutay H, Ghoshal K, et al. Protein tyrosine phosphatase receptor-type O (PTPRO) exhibits characteristics of a candidate tumor suppressor in human lung cancer. Proc Natl Acad Sci U S A. 2004;101:13844–9. doi: 10.1073/pnas.0405451101. Epub 2004 Sep 8. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 14.Aguiar RC, Yakushijin Y, Kharbanda S, et al. PTPROt: an alternatively spliced and developmentally regulated B-lymphoid phosphatase that promotes G0/G1 arrest. Blood. 1999;94:2403–13. [PubMed] [Google Scholar]
- 15.Wendel-Hansen V, Sallstrom J, De Campos-Lima PO, et al. Epstein-Barr virus (EBV) can immortalize B-cll cells activated by cytokines. Leukemia. 1994;8:476–84. [PubMed] [Google Scholar]
- 16.Chomczynski P, Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987;162:156–9. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- 17.Majumder S, Ghoshal K, Li Z, Jacob ST. Hypermethylation of metallothionein-I promoter and suppression of its induction in cell lines overexpressing the large subunit of Ku protein. J Biol Chem. 1999;274:28584–9. doi: 10.1074/jbc.274.40.28584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Majumder S, Ghoshal K, Li Z, Bo Y, Jacob ST. Silencing of metallothionein-I gene in mouse lymphosarcoma cells by methylation. Oncogene. 1999;18:6287–95. doi: 10.1038/sj.onc.1203004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ghoshal K, Majumder S, Li Z, Dong X, Jacob ST. Suppression of metallothionein gene expression in a rat hepatoma because of promoter-specific DNA methylation. J Biol Chem. 2000;275:539–47. doi: 10.1074/jbc.275.1.539. [DOI] [PubMed] [Google Scholar]
- 20.Majumder S, Ghoshal K, Summers D, et al. Chromium(VI) down-regulates heavy metal-induced metallothionein gene transcription by modifying trans-activation potential of the key transcription factor, metal-responsive transcription factor 1. J Biol Chem. 2003;278:26216–26. doi: 10.1074/jbc.M302887200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Majumder S, Ghoshal K, Gronostajski RM, Jacob ST. Downregulation of constitutive and heavy metal-induced metallothionein-I expression by nuclear factor I. Gene Expr. 2001;9:203–15. doi: 10.3727/000000001783992588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ghoshal K, Datta J, Majumder S, et al. 5-Aza-deoxycytidine induces selective degradation of DNA methyltransferase 1 by a proteasomal pathway that requires the KEN box, bromo-adjacent homology domain, and nuclear localization signal. Mol Cell Biol. 2005;25:4727–41. doi: 10.1128/MCB.25.11.4727-4741.2005. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Motiwala T, Jacob ST. Role of protein tyrosine phosphatases in cancer. Prog Nucleic Acid Res Mol Biol. 2006;81:297–329. doi: 10.1016/S0079-6603(06)81008-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Tonks NK. Protein tyrosine phosphatases: from genes, to function, to disease. Nat Rev Mol Cell Biol. 2006;7:833–46. doi: 10.1038/nrm2039. [DOI] [PubMed] [Google Scholar]
- 25.Ostman A, Hellberg C, Bohmer FD. Protein-tyrosine phosphatases and cancer. Nat Rev Cancer. 2006;6:307–20. doi: 10.1038/nrc1837. [DOI] [PubMed] [Google Scholar]
- 26.Laytragoon-Lewin N, Rossmann ED, Castro J, Mellstedt H. Significance of phosphotyrosine proteins, Bcl-2 and p53 for apoptosis in resting B-chronic lymphocytic leukemia (CLL) cells. Int J Cancer. 2002;97:344–8. doi: 10.1002/ijc.1616. [DOI] [PubMed] [Google Scholar]
- 27.Fais F, Ghiotto F, Hashimoto S, et al. Chronic lymphocytic leukemia B cells express restricted sets of mutated and unmutated antigen receptors. J Clin Invest. 1998;102:1515–25. doi: 10.1172/JCI3009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wharram BL, Goyal M, Gillespie PJ, et al. Altered podocyte structure in GLEPP1 (Ptpro)-deficient mice associated with hypertension and low glomerular filtration rate. J Clin Invest. 2000;106:1281–90. doi: 10.1172/JCI7236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Stepanek L, Stoker AW, Stoeckli E, Bixby JL. Receptor tyrosine phosphatases guide vertebrate motor axons during development. J Neurosci. 2005;25:3813–23. doi: 10.1523/JNEUROSCI.4531-04.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Beltran PJ, Bixby JL, Masters BA. Expression of PTPRO during mouse development suggests involvement in axonogenesis and differentiation of NT-3 and NGF-dependent neurons. J Comp Neurol. 2003;456:384–95. doi: 10.1002/cne.10532. [DOI] [PubMed] [Google Scholar]
- 31.Jacob ST, Motiwala T. Epigenetic regulation of protein tyrosine phosphatases: potential molecular targets for cancer therapy. Cancer GeneTher. 2005;12:665–72. doi: 10.1038/sj.cgt.7700828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Frank DA, Mahajan S, Ritz J. B lymphocytes from patients with chronic lymphocytic leukemia contain signal transducer and activator of transcription (STAT) 1 and STAT3 constitutively phosphorylated on serine residues. J Clin Invest. 1997;100:3140–8. doi: 10.1172/JCI119869. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ringshausen I, Schneller F, Bogner C, et al. Constitutively activated phosphatidylinositol-3 kinase (PI-3K) is involved in the defect of apoptosis in B-CLL: association with protein kinase Cy. Blood. 2002;100:3741–8. doi: 10.1182/blood-2002-02-0539. [DOI] [PubMed] [Google Scholar]
- 34.Crespo M, Bosch F, Villamor N, et al. ZAP-70 expression as a surrogate for immunoglobulin-variable-region mutations in chronic lymphocytic leukemia. N Engl J Med. 2003;348:1764–75. doi: 10.1056/NEJMoa023143. [DOI] [PubMed] [Google Scholar]
- 35.Durig J, Nuckel H, Cremer M, et al. ZAP-70 expression is a prognostic factor in chronic lymphocytic leukemia. Leukemia. 2003;17:2426–34. doi: 10.1038/sj.leu.2403147. [DOI] [PubMed] [Google Scholar]
- 36.Contri A, Brunati AM, Trentin L, et al. Chronic lymphocytic leukemia B cells contain anomalous Lyn tyrosine kinase, a putative contribution to defective apoptosis. J Clin Invest. 2005;115:369–78. doi: 10.1172/JCI22094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Chen L, Juszczynski P, Takeyama K, Aguiar RC, Shipp MA. Protein tyrosine phosphatase receptor-type O truncated (PTPROt) regulates SYK phosphorylation, proximal B-cell-receptor signaling, and cellular proliferation. Blood. 2006;108:3428–33. doi: 10.1182/blood-2006-03-013821. [DOI] [PubMed] [Google Scholar]