One can think of the chromosomal translocation process as consisting of two DNA double-strand breaks (DSBs) followed by a joining of the resulting four DNA ends in the new (i.e., translocated) configuration 1. Two recent papers provide major new insights regarding the joining phase 2,3. Both papers use the most sophisticated approaches yet devised for how DNA ends are joined in chromosomal translocations.
Both studies find that chromosomal translocations are more common when NHEJ (nonhomologous DNA end joining) proteins are missing 2,3. The richness of the data in these papers, which is quite well-discussed within them (the reader is referred there), drives consideration of an even wider range of mechanistic possibilities than either paper has space to discuss. Both studies suggest that NHEJ suppresses chromosomal translocations. In addition to this direct interpretation, another possibility arises out of the fact that the two DSBs, on the two different chromosomes, do not usually occur at the same time (except in the less common case of off-target V(D)J recombination, where the two sites are enzymatically paired) 1. For most translocations, the DSB at one location has a high probability of being rejoined to restore the original chromosomal configuration prior to the time when a DSB at any other location is created. That is, only rarely are the two DSBs generated concurrently and with sufficient longevity to exist within the same time window. If the rejoining of either DSB is slowed, then this widens the time window before closure, thereby increasing the chance that both DSBs will be open at the same time and increasing the chance of a translocation. Hence, the increase in translocation frequency when NHEJ is inefficient, due to a missing or mutant component, may reflect increased encounters of concurrent DSBs. In short, slowed NHEJ would be expected to increase the time of overlap for which two breaks would be unrepaired, thereby increasing the chance of translocation. This interpretation would be equivalent to the impression that NHEJ suppresses translocations but might be stated slightly differently as, NHEJ reduces the temporal opportunity for translocations by reducing the time window during which two DSBs exist.
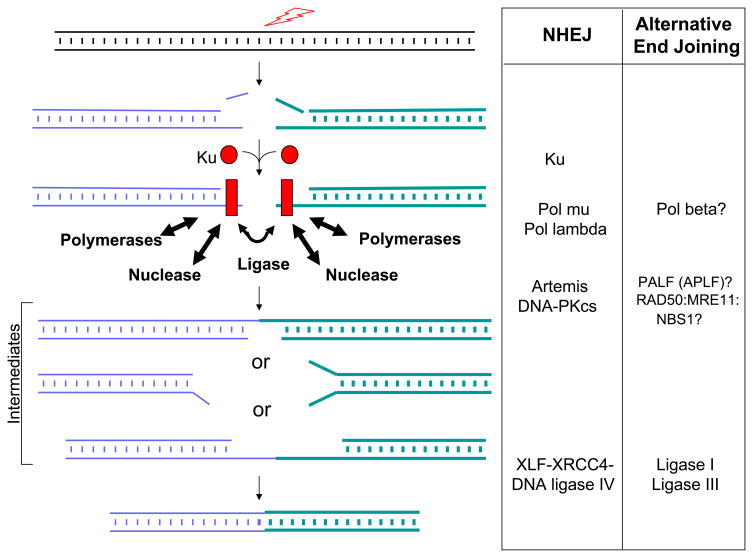
Both studies demonstrate once again that translocations can occur when one or more key NHEJ components are missing, verifying that some form of end joining can occur even under such adverse circumstances (Figure 1). This form of end joining has been given several designations in eukaryotes: alternative NHEJ (or simply, alternative EJ), back-up NHEJ, and microhomology-mediated NHEJ 4–7. However, there is little certainty about which enzymes participate in these end joining events. The relative kinetics of NHEJ versus any of these alternative end joining pathways is also a subject for continuing investigation. The earliest time points for end joining assays (~24 hrs) in cells lacking ligase IV suggest that alternative pathways are about 10-fold slower than NHEJ 4,8. In wild type cells, the fold difference between NHEJ and all alternative end joining pathways might be greater than 10-fold if the alternative9 pathways are employed only when the NHEJ pathway is mutated or missing one or more components. That is, there may be some DSB accumulation in cells missing the NHEJ pathway, and the joining of these accumulated DSBs may occur more slowly by a less efficient alternative end joining pathway. (There is a precedent for a backup NHEJ in prokaryotes, illustrating the importance of repairing DSBs throughout evolution10,11.)
Figure 1.
Components of NHEJ and Candidates for Alternative Pathways. In yeast, plants, and invertebrates, there is no Artemis or DNA-PKcs to serve as a nuclease complex, and RAD50:MRE11:XRS1 may play an equivalent role (XRS1 is the homologue of human NBS1). POL4 in yeast serves the role of pol mu and pol lambda in vertebrates, and these, along with pol beta, are all members of the POL X family of polymerases. The reader is referred elsewhere for discussions of homologues across prokaryotes and eukaryotes (Refs. 7 & 11).
Despite the richness of insight from these two papers 2,3 and earlier outstanding ones from these and other groups, the field lacks detail on the precise enzymatic components that function in the alternative end joining pathways. One can consider that end joining relies on a nuclease to remove damaged DNA, a polymerase to fill-in, and a ligase to join. There is mounting evidence that the RAD50:MRE11:NBS1 complex may function in a subset of alternative end joining events, but the rules governing such possible participation are unclear, nor is it clear what role this complex might actually serve 12–15.
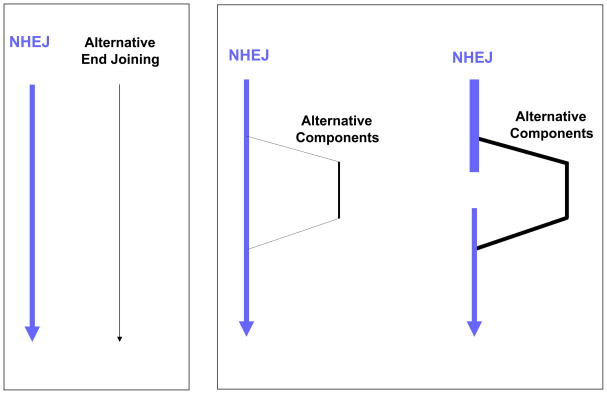
A minority opinion (this author) is that NHEJ, even without Ku and the XRCC4:DNA ligase IV complex, might still function with substitutions for the ‘first-string players’ (Figure 2). Other nucleases might substitute for the Artemis:DNA-PKcs nuclease, and ligase I or III might substitute for ligase IV7. It is clear that DNA ligases I and III can substitute for XRCC4:DNA ligase IV in purified biochemical end joining systems9,16, and there is even some in vivo data supporting the role of DNA ligase III17. Since these are the only three ligases in mammalian cells18, there only two choices for the back-up ligase when DNA ligase IV is missing. Ku does not have an enzymatic function but rather serves to improve the binding affinity of the polymerases (pol mu and pol lambda), the nuclease (Artemis:DNA-PKcs), and the XRCC4:DNA ligase IV complex 7. In both biochemical and in vivo systems, incompatible end joining can occur without Ku 9,19, but the joining is much less efficient9.
Figure 2.
Two Possible Relationships Between NHEJ and Alternative End Joining Pathways. In the left box, NHEJ and alternative end joining are depicted as two independent pathways. In the right box, NHEJ is viewed as the ‘first-string’ enzymes, with back-up enzymes filling-in when one or more primary NHEJ enzymes or components are mutated, missing (right side of right box), or perhaps even overwhelmed with a large number of DSBs.
There are several other interesting aspects to these studies. In the Jasin paper, they find junctional additions containing templated direct repeats or inverted repeats derived from either of the DNA strands (top or bottom) from any of the four DNA ends involved in the two DSBs 3. Junctional additions containing direct or inverted repeats from the adjacent DNA ends have been seen at human chromosomal translocation junctions and are called T-nucleotides20. T-nucleotides are suspected to be due to pol mu or pol lambda, either of which can slip on a template21,22. More importantly, the Jasin paper finds T-nucleotides from more than one genomic location3. They point out that this is likely one more facet to the iterative nature of NHEJ7. T-nucleotides from multiple and diverse genomic locations hint that DNA ends in a chromosomal translocation may sample a large fraction of the nuclear space in search of a partner to which they can join.
The participation of pol mu may explain one other perplexing aspect of these alternative end joinings (joins formed in the absence of Ku, XRCC4, or DNA ligase IV). Though alternative end joining junctions typically average ~2 nts of terminal microhomology, there can be a significant subset (0 to 30%) of junctions that have no microhomology. Yet joins by ligase I and III appear to rely more heavily on terminal microhomology in biochemical systems9,16. Thus, how can joining by ligase I or III occur without microhomology. As mentioned in the Jasin paper, the TdT-like activity of pol mu may create terminal microhomology, and this polymerase-generated microhomology would not score as microhomology (because it is equivalent to fill-in synthesis)7. In addition, pol mu may also have some ability to polymerize from one DNA end into another, if the receiving end is a 5′-overhang23. Like the TdT-like additions, this polymerase-generated microhomology would also be inapparent (‘occult’ microhomology, as some have called it24).
The detailed characterization of end joinings when NHEJ has been disabled or handicapped will be critical for determining the enzymes that participate in the alternative pathway(s). The Jasin and Alt systems continue to provide the experimental platforms for such continued important studies.
References
- 1.Tsai AG, Lieber MR. Mechanisms of chromosomal rearrangement in the human genome. BMC Genomics. 2010;11 (Suppl 1):S1. doi: 10.1186/1471-2164-11-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Boboila C, et al. Alternative end-joining catalyzes robust IgH locus deletions and translocations in the combined absence of ligase 4 and Ku70. Proc Natl Acad Sci U S A. 2010;107:3034–9. doi: 10.1073/pnas.0915067107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Simsek D, Jasin M. Alternative end-joining is suppressed by the canonical NHEJ component Xrcc4 ligase IV during chromosomal translocation formation. Nature Structure & Molecular Biology. 2010 doi: 10.1038/nsmb.1773. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yan CT, et al. IgH class switching and translocations use a robust non-classical end-joining pathway. Nature. 2007;449:478–482. doi: 10.1038/nature06020. [DOI] [PubMed] [Google Scholar]
- 5.Corneo B, et al. A mutation in RAG2 reveals robust, error-prone alternative end joining. Nature. 2007;449:483–486. doi: 10.1038/nature06168. [DOI] [PubMed] [Google Scholar]
- 6.Ma JL, Kim EM, Haber JE, Lee SE. Yeast Mre11 and Rad1 proteins define a Ku-independent mechanism to repair double-strand breaks lacking overlapping end sequences. Mol Cell Biol. 2003;23:8820–8828. doi: 10.1128/MCB.23.23.8820-8828.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lieber MR. The Mechanism of Double-Strand DNA Break Repair by the Nonhomologous DNA End-Joining Pathway. Annu Rev Biochem. 2010 doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Han L, Yu K. Altered kinetics of nonhomologous end joining and class switch recombination in ligase IV--deficient B cells. J Exp Med. 2008;205:2745–53. doi: 10.1084/jem.20081623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gu J, et al. XRCC4:DNA ligase IV can ligate incompatible DNA ends and can ligate across gaps. Embo J. 2007;26:1010–23. doi: 10.1038/sj.emboj.7601559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Aniukwu J, Glickman MS, Shuman S. The pathways and outcomes of mycobacterial NHEJ depend on the structure of the broken DNA ends. Genes Dev. 2008;22:512–27. doi: 10.1101/gad.1631908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gu J, Lieber MR. Mechanistic flexibility as a conserved theme across 3 billion years of nonhomologous DNA end-joining. Genes Dev. 2008;22:411–5. doi: 10.1101/gad.1646608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deriano L, Stracker TH, Baker A, Petrini JH, Roth DB. Roles for NBS1 in alternative nonhomologous end-joining of V(D)J recombination intermediates. Mol Cell. 2009;34:13–25. doi: 10.1016/j.molcel.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rass E, et al. Role of Mre11 in chromosomal nonhomologous end joining in mammalian cells. Nat Struct Mol Biol. 2009;16:819–24. doi: 10.1038/nsmb.1641. [DOI] [PubMed] [Google Scholar]
- 14.Xie A, Kwok A, Scully R. Role of mammalian Mre11 in classical and alternative nonhomologous end joining. Nat Struct Mol Biol. 2009;16:814–8. doi: 10.1038/nsmb.1640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dinkelmann M, et al. Multiple functions of MRN in end-joining pathways during isotype class switching. Nat Struct Mol Biol. 2009;16:808–13. doi: 10.1038/nsmb.1639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen L, Trujillo K, Sung P, Tomkinson AE. Interactions of the DNA ligase IV-XRCC4 complex with DNA ends and the DNA-dependent protein kinase. J Biol Chem. 2000;275:26196–205. doi: 10.1074/jbc.M000491200. [DOI] [PubMed] [Google Scholar]
- 17.Wang H, et al. DNA ligase III as a candidate component of backup pathways of nonhomologous end joining. Cancer Res. 2005;65:4020–30. doi: 10.1158/0008-5472.CAN-04-3055. [DOI] [PubMed] [Google Scholar]
- 18.Tomkinson AE, Vijayakumar S, Pascal JM, Ellenberger T. DNA ligases: structure, reaction mechanism, and function. Chem Rev. 2006;106:687–699. doi: 10.1021/cr040498d. [DOI] [PubMed] [Google Scholar]
- 19.Weinstock DM, Brunet E, Jasin M. Formation of NHEJ-derived reciprocal chromosomal translocations does not require Ku70. Nat Cell Biol. 2007;9:978–81. doi: 10.1038/ncb1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jaeger U, et al. Follicular lymphomas BCL-2/IgH junctions contain templated nucleotide insertions: novel insights into the mechanism of t(14;18) translocation. Blood. 2000;95:3520–3529. [PubMed] [Google Scholar]
- 21.Tippin B, Kobayashi S, Bertram JG, Goodman MF. To slip or skip, visualizing frameshift mutation dynamics for error-prone DNA polymerases. J Biol Chem. 2004;279:5360–5368. doi: 10.1074/jbc.M408600200. [DOI] [PubMed] [Google Scholar]
- 22.Blanca G, et al. Human DNA polymerases lambda and beta show different efficiencies of translesion DNA synthesis past abasic sites and alternative mechanisms for frameshift generation. Biochemistry. 2004;43:11605–15. doi: 10.1021/bi049050x. [DOI] [PubMed] [Google Scholar]
- 23.NickMcElhinny SA, et al. A gradient of template dependence defines distinct biological roles for family x polymerases in nonhomologous end joining. Mol Cell. 2005;19:357–366. doi: 10.1016/j.molcel.2005.06.012. [DOI] [PubMed] [Google Scholar]
- 24.Komori T, Okada A, Stewart V, Alt F. Lack of N regions in antigen receptor variable region genes of TdT-deficient lymphocytes. Science. 1993;261:1171–1175. doi: 10.1126/science.8356451. [DOI] [PubMed] [Google Scholar]