Abstract
Recent advances in mass spectrometry (MS) have provided means for large-scale phosphoproteomic profiling of specific tissues. Here, we report results from large-scale tandem MS [liquid chromatography (LC)-MS/MS]-based phosphoproteomic profiling of biochemically isolated membranes from the renal cortex, with focus on transporters and regulatory proteins. Data sets were filtered (by target-decoy analysis) to limit false-positive identifications to <2%. A total of 7,125 unique nonphosphorylated and 743 unique phosphorylated peptides were identified. Among the phosphopeptides identified were sites on transporter proteins, i.e., solute carrier (Slc, n = 63), ATP-binding cassette (Abc, n = 4), and aquaporin (Aqp, n = 3) family proteins. Database searches reveal that a majority of the phosphorylation sites identified in transporter proteins were previously unreported. Most of the Slc family proteins are apical or basolateral transporters expressed in proximal tubule cells, including proteins known to mediate transport of glucose, amino acids, organic ions, and inorganic ions. In addition, we identified potentially important phosphorylation sites for transport proteins from distal nephron segments, including the bumetanide-sensitive Na-K-2Cl cotransporter (Slc12a1 or NKCC2) at Ser87, Thr101, and Ser126 and the thiazide-sensitive Na-Cl cotransporter (Slc12a3 or NCC) at Ser71 and Ser124. A subset of phosphorylation sites in regulatory proteins coincided with known functional motifs, suggesting specific regulatory roles. An online database from this study (http://dir.nhlbi.nih.gov/papers/lkem/rcmpd/) provides a resource for future studies of transporter regulation.
Keywords: mass spectrometry, kidney, thiazide, sodium-chloride cotransporter
new developments in mass spectrometry (MS) have led to rapid progress in phosphoproteomic profiling of various tissues and cell types (17, 32, 51). In the kidney, studies of the phosphoproteome of inner medullary collecting duct cells (3, 22) and cultured cortical collecting duct (mpkCCD) cells (41), as well as native medullary thick ascending limb cells (18), have yielded new hypotheses about the vasopressin-mediated regulation of solute and water transport (20, 31). Here we extend the analysis to membrane proteins in the renal cortex. The renal cortex contains proximal tubules, cortical thick ascending limbs, distal convoluted tubules, connecting tubules, and cortical collecting ducts, as well as glomeruli. Among these, the proximal tubule is, on a volume basis, the dominant structure.
A biochemical protocol for isolation of cortical membranes by magnesium precipitation has been described (5) and extensively utilized to study transport function of the proximal tubule and its regulation. This technique has been used chiefly to study apical plasma membrane (“brush border”) proteins of the proximal tubule, which are highly enriched in the isolation process. Prior studies include general proteomic profiling (7, 49), but not phosphoproteomics. With the magnesium precipitation method, however, other types of membranes, including proximal tubule basolateral membranes (7, 47) and membranes from distal nephron segments, are moderately enriched as well. Thus membrane isolation from renal cortex potentially allows large-scale identification of phosphorylation sites in physiologically important transporters and associated regulatory proteins in the apical and basolateral plasma membranes of epithelial cells from proximal and distal nephron. In this study, we apply tandem MS [liquid chromatography (LC)-MS/MS] methods similar to those used in our prior studies of collecting duct (22) and thick ascending limb of Henle (18) to carry out large-scale identification of phosphorylation sites in renal cortical membrane proteins isolated by MgCl2 precipitation. A total of 743 unique phosphorylation peptides were identified. This study presents many previously unreported sites on a number of transporters critical to renal transport physiology and provides a database resource for future studies of renal epithelia and related epithelia in other tissues.
METHODS
Animals.
Pathogen-free 6- to 8-wk-old male Sprague-Dawley rats (Taconic Farms, Germantown, NY) were maintained on drinking water and ad libitum rat chow (NIH-07, Zeigler, Gardners, PA) in the Small Animal Facility of the Intramural Research Program at the National Institutes of Health. All experiments followed the animal protocol H-0110 as approved by the Animal Care and Use Committee of the National Heart, Lung, and Blood Institute.
Antibodies (listed by gene symbol).
Rabbit polyclonal antibodies against aquaporin-1 and -2 [Aqp1 (46) and Aqp2 (20)], Na/K pump Na-K-ATPase α1-subunit [Atp1a1 (37)], thiazide-sensitive Na-Cl cotransporter NCC [Slc12a3 (28)], bumetanide-sensitive Na-K-2Cl cotransporter NKCC2 [Slc12a1 (35)], ATP-binding cassette subfamily C, member 4 [Abcc4 (16)], and Na-Pi cotransporter NaPi2 [Slc34a1 (27)] were generated in our laboratory.1 The species-specific secondary antibodies conjugated with fluorophores were obtained from Rockland Immunochemicals (Gilbertsville, PA).
Homogenization and differential centrifugation.
Cortices from two rat kidneys were dissected, pooled, and homogenized for 15 s three times in 10 ml of cold sucrose buffer [10 mM triethanolamine, 250 mM sucrose, pH 7.6, 1× Complete Mini Protein inhibitor cocktail (Roche, Indianapolis, IN), and 1× HALT phosphatase inhibitor (Pierce, Rockford, IL)] and then subjected to differential centrifugation (47). Twenty milliliters of ultrapure water and 350 μl of MgCl2 were added to give a concentration of 12 mM to the homogenate, which was then vortexed. A Sorvall RC2-B refrigerated centrifuge was used to process the samples at 4,500 rpm for 15 min at 4°C. The supernatant was removed and further centrifuged at 16,000 rpm for 30 min. The resulting pellet was resuspended in 20 ml of 5 mM triethanolamine and 125 mM sucrose solution with the addition of 240 μl of 1 M MgCl2. The supernatant was centrifuged at 4,500 rpm for 15 min and again at 16,000 rpm for 30 min. The remaining pellet was resuspended in 20 ml of 5 mM triethanolamine and 125 mM sucrose solution and centrifuged at 16,000 rpm for 30 min. The final pellet contained the membrane fraction used for LC-MS/MS analysis. The samples were resuspended in 250 μl of Laemmli buffer (1.5% SDS, 10 mM Tris, pH 6.8) to a concentration of 1–2 μg/μl, while the samples for phosphoproteomic analysis were resuspended in 250 μl of 8 M urea, 75 mM NaCl, and 50 mM Tris·HCl. The protein concentration of the samples was determined by the bicinchoninic acid method (Pierce).
Immunoblot analysis.
Immunoblotting followed procedures described by Pisitkun et al. (38). Protein (20 μg) in Laemmli buffer was loaded onto a 4–20% gradient SDS-polyacrylamide gel, and electrophoresis was performed at 200 V. Proteins were then transferred onto a nitrocellulose membrane (0.2-μm pore size) under 250 mA for 1 h. After 1 h of incubation in Odyssey blocking buffer (LI-COR, Lincoln, NE), primary antibody was added to the membrane, which was incubated overnight. The membrane was washed four times using 1× PBS with 0.1% Tween 20 and then incubated for 1 h in secondary antibody. The membrane was washed four times with 1× PBS with 0.1% Tween 20 and, finally, rinsed with 1× PBS. The protein bands on the membrane were scanned using the LI-COR Odyssey scanner and further analyzed with Odyssey software version 2.1.
In-gel trypsin digestion for proteomic analysis.
Protein (∼170 μg) was loaded onto a 4–15% gradient SDS-polyacrylamide mini gel. Gel electrophoresis was performed for 70 min at 200 V. The gel was rinsed three times with water, stained with Imperial protein stain Pierce, and then destained for 1 h in LC-MS-grade water. The gel was cut into 35 slices, each ∼1–2 mm thick. Slices were further diced and placed into a 1.5-ml Eppendorf tube and destained with 25 mM ammonium bicarbonate in 50% acetonitrile (ACN). As previously described, the gel slices were reduced with 10 mM DTT in 25 mM ammonium bicarbonate and alkylated with 55 mM iodoacetamide and later washed with 25 mM ammonium bicarbonate and dehydrated with 25 mM ammonium bicarbonate in 50% ACN. Samples were digested with trypsin and incubated at 37°C overnight. The digested solution was transferred, and the remaining peptides were extracted from the gel pieces using 50% ACN and 0.5% formic acid and dried in vacuo.
In-solution trypsin digestion for phosphoproteomic analysis.
Reduction, alkylation, and trypsinization were performed as previously described (22) with modifications. To 100 μl (∼1.5 mg of protein) of solution, 5 μl of 200 mM DTT in ammonium bicarbonate were added. After the solution was incubated for 1 h at 37°C, 20 μl of 200 mM iodoacetamide were added for 1 h; then an additional 20 μl of 200 mM DTT were added to quench unreacted iodoacetamide. Samples were diluted to <1 M urea with 50 mM ammonium bicarbonate and digested overnight at 37°C with trypsin [1:30 (wt/wt)]. Samples were acidified with 5 μl of 100% formic acid and spun at 16,000 rpm for 20 min at 4°C, and the supernatant was saved. Samples were desalted using Oasis 1cc HLB columns (Waters, Milford, MA) and resuspended in strong cation-exchange (SCX) solvent A.
SCX chromatography and immobilized metal affinity chromatography.
The dried peptides were resuspended in 300 μl of solvent A (5 mM KH2PO4, 25% acetonitrile, pH 2.7) and injected onto a PolySULFOETHYL A SCX column (4.6 mm ID × 20 cm long, 5-μm particle size, 300-Å pore size). SCX chromatography was carried out on an Agilent HP1100 system at a flow rate of 1 ml/min using the following gradient: 0% solvent B (5 mM KH2PO4, 25% ACN, and 350 mM KCl, pH 2.7) for 2 min, 0–20% solvent B for 40 min, 20–100% solvent B for 5 min, and 100% solvent B for 5 min. UV absorbance at 214 nm was monitored while fractions were collected at 1.5-ml intervals. All 20 fractions were dried down by Speed Vac and desalted on 1cc HLB cartridges (Waters) prior to phosphopeptide enrichment, which was performed as described elsewhere (22) using immobilized metal affinity chromatography (IMAC) with a Ga3+ matrix (Phosphopeptide Isolation Kit, Pierce). Before analysis by LC-MS/MS, samples were desalted using C18 ZipTip pipette tips (Millipore) and resuspended in 0.1% formic acid.
LC-MS/MS analysis on a linear ion trap mass spectrometer.
LC-MS/MS analysis was performed as described previously (22). Briefly, isolated phosphopeptide samples were analyzed on a nanoflow system (model 1100, Agilent Technologies, Palo Alto, CA) connected to a Finnigan linear ion trap mass spectrometer (LTQ-FT, Thermo Electron, San Jose, CA) equipped with a nanoelectrospray ion source. The LTQ was used for the full MS scan and subsequent spectra (MS2 and MS3). An MS3 scan was triggered when the presence of a neutral loss peak (−98, −49, or −32.7 mass-to-charge ratio from precursor ion) was detected in the MS2 scan.
Data searching, scoring, and quantification.
Searches were performed using the latest version of the rat RefSeq database (National Center for Biotechnology Information) with concatenated forward and reverse sequences to allow for target-decoy analysis. The database also contained sequences for common MS contaminants, such as human keratin and porcine trypsin. Spectra were searched with SEQUEST (11), InsPecT (44), and the Open Mass Spectrometry Search Algorithm (OMSSA) (13). MS searches were aided by the high-performance computational capabilities of the Biowulf Linux cluster at the National Institutes of Health (http://biowulf.nih.gov). False discovery rate was filtered and determined as described previously (23). All data sets were filtered for a false discovery rate of <2%. Phosphorylation site localization was performed using Ascore (4) and PhosphoScore (42) for the SEQUEST data and Phosphate Localization Score for the InsPecT data (1).
Miscellaneous computational analysis.
Phosphoproteins were classified through PANTHER [Protein ANalysis THrough Evolutionary Relationships (www.pantherdb.org)]. Transmembrane prediction using hidden Markov models [TMHMM (http://www.cbs.dtu.dk/services/TMHMM-2.0/)] was used to predict transmembrane helices in proteins. The PDZ domain in phosphoproteins was identified by string searching the corresponding RefSeq records. We determined conservation of phosphorylated amino acids among multiple mammalian species using the HomoloGene database (release 64: http://www.ncbi.nlm.nih.gov/homologene) to obtain amino acid sequences for orthologous proteins and aligning these sequences using ClustalW2 (http://www.ebi.ac.uk/Tools/clustalw2/). The online database PhosphoSite (http://www.phosphosite.org/) was used to determine whether phosphorylation sites identified in this study had been previously described. The online utility Motif-X (43) was used to classify the identified phosphorylation sites with regard to patterns of neighboring amino acids.
RESULTS
Enrichment of renal cortical membrane proteins by MgCl2 precipitation.
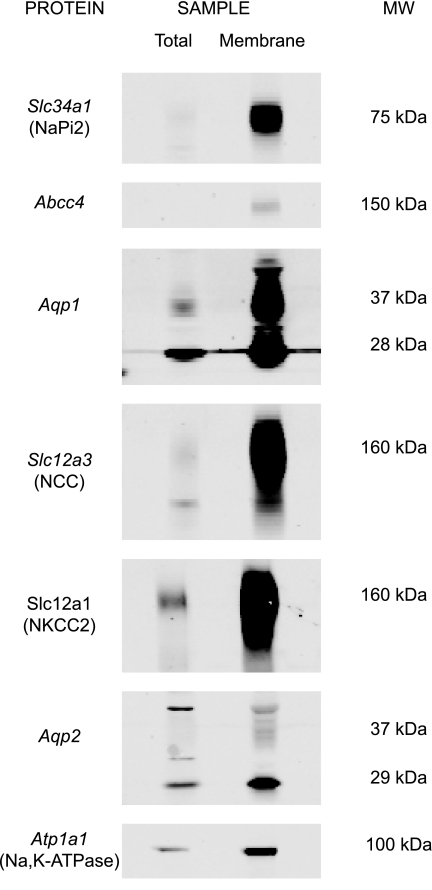
Before LC-MS/MS phosphoproteomic profiling, immunoblotting was utilized to confirm enrichment of cortical membrane proteins (Fig. 1). The whole cortex homogenate (“total”) and cortical membrane fraction (“membrane”) were prepared according to procedures described in methods and then analyzed by immunoblotting for proteins known to be expressed in cortical nephron segments. Membrane proteins expressed in proximal tubule, including Slc34a1, Abcc4 (a cAMP transporter), and Aqp1 (the water channel aquaporin-1), were more abundant in the cortical membrane fraction than total homogenate. A number of membrane proteins from other nephron segments, including Slc12a3 (the thiazide-sensitive Na-Cl cotransporter NCC found in distal convoluted tubule), Slc12a1 (the bumetanide-sensitive Na-K-2Cl cotransporter NKCC2 found in thick ascending limb), and Aqp2 (the vasopressin-regulated water channel aquaporin-2 found in connecting tubule and cortical collecting duct), also were enriched in the cortical membrane fraction. In addition, Atp1a1 (the Na/K pump Na-K-ATPase α1-subunit found in all renal tubule segments) was enriched in the membrane fraction. These results show that the described membrane isolation technique succeeds at enriching proximal tubule membrane proteins and membrane proteins from other nephron segments.
Fig. 1.
Immunoblot analysis of cortical membrane marker proteins. After membrane fractions were isolated from rat cortex by MgCl2 precipitation, 20 μg of protein were immunoblotted for various integral membrane proteins known to be expressed in renal cortical nephron segments indicated by gene symbols: Slc34a1 (proximal tubule), Abcc4 (proximal tubule), Aqp1 (proximal tubule), Slc12a3 (distal convoluted tubule), Slc12a1 (cortical thick ascending limb), Aqp2 (connecting tubule and cortical collecting duct), and Atp1a1 (Na-K-ATPase α1-subunit, all segments). All marker proteins were enriched in the cortical membrane fraction (“membrane”) compared with whole rat cortex (“total”).
Phosphoproteomic profiling of rat cortical membrane fractions.
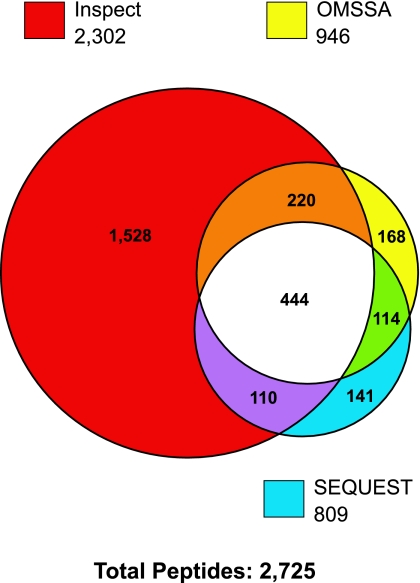
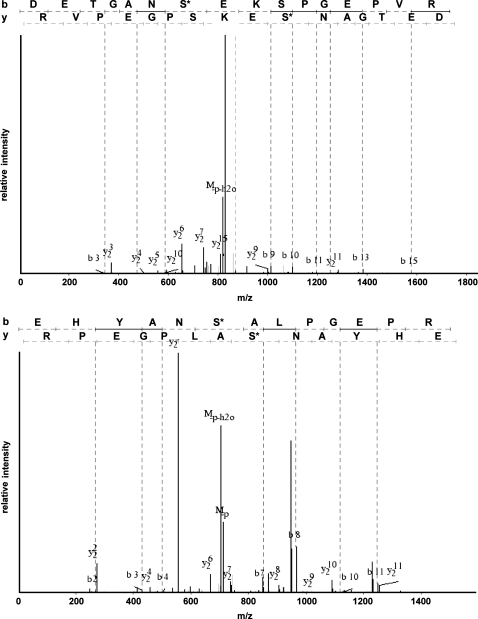
Rat cortical membrane fractions were subjected to in-gel proteomic and in-solution phosphoproteomic analysis as described in methods. Peptide samples were analyzed on a Thermo LTQ mass spectrometer, and the resulting spectra were searched using three different search algorithms, SEQUEST, InsPecT, and OMSSA, to ensure the highest possible yield of phosphopeptide identifications (3, 45). Final data sets were filtered using the target-decoy approach (9, 10) to limit false-positive identifications to <2%. Figure 2 shows a Venn diagram of phosphopeptides identified from each of the three search algorithms. A total of 2,725 phosphopeptides were identified by the combination of the three search algorithms. InsPecT identified the largest number of phosphopeptides (2,302), with the highest number of identifications that were not shared with either of the other two search algorithms (1,528). After redundant peptide identifications were filtered out, 743 unique phosphorylated peptides remained (see Supplemental Table S1 in Supplemental Material for this article, available online at the Journal website). Almost half of the phosphorylation sites identified in membrane transporters were not found on the PhosphoSite database and are therefore considered “previously unidentified” or “novel.” Annotated phosphopeptide data from all searches are accessible online (http://dir.nhlbi.nih.gov/papers/lkem/rcmpd/). Examples of phosphopeptide spectra (Ser71 and Ser124 of Slc12a3, the thiazide-sensitive cotransporter) are shown in Fig. 3.
Fig. 2.
Numbers of phosphopeptides identified in rat renal cortical membrane samples. Individual circles in Venn diagram show the number of phosphopeptides identified from the same spectra using each of three search algorithms (SEQUEST, InsPecT, and OMSSA). A false discovery rate of <2% was specified for each of the searches.
Fig. 3.
Typical spectra showing sites detected in the thiazide-sensitive Na-Cl cotransporter of the renal distal convoluted tubule (Slc12a3). Top: peptide containing phosphorylated Ser124. Bottom: peptide containing phosphorylated Ser71. m/z, Mass-to-charge ratio.
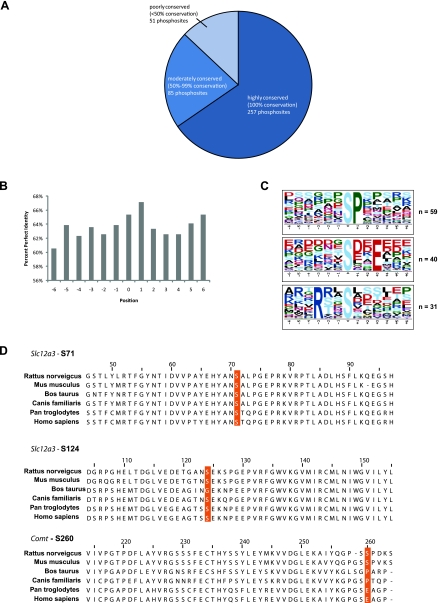
We analyzed the degree of conservation of the identified phosphorylation sites among mammalian species other than rat. For this we used the entries in Supplemental Table S1, which also were found on the HomoloGene database (see methods). Among the species examined, 65% of sites proved to be 100% conserved, while 22% (50–99% of species) were moderately conserved and 13% (<50% of species) were poorly conserved (Fig. 4A). We interpret these data to indicate that most of the detected sites are compatible with phosphorylation events that have regulatory significance in multiple species. However, as demonstrated in Fig. 4B, the mean level of conservation of neighboring amino acids appears to be equally high, indicating that the variability of the phosphorylation sites among species may simply be a reflection of the background variability of the primary structure. On the other hand, the 13% of phosphorylation sites that proved to be poorly conserved among mammalian species can probably be regarded as having questionable functional significance. Figure 4C shows sequence logos summarizing detectable amino acid patterns in the sequence flanking the phosphorylation sites that were 100% conserved [analyzed using Motif-X (http://motif-x.med.harvard.edu/)]. Figure 4D gives examples of each of the three levels of conservation listed in Fig. 4A. The site corresponding to Ser71 of Slc12a3 was totally conserved among species. The site corresponding to Ser124 of Slc12a3 was conserved among five of six of the species examined. Finally, Ser260 of Comt (catechol O-methyltransferase) was conserved in only two of six species.
Fig. 4.
Conservation of phosphorylation sites detected by mass spectrometry (MS) in rat renal cortical membrane samples. A: Venn diagram showing number of sites that are highly conserved, moderately conserved, or poorly conserved among mammalian species. B: percent perfect identity (100% conservation) of neighboring amino acids of phosphorylated residue (position 0). C: sequence logos showing overrepresented phosphorylation motifs extracted from sites that are 100% conserved among species examined. Position 0 represents phosphorylated residue. Three classes of phosphorylation motif were found: proline-directed motif (top), acidophilic phosphorylation motif (middle), and basophilic phosphorylation motif (bottom). Motif-x software was used for this analysis. D: examples of highly conserved (top), moderately conserved (middle), or poorly conserved (bottom) phosphorylation sites with alignments.
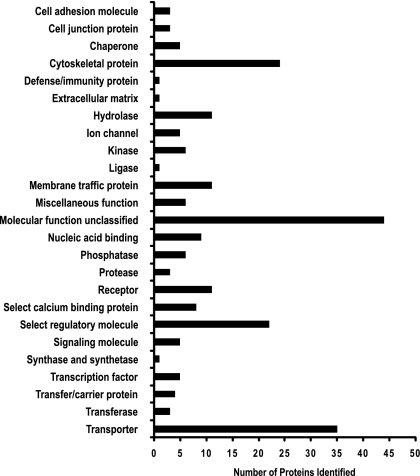
We next used the online classification system PANTHER to classify all the identified cortical membrane phosphoproteins in our data set. If we disregard unclassified identifications, the three most predominant “molecular function” terms were “transporter,” “select regulatory molecule,” and “cytoskeletal protein” (Fig. 5). Detailed descriptions of proteins in these categories are included in Tables 1 and 2. Table 1 includes transporters from Slc (n = 30), Abc (n = 4), and ATPase (n = 5) family members. Most of these proteins are well-known apical or basolateral transporters expressed in proximal tubule cells, including proteins known to mediate transport of glucose, amino acids, organic ions, and inorganic ions (Figs. 6 and 7). Table 1 also includes phosphopeptides for transport proteins from other nephron segments, including NKCC2 (Slc12a1) and NCC (Slc12a3). For NKCC2, two sites (Thr101 and Ser126) had been previously identified (14, 18) and the third, Ser87, is previously unreported to our knowledge. For NCC, one site (Ser71) had been previously identified (33), while the other site (Ser124) was not previously identified. Interestingly, the sequences surrounding Ser71 [E H Y A N S* A L P G E P (conserved amino acids underlined)] and Ser124 (E T G A N S* E K S P G E P) of NCC are similar (Fig. 3), suggesting that the same kinase may phosphorylate both. Ser71 of Slc12a3 has been demonstrated to be a phosphoacceptor site for the Ser/Thr kinases STE20/SPS1-related proline/alanine-rich kinase (SPAK) and odd-skipped related 1 (OSR1) (33, 53, 52).
Fig. 5.
Classification of cortical membrane phosphoproteins identified by tandem MS (LC-MS/MS). Histogram shows number of phosphoproteins (horizontal axis) associated with a particular PANTHER classification term (vertical axis).
Table 1.
Identification of phosphorylation sites of transport proteins
| Protein Name | Gene Symbol | Accession No. | Peptide Sequence | Phosphorylation Site | Relative Location |
|---|---|---|---|---|---|
| Solute carriers | |||||
| Excitatory amino acid transporter 3 | Slc1a1 | NP_037164 | ALEPTILDNEDS*DTKK | S496† | COOH-terminal tail |
| SDTIS*FTQTSQF | S516 | COOH-terminal tail | |||
| Proton myo-inositol cotransporter | Slc2a13 | NP_598295 | EAS*AAGPIICR | S277† | Loop |
| Neutral and basic amino acid transport protein rBAT | Slc3a1 | NP_058912 | DKRDS*IQMSMK | S10† | NH2-terminal tail |
| DSIQMS*MK | S14† | NH2-terminal tail | |||
| Band 3 anion transport protein | Slc4a1 | NP_036783 | VLEIPDRDS* | S18 | NH2-terminal tail |
| Electrogenic Na-bicarbonate cotransporter 1 | Slc4a4 | NP_445876 | MFSNPDNGS*PAMTHR | S245 | NH2-terminal tail |
| NLTSSS*LNDISDKPEK | S257 | NH2-terminal tail | |||
| KKGS*LDSDNDDSDCPYSEK | S1026 | COOH-terminal tail | |||
| KGSLDS*DNDDSDCPYSEK | S1029 | COOH-terminal tail | |||
| KKGS*LDS*DNDDSDCPYSEK | S1026, S1029 | COOH-terminal tail | |||
| Na-glucose cotransporter 2 | Slc5a2 | NP_072112 | S*GSGSPPPTTEEVAATTR | S619† | Loop |
| SGS*GSPPPTTEEVAATTR | S621† | Loop | |||
| SGSGS*PPPTTEEVAATTR | S623† | Loop | |||
| Na-dependent neutral amino acid transporter B(0)AT3 | Slc6a18 | NP_058859 | NTHLESALKPQES*R | S612‡ | COOH-terminal tail |
| Na-dependent neutral amino acid transporter B(0)AT1 | Slc6a19 | NP_001034811 | ALSTAS*VNGDLKN | S627† | COOH-terminal tail |
| Y + L amino acid transporter 1 | Slc7a7 | NP_112631 | DEADGS*AQGDGAGPAAEQVK | S21 | NH2-terminal tail |
| Large neutral amino acid transporter small subunit 2 | Slc7a8 | NP_445894 | NHPDRGS*DTSPEAEASSGGGGVALK | S20† | NH2-terminal tail |
| B(0,+)-type amino acid transporter 1 | Slc7a9 | NP_446381 | EDEKS*VHSTEPK | S15† | NH2-terminal tail |
| EDEKS*VHS*TEPK | S15†, S18† | NH2-terminal tail | |||
| EDEKS*VHST*EPK | S15†, T19† | NH2-terminal tail | |||
| Na/H exchanger 3 | Slc9a3 | NP_036786 | RGS*LAFIR | S552 | COOH-terminal tail |
| HELT*PNEDEKQDK | T639† | COOH-terminal tail | |||
| EKDLELS*EPE | S716 | COOH-terminal tail | |||
| VQIPNS*PSNFR | S791† | COOH-terminal tail | |||
| VQIPNSPS*NFR | S793† | COOH-terminal tail | |||
| DGPEEQLQPAS*PESTHM | S825† | COOH-terminal tail | |||
| Solute carrier family 12 member 1 | Slc12a1 | NP_062007 | TDTTFHAYDS*HTNTY | S87† | NH2-terminal tail |
| YLQTFGHNT*MDAVPK | T101 | NH2-terminal tail | |||
| S*LQEIHEQLAK | S126 | NH2-terminal tail | |||
| Solute carrier family 12 member 3 | Slc12a3 | NP_062218 | EHYANS*ALPGEPR | S71 | NH2-terminal tail |
| DETGANS*EKSPGEPVR | S124† | NH2-terminal tail | |||
| Monocarboxylate transporter 1 | Slc16a1 | NP_036848 | LKS*KES*LQEAGK | S210, S213 | Loop |
| SKES*LQEAGK | S213 | Loop | |||
| DGKEDETST*DVDEKPK | T461 | COOH-terminal tail | |||
| ETQSPAPLQNSSGDPAEEES*PV | S492 | COOH-terminal tail | |||
| Sialin | Slc17a5 | NP_001009713 | AGNDDEESS*DSTPLLPSAR | S17† | NH2-terminal tail |
| Solute carrier organic anion transporter family member 1A3 | Slc21a4 | NP_110464 | INS*SEMEIAEMK | S633‡ | COOH-terminal tail |
| INSS*EMEIAEMK | S634‡ | COOH-terminal tail | |||
| Solute carrier family 22 member 2 | Slc22a2 | NP_113772 | ENLPPSQASRPS*AK | S553† | COOH-terminal tail |
| Solute carrier family 22 member 4 | Slc22a4 | NP_071606 | KST*VSMDREENPK | T537† | COOH-terminal tail |
| STVS*MDREENPK | S539† | COOH-terminal tail | |||
| Solute carrier family 22 member 5 | Slc22a5 | NP_062142 | QWQIQS*QTR | S537† | COOH-terminal tail |
| TQKDGGES*PTVLK | S548† | COOH-terminal tail | |||
| Solute carrier family 22 member 12 | Slc22a12 | NP_001030115 | VTHDIAGGS*VLK | S546† | COOH-terminal tail |
| Solute carrier family 23 member 1 | Slc23a1 | NP_059011 | QHECPDS*AGTSTR | S18† | NH2-terminal tail |
| QHECPDS*AGTS*TR | S18†, S22† | NH2-terminal tail | |||
| DAPDNTET*GSVCTKV | T597† | COOH-terminal tail | |||
| TENQPAVLEDAPDNTETGS*VCTK | S599 | COOH-terminal tail | |||
| Solute carrier family 23 member 2 | Slc23a2 | NP_059012 | ETLDSTGS*LDPQR | S25 | NH2-terminal tail |
| Sulfate anion transporter 1 | Slc26a1 | NP_071623 | GTEVGVS*NR | S586 | COOH-terminal tail |
| Na-dependent phosphate transport protein 2A | Slc34a1 | NP_037162 | AVS*PLPVR | S14† | NH2-terminal tail |
| EELPPAT*PS*PR | T621†, S623† | COOH-terminal tail | |||
| VFLEELPPATPS*PR | S623† | COOH-terminal tail | |||
| Na-dependent phosphate transport protein 2C | Slc34a3 | NP_647554 | GETPQGS*SEECDLSGSCTER | S285† | Loop |
| GETPQGSS*EECDLSGSCTER | S286† | Loop | |||
| Large neutral amino acid transporter small subunit 4 | Slc43a2 | NP_001099282 | RLS*VGSSMR | S274 | Loop |
| Solute carrier organic anion transporter family member 1A1 | Slco1a1 | NP_058807 | ESEHTDVHGS*PQVENDGELK | S657† | COOH-terminal tail |
| Solute carrier organic anion transporter family member 4C1 | Slco4c1 | NP_001002024 | GVENPAFVPS*SPDTPRR | S15† | NH2-terminal tail |
| GVENPAFVPSS*PDTPR | S16† | NH2-terminal tail | |||
| S*ASPSQVEVSAVASR | S24† | NH2-terminal tail | |||
| S*ASPS*QVEVSAVASR | S24†, S28† | NH2-terminal tail | |||
| Predicted: similar to solute carrier family 7 (cationic amino acid transporter, y+ system), member 12 | LOC688389 | XP_001066729 | DEQDKNLCS*E | S553‡ | COOH-terminal tail |
| ATP binding cassettes | |||||
| ATP-binding cassette, subfamily B (MDR/TAP), member 1A | Abcb1a | NP_596892 | DSGSS*LIR | S653 | Loop |
| ATP-binding cassette subfamily G member 2 | Abcg2 | NP_852046 | DHVLVPMS*QR | S13‡ | NH2-terminal tail |
| Multidrug resistance-associated protein 4 | Abcc4 | NP_596902 | ENEEAEPSPVPGT*PTLR | T646 | Loop |
| Similar to ATP-binding cassette, subfamily G (white), member 3 | LOC360997 | NP_001032282 | RHS*DLPETNTS | S18‡ | NH2-terminal tail |
| ATPase | |||||
| V-type proton ATPase 116-kDa subunit a isoform 4 | Atp6v0a4 | NP_001100061 | SQLQS*FTIHE | S645† | Loop |
| V-type proton ATPase subunit H | Atp6v1 h | NP_001013951 | QEMLQTEGS*QCAK | S71† | Non-transmembrane protein |
| Predicted: ATPase, aminophospholipid transporter (APLT), class I, type 8A, member 1 | Atp8a1 | XP_001070245 | TDDVSEKTS*LADQEEVR | S29 | NH2-terminal tail |
| Probable phospholipid-transporting ATPase IH | Atp11a | NP_001100794 | DS*FSGLSTDMH | S747 | Loop |
| Predicted: similar to probable cation-transporting ATPase 13A3 (ATPase family homolog upregulated in senescence cells 1) | LOC292449 | XP_001067586 | HDS*LEDLQVTR | S744‡ | Loop |
Phosphorylated amino acid.
Novel phosphorylation site.
Protein not in www.phosphosite.org database.
Table 2.
Identification of phosphorylation sites of cytoskeletal proteins
| Protein Name | Gene Symbol | Accession No. | Peptide Sequence | Phosphorylation Site |
|---|---|---|---|---|
| Actin-binding LIM protein 1 | Ablim1 | NP_001037859 | TLS*PTPSAEGFQDGR | S115 |
| A-adducin | Add1 | NP_058686 | QKGS*EENLDETR | S586 |
| Whirlin | Dfnb31 | NP_851602 | FDHLVLRREIES* | S501† |
| Cortactin isoform B | Cttn | NP_068640 | DRPPSS*PIYEDAAPLK | S381 |
| Espin | Espn | NP_062568 | ARS*PTPPASGP | S688† |
| ES*PQPAVS*PGPSR | S674†, S680† | |||
| ESPQPAVS*PGPSR | S680† | |||
| Keratin, type II cytoskeletal 80 | Krt80 | NP_001008815 | TT*AS*KSGLS*K | T412†, S414†, S419† |
| LIM domain only protein 7 | Lmo7 | NP_001001515 | EVSRS*PDQFSDM@R | S1061† |
| SHS*PSM@SQSGSQLR | S1639 | |||
| SRS*TTELNDPLIEK | S1476 | |||
| Microtubule-associated protein 4 | Map4 | NP_001019449 | DVS*PSPETETAK | S520 |
| Myosin-9 | Myh9 | NP_037326 | KGTGDCS*DEEVDGKADGADAK | S1944 |
| Septin-2 | Sept2 | NP_476489 | DAES*DEDEDFKEQTR | S218 |
| Spectrin β-chain, brain 1 | Sptbn1 | NP_001013148 | ESS*PVPS*PTSDR | S2159, S2163 |
| GDQVSQNGLPAEQGS*PR | S2132 | |||
| RPPS*PEPSAK | S2097 | |||
| TSS*KES*SPVPSPTSDRK | S2155, S2158 | |||
| TSS*KESS*PVPS*PTSDRK | S2155, S2159, S2163 | |||
| TSS*KESS*PVPSPTSDRK | S2155, S2159 | |||
| Myosin 18a | LOC360570 | NP_001165608 | HNYLS*DS*DTEAK | S2041, S2043 |
| HNYLSDS*DTEAK | S2043 | |||
| LIM domain only protein 7 | RGD1305269 | XP_223397 | GSSDGRGS*DSESDLPHR | S374‡ |
| Predicted: myosin VI-like | Myo6 | XP_001061392 | GT*VIKVPLK | T405 |
| Predicted: myosin VIIB-like | Myo7b | XP_001059493 | S*SLTGSSVMR | S1035† |
| Predicted: InaD-like (Drosophila) isoform 2 | Inadl | XP_001055452 | DDEAS*VDEPR | S647 |
| KTS*LSASPFEQPSSR | S455† |
Phosphorylated amino acid.
Novel phosphorylation site.
Protein not in www.phosphosite.org database.
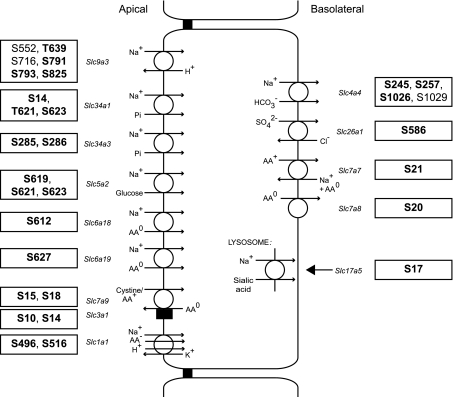
Fig. 6.
Schematic of a proximal tubule cell showing identified phosphorylation sites in transporters involved in inorganic ion, glucose, and amino acid transport. Novel phosphorylation sites are in bold. Gene symbols for the various transporters are in italics. Substrates and substrate stoichiometry are summarized from http://www.bioparadigms.org/.
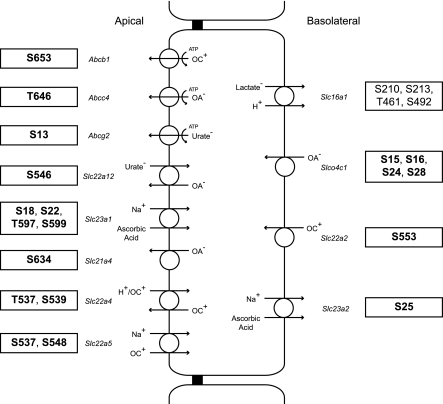
Fig. 7.
Schematic of a proximal tubule cell showing identified phosphorylation sites in transporters involved in organic anion and cation transport. Novel phosphorylation sites are in bold. Gene symbols are in italics. Substrates and substrate stoichiometry for Slc family members are summarized from http://www.bioparadigms.org/.
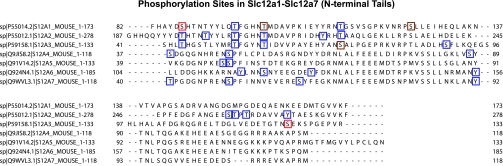
The phosphorylation sites identified in Slc12a1 and Slc12a3 in this study are shown in Fig. 8, along with all sites that have been previously reported on the PhosphoSite database for Slc12a1 through Slc12a7, aligned to show corresponding amino acids in the different paralogs. The novel Ser87 site in Slc12a1 (NKCC2) corresponds to threonines previously demonstrated to be phosphorylated in Slc12a2 (48) and Slc12a3 (39), supporting the possibility of a single functional role common to this site in the three paralogs. The corresponding sites in Slc12a3 (Thr44) and Slc12a2 (Thr197) are believed to be phosphoacceptor sites for the Ser/Thr kinases SPAK and OSR1 (39, 48). Therefore, it appears likely that Ser87 of Slc12a1 is similarly targeted by these kinases. The novel Ser124 site in Slc12a3 corresponds to a known tyrosine phosphorylation site in Slc12a2 (25), but the corresponding site in Slc12a1 is a lysine, i.e., not a potential phosphorylation target.
Fig. 8.
Known phosphorylation sites in distal portions of NH2-terminal tails of Slc12 member Cl-cation cotransporters. Mouse sequences are shown to provide a common reference for data from rat, mouse, and human. Phosphorylation sites identified in Slc12a1 and Slc12a3 in this study are indicated by red boxes (novel sites) or brown boxes (previously demonstrated sites). Blue boxes indicate sites demonstrated in prior studies and not identified in renal cortical membranes in this study. Previously demonstrated sites are culled from the PhosphoSite database.
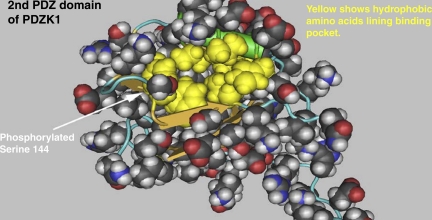
Other proteins relevant to our study include those with PANTHER classification terms “transmembrane” (Table 3) and “PDZ domain” (Table 4). Another way to assess possible functional relevance of the identified phosphorylation sites is to cross-reference the sites to the locations of known functional domains in the phosphorylated proteins (Table 5). These data can be used to formulate hypotheses about the sites' functional roles, as illustrated in Fig. 9. Figure 9 shows the location of the Ser144 phosphorylation site in the regulatory/scaffolding protein Pdzk1, also known as Na/H exchanger regulatory factor 3 (NHERF-3), with respect to the three-dimensional structure of the second PDZ domain of Pdzk1 determined by solution NMR (pdb 2EDZ A). The phosphorylated amino acid is located at the mouth of the PDZ ligand binding pocket, suggesting that phosphorylation could play a regulatory role in protein-protein interactions.
Table 3.
Identification of phosphorylation sites of transmembrane proteins
| Protein Name | Gene Symbol | Accession No. | Peptide Sequence | Phosphorylation Site | Relative Location |
|---|---|---|---|---|---|
| Acetyl-coenzyme A synthetase, cytoplasmic | Acss2 | NP_001101263 | RGWS*PPPEVR | S30 | |
| Aquaporin-1 | Aqp1 | NP_036910 | EYDLDADDINS*R | S262 | COOH terminus |
| Aquaporin-2 | Aqp2 | NP_037041 | RQS*VELHS*PQSLPR | S256, S261 | COOH terminus |
| Baculoviral IAP repeat-containing protein 6 | Birc6 | NP_001164067 | LEGDSDDLLEDS*DSEEHSR | S631 | |
| C5a anaphylatoxin chemotactic receptor | C5ar1 | NP_446071 | NVLSEDS*LGR | S329 | COOH terminus |
| Calnexin precursor | Canx | NP_742005 | AEEDEILNRS*PR | S582 | COOH terminus |
| QKS*DAEEDGGTGS*QDEEDSKPK | S553, S563 | COOH terminus | |||
| QKSDAEEDGGTGS*QDEEDSKPK | S563 | COOH terminus | |||
| Carbonic anhydrase 14 | Car14 | NP_001103125 | S*VVFTSAR | S325‡ | |
| Phosphoprotein associated with glycosphingolipid-enriched microdomains 1 | Cbp | NP_071589 | FSS*LSYK | S293† | |
| SSSS*CNDLYATVK | S346† | ||||
| Phosphatidate cytidylyltransferase 2 | Cds2 | NP_446095 | LDGETAS*DSESR | S32 | NH2 terminus |
| Catechol O-methyltransferase | Comt | NP_036663 | AIYQGPSS*PDKS | S260 | COOH terminus |
| Crumbs protein homolog 3 | Crb3 | NP_001020832 | QTEGTY*RPSSEEQVGAR | Y87† | |
| Glutamate carboxypeptidase 2 | Folh1 | NP_476533 | DSDS*AEALGRR | S10† | NH2 terminus |
| Predicted: G protein-coupled receptor, family C, group 5, member C isoform 1 | Gprc5c | XP_001081657 | SEGAY*DVILPR | Y474 | COOH terminus |
| AEDM@Y*M@VQSHQVATPTK | Y501 | COOH terminus | |||
| Cation-independent mannose-6-phosphate receptor | Igf2r | NP_036888 | DDS*DEDLLHI | S2472 | COOH terminus |
| Integrin-α4 | Itga4 | NP_001101207 | RDS*WSYVNSK | S1025 | COOH terminus |
| Low-density lipoprotein receptor-related protein 2 precursor | Lrp2 | NP_110454 | DAVAVAPPPS*PSLPAK | S4624 | COOH terminus |
| DTANLVKEDS*DV | S4658† | COOH terminus | |||
| E3 ubiquitin-protein ligase MARCH8 | March8 | NP_001101352 | TSVTPS*NQDICR | S71† | |
| Microfibrillar-associated protein 3-like precursor | Mfap3l | NP_001012049 | DASS*LHEQPQQIAIK | S307† | |
| Neprilysin | Mme | NP_036740 | GRS*ESQM@DITDINAPK | S4† | NH2 terminus |
| SES*QM@DITDINAPKPK | S6 | NH2 terminus | |||
| Protein LYRIC | Mtdh | NP_596889 | SQEPISNDQKDS*DDDKEK | S425 | COOH terminus |
| Cadherin-related family member 5 precursor | Mucdhl | NP_612534 | AKWS*PTSNR | S729† | COOH terminus |
| SSS*PTPPSSMPPS*PQPK | S756†, S766† | COOH terminus | |||
| T*PPSSM@PPS*PQPK | T758†, S766† | COOH terminus | |||
| TVQAGDS*PSAVR | S783 | COOH terminus | |||
| TADVDSASASGSEGS*DDDDDPDQKK | S839† | COOH terminus | |||
| Predicted: osteopetrosis-associated transmembrane protein 1-like | LOC100362414 | XP_002729028 | SSTS*FANIQENST | S329 | COOH terminus |
| PDZK1-interacting protein 1 | Pdzk1ip1 | NP_569085 | YSS*M@ASGFR | S85† | COOH terminus |
| Plexin domain-containing protein 2 | Plxdc2 | NP_001101892 | RGS*GHPAYAEVEPVGEK | S507 | |
| Lipid phosphate phosphohydrolase 3 | Ppap2b | NP_620260 | NGGS*PALNNNPR | S19 | NH2 terminus |
| Receptor-type tyrosine-protein phosphatase C isoform 4 | Ptprc | NP_612516 | NRSS*NVVPYDFNR | S944 | COOH terminus |
| ANS*QDKIEFHNEVDGAK | S1209† | COOH terminus | |||
| Roundabout 2 | Robo2 | NP_115289 | KAEILRGS*HQR | S1446† | |
| Secretory carrier membrane protein 3 | Scamp3 | NP_113912 | KLS*PAEPK | S147† | NH2 terminus |
| Na channel subunit β2 precursor | Scn2b | NP_037009 | Y*DVS*VTLK | Y108†, S111† | NH2 terminus |
| Serine incorporator 1 | Serinc1 | NP_891996 | SDGS*LDDGEGVHR | S364 | Loop |
| Predicted: sphingosine-1-phosphate phosphatase 1 | Sgpp1 | XP_001080791 | RNS*LTGEEGELAK | S200 | NH2 terminus |
| Syntaxin-4 | Stx4 | NP_112387 | QGDNIS*DDEDEVR | S15 | NH2 terminus |
| Syntaxin-7 | Stx7 | NP_068641 | SRVS*GGFPEDSSK | S129 | NH2 terminus |
| Syntaxin-8 | Stx8 | NP_113844 | NEGS*EPDLIR | S102† | NH2 terminus |
| Taste receptor type 2 member 135 | Tas2r135 | NP_001020233 | T*LSAHNK | T60‡ | |
| Transmembrane 7 superfamily member 3 precursor | Tm7 sf3 | NP_001011970 | EQPAGERT*PLLL | T561† | |
| Transmembrane protein 9 | Tmem9 | NP_001099423 | S*M@AAAAASIGGPR | S76 | COOH terminus |
| Transmembrane protein 51 | Tmem51 | NP_001102743 | AETETS*PGHAPDR | S178† | COOH terminus |
| EEDS*QEEEEEASSR | S114 | COOH terminus | |||
| Transmembrane protein 106A | Tmem106a | NP_001020138 | S*HSCVPCEK | S41† | NH2 terminus |
| Transmembrane protein 116 | Tmem116 | NP_001153097 | DTQT*PLLCSQK | T317‡ | |
| Transmembrane protein 192 | Tmem192 | NP_001014163 | LLELATQPART* | T266† | |
| Two-pore Ca channel protein 1 | Tpcn1 | NP_647548 | TKS*DLSLK | S767 | COOH terminus |
| Vesicle-associated membrane protein 8 | Vamp8 | NP_114015 | DLEAT*SEHFK | T53 | NH2 terminus |
| Hypothetical protein LOC300783 | RGD1565496 | NP_001100301 | DES*DAEM@ELR | S114‡ | NH2 terminus |
| Hypothetical protein LOC311536 | RGD1304644 | NP_001032275 | DTEDTGS*VGAPVSSPGK | S269‡ | Loop |
| Protein ITFG3 | Itfg3 | NP_001009701 | NEDKKS*QENLGNLPK | S21 | NH2 terminus |
| Predicted: rCG24298-like | Pcdh24 | XP_001070261 | DLPHNPPES* | S1274‡ | COOH terminus |
| Predicted: olfactory receptor Olfr1291-like | RGD1566059 | XP_001080280 | WPDT*WER | T18‡ | NH2 terminus |
| Predicted: Leu-rich repeats and immunoglobulin-like domains 3 | Lrig3 | XP_001055013 | S*IPWPHS*R | S1132‡, S1138‡ |
Phosphorylated amino acid.
Methionine oxidation.
Novel phosphorylation site.
Protein not in www.phosphosite.org database.
Table 4.
Identification of phosphorylation sites of PDZ domain proteins
| Protein Name | Gene Symbol | Accession No. | Peptide Sequence | Phosphorylation Site |
|---|---|---|---|---|
| Whirlin | Dfnb31 | NP_851602 | FDHLVLRREIES* | S501† |
| LIM domain only protein 7 | Lmo7 | NP_001001515 | EVSRS*PDQFSDMR | S1061† |
| SRS*TTELNDPLIEK | S1476 | |||
| SHS*PSMSQSGSQLR | S1639 | |||
| p55 protein | LOC652956 | NP_001032748 | TNGS*VDLGEEEEAAR | S57† |
| Membrane protein, palmitoylated 5 (MAGUK p55 subfamily member 5) | Mpp5 | NP_001101504 | KQELDLNSS*MR | S84† |
| Na/H exchange regulatory cofactor NHE-RF3 | Pdzk1 | NP_113900 | DQS*PREPALNEK | S108† |
| EGNS*FGFSLK | S144† | |||
| S*NGYGFYLR | S250† | |||
| SQELPNGS*VK | S348† | |||
| TLSAAS*HSSSNSE | S512 | |||
| TLSAAS*HSS*SNS*EDTVM | S512, S515†, S518 | |||
| TLSAAS*HSSS*NS*EDTVM | S512, S516, S518 | |||
| TLSAASHS*SSNS*EDTVM | S514, S518 | |||
| TLSAASHS*S*SNS*EDTVM | S514, S515†, S518 | |||
| TLSAASHSS*S*NSEDTVM | S515†, S516 | |||
| TLSAASHSS*SNS*EDTVM | S515†, S518 | |||
| TLSAASHSSS*NS*EDTVM | S516, S518 | |||
| Rap guanine nucleotide exchange factor 2 | Rapgef2 | NP_001101154 | SETS*PVAPR | S820 |
| Signal-induced proliferation-associated 1-like protein 1 | Sipa1l1 | NP_647546 | LIDLES*PTPESQK | S1567 |
| Na/H exchange regulatory cofactor NHE-RF1 | Slc9a3r1 | NP_067605 | NGYGFNLHS*DK | S167† |
| AS*ESPRPALAR | S275 | |||
| LVEPASES*PRPALAR | S277 | |||
| S*ASSDTSEELNAQ | S285 | |||
| S*AS*SDTSEELNAQDSPK | S285, S287 | |||
| S*ASSDT*SEELNAQDSPK | S285, T290 | |||
| S*ASSDTS*EELNAQDSPK | S285, S291 | |||
| SAS*SDTSEELNAQDSPKR | S287 | |||
| SAS*S*DTSEELNAQDSPK | S287, S288 | |||
| SAS*SDT*SEELNAQDSPK | S287, T290 | |||
| SAS*SDTS*EELNAQDSPK | S287, S291 | |||
| SAS*SDTSEELNAQDS*PKR | S287, S299 | |||
| SAS*SDTS*EELNAQDS*PK | S287, S291, S299 | |||
| SASS*DTSEELNAQDSPK | S288 | |||
| SASS*DT*SEELNAQDSPK | S288, T290 | |||
| SASS*DTS*EELNAQDSPK | S288, S291 | |||
| SS*DTSEELNAQDS*PKR | S288, S299 | |||
| SASS*DTS*EELNAQDS*PKR | S288, S291, S299 | |||
| SASSDT*SEELNAQDSPK | T290 | |||
| SASSDT*S*EELNAQDSPK | T290, S291 | |||
| SSDT*SEELNAQDS*PKR | T290, S299 | |||
| SASSDT*S*EELNAQDS*PKR | T290, S291, S299 | |||
| SSDTS*EELNAQDSPK | S291 | |||
| SASSDTS*EELNAQDS*PKR | S291, S299 | |||
| SASSDTSEELNAQDS*PK | S299 | |||
| Na/H exchange regulatory cofactor NHE-RF2 | Slc9a3r2 | NP_446263 | SDLPGS*EKDNEDGSAWK | S280† |
| Tight junction protein ZO-2 | Tjp2 | NP_446225 | RQQYS*DQEYHSSTEK | S404 |
| DGS*PPPAFKPEPPK | S966 | |||
| Usher syndrome 1C | Ush1c | NP_997686 | EMEQIS*EEEEK | S364† |
| Myosin 18a | LOC360570 | NP_001165608 | HNYLS*DS*DTEAK | S2041, S2043 |
| HNYLSDS*DTEAK | S2043 | |||
| Predicted: InaD-like (Drosophila) isoform 2 | Inadl | XP_001055452 | KTS*LSASPFEQPSSR | S455† |
| DDEAS*VDEPR | S647 |
Phosphorylated amino acid.
Novel phosphorylation site.
Protein not in www.phosphosite.org database.
Table 5.
Functional motifs
| Protein Name | Accession No. | Phosphorylation Site | Region Site |
|---|---|---|---|
| Glucosamine-fructose-6-phosphate aminotransferase (isomerizing) 1 | NP_001005879 | S239 | Glutamine amidotransferases class-II (Gn-AT)_GFAT-type; cd00714, PLN02981 |
| Protein TSSC1 | NP_001012192 | S320 | WD40 domain; cl02567 |
| DNA polymerase-λ | NP_001014190 | Y565 | Nucleotidyltransferase (NT) domain of family X DNA polymerases; cd00141 |
| RISC-loading complex subunit TARBP2 | NP_001030113 | T67 | Double-stranded RNA binding motif; cd00048 |
| RISC-loading complex subunit TARBP2 | NP_001030113 | T74 | Double-stranded RNA binding motif; cd00048 |
| Ras-related GTP-binding protein C | NP_001041649 | S94 | Switch I region |
| CAD protein | NP_001099180 | Y2103 | Asp/Orn carbamoyltransferase, Asp/Orn binding domain; pfam00185 |
| Formin-like protein 1 | NP_001099316 | S965 | Formin homology 2 domain; smart00498 |
| Hypothetical protein LOC300783 | NP_001100301 | S114 | p23_like domain; cd06465 |
| Hypothetical protein LOC498014 | NP_001102520 | S17 | NTR_like domain; cl02512 |
| Hypothetical protein LOC498014 | NP_001102520 | Y20 | NTR_like domain; cl02512 |
| Rho guanine nucleotide exchange factor 7 isoform b | NP_001106994 | S340 | Pleckstrin homology-like domain; cl00273 |
| Fructose-1,6-bisphosphatase 1 | NP_036690 | Y216 | Active site |
| Protein kinase Cβ isoform 1 | NP_036845 | T641 | Turn motif phosphorylation site |
| Dual-specificity tyrosine phosphorylation-regulated kinase 1A | NP_036923 | Y321 | Ser/Thr protein kinases, catalytic domain; smart00220 |
| Na channel subunit-β2 precursor | NP_037009 | S111 | Immunoglobulin domain; cl11960 |
| Na channel subunit-β2 precursor | NP_037009 | Y108 | Immunoglobulin domain; cl11960 |
| Utrophin | NP_037202 | S2602 | Spectrin repeats; cd00176 |
| Afadin | NP_037349 | S1090 | PDZ domain; cd00992 |
| Glycogen synthase kinase-3α | NP_059040 | Y279 | Protein kinase domain; pfam00069; protein kinases, catalytic domain; cl09925 |
| Solute carrier family 12 member 1 | NP_062007 | S116 | Amino acid permease NH2-terminal; pfam08403 |
| Solute carrier family 12 member 1 | NP_062007 | S126 | Amino acid permease NH2-terminal; pfam08403 |
| Solute carrier family 12 member 1 | NP_062007 | S87 | Amino acid permease NH2-terminal; pfam08403 |
| Solute carrier family 12 member 1 | NP_062007 | T101 | Amino acid permease NH2-terminal; pfam08403 |
| Solute carrier family 12 member 1 | NP_062007 | T114 | Amino acid permease NH2-terminal; pfam08403 |
| Solute carrier family 12 member 3 | NP_062218 | S71 | Amino acid permease NH2-terminal; pfam08403 |
| 3-Hydroxyanthranilate 3,4-dioxygenase | NP_064461 | S9 | Cupin domain; cl09118 |
| Na/H exchange regulatory cofactor NHE-RF1 | NP_067605 | S167 | PDZ domain found; cd00992 |
| Syntaxin-7 | NP_068641 | S126 | Syntaxin NH2-terminus domain; cd00179 |
| Syntaxin-7 | NP_068641 | S129 | Linker region; syntaxin NH2-terminus domain; cd00179 |
| Sulfate anion transporter 1 | NP_071623 | S586 | Sulfate transporter and anti-σ factor antagonist domain of SulP-like sulfate transporters; cd07042 |
| Protein kinase Cξ | NP_071952 | T560 | Turn motif phosphorylation site; catalytic domain of the protein Ser/Thr kinase, atypical protein kinase C; cd05588 |
| Synaptosomal-associated protein 23 | NP_073180 | S109 | SNAP-25 family; pfam00835 |
| Synaptosomal-associated protein 23 | NP_073180 | S110 | SNAP-25 family; pfam00835 |
| Synaptosomal-associated protein 23 | NP_073180 | S20 | Soluble NSF (N-ethylmaleimide-sensitive fusion protein)-Attachment protein (SNAP) REceptor domain; cl00152 |
| Synaptosomal-associated protein 23 | NP_073180 | S23 | Soluble NSF (N-ethylmaleimide-sensitive fusion protein)-attachment protein (SNAP) receptor domain; cl00152 |
| Synaptosomal-associated protein 23 | NP_073180 | S95 | SNAP-25 family; pfam00835 |
| Na/H exchange regulatory cofactor NHE-RF3 | NP_113900 | S144 | PDZ domain; cd00992 |
| Na/H exchange regulatory cofactor NHE-RF3 | NP_113900 | S250 | PDZ domain; cd00992 |
| Vesicle-associated membrane protein 8 | NP_114015 | T53 | Synaptobrevin; pfam00957 |
| Protein kinase Cι | NP_114448 | T564 | Turn motif phosphorylation site; catalytic domain of the protein Ser/Thr kinase, atypical protein kinase C; cd05588 |
| Electrogenic Na-bicarbonate cotransporter 1 | NP_445876 | S245 | Band 3 cytoplasmic domain; pfam07565 |
| Electrogenic Na-bicarbonate cotransporter 1 | NP_445876 | S257 | Band 3 cytoplasmic domain; pfam07565 |
| Electrogenic Na-bicarbonate cotransporter 1 | NP_445876 | T249 | Band 3 cytoplasmic domain; pfam07565 |
| Band 4.1-like protein 3 | NP_446379 | S446 | FERM adjacent (FA); pfam08736 |
| A-kinase anchor protein 12 isoform-α | NP_476444 | S614 | WSK motif; pfam03832 |
| A-kinase anchor protein 12 isoform-α | NP_476444 | S616 | WSK motif; pfam03832 |
| Brain-specific angiogenesis inhibitor 1-associated protein 2 | NP_476544 | S396 | Src homology 3 domains; cd00174 |
| Protein kinase Cδ | NP_579841 | S662 | Protein kinases, catalytic domain; cl09925 |
| Receptor-type tyrosine-protein phosphatase C isoform 4 | NP_612516 | S944 | Protein tyrosine phosphatase, catalytic domain; smart00194, cd00047 |
| Spectrin α-chain, brain | NP_741984 | S1217 | Spectrin repeats; cd00176 |
| Predicted: myosin VIIB-like | XP_001059493 | S1035 | MyTH4 domain; cl02480 |
| Predicted: myosin VIIB-like | XP_001059493 | S1036 | MyTH4 domain; cl02480 |
| Predicted: myosin VIIB-like | XP_001059493 | T1038 | MyTH4 domain; cl02480 |
| Predicted: myosin VI-like | XP_001061392 | T405 | Myosin head (motor domain); pfam00063; Myosin motor domain, type VI myosins; cd01382 |
| Predicted: erythrocyte membrane protein band 4.1 | XP_001063302 | S611 | SAB domain; pfam04382 |
Fig. 9.
Space-filling diagram showing structure of 2nd PDZ domain in the scaffold protein Pdzk1 [Na/H exchanger regulatory factor 3 (NHERF-3)] from mouse indicating location of a novel phosphorylation site found in this study. Structure was downloaded from National Center for Biotechnology Information (NCBI) Structure Database (pdb 2EDZ A) and displayed via Cn3D Viewer from the NCBI. This is one of many sites demonstrated within known functional domains (see Table 5).
Analysis of nonphosphorylated peptides in the rat renal cortical membrane fraction.
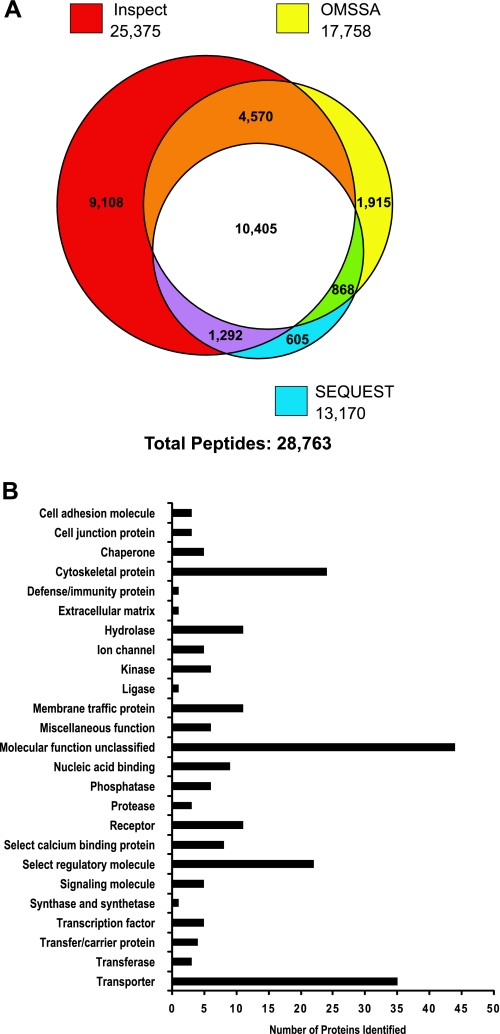
We also carried out LC-MS/MS analysis of the flow-through samples from the IMAC columns (see above) to identify the nonphosphorylated peptides in the renal cortical membrane fraction. Searches of the spectral data using SEQUEST, InsPecT, and OMSSA identified a total of 28,763 peptides (Fig. 10A). Sorting of these data gave a total of 7,170 unique nonphosphorylated peptides corresponding to 1,817 unique proteins (see Supplemental Table S3). We used the online classification system PANTHER to classify all the identified cortical membrane proteins (Fig. 10B). The most frequent identifiers were transporter, cytoskeletal protein, and select regulatory molecule, in accord with expectations from the use of a cortical membrane fraction for the analysis. The transporter proteins identified are summarized in Supplemental Table S4.
Fig. 10.
LC-MS/MS analysis of nonphosphorylated peptides from rat cortical membrane fraction obtained from flow-through fraction of immobilized metal ion affinity chromatography (IMAC) procedure. A: numbers of peptides identified. Individual circles in Venn diagram show the number of phosphopeptides identified using each of three search algorithms (SEQUEST, InsPecT, and OMSSA). A false discovery rate of <2% was used for each of the searches. B: classification of cortical membrane proteins identified by LC-MS/MS. Histogram shows number of proteins (horizontal axis) associated with a particular PANTHER classification term (vertical axis).
DISCUSSION
Systems biology and epithelial cell physiology.
Physiology is largely an integrative science: knowledge about objects at one level of integration is used to understand function at a higher level of integration (an approach popularly referred to as “systems biology”). Much of cell physiology seeks to understand function of the whole cell in terms of the proteins and other molecules contained in the cell. Step 1 of this process is enumeration of the parts (chiefly proteins) and their properties; step 2 is assembly of this information into various types of models, including mathematical models that can make functional predictions that can be tested experimentally (see Approaches to addressing the functional significance of demonstrated phosphorylation sites). For epithelial cell biology, step 1 began in earnest in the late 1980s, when investigators began to clone cDNAs corresponding to their favorite proteins [e.g., by expression cloning in Xenopus oocytes (19)], generating sequence information and tools that led to an enormous expansion of physiological knowledge. Step 1 has been greatly aided by the advent of the single-species genomic sequencing projects that came to fruition around the turn of the century, spawning large-scale methodologies such as protein MS, expression microarrays, and deep-sequencing of nucleic acid elements. In this study, we apply one of these techniques, protein MS, to the large-scale identification of phosphorylation sites in membrane proteins isolated from the epithelia of the renal cortex.
The renal cortex contains a number of epithelial cell types, including proximal tubule cells, thick ascending limb cells, distal convoluted tubule cells, connecting tubule cells, principal cells of the cortical collecting duct, and various types of intercalated cells, as well as podocytes in the glomerulus. Because of the volumetric dominance of proximal tubule cells, we expected, in the present study, to identify phosphorylation sites mainly, but not exclusively, in proximal tubule proteins, a prediction that was borne out by the data. Since the most visible function of the proximal tubule is transport of a myriad of molecules, we expected to identify a large number of phosphorylation sites in transporter proteins, a prediction that was also confirmed by the data (see Figs. 6 and 7 for an enumeration of the substrates). Finally, since transport is strongly regulated in the kidney, we expected to see phosphorylation sites in membrane-associated regulatory molecules such as PDZ domain-containing proteins, an expectation that was fulfilled. Overall, the study identified a large number of phosphorylation sites, most of which were not found in online database searches and are therefore labeled “novel” on the basis of that criterion. The entire database of phosphorylation sites found in this study can be downloaded or browsed at the National Heart, Lung, and Blood Institute Intramural Research website (http://dir.nhlbi.nih.gov/papers/lkem/rcmpd/).
Phosphoproteomics.
Protein phosphorylation is one of the major ways that signaling networks are altered to effect regulation of the cellular functions of cells, including transport in renal tubule cells (21, 24). The level of protein phosphorylation at a given site is dependent on a balance between the action of individual kinases to phosphorylate the site and individual phosphatases to dephosphorylate it. Phosphorylation is critical not only to regulation of normal function but, also, a variety of pathophysiological processes, including various forms of cancer (8), hypertension (26), and diabetes (2). Therefore, it is valuable to document, on a large scale, the repertoire of phosphorylation sites in renal transporters and associated regulatory proteins. The advent of high-resolution tandem mass spectrometers and sophisticated biochemical separations methodologies upstream from the MS, including phosphopeptide enrichment by IMAC, has facilitated the identification of thousands of phosphorylation sites in multiple cell and tissue types (6). Such studies in the kidney have focused primarily on renal medullary nephron segments (3, 18, 22), urinary exosomes (16), and cell culture models (40, 41). The present study, therefore, provides unique information on phosphorylation sites in membrane proteins in the kidney cortex.
Cutillas et al. (7) and Walmsley et al. (49) reported on the general proteome of membrane fractions from the renal cortex, identifying 251 and 271 proteins, respectively. Our ancillary analysis of nonphosphorylated peptides isolated from the flow-through fraction from the IMAC-enrichment step of the phosphoproteomic workflow included 1,817 proteins, many of which were not identified in the prior studies (Fig. 10; see Supplemental Table S3), significantly extending the known cortical membrane proteome. Although a majority of phosphoproteins identified appear to originate from proximal tubule cells, a significant number of very abundant phosphoproteins from other nephron segments, including the bumetanide-sensitive Na-K-2Cl cotransporter of the thick ascending limb of Henle, the thiazide-sensitive Na-Cl cotransporter of the distal convoluted tubule, and aquaporin-2, a water channel expressed in the connecting tubule and cortical collecting duct, were identified. Interesting, although glomerular membranes may have been expected to be included in the preparation, glomerulus-specific membrane proteins, such as nephrin and podocin, were not found in the list of phosphoproteins identified, possibly because of the volumetric dominance of tubular elements in the renal cortex relative to glomerular elements.
Approaches to addressing the functional significance of demonstrated phosphorylation sites.
Step 2 of the process of understanding function of the whole cell in terms of the proteins and other molecules contained in the cell (see Systems biology and epithelial cell physiology) is the use of information about individual proteins accumulated from all sources to understand the integrated function of epithelial cells. The challenge is to combine knowledge from all types of studies, including reductionist studies, to understand the integrated system, in this case, the individual epithelia that make up the renal cortex. Major progress in this area will require development of computational methods, including mathematical modeling of the type pioneered by Weinstein (50), which uses mass balance equations to show the functional interrelationship between transporters. It will also require further studies at a reductionist level to ascertain the functional consequences of the individual phosphorylation events demonstrated in the present study. In this study, we have used two bioinformatic approaches to address the possible functional significance of individual phosphorylation events: 1) analysis of conservation of the phosphorylated amino acids among mammalian species and 2) analysis of relationships of phosphorylation sites to known functional motifs.
Analysis of phosphorylation site conservation.
One criterion that has been proposed for functional significance of a given phosphorylation site is evolutionary conservation (29); i.e., a given site is considered more likely to be functional if the phosphorylated amino acid (serine, threonine, or tyrosine) is conserved in other species. We analyzed all sites identified in rat renal cortical membrane proteins in relation to the orthologous proteins in other mammalian species reported in the HomoloGene database (see Supplemental Table S2). Among the phosphorylated amino acids reported in this study, 65% showed complete conservation among all species. However, the finding of a similar level of conservation of the amino acids immediately surrounding the phosphorylation sites indicates that the level of phosphorylation site conservation may be a consequence of factors beyond the functional role of the phosphorylation events (Fig. 4B). Nevertheless, the 13% of phosphorylation sites identified that were poorly conserved among mammals other than rats raises doubt about the functional importance of these sites in rat.
Analysis of relationships of phosphorylation sites to known functional motifs.
We also scanned the RefSeq records of all phosphorylated proteins identified in the present study to identify specialized functional regions that contain the identified phosphorylation sites. As shown in Table 5, the phosphorylated functional regions identified included the second PDZ domain in the scaffolding protein Pdzk1 (or NHERF-3). Each identified phosphorylation site yields an independent hypothesis for further experimentation. For example, as shown in Fig. 9, the phosphorylated serine in the second PDZ domain in Pdzk1 lies at the mouth of the PDZ-ligand binding pocket in a position that could be hypothesized to affect the binding of this domain to its ligands. Pdzk1 is strongly expressed in the renal proximal tubule (15) and complexes with a number of proteins, including Slc34a1 (NaPi-IIa), Slc17a1 (NaPi-I), Slc9a3 (NHE-3), Slc22a4 (OCTN1), Slc26a6 (CFEX), and Slc22a12 (URAT1), presumably providing a key element of regulation.
Phosphorylation sites in the Slc12 family of cation-Cl cotransporters, including the thiazide-sensitive Na-Cl cotransporter of the renal distal convoluted tubule.
A particularly important family of transporters are those in the Slc12 group of cation-Cl cotransporters (12). We identified phosphorylation sites in two members of this family, i.e., Slc12a1 (the bumetanide-sensitive Na-K-Cl cotransporter of the thick ascending limb) and Slc12a3 (the thiazide-sensitive Na-Cl cotransporter of the distal convoluted tubule), including a previously unidentified site in each. The Na-K-Cl cotransporter of the thick ascending limb (NKCC2) plays a central role in regulation of water balance through its role in the urinary concentrating mechanism (as the driving force for countercurrent multiplier process) and in dilution of the tubule fluid (30). The thiazide-sensitive Na-Cl cotransporter plays a critical role in the regulation of systemic blood pressure and is a target for regulation by aldosterone (28) and vasopressin (34, 36). Because of their physiological importance, these transporters are the focus of concerted research in a number of laboratories worldwide. A future goal will be to identify the network of kinases and phosphatases that regulate these phosphorylation events as a function of physiological signals.
Similarly, a large number of phosphorylation sites were identified in proximal tubule transporters, many of which are classified as novel. An important goal for the future will be to identify which of these are regulated in response to physiological stimuli, such as changes in parathyroid hormone and angiotensin II levels, and to identify the kinases that are responsible for regulated phosphorylation. A partial clue relevant to the latter goal is the analysis of sequences surrounding the detected phosphorylation sites (Fig. 4C), which showed that most of the sites correspond to acidophilic motifs (dominated by aspartic acid and glutamic acid moieties downstream from the site), basophilic motifs (dominated by arginines and lysines upstream from the site), and so-called proline-directed sites (dominated by prolines up- and downstream from the phosphorylation site) (41).
GRANTS
This study was supported by the Intramural Budget of the NHLBI (Project Z01-HL-001285). Mass spectrometry was done in the NHLBI Proteomics Core Facility (Marjan Gucek, Director). M. Feric was a student intern from the University of Maryland Department of Chemical Engineering (e-mail: mferic@princeton.edu). B. Zhao was a student intern from the University of Michigan in the National Institutes of Health Biomedical Engineering Summer Internship Program, funded by the National Institute of Biomedical Imaging and Bioengineering (e-mail: zhaob@umich.edu).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the authors.
Supplementary Material
ACKNOWLEDGMENTS
The authors thank Prof. Francois Verrey (Institute of Physiology, University of Zurich) for advice concerning transporter substrates and substrate stoichiometry and Guozhong Ma [National Heart, Lung, and Blood Institute (NHLBI)] for help in setting up the Renal Cortical Membrane Phosphoproteome Database.
Footnotes
By convention, all official gene symbols are italicized, whether they refer to proteins or their genes of origin.
REFERENCES
- 1. Albuquerque CP, Smolka MB, Payne SH, Bafna V, Eng J, Zhou H. A multidimensional chromatography technology for in-depth phosphoproteome analysis. Mol Cell Proteomics 7: 1389–1396, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Balaban RS. The mitochondrial proteome: a dynamic functional program in tissues and disease states. Environ Mol Mutagen 51: 352–359, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Bansal AD, Hoffert JD, Pisitkun T, Hwang S, Chou CL, Boja ES, Wang G, Knepper MA. Phosphoproteomic profiling reveals vasopressin-regulated phosphorylation sites in collecting duct. J Am Soc Nephrol 21: 303–315, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Beausoleil SA, Villen J, Gerber SA, Rush J, Gygi SP. A probability-based approach for high-throughput protein phosphorylation analysis and site localization. Nat Biotechnol 24: 1285–1292, 2006 [DOI] [PubMed] [Google Scholar]
- 5. Biber J, Stieger B, Haase W, Murer H. A high yield preparation for rat kidney brush border membranes. Different behaviour of lysosomal markers. Biochim Biophys Acta 647: 169, 1981 [DOI] [PubMed] [Google Scholar]
- 6. Choudhary C, Mann M. Decoding signalling networks by mass spectrometry-based proteomics. Nat Rev Mol Cell Biol 11: 427–439, 2010 [DOI] [PubMed] [Google Scholar]
- 7. Cutillas PR, Biber J, Marks J, Jacob R, Stieger B, Cramer R, Waterfield M, Burlingame AL, Unwin RJ. Proteomic analysis of plasma membrane vesicles isolated from the rat renal cortex. Proteomics 5: 101–112, 2005 [DOI] [PubMed] [Google Scholar]
- 8. Del Rosario AM, White FM. Quantifying oncogenic phosphotyrosine signaling networks through systems biology. Curr Opin Genet Dev 20: 23–30, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Elias JE, Gibbons FD, King OD, Roth FP, Gygi SP. Intensity-based protein identification by machine learning from a library of tandem mass spectra. Nat Biotechnol 22: 214–219, 2004 [DOI] [PubMed] [Google Scholar]
- 10. Elias JE, Gygi SP. Target-decoy search strategy for increased confidence in large-scale protein identifications by mass spectrometry. Nat Methods 4: 207–214, 2007 [DOI] [PubMed] [Google Scholar]
- 11. Eng JK, McCormack AL, Yates JR., 3rd An approach to correlate tandem mass spectral data of peptides with amino acid sequences in a protein database. J Am Soc Mass Spectrom 5: 976–989, 1994 [DOI] [PubMed] [Google Scholar]
- 12. Gamba G. Molecular physiology and pathophysiology of electroneutral cation-chloride cotransporters. Physiol Rev 85: 423–493, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Geer LY, Markey SP, Kowalak JA, Wagner L, Xu M, Maynard DM, Yang X, Shi W, Bryant SH. Open mass spectrometry search algorithm. J Proteome Res 3: 958–964, 2004 [DOI] [PubMed] [Google Scholar]
- 14. Gimenez I, Forbush B. Short-term stimulation of the renal Na-K-Cl cotransporter (NKCC2) by vasopressin involves phosphorylation and membrane translocation of the protein. J Biol Chem 278: 26946–26951, 2003 [DOI] [PubMed] [Google Scholar]
- 15. Gisler SM, Pribanic S, Bacic D, Forrer P, Gantenbein A, Sabourin LA, Tsuji A, Zhao ZS, Manser E, Biber J, Murer I. HPDZK1: a major scaffolder in brush borders of proximal tubular cells. Kidney Int 64: 1733–1745, 2003 [DOI] [PubMed] [Google Scholar]
- 16. Gonzales PA, Pisitkun T, Hoffert JD, Tchapyjnikov D, Star RA, Kleta R, Wang NS, Knepper MA. Large-scale proteomics and phosphoproteomics of urinary exosomes. J Am Soc Nephrol 20: 363–379, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Gruhler A, Olsen JV, Mohammed S, Mortensen P, Faergeman NJ, Mann M, Jensen ON. Quantitative phosphoproteomics applied to the yeast pheromone signaling pathway. Mol Cell Proteomics 4: 310–327, 2005 [DOI] [PubMed] [Google Scholar]
- 18. Gunaratne R, Braucht DW, Rinschen MM, Chou CL, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals cAMP/vasopressin-dependent signaling pathways in native renal thick ascending limb cells. Proc Natl Acad Sci USA 107: 15653–15658, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hediger MA, Coady MJ, Ikeda TS, Wright EM. Expression cloning and cDNA sequencing of the Na+/glucose co-transporter. Nature 330: 379–381, 1987 [DOI] [PubMed] [Google Scholar]
- 20. Hoffert JD, Fenton RA, Moeller HB, Simons B, Tchapyjnikov D, McDill BW, Yu MJ, Pisitkun T, Chen F, Knepper MA. Vasopressin-stimulated increase in phosphorylation at Ser269 potentiates plasma membrane retention of aquaporin-2. J Biol Chem 283: 24617–24627, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Hoffert JD, Knepper MA. Taking aim at shotgun phosphoproteomics. Anal Biochem 375: 1–10, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Hoffert JD, Pisitkun T, Wang G, Shen RF, Knepper MA. Quantitative phosphoproteomics of vasopressin-sensitive renal cells: regulation of aquaporin-2 phosphorylation at two sites. Proc Natl Acad Sci USA 103: 7159–7164, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Hoffert JD, Wang G, Pisitkun T, Shen RF, Knepper MA. An automated platform for analysis of phosphoproteomic datasets: application to kidney collecting duct phosphoproteins. J Proteome Res 6: 3501–3508, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Hoffert JD, Chou CL, Knepper MA. Aquaporin-2 in the “-omics” era. J Biol Chem 284: 14683–14687, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Jorgensen C, Sherman A, Chen GI, Pasculescu A, Poliakov A, Hsiung M, Larsen B, Wilkinson DG, Linding R, Pawson T. Cell-specific information processing in segregating populations of Eph receptor ephrin-expressing cells. Science 326: 1502–1509, 2009 [DOI] [PubMed] [Google Scholar]
- 26. Kahle KT, Rinehart J, Giebisch G, Gamba G, Hebert SC, Lifton RP. A novel protein kinase signaling pathway essential for blood pressure regulation in humans. Trends Endocrinol Metab 19: 91–95, 2008 [DOI] [PubMed] [Google Scholar]
- 27. Kim GH, Martin SW, Fernandez-Llama P, Masilamani S, Packer RK, Knepper MA. Long-term regulation of renal Na-dependent cotransporters and ENaC: response to altered acid-base intake. Am J Physiol Renal Physiol 279: F459–F467, 2000 [DOI] [PubMed] [Google Scholar]
- 28. Kim GH, Masilamani S, Turner R, Mitchell C, Wade JB, Knepper MA. The thiazide-sensitive Na-Cl cotransporter is an aldosterone-induced protein. Proc Natl Acad Sci USA 95: 14552–14557, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Levy ED, Landry CR, Michnick SW. Cell signaling. Signaling through cooperation. Science 328: 983–984, 2010 [DOI] [PubMed] [Google Scholar]
- 30. Masilamani S, Knepper MA, Burg MB. Urine concentration and dilution. In: The Kidney, edited by Brenner BM. Philadelphia: Saunders, 2000, p. 595–636 [Google Scholar]
- 31. Moeller HB, MacAulay N, Knepper MA, Fenton RA. Role of multiple phosphorylation sites in the COOH-terminal tail of aquaporin-2 for water transport: evidence against channel gating. Am J Physiol Renal Physiol 296: F649–F657, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Molina H, Horn DM, Tang N, Mathivanan S, Pandey A. Global proteomic profiling of phosphopeptides using electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci USA 104: 2199–2204, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Moriguchi T, Urushiyama S, Hisamoto N, Iemura S, Uchida S, Natsume T, Matsumoto K, Shibuya H. WNK1 regulates phosphorylation of cation-chloride-coupled cotransporters via the STE20-related kinases, SPAK and OSR1. J Biol Chem 280: 42685–42693, 2005 [DOI] [PubMed] [Google Scholar]
- 34. Mutig K, Saritas T, Uchida S, Kahl T, Borowski T, Paliege A, Bohlick A, Bleich M, Shan Q, Bachmann S. Short-term stimulation of the thiazide-sensitive Na+-Cl− cotransporter by vasopressin involves phosphorylation and membrane translocation. Am J Physiol Renal Physiol 298: F502–F509, 2010 [DOI] [PubMed] [Google Scholar]
- 35. Nielsen S, Maunsbach AB, Ecelbarger CA, Knepper MA. Ultrastructural localizatioon of Na-K-2Cl cotransporter in thick ascending limb and macula densa of rat kidney. Am J Physiol Renal Physiol 275: F885–F893, 1998 [DOI] [PubMed] [Google Scholar]
- 36. Pedersen NB, Hofmeister MV, Rosenbaek LL, Nielsen J, Fenton RA. Vasopressin induces phosphorylation of the thiazide-sensitive sodium chloride cotransporter in the distal convoluted tubule. Kidney Int 78: 160–169, 2010 [DOI] [PubMed] [Google Scholar]
- 37. Pisitkun T, Bieniek J, Tchapyjnikov D, Wang G, Wu WW, Shen RF, Knepper MA. High-throughput identification of IMCD proteins using LC-MS/MS. Physiol Genomics 25: 263–276, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Pisitkun T, Jacob V, Schleicher SM, Chou CL, Yu MJ, Knepper MA. Akt and ERK1/2 pathways are components of the vasopressin signaling network in rat native IMCD. Am J Physiol Renal Physiol 295: F1030–F1043, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Richardson C, Rafiqi FH, Karlsson HK, Moleleki N, Vandewalle A, Campbell DG, Morrice NA, Alessi DR. Activation of the thiazide-sensitive Na+-Cl− cotransporter by the WNK-regulated kinases SPAK and OSR1. J Cell Sci 121: 675–684, 2008 [DOI] [PubMed] [Google Scholar]
- 40. Rinehart J, Maksimova YD, Tanis JE, Stone KL, Hodson CA, Zhang J, Risinger M, Pan W, Wu D, Colangelo CM, Forbush B, Joiner CH, Gulcicek EE, Gallagher PG, Lifton RP. Sites of regulated phosphorylation that control K-Cl cotransporter activity. Cell 138: 525–536, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Rinschen MM, Yu MJ, Wang G, Boja ES, Hoffert JD, Pisitkun T, Knepper MA. Quantitative phosphoproteomic analysis reveals vasopressin V2-receptor-dependent signaling pathways in renal collecting duct cells. Proc Natl Acad Sci USA 107: 3882–3887, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Ruttenberg BE, Pisitkun T, Knepper MA, Hoffert JD. PhosphoScore: an open-source phosphorylation site assignment tool for MSn data. J Proteome Res 7: 3054–3059, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23: 1391–1398, 2005 [DOI] [PubMed] [Google Scholar]
- 44. Tanner S, Shu H, Frank A, Wang LC, Zandi E, Mumby M, Pevzner PA, Bafna V. InsPecT: identification of posttranslationally modified peptides from tandem mass spectra. Anal Chem 77: 4626–4639, 2005 [DOI] [PubMed] [Google Scholar]
- 45. Tchapyjnikov D, Li Y, Pisitkun T, Hoffert JD, Yu MJ, Knepper MA. Proteomic profiling of nuclei from native renal inner medullary collecting duct cells using LC-MS/MS. Physiol Genomics 40: 167–183, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Terris J, Ecelbarger CA, Nielsen S, Knepper MA. Long-term regulation of four renal aquaporins in rat. Am J Physiol Renal Fluid Electrolyte Physiol 271: F414–F422, 1996 [DOI] [PubMed] [Google Scholar]
- 47. Turban S, Beutler KT, Morris RG, Masilamani S, Fenton RA, Knepper MA, Packer RK. Long-term regulation of proximal tubule acid-base transporter abundance by angiotensin II. Kidney Int 70: 660–668, 2006 [DOI] [PubMed] [Google Scholar]
- 48. Vitari AC, Thastrup J, Rafiqi FH, Deak M, Morrice NA, Karlsson HK, Alessi DR. Functional interactions of the SPAK/OSR1 kinases with their upstream activator WNK1 and downstream substrate NKCC1. Biochem J 397: 223–231, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Walmsley SJ, Broeckling C, Hess A, Prenni J, Curthoys NP. Proteomic analysis of brush-border membrane vesicles isolated from purified proximal convoluted tubules. Am J Physiol Renal Physiol 298: F1323–F1331, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Weinstein AM. A mathematical model of rat ascending Henle limb. I. Cotransporter function. Am J Physiol Renal Physiol 298: F512–F524, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Wolf-Yadlin A, Hautaniemi S, Lauffenburger DA, White FM. Multiple reaction monitoring for robust quantitative proteomic analysis of cellular signaling networks. Proc Natl Acad Sci USA 104: 5860–5865, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Yang SS, Lo YF, Wu CC, Lin SW, Yeh CJ, Chu P, Sytwu HK, Uchida S, Sasaki S, Lin SH. SPAK-knockout mice manifest Gitelman syndrome and impaired vasoconstriction. J Am Soc Nephrol 21: 1868–1877, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Yang SS, Morimoto T, Rai T, Chiga M, Sohara E, Ohno M, Uchida K, Lin SH, Moriguchi T, Shibuya H, Kondo Y, Sasaki S, Uchida S. Molecular pathogenesis of pseudohypoaldosteronism type II: generation and analysis of a Wnk4D561A/+ knockin mouse model. Cell Metab 5: 331–344, 2007 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.