Abstract
Hepcidin negatively regulates systemic iron homeostasis in response to inflammation and elevated serum iron. Conversely, hepcidin expression is diminished in response to hypoxia, oxidative stress, and increased erythropoietic demand, though the molecular intermediates involved are incompletely understood. To address this, we have investigated hypoxic hepcidin regulation in HuH7 hepatoma cells either cultured alone or cocultured with activated THP-1 macrophages. HuH7 hepcidin mRNA expression was determined using quantitative polymerase chain reaction (Q-PCR). Hepcidin promoter activity was measured using luciferase reporter constructs containing a 0.9 kb fragment of the wild-type human hepcidin promoter, and constructs containing mutations in bone morphogenetic protein (BMP)/SMAD4, signal transducer and activator of transcription 3 (STAT3), CCAAT/enhancer-binding protein (C/EBP), and E-box-responsive elements. Hepatic expression of bone morphogenetic proteins BMP2 and BMP6 and the BMP inhibitor noggin was determined using Q-PCR, and the protein expression of hemojuvelin (HJV), pSMAD 1/5/8, and SMAD4 was determined by western blotting. Following exposure to hypoxia or H2O2, hepcidin mRNA expression and promoter activity increased in HuH7 cells monocultures but were decreased in HuH7 cells cocultured with THP-1 macrophages. This repression was attenuated by mutation of the BMP/SMAD4-response element, suggesting that modulation of SMAD signaling mediated the response to hypoxia. No changes in hepatocyte BMP2, BMP6 or noggin mRNA, or protein expression of HJV or pSMAD 1/5/8 were detected. However, treatment with hypoxia caused a marked decrease in nuclear and cytosolic SMAD4 protein and SMAD4 mRNA expression in cocultured HuH7 cells. Together these data indicate that hypoxia represses hepcidin expression through inhibition of BMP/SMAD signaling.
Keywords: hepatocytes, macrophages, oxidative stress
the liver-expressed peptide hepcidin (23) has emerged as a major regulator of systemic iron homeostasis. During inflammation, hepcidin is secreted into the serum, where it blocks iron release from duodenal enterocytes and reticuloendothelial macrophages, causing hypoferremia (8, 10, 12, 20, 36). Hepcidin transcription is also sensitive to systemic iron levels, body iron stores, erythropoietic factors, and hypoxia in vivo (14, 29, 33), and the regulated production and release of the peptide plays a central role in the maintenance of iron homeostasis. This is supported by the inappropriate regulation of hepcidin expression seen in transgenic animal models of iron overload that lack functional genes for HFE, hemojuvelin (HJV), or transferrin receptor 2 (1, 6, 28, 31).
The systemic and intracellular mechanisms that underlie negative regulation of hepcidin by hypoxia and oxidative stress are incompletely understood. Studies in rodent models have demonstrated dramatic effects of hypoxia on iron homeostasis: increased intestinal iron absorption (24, 25, 30, 37) and decreased expression of hepcidin (24, 29, 32). The molecular intermediates that signal the effects of hypoxia on hepcidin expression are unclear, but possible factors include erythropoietin (16, 34), hypoxia-inducible factors (HIFs) (32), and prolyl hydroxylases (5). While the hepcidin promoter contains putative HIF-binding sites (4, 42), recent evidence has demonstrated that the effects of hypoxia on hepcidin expression are not mediated directly by HIF-promoter binding (42). Reactive oxygen species produced as a consequence of hypoxia have also been implicated in the transcriptional repression of hepcidin and appear to operate via inhibitory effects on signal transducer and activator of transcription 3 (STAT3) and CCAAT/enhancer-binding protein (C/EBP) signaling (9). Furthermore, there is evidence that hypoxia inhibits the HJV/bone morphogenetic protein (BMP) signaling cascade and thereby decreases hepcidin expression (39). In the present study we have further investigated the roles of these pathways in the hypoxic regulation of hepcidin expression.
Experiments were performed using HuH7 hepatoma cells as a hepatocyte model. Cells either were used alone as monocultures or were cocultured with THP-1 macrophages. We have recently characterized this coculture model (26) and have shown along with others (17) that it ensures an appropriate hepatocyte hepcidin response to a number of stimuli.
METHODS
Cell culture.
HuH7 human hepatoma cells were grown in DMEM containing 10% fetal bovine serum and were used for experiments at 80% confluence. THP-1 cells, grown in RPMI 1640 medium containing 10% fetal bovine serum, were seeded at 106 cells per well on Transwell filters and treated overnight with phorbol myristate acetate (100 nmol/l) to induce differentiation and filter attachment. After differentiation, cells were washed and incubated in fresh media for 24 h before experimentation.
Coculture and treatments.
HuH7 hepatoma cells were seeded at a density of 0.5 × 106 cells per well in six-well plates and grown for 48 h. On the day of experiment, HuH7 cells were washed and given fresh medium and Transwell inserts containing differentiated THP-1 macrophages were overlaid onto six-well plates containing HuH7 cells. In some experiments, HuH7 cells were exposed either to conditioned medium from THP-1 macrophages cultured under normoxic condition or to IL-1β [10 ng/ml; this concentration was chosen to be consistent with our previous studies using this coculture model; (26)].
For hypoxia treatments, cell cultures were placed in a sealed Perspex chamber. The chamber was then flushed with air containing 1% O2, 5% CO2 and 94% N2 and then sealed and incubated at 37°C for 24 h. In some experiments, cells were cultured under normoxic conditions in the presence of the oxidative stress agent H2O2 (100 μM). On the basis of Trypan blue exclusion studies, there was no effect of either treatment on cell viability (data not shown).
Real-time quantitative PCR.
Total RNA was isolated from HuH7 or THP-1 cells using TRIzol reagent (Invitrogen). Following first-strand synthesis, expression levels of hepcidin, BMP2, BMP6, noggin, SMAD4, SMAD7, and 18S (used as a housekeeper gene) mRNA were analyzed by real-time PCR using an ABI Prism 7000HT PCR cycler with gene-specific primers (15, 21, 26) and a Quanti-Tect SYBR Green PCR kit (Qiagen), according to the manufacturer's protocol. Quantitative measurements of each gene were derived from a standard curve constructed from known concentrations of PCR product.
Cell transfection and luciferase reporter assays.
Full details concerning the generation and utilization of hepcidin promoter plasmid constructs have been described previously (26). Briefly, HuH7 cells were transfected with the wild-type, STAT3 mutant, C/EBP mutant, E-box mutant, BMP/SMAD4 mutant hepcidin-luciferase constructs or the empty pGL3-basic vector, using Fugene6 (Roche) according to the manufacturer's instructions. As a normalization control, the pRL-SV40 Renilla luciferase plasmid (Promega) was cotransfected alongside the hepcidin constructs in a 1:50 ratio. After 24 h, cells were exposed to H2O2 or hypoxia for a further 24 h and luciferase activity was determined in triplicate using the Promega Dual Luciferase Reporter Assay, according to the manufacturer's instructions.
Preparation of nuclear protein extracts.
HuH7 cells were scraped off plates, washed in ice-cold PBS, and lysed in sucrose buffer (in mM: 320 sucrose, 3 CaCl2 2 Mg acetate, 0.1 EDTA, 1 DTT, and 0.5 PMSF) containing 0.5% NP-40. After centrifugation at 1,500 g for 5 min, the cytosolic fraction (supernatant) was removed and stored at −80°C. The nuclear pellet was washed in sucrose buffer without NP-40 and centrifuged at 1,500 g for a further 5 min. After the pellet was dried, nuclei were resuspended in sucrose buffer. Approximately 0.6 volumes of high salt buffer (20 mM HEPES, pH 7.9, 1.5 mM MgCl2 800 mM KCl, 0.2 mM EDTA, 1 mM DTT, 0.5 mM PMSF, 25% glycerol, and 0.5% NP-40) were then added in 0.2 volume aliquots until nuclei were lysed.
Western blot analysis.
HuH7 cells were harvested into ice-cold lysis buffer (PBS containing 1% sodium dodecyl sulfate and 10 mg/l protease inhibitor cocktail) and homogenized by repeated passing through at 25-gauge needle. Protein samples (40 μg) were solubilized in sample loading buffer and subjected to polyacrylamide gel electrophoresis. Following immobilization on nitrocellulose, the proteins were exposed to commercially available anti-phospho-SMAD 1/5/8 antibody, SMAD4 antibody (1:1,000 dilution, Cell Signaling Technology), or HJV antibody (1:1,000 dilution, anti-RMGc, Santa Cruz; Supplemental Fig. S2; Supplemental Material for this article is provided in an online supplement at the Journal website). Cross-reactivity was observed using a horseradish peroxidase-linked secondary antibody (Dako) and ECL Plus (GE Healthcare). Band densities were semiquantified using Scion Image software (Scion, Frederick, MD). At the end of the experiment, the nitrocellulose membranes were stripped (Western Stripping Buffer, Perbio Science) and reprobed with antibodies to actin (1:2,000 dilution, Sigma-Aldrich), which acted as a loading control. In some experiments, β-tubulin (1:1,000 dilution, Cell Signaling Technology) was used as a cytosolic protein marker.
Statistical analysis.
Data are presented as means ± SE. Statistical differences (P < 0.05) among groups were determined by one-way ANOVA, followed by Tukey's post hoc test when F-test was significant at P < 0.05.
RESULTS
Hypoxia causes repression of hepcidin in HuH7 cells cultured in the presence of THP-1 macrophages.
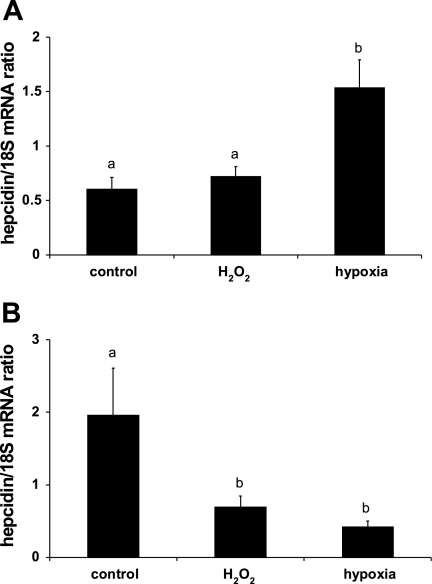
Hepcidin mRNA expression in monocultured HuH7 hepatoma cells was unchanged following exposure to 100 μM H2O2 but was increased in cells cultured under hypoxic conditions (Fig. 1A). In contrast, when HuH7 cells were cocultured with THP-1 macrophages, treatment with both H2O2 and hypoxia decreased hepcidin mRNA to 17 ± 6% and 29 ± 6% (P < 0.001) of the control, respectively (Fig. 1B).
Fig. 1.
Hypoxia represses hepcidin expression in HuH7/THP-1 cell cocultures. HuH7 cells either cultured alone (A) or cocultured with THP-1 macrophages (B) were exposed to hypoxia (1% O2) or 100 μM H2O2. In HuH7 monocultures, hypoxia significantly increased hepcidin expression, whereas in the coculture model, HuH7 hepcidin levels were significantly decreased by both hypoxia and H2O2 treatment. Data are presented as hepcidin/18S ratios and are means ± SE of 6–12 observations in each group. Different letters above data bars indicate that these groups are significantly different (P < 0.05).
Similar observations were also made in a second human hepatoma cell line (HepG2 cells); hypoxia increased hepcidin expression in monocultured HepG2 cells but significantly repressed hepcidin expression in THP-1-stimulated HepG2 cells (Supplemental Fig. S1).
Oxidative repression of HuH7 hepcidin mRNA is reconstituted in the presence of conditioned medium from THP-1 macrophages.
The decrease in hepcidin mRNA observed in the coculture model (Fig. 1B) could arise from direct effects of hypoxia or H2O2 on either the THP-1 cells or the HuH7 cells or both. We have previously demonstrated that the generation and release of cytokines, particularly IL-1β, by activated THP-1 cells are required for induction of hepcidin mRNA in our coculture system (26). In the present study the expression of IL-1β mRNA in THP-1 cells was significantly decreased by treatment with H2O2 (−43.6 ± 8.5%, P < 0.05) and hypoxia (−53.8 ± 9.0%, P < 0.05), suggesting that suppression of cytokine production by THP-1 cells may be involved in the attenuated hepcidin response.
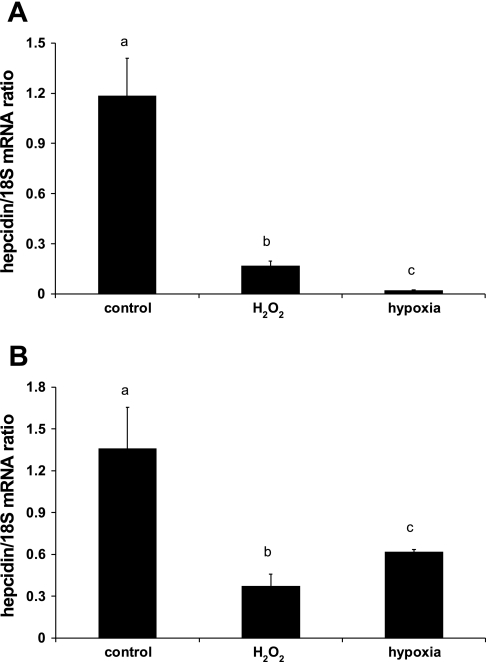
To determine whether there was also a direct influence of hypoxia or oxidative stress on HuH7 cells, we prestimulated HuH7 monocultures with conditioned medium derived from activated THP-1 macrophages grown under normoxic conditions. HuH7 cells were subsequently treated with H2O2 or hypoxia for 24 h. Under both of these conditions, hepcidin mRNA was significantly repressed (Fig. 2A). Furthermore, monocultured HuH7 cells prestimulated with IL-1β and subsequently exposed to 100 μM H2O2 or 1% O2 for 24 h also displayed a significant decrease in hepcidin mRNA (Fig. 2B).
Fig. 2.
Hypoxic repression of hepcidin mRNA is reconstituted in the presence of THP-1-conditioned medium. HuH7 monocultures were exposed to conditioned medium from activated THP-1 macrophages cultured under normoxic conditions (A) or were treated with IL-1β (10 ng/ml) (B) before exposure to hypoxia (1% O2) or 100 μM H2O2 for 24 h. Hepcidin mRNA (expressed as hepcidin/18S ratio) was significantly decreased in all groups compared with controls. Data are means ± SE of 6 observations in each group. Different letters above data bars indicate that these groups are significantly different (P < 0.05).
Mutation of proximal SMAD4-binding site on the hepcidin promoter prevents repression by hypoxia.
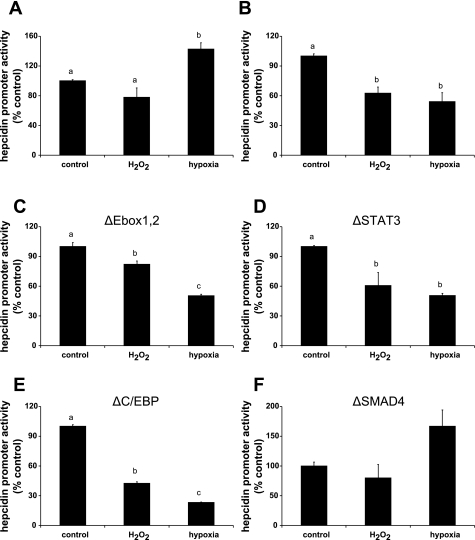
To identify the transcriptional mechanisms by which hypoxia and H2O2 regulate hepcidin expression, HuH7 hepatoma cells were transfected with a luciferase construct containing 0.9 kb of the wild-type hepcidin promoter. Consistent with the quantitative PCR (Q-PCR) experiments (Fig. 1), luciferase reporter activity in HuH7 monocultures was significantly increased by hypoxia (Fig. 3A), whereas in the coculture system it was significantly decreased (Fig. 3B) compared with the untreated control groups. Hepcidin expression was also decreased by H2O2 treatment in the coculture experiments (Fig. 3B).
Fig. 3.
Human hepcidin promoter activity is repressed by hypoxia and oxidative stress. HuH7 cells were transfected with luciferase reporter constructs and 24 h later were exposed to hypoxia (1% O2) or 100 μM H2O2 for 24 h in the absence (A) or presence (B–F) of activated THP-1 cells. Luciferase activity was measured after 24 h. Hepcidin promoter activity was repressed by hypoxia and H2O2 treatments in HuH7 cocultures transfected with the wild-type (B) and E-box (C)-, signal transducer and activator of transcription 3 (STAT3) (D)-, and CCAAT/enhancer-binding protein (C/EBP)-mutant (E) constructs. No effect of hypoxia and H2O2 treatments was observed in HuH7 monocultures transfected with the wild-type promoter (A) or in cocultures transfected with bone morphogenetic protein (BMP)/SMAD mutant promoter (F). Data are means ± SE of 4–6 observations in each group and are expressed as a percentage of the control group for each experiment. Different letters above data bars indicate that these groups are significantly different (P < 0.05).
To investigate further the mechanism for the transcriptional repression of hepcidin expression by hypoxia and H2O2 in the HuH7/THP-1 coculture system, we transfected HuH7 cells with mutant [ΔSTAT3, ΔC/EBP, ΔE-box, ΔSMAD4; (26)] luciferase constructs. Suppression of hepcidin promoter activity was sustained in the presence of mutations in the STAT3 (Fig. 3D) and C/EBP (Fig. 3E) response elements, and with the reporter construct containing mutations in the E-boxes (Fig. 3C). However, mutation of the SMAD4 response element prevented H2O2- and hypoxia-mediated repression of hepcidin promoter activity in cocultured HuH7 cells (Fig. 3F).
Hypoxia diminishes BMP signaling in HuH7 cells.
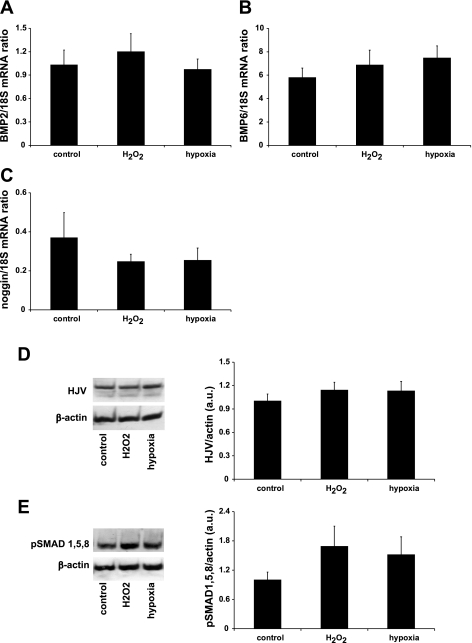
Given the necessity of the SMAD4 response element for regulation of hepcidin by oxidative stress and hypoxia, we tested whether regulation of hepatic expression of BMPs may be responsible for hepcidin repression. However, mRNA expression of BMP2, BMP6, and the endogenous BMP inhibitor noggin was not regulated following treatment with H2O2 or hypoxia (Fig. 4, A–C).
Fig. 4.
Endogenous expression of BMPs and receptor-activated SMAD 1/5/8 (R-SMADs) in HuH7 cells is not altered by hypoxia. mRNA expression levels of the endogenous BMP/hemojuvelin (HJV) receptor ligands BMP2 (A) and BMP6 (B) together with the BMP receptor inhibitor noggin (C) were not altered by treatment with H2O2 or hypoxia in HuH7 cocultures. In addition, exposure to hypoxia (1% O2) or 100 μM H2O2 did not alter the protein expression of HJV (D), the coreceptor required for BMP-mediated regulation of hepcidin expression, or phospho-SMAD 1/5/8 (E) activated by BMP binding to the BMP receptor/HJV complex. AU, arbitrary units. Data are presented as means ± SE of 4–6 observations.
Next, we determined whether hypoxia might directly influence the BMP/hepcidin signaling pathway. HJV is a coreceptor that is required for the initiation of the BMP receptor signaling cascade. However, Western blotting revealed that HJV protein levels were unaltered by H2O2 or hypoxia (Fig. 4D). Similarly, there was no significant change in the phosphorylation of HJV/BMP receptor-activated SMAD 1/5/8 in either treatment group (Fig. 4E).
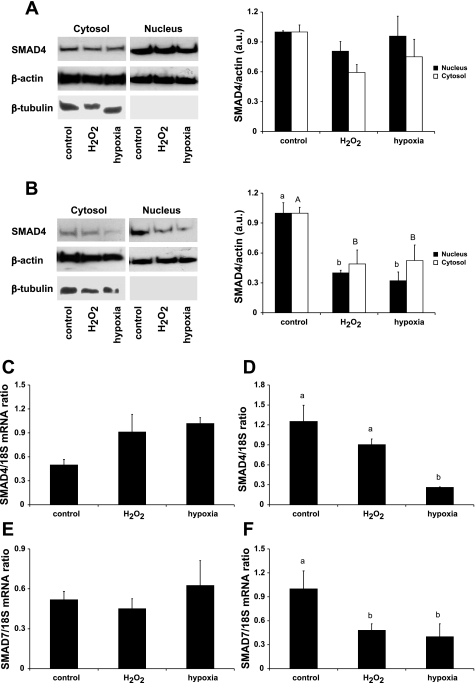
The transcription factor SMAD4 is a co-SMAD that dimerizes with receptor-activated SMADs and enhances BMP and transforming growth factor-β signaling. We detected SMAD4 protein in nuclear and cytosolic extracts from HuH7 cells (cytosolic extracts were positive for the marker protein β-tubulin; nuclear extracts were β-tubulin negative; Fig. 5). In HuH7 monocultures there were no significant changes in either nuclear or cytosolic SMAD4 protein following treatment with H2O2 or hypoxia (Fig. 5A). However, both nuclear (P < 0.001) and cytosolic (P < 0.05) SMAD4 protein was significantly decreased by H2O2 and hypoxia in THP-1-stimulated HuH7 cells compared with control (Fig. 5B). SMAD4 mRNA in HuH7 monoculture was not significantly affected by either treatment (Fig. 5C); however, in THP-1-stimulated HuH7 cells, SMAD4 mRNA was significantly decreased by hypoxia (Fig. 5D).
Fig. 5.
Hypoxia suppresses the expression of SMAD4 and SMAD7 in THP-1-stimulated HuH7 cells. Protein (A) and mRNA (C) expression levels of SMAD4, the coactivator of BMP signaling, were not altered by hypoxia or H2O2 in HuH7 monocultures. However, in THP-1-stimulated HuH7 cells, SMAD4 protein levels in both nuclear and cytosolic fractions were significantly decreased by hypoxia and oxidative stress (B). SMAD4 mRNA was also significantly decreased by hypoxia in THP-1-stimulated HuH7 cells (D). In addition, expression of the SMAD4-regulated inhibitory factor SMAD7 was decreased by both hypoxia and H2O2 in THP-1-stimulated HuH7 cells (F) but not in HuH7 monocultures (E). Data are means ± SE of 4–6 observations in each group. Different letters above data bars indicate that these groups are significantly different (P < 0.05). In B, lowercase letters denote differences in nuclear fractions and uppercase letters denote differences in cytosolic fractions.
SMAD7 is a SMAD4-regulated gene (18) and has recently been identified as a negative regulator of hepcidin expression in hepatoma cells (27). We investigated whether SMAD7 expression might be upregulated by hypoxia in THP-1-stimulated HuH7 cells and thereby mediate the inhibition of hepcidin. However, while there was no effect of hypoxia or H2O2 on SMAD7 mRNA levels in monocultured cells (Fig. 5E), SMAD7 mRNA was significantly decreased by both hypoxia and H2O2 in HuH7 cocultures (Fig. 5F).
DISCUSSION
Hepcidin is a major regulator of systemic iron homeostasis and as such is responsive to changes in iron status (14, 33), erythropoietic requirements (13, 29), and hypoxia (24, 29, 32). In hepatic cell lines, hypoxia is linked to suppression of hepcidin expression (5, 9); however, this finding is not consistent and recent studies suggest that hypoxia increases hepcidin expression in HuH7 hepatoma cells (42). We have recently characterized a HuH7 cell/THP-1 macrophage coculture system in which cross talk between macrophages and hepatoma cell lines leads to the induction of hepcidin mRNA (26). In the present study we further explored the influence of macrophages on the regulation of hepcidin in the context of known negative stimuli, namely, hypoxia and oxidative stress (induced by H2O2). Interestingly, we found differential effects of hypoxia on hepcidin expression in our studies: an increase in expression in monocultures of HuH7 cells—consistent with the recent findings of Volke et al. (42)—but a significant decrease in hepcidin mRNA in the HuH7/THP-1 coculture model.
As shown in our previous study (26) and by others (22), activated THP-1 macrophages secrete a number of inflammatory cytokines including IL-1β and IL-6, which can subsequently stimulate hepcidin expression. We hypothesized that decreased macrophage production of these cytokines may contribute to hepatic suppression of hepcidin by hypoxia and oxidative stress. In preliminary experiments we observed a significant decrease in macrophage IL-1β mRNA following hypoxia and oxidative stress; however, it was clear from subsequent studies that the decrease in THP-1 IL-1β production alone could not account for the inhibitory effects on HuH7 cell hepcidin mRNA in the coculture model. When HuH7 cell monocultures were prestimulated with conditioned media derived from THP-1 cells grown under normoxic conditions, we also observed a dramatic hypoxic and oxidative suppression of hepcidin mRNA. Interestingly, we observed the same significant suppression of hepcidin mRNA in HuH7 monocultures prestimulated with IL-1β and subsequently exposed to hypoxia or H2O2. Hence it appears, in our in vitro model at least, that macrophage-derived factors are required to prime basal hepatic hepcidin expression, but that hypoxia and oxidative stress act directly on the stimulated HuH7 cells to suppress hepcidin mRNA. It remains unclear whether macrophage/hepatocyte cross talk is equally important in the regulation of hepcidin expression in vivo.
Next we focused on the transcriptional regulation of hepatocyte hepcidin expression. A number of putative transcription factor-binding sites have been identified in the proximal 0.9 kb of the human hepcidin promoter, including characterized response elements for STAT3 (41), SMAD4 (40), and members of the CCAAT enhancer-binding protein family (C/EBP) (11), as well as two E-boxes (4) which bind a number of basic helix-loop-helix factors and potentially HIFs (32). Using hepcidin promoter-luciferase reporter constructs containing mutations in these transcription factor-binding sites, we showed that the proximal SMAD4-binding site is required for the repression of hepcidin in this model. Within the hepatocyte the SMAD pathway is essential for the transmission of BMP signals and the regulation of hepcidin gene transcription (3, 7, 43). Importantly, an intact BMP/SMAD signaling pathway is essential for maintaining basal hepcidin expression (26, 40). We therefore hypothesized that a decrease in hepatic BMP/SMAD signaling may offer an explanation for oxidative and hypoxic hepcidin repression. There were no changes in hepatic BMP2, BMP6, or noggin levels, suggesting that regulation of endogenous hepatic ligands and inhibitors of this signaling pathway is not involved in this mechanism. Additionally, membrane levels of HJV, the coreceptor required for the initiation of the BMP receptor signaling cascade, were unaltered by exposure to H2O2 and hypoxia, and, furthermore, no significant changes in phosphorylation of the receptor SMAD 1/5/8 complex were observed.
We therefore investigated the effect of hypoxia and oxidative stress on expression of SMAD4, commonly referred to as the co-SMAD, which forms complexes with receptor SMADs to elicit their transcriptional response (44). SMAD4 mRNA was increased by hypoxia in monocultured HuH7 cells, but this did not reach statistical significance (P = 0.07). However, in THP-1-stimulated HuH7 cells, SMAD4 mRNA was significantly decreased by hypoxia. SMAD4 protein (both nuclear and cytosolic) was also decreased in the THP-1-stimulated HuH7 cells following exposure to hypoxia and H2O2, suggesting that the decreased expression of SMAD4 might mediate the hypoxic repression of hepcidin expression in the coculture model.
SMAD4 is involved in chromatin remodeling, stabilizing transcriptional complexes, and recruiting coactivators (44). The complete blunting of hepcidin to all inflammatory cytokines and BMPs in SMAD4-knockout mice (43) suggests that decreased SMAD4 may be a central mechanism of hepcidin repression. Hypoxia and reactive oxygen species have been implicated in both the induction and the inhibition of receptor SMADs (2, 35, 38, 46) and SMAD4 nuclear localization (19). Furthermore, inhibitory SMADs and a number of E3 ubiquitin ligases are implicated in this process because they target SMAD4 for proteolytic degradation (44, 45). Further studies are needed to determine the precise mechanisms involved in the regulation of SMAD4 in our model.
In conclusion, hepcidin expression is decreased by hypoxia and oxidative stress in HuH7 hepatocytes (Fig. 6), but only when they are cocultured with conditioned medium from activated THP-1 macrophages. IL-1β appears to be the main mediator of this effect. Because hepatocytes are normally exposed to a stream of cytokines from Kupffer cells and circulating macrophages, our model may be physiologically relevant for the study of hepatic hepcidin expression. Under these conditions we have identified a transcriptional mechanism that, following stimulation, represents an intracellular hepatic regulatory circuit for direct SMAD4-dependent repression of hepcidin transcription.
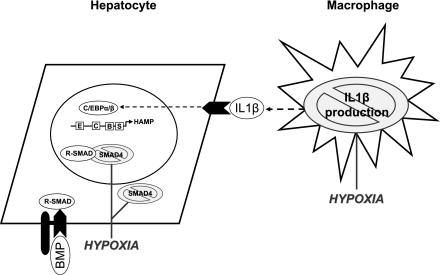
Fig. 6.
Cross talk between macrophages and hepatocytes and the regulation of hepcidin expression. Data from this study and from previous work by our group (26) suggest that cross talk between macrophages and hepatocytes is important in controlling the production and release of hepcidin, the central regulator of iron homeostasis. These effects appear to be mediated by macrophage production of the proinflammatory cytokine IL-1β, which activates the C/EBP pathway in HuH7 cells. In the context of hypoxia, we observed a decrease in macrophage production of IL-1β. In addition, there are direct effects of hypoxia on SMAD4 protein levels in the cytosol and nucleus of HuH7 cells. Together, these mechanisms serve to abrogate the BMP/SMAD signaling pathway that is crucial for the regulation of hepcidin expression. HAMP, hepcidin antimicrobial peptide; B, BMP/SMAD-responsive element; C, CCAAT enhancer-binding protein responsive element; E, E-box elements; S, STAT3-responsive element.
GRANTS
This work was funded by the Biotechnology and Biological Sciences Research Council. P. Matak was funded by a studentship from the Medical Research Council. K. Pourvali is funded by a studentship from the Shaheed Beheshti Medical University, Tehran, Iran.
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
REFERENCES
- 1. Ahmad KA, Ahmann JR, Migas MC, Waheed A, Britton RS, Bacon BR, Sly WS, Fleming RE. Decreased liver hepcidin expression in the Hfe knockout mouse. Blood Cells Mol Dis 29: 361–366, 2002 [DOI] [PubMed] [Google Scholar]
- 2. Akman HO, Zhang H, Siddiqui MA, Solomon W, Smith EL, Batuman OA. Response to hypoxia involves transforming growth factor-beta2 and Smad proteins in human endothelial cells. Blood 98: 3324–3331, 2001 [DOI] [PubMed] [Google Scholar]
- 3. Babitt JL, Huang FW, Wrighting DM, Xia Y, Sidis Y, Samad TA, Campagna JA, Chung RT, Schneyer AL, Woolf CJ, Andrews NC, Lin HY. Bone morphogenetic protein signaling by hemojuvelin regulates hepcidin expression. Nat Genet 38: 531–539, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Bayele HK, McArdle H, Srai SK. Cis and trans regulation of hepcidin expression by upstream stimulatory factor. Blood 108: 4237–4245, 2006 [DOI] [PubMed] [Google Scholar]
- 5. Braliou GG, Verga Falzacappa MV, Chachami G, Casanovas G, Muckenthaler MU, Simos G. 2-Oxoglutarate-dependent oxygenases control hepcidin gene expression. J Hepatol 48: 801–810, 2008 [DOI] [PubMed] [Google Scholar]
- 6. Bridle KR, Frazer DM, Wilkins SJ, Dixon JL, Purdie DM, Crawford DH, Subramaniam VN, Powell LW, Anderson GJ, Ramm GA. Disrupted hepcidin regulation in HFE-associated haemochromatosis and the liver as a regulator of body iron homoeostasis. Lancet 361: 669–673, 2003 [DOI] [PubMed] [Google Scholar]
- 7. Casanovas G, Mleczko-Sanecka K, Altamura S, Hentze MW, Muckenthaler MU. Bone morphogenetic protein (BMP)-responsive elements located in the proximal and distal hepcidin promoter are critical for its response to HJV/BMP/SMAD. J Mol Med 87: 471–480, 2009 [DOI] [PubMed] [Google Scholar]
- 8. Chaston T, Chung B, Mascarenhas M, Marks J, Patel B, Srai SK, Sharp P. Evidence for differential effects of hepcidin in macrophages and intestinal epithelial cells. Gut 57: 374–382, 2008 [DOI] [PubMed] [Google Scholar]
- 9. Choi SO, Cho YS, Kim HL, Park JW. ROS mediate the hypoxic repression of the hepcidin gene by inhibiting C/EBPalpha and STAT-3. Biochem Biophys Res Commun 356: 312–317, 2007 [DOI] [PubMed] [Google Scholar]
- 10. Chung B, Chaston T, Marks J, Srai SK, Sharp PA. Hepcidin decreases iron transporter expression in vivo in mouse duodenum and spleen and in vitro in THP-1 macrophages and intestinal Caco-2 cells. J Nutr 139: 1457–1462, 2009 [DOI] [PubMed] [Google Scholar]
- 11. Courselaud B, Pigeon C, Inoue Y, Inoue J, Gonzalez FJ, Leroyer P, Gilot D, Boudjema K, Guguen-Guillouzo C, Brissot P, Loreal O, Ilyin G. C/EBPalpha regulates hepatic transcription of hepcidin, an antimicrobial peptide and regulator of iron metabolism. Cross-talk between C/EBP pathway and iron metabolism. J Biol Chem 277: 41163–41170, 2002 [DOI] [PubMed] [Google Scholar]
- 12. Delaby C, Pilard N, Goncalves AS, Beaumont C, Canonne-Hergaux F. Presence of the iron exporter ferroportin at the plasma membrane of macrophages is enhanced by iron loading and down-regulated by hepcidin. Blood 106: 3979–3984, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Frazer DM, Inglis HR, Wilkins SJ, Millard KN, Steele TM, McLaren GD, McKie AT, Vulpe CD, Anderson GJ. Delayed hepcidin response explains the lag period in iron absorption following a stimulus to increase erythropoiesis. Gut 53: 1509–1515, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Frazer DM, Wilkins SJ, Becker EM, Vulpe CD, McKie AT, Trinder D, Anderson GJ. Hepcidin expression inversely correlates with the expression of duodenal iron transporters and iron absorption in rats. Gastroenterology 123: 835–844, 2002 [DOI] [PubMed] [Google Scholar]
- 15. Haudenschild DR, Palmer SM, Moseley TA, You Z, Reddi AH. Bone morphogenetic protein (BMP)-6 signaling and BMP antagonist noggin in prostate cancer. Cancer Res 64: 8276–8284, 2004 [DOI] [PubMed] [Google Scholar]
- 16. Huang H, Constante M, Layoun A, Santos MM. Contribution of STAT3 and SMAD4 pathways to the regulation of hepcidin by opposing stimuli. Blood 113: 3593–3599, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Jacolot S, Ferec C, Mura C. Iron responses in hepatic, intestinal and macrophage/monocyte cell lines under different culture conditions. Blood Cells Mol Dis 41: 100–108, 2008 [DOI] [PubMed] [Google Scholar]
- 18. Kautz L, Meynard D, Monnier A, Darnaud V, Bouvet R, Wang RH, Deng C, Vaulont S, Mosser J, Coppin H, Roth MP. Iron regulates phosphorylation of Smad1/5/8 and gene expression of Bmp6, Smad7, Id1, and Atoh8 in the mouse liver. Blood 112: 1503–1509, 2008 [DOI] [PubMed] [Google Scholar]
- 19. Kim YK, Bae GU, Kang JK, Park JW, Lee EK, Lee HY, Choi WS, Lee HW, Han JW. Cooperation of H2O2-mediated ERK activation with Smad pathway in TGF-beta1 induction of p21WAF1/Cip1. Cell Signal 18: 236–243, 2006 [DOI] [PubMed] [Google Scholar]
- 20. Knutson MD, Oukka M, Koss LM, Aydemir F, Wessling-Resnick M. Iron release from macrophages after erythrophagocytosis is up-regulated by ferroportin 1 overexpression and down-regulated by hepcidin. Proc Natl Acad Sci USA 102: 1324–1328, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Kochanowska I, Chaberek S, Wojtowicz A, Marczynski B, Wlodarski K, Dytko M, Ostrowski K. Expression of genes for bone morphogenetic proteins BMP-2, BMP-4 and BMP-6 in various parts of the human skeleton. BMC Musculoskelet Disord 8: 128, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Kohro T, Tanaka T, Murakami T, Wada Y, Aburatani H, Hamakubo T, Kodama T. A comparison of differences in the gene expression profiles of phorbol 12-myristate 13-acetate differentiated THP-1 cells and human monocyte-derived macrophage. J Atheroscler Thromb 11: 88–97, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Krause A, Neitz S, Magert HJ, Schulz A, Forssmann WG, Schulz-Knappe P, Adermann K. LEAP-1, a novel highly disulfide-bonded human peptide, exhibits antimicrobial activity. FEBS Lett 480: 147–150, 2000 [DOI] [PubMed] [Google Scholar]
- 24. Leung PS, Srai SK, Mascarenhas M, Churchill LJ, Debnam ES. Increased duodenal iron uptake and transfer in a rat model of chronic hypoxia is accompanied by reduced hepcidin expression. Gut 54: 1391–1395, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Mastrogiannaki M, Matak P, Keith B, Simon MC, Vaulont S, Peyssonnaux C. HIF-2alpha, but not HIF-1alpha, promotes iron absorption in mice. J Clin Invest 119: 1159–1166, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Matak P, Chaston TB, Chung B, Srai SK, McKie AT, Sharp PA. Activated macrophages induce hepcidin expression in HuH7 hepatoma cells. Haematologica 94: 773–780, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Mleczko-Sanecka K, Casanovas G, Ragab A, Breitkopf K, Muller A, Boutros M, Dooley S, Hentze MW, Muckenthaler MU. SMAD7 controls iron metabolism as a potent inhibitor of hepcidin expression. Blood 115: 2657–2665, 2009 [DOI] [PubMed] [Google Scholar]
- 28. Nemeth E, Roetto A, Garozzo G, Ganz T, Camaschella C. Hepcidin is decreased in TFR2 hemochromatosis. Blood 105: 1803–1806, 2005 [DOI] [PubMed] [Google Scholar]
- 29. Nicolas G, Chauvet C, Viatte L, Danan JL, Bigard X, Devaux I, Beaumont C, Kahn A, Vaulont S. The gene encoding the iron regulatory peptide hepcidin is regulated by anemia, hypoxia, and inflammation. J Clin Invest 110: 1037–1044, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. O'Riordan DK, Debnam ES, Sharp PA, Simpson RJ, Taylor EM, Srai SK. Mechanisms involved in increased iron uptake across rat duodenal brush-border membrane during hypoxia. J Physiol 500: 379–384, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Papanikolaou G, Samuels ME, Ludwig EH, MacDonald ML, Franchini PL, Dube MP, Andres L, MacFarlane J, Sakellaropoulos N, Politou M, Nemeth E, Thompson J, Risler JK, Zaborowska C, Babakaiff R, Radomski CC, Pape TD, Davidas O, Christakis J, Brissot P, Lockitch G, Ganz T, Hayden MR, Goldberg YP. Mutations in HFE2 cause iron overload in chromosome 1q-linked juvenile hemochromatosis. Nat Genet 36: 77–82, 2004 [DOI] [PubMed] [Google Scholar]
- 32. Peyssonnaux C, Zinkernagel AS, Schuepbach RA, Rankin E, Vaulont S, Haase VH, Nizet V, Johnson RS. Regulation of iron homeostasis by the hypoxia-inducible transcription factors (HIFs). J Clin Invest 117: 1926–1932, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Pigeon C, Ilyin G, Courselaud B, Leroyer P, Turlin B, Brissot P, Loreal O. A new mouse liver-specific gene, encoding a protein homologous to human antimicrobial peptide hepcidin, is overexpressed during iron overload. J Biol Chem 276: 7811–7819, 2001 [DOI] [PubMed] [Google Scholar]
- 34. Pinto JP, Ribeiro S, Pontes H, Thowfeequ S, Tosh D, Carvalho F, Porto G. Erythropoietin mediates hepcidin expression in hepatocytes through EPOR signaling and regulation of C/EBPalpha. Blood 111: 5727–5733, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Pistollato F, Chen HL, Rood BR, Zhang HZ, D'Avella D, Denaro L, Gardiman M, te Kronnie G, Schwartz PH, Favaro E, Indraccolo S, Basso G, Panchision DM. Hypoxia and HIF1alpha repress the differentiative effects of BMPs in high-grade glioma. Stem Cells 27: 7–17, 2009 [DOI] [PubMed] [Google Scholar]
- 36. Rivera S, Nemeth E, Gabayan V, Lopez MA, Farshidi D, Ganz T. Synthetic hepcidin causes rapid dose-dependent hypoferremia and is concentrated in ferroportin-containing organs. Blood 106: 2196–2199, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shah YM, Matsubara T, Ito S, Yim SH, Gonzalez FJ. Intestinal hypoxia-inducible transcription factors are essential for iron absorption following iron deficiency. Cell Metab 9: 152–164, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Shi YF, Fong CC, Zhang Q, Cheung PY, Tzang CH, Wu RS, Yang M. Hypoxia induces the activation of human hepatic stellate cells LX-2 through TGF-beta signaling pathway. FEBS Lett 581: 203–210, 2007 [DOI] [PubMed] [Google Scholar]
- 39. Silvestri L, Pagani A, Nai A, De Domenico I, Kaplan J, Camaschella C. The serine protease matriptase-2 (TMPRSS6) inhibits hepcidin activation by cleaving membrane hemojuvelin. Cell Metab 8: 502–511, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Verga Falzacappa MV, Casanovas G, Hentze MW, Muckenthaler MU. A bone morphogenetic protein (BMP)-responsive element in the hepcidin promoter controls HFE2-mediated hepatic hepcidin expression and its response to IL-6 in cultured cells. J Mol Med 86: 531–540, 2008 [DOI] [PubMed] [Google Scholar]
- 41. Verga Falzacappa MV, Vujic SM, Kessler R, Stolte J, Hentze MW, Muckenthaler MU. STAT3 mediates hepatic hepcidin expression and its inflammatory stimulation. Blood 109: 353–358, 2007 [DOI] [PubMed] [Google Scholar]
- 42. Volke M, Gale DP, Maegdefrau U, Schley G, Klanke B, Bosserhoff AK, Maxwell PH, Eckardt KU, Warnecke C. Evidence for a lack of a direct transcriptional suppression of the iron regulatory peptide hepcidin by hypoxia-inducible factors. PLoS One 4: e7875, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Wang RH, Li C, Xu X, Zheng Y, Xiao C, Zerfas P, Cooperman S, Eckhaus M, Rouault T, Mishra L, Deng CX. A role of SMAD4 in iron metabolism through the positive regulation of hepcidin expression. Cell Metab 2: 399–409, 2005 [DOI] [PubMed] [Google Scholar]
- 44. Wrana JL. Regulation of Smad activity. Cell 100: 189–192, 2000 [DOI] [PubMed] [Google Scholar]
- 45. Yamashita M, Ying SX, Zhang GM, Li C, Cheng SY, Deng CX, Zhang YE. Ubiquitin ligase Smurf1 controls osteoblast activity and bone homeostasis by targeting MEKK2 for degradation. Cell 121: 101–113, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhang H, Akman HO, Smith EL, Zhao J, Murphy-Ullrich JE, Batuman OA. Cellular response to hypoxia involves signaling via Smad proteins. Blood 101: 2253–2260, 2003 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.