Abstract
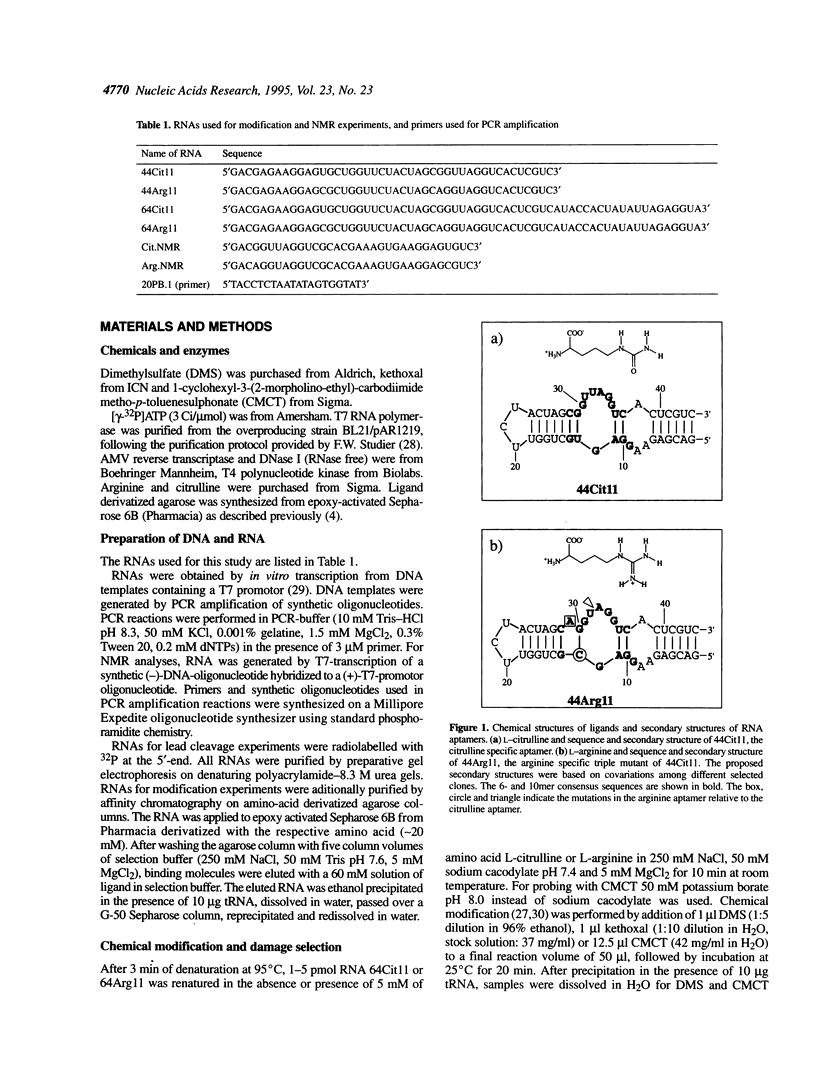
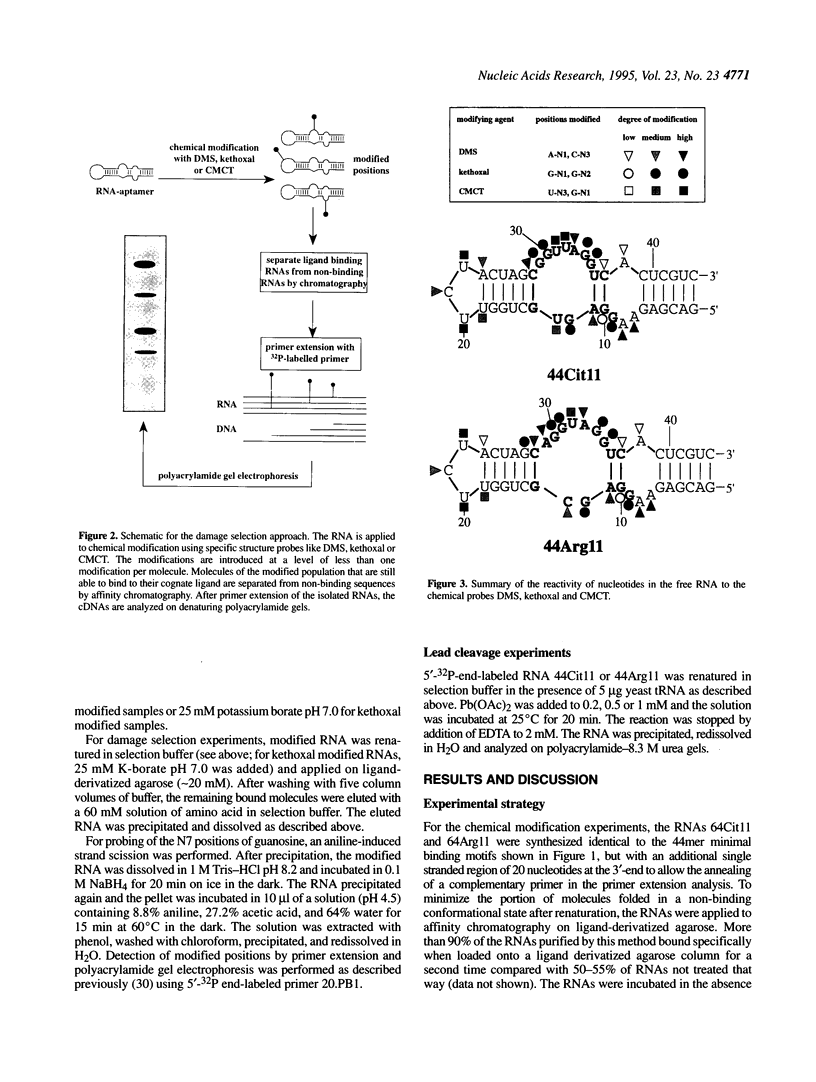
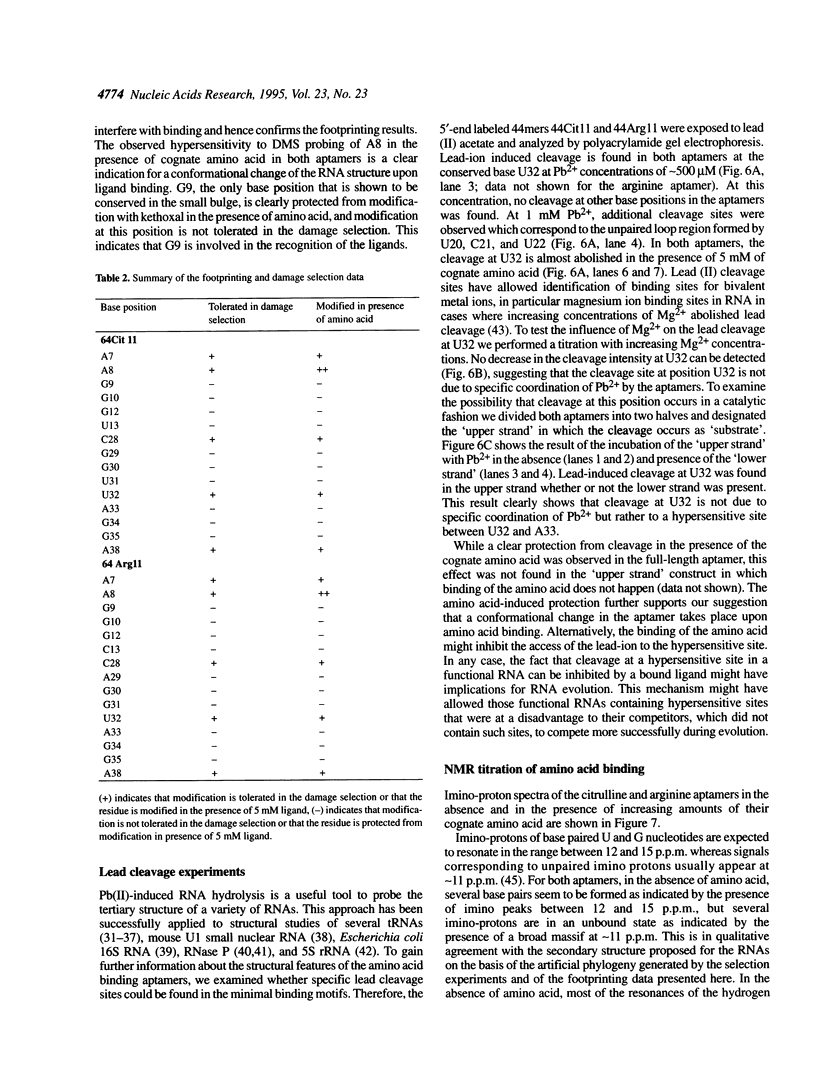
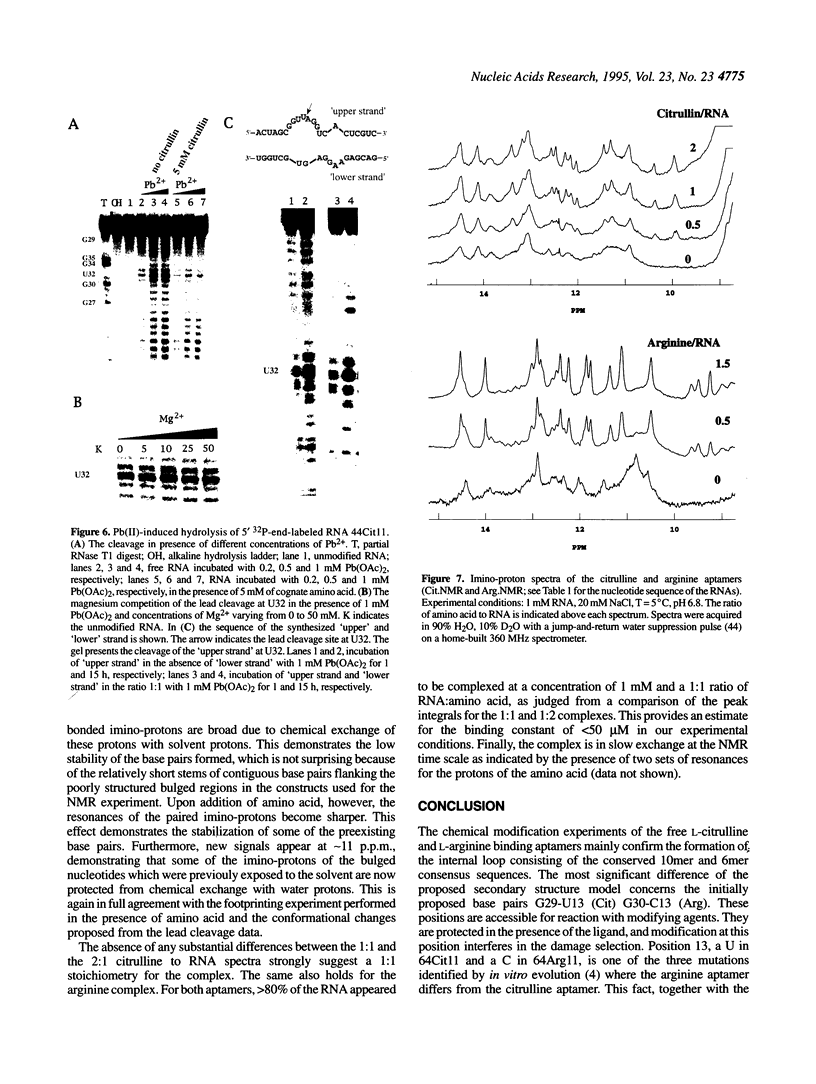
In a recent study, an RNA aptamer for the specific recognition of the amino acid L-arginine was evolved from an in vitro selected L-citrulline binding parent sequence [M. Famulok (1994) J. Am. Chem. Soc. 116, 1698-1706]. We have now carried out a structural analysis of these aptamers by using chemical modification experiments. Footprinting experiments and a damage selection approach were performed to identify those positions protected from modification in the presence of the amino acids and modifications that interfere with the binding of the ligand. It is shown that of the two bulged regions present in both aptamers one can be modified without loss of binding activity whereas in the other bulge nearly every position is shown to be involved in the recognition of the ligands. This might be indicative for non-canonical base pairing to occur within the non-Watson-Crick paired regions which might be stabilized by the complexed amino acid. Binding to the cognate amino acid significantly enhances the conformational stability of the RNA. We also tested the sensitivity of both aptamers towards lead (II) ion induced cleavage and identified a hypersensitive cleavage site within the invariant bulged region. Lead cleavage is inhibited by the complexed amino acid, indicating a conformational change of the aptamer upon ligand binding. NMR titration data obtained with both aptamers and their cognate ligands confirm the proposed conformational changes and indicate the formation of a 1:1 complex of RNA:amino acid.
Full text
PDF



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Behlen L. S., Sampson J. R., DiRenzo A. B., Uhlenbeck O. C. Lead-catalyzed cleavage of yeast tRNAPhe mutants. Biochemistry. 1990 Mar 13;29(10):2515–2523. doi: 10.1021/bi00462a013. [DOI] [PubMed] [Google Scholar]
- Bock L. C., Griffin L. C., Latham J. A., Vermaas E. H., Toole J. J. Selection of single-stranded DNA molecules that bind and inhibit human thrombin. Nature. 1992 Feb 6;355(6360):564–566. doi: 10.1038/355564a0. [DOI] [PubMed] [Google Scholar]
- Ciesiolka J., Hardt W. D., Schlegl J., Erdmann V. A., Hartmann R. K. Lead-ion-induced cleavage of RNase P RNA. Eur J Biochem. 1994 Jan 15;219(1-2):49–56. doi: 10.1111/j.1432-1033.1994.tb19913.x. [DOI] [PubMed] [Google Scholar]
- Ciesiołka J., Lorenz S., Erdmann V. A. Structural analysis of three prokaryotic 5S rRNA species and selected 5S rRNA--ribosomal-protein complexes by means of Pb(II)-induced hydrolysis. Eur J Biochem. 1992 Mar 1;204(2):575–581. doi: 10.1111/j.1432-1033.1992.tb16670.x. [DOI] [PubMed] [Google Scholar]
- Ciesiołka J., Wrzesinski J., Górnicki P., Podkowiński J., Krzyzosiak W. J. Analysis of magnesium, europium and lead binding sites in methionine initiator and elongator tRNAs by specific metal-ion-induced cleavages. Eur J Biochem. 1989 Dec 8;186(1-2):71–77. doi: 10.1111/j.1432-1033.1989.tb15179.x. [DOI] [PubMed] [Google Scholar]
- Connell G. J., Illangesekare M., Yarus M. Three small ribooligonucleotides with specific arginine sites. Biochemistry. 1993 Jun 1;32(21):5497–5502. doi: 10.1021/bi00072a002. [DOI] [PubMed] [Google Scholar]
- Davanloo P., Rosenberg A. H., Dunn J. J., Studier F. W. Cloning and expression of the gene for bacteriophage T7 RNA polymerase. Proc Natl Acad Sci U S A. 1984 Apr;81(7):2035–2039. doi: 10.1073/pnas.81.7.2035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ehresmann C., Baudin F., Mougel M., Romby P., Ebel J. P., Ehresmann B. Probing the structure of RNAs in solution. Nucleic Acids Res. 1987 Nov 25;15(22):9109–9128. doi: 10.1093/nar/15.22.9109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. In vitro selection of RNA molecules that bind specific ligands. Nature. 1990 Aug 30;346(6287):818–822. doi: 10.1038/346818a0. [DOI] [PubMed] [Google Scholar]
- Ellington A. D., Szostak J. W. Selection in vitro of single-stranded DNA molecules that fold into specific ligand-binding structures. Nature. 1992 Feb 27;355(6363):850–852. doi: 10.1038/355850a0. [DOI] [PubMed] [Google Scholar]
- Gold L. Oligonucleotides as research, diagnostic, and therapeutic agents. J Biol Chem. 1995 Jun 9;270(23):13581–13584. doi: 10.1074/jbc.270.23.13581. [DOI] [PubMed] [Google Scholar]
- Gold L., Polisky B., Uhlenbeck O., Yarus M. Diversity of oligonucleotide functions. Annu Rev Biochem. 1995;64:763–797. doi: 10.1146/annurev.bi.64.070195.003555. [DOI] [PubMed] [Google Scholar]
- Gornicki P., Baudin F., Romby P., Wiewiorowski M., Kryzosiak W., Ebel J. P., Ehresmann C., Ehresmann B. Use of lead(II) to probe the structure of large RNA's. Conformation of the 3' terminal domain of E. coli 16S rRNA and its involvement in building the tRNA binding sites. J Biomol Struct Dyn. 1989 Apr;6(5):971–984. doi: 10.1080/07391102.1989.10506525. [DOI] [PubMed] [Google Scholar]
- Huizenga D. E., Szostak J. W. A DNA aptamer that binds adenosine and ATP. Biochemistry. 1995 Jan 17;34(2):656–665. doi: 10.1021/bi00002a033. [DOI] [PubMed] [Google Scholar]
- Jenison R. D., Gill S. C., Pardi A., Polisky B. High-resolution molecular discrimination by RNA. Science. 1994 Mar 11;263(5152):1425–1429. doi: 10.1126/science.7510417. [DOI] [PubMed] [Google Scholar]
- Jensen K. B., Green L., MacDougal-Waugh S., Tuerk C. Characterization of an in vitro-selected RNA ligand to the HIV-1 Rev protein. J Mol Biol. 1994 Jan 7;235(1):237–247. doi: 10.1016/s0022-2836(05)80030-0. [DOI] [PubMed] [Google Scholar]
- Joyce G. F. In vitro evolution of nucleic acids. Curr Opin Struct Biol. 1994;4:331–336. doi: 10.1016/s0959-440x(94)90100-7. [DOI] [PubMed] [Google Scholar]
- Klug S. J., Famulok M. All you wanted to know about SELEX. Mol Biol Rep. 1994;20(2):97–107. doi: 10.1007/BF00996358. [DOI] [PubMed] [Google Scholar]
- Krzyzosiak W. J., Marciniec T., Wiewiorowski M., Romby P., Ebel J. P., Giegé R. Characterization of the lead(II)-induced cleavages in tRNAs in solution and effect of the Y-base removal in yeast tRNAPhe. Biochemistry. 1988 Jul 26;27(15):5771–5777. doi: 10.1021/bi00415a056. [DOI] [PubMed] [Google Scholar]
- Lato S. M., Boles A. R., Ellington A. D. In vitro selection of RNA lectins: using combinatorial chemistry to interpret ribozyme evolution. Chem Biol. 1995 May;2(5):291–303. doi: 10.1016/1074-5521(95)90048-9. [DOI] [PubMed] [Google Scholar]
- Lauhon C. T., Szostak J. W. RNA aptamers that bind flavin and nicotinamide redox cofactors. J Am Chem Soc. 1995 Feb 1;117(4):1246–1257. doi: 10.1021/ja00109a008. [DOI] [PubMed] [Google Scholar]
- Lorsch J. R., Szostak J. W. In vitro selection of RNA aptamers specific for cyanocobalamin. Biochemistry. 1994 Feb 1;33(4):973–982. doi: 10.1021/bi00170a016. [DOI] [PubMed] [Google Scholar]
- Macaya R. F., Schultze P., Smith F. W., Roe J. A., Feigon J. Thrombin-binding DNA aptamer forms a unimolecular quadruplex structure in solution. Proc Natl Acad Sci U S A. 1993 Apr 15;90(8):3745–3749. doi: 10.1073/pnas.90.8.3745. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Majerfeld I., Yarus M. An RNA pocket for an aliphatic hydrophobe. Nat Struct Biol. 1994 May;1(5):287–292. doi: 10.1038/nsb0594-287. [DOI] [PubMed] [Google Scholar]
- Marciniec T., Ciesiołka J., Wrzesinski J., Krzyzosiak W. J. Identification of the magnesium, europium and lead binding sites in E. coli and lupine tRNAPhe by specific metal ion-induced cleavages. FEBS Lett. 1989 Jan 30;243(2):293–298. doi: 10.1016/0014-5793(89)80148-6. [DOI] [PubMed] [Google Scholar]
- Milligan J. F., Uhlenbeck O. C. Synthesis of small RNAs using T7 RNA polymerase. Methods Enzymol. 1989;180:51–62. doi: 10.1016/0076-6879(89)80091-6. [DOI] [PubMed] [Google Scholar]
- Morris K. N., Tarasow T. M., Julin C. M., Simons S. L., Hilvert D., Gold L. Enrichment for RNA molecules that bind a Diels-Alder transition state analog. Proc Natl Acad Sci U S A. 1994 Dec 20;91(26):13028–13032. doi: 10.1073/pnas.91.26.13028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Otzen D. E., Barciszewski J., Clark B. F. Altered lead(II)-cleavage pattern of free Phe-tRNAPhe and Phe-tRNAPhe in ternary complex with EF-Tu:GTP. Biochem Mol Biol Int. 1993 Sep;31(1):95–103. [PubMed] [Google Scholar]
- Pan T., Gutell R. R., Uhlenbeck O. C. Folding of circularly permuted transfer RNAs. Science. 1991 Nov 29;254(5036):1361–1364. doi: 10.1126/science.1720569. [DOI] [PubMed] [Google Scholar]
- Prudent J. R., Uno T., Schultz P. G. Expanding the scope of RNA catalysis. Science. 1994 Jun 24;264(5167):1924–1927. doi: 10.1126/science.8009223. [DOI] [PubMed] [Google Scholar]
- Sampson J. R., Sullivan F. X., Behlen L. S., DiRenzo A. B., Uhlenbeck O. C. Characterization of two RNA-catalyzed RNA cleavage reactions. Cold Spring Harb Symp Quant Biol. 1987;52:267–275. doi: 10.1101/sqb.1987.052.01.032. [DOI] [PubMed] [Google Scholar]
- Sassanfar M., Szostak J. W. An RNA motif that binds ATP. Nature. 1993 Aug 5;364(6437):550–553. doi: 10.1038/364550a0. [DOI] [PubMed] [Google Scholar]
- Schultze P., Macaya R. F., Feigon J. Three-dimensional solution structure of the thrombin-binding DNA aptamer d(GGTTGGTGTGGTTGG). J Mol Biol. 1994 Feb 4;235(5):1532–1547. doi: 10.1006/jmbi.1994.1105. [DOI] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Streicher B., von Ahsen U., Schroeder R. Lead cleavage sites in the core structure of group I intron-RNA. Nucleic Acids Res. 1993 Jan 25;21(2):311–317. doi: 10.1093/nar/21.2.311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varani G., Tinoco I., Jr RNA structure and NMR spectroscopy. Q Rev Biophys. 1991 Nov;24(4):479–532. doi: 10.1017/s0033583500003875. [DOI] [PubMed] [Google Scholar]
- Wallis M. G., von Ahsen U., Schroeder R., Famulok M. A novel RNA motif for neomycin recognition. Chem Biol. 1995 Aug;2(8):543–552. doi: 10.1016/1074-5521(95)90188-4. [DOI] [PubMed] [Google Scholar]
- Wang K. Y., McCurdy S., Shea R. G., Swaminathan S., Bolton P. H. A DNA aptamer which binds to and inhibits thrombin exhibits a new structural motif for DNA. Biochemistry. 1993 Mar 2;32(8):1899–1904. doi: 10.1021/bi00059a003. [DOI] [PubMed] [Google Scholar]
- Wang Y., Rando R. R. Specific binding of aminoglycoside antibiotics to RNA. Chem Biol. 1995 May;2(5):281–290. doi: 10.1016/1074-5521(95)90047-0. [DOI] [PubMed] [Google Scholar]
- Zito K., Hüttenhofer A., Pace N. R. Lead-catalyzed cleavage of ribonuclease P RNA as a probe for integrity of tertiary structure. Nucleic Acids Res. 1993 Dec 25;21(25):5916–5920. doi: 10.1093/nar/21.25.5916. [DOI] [PMC free article] [PubMed] [Google Scholar]