Abstract
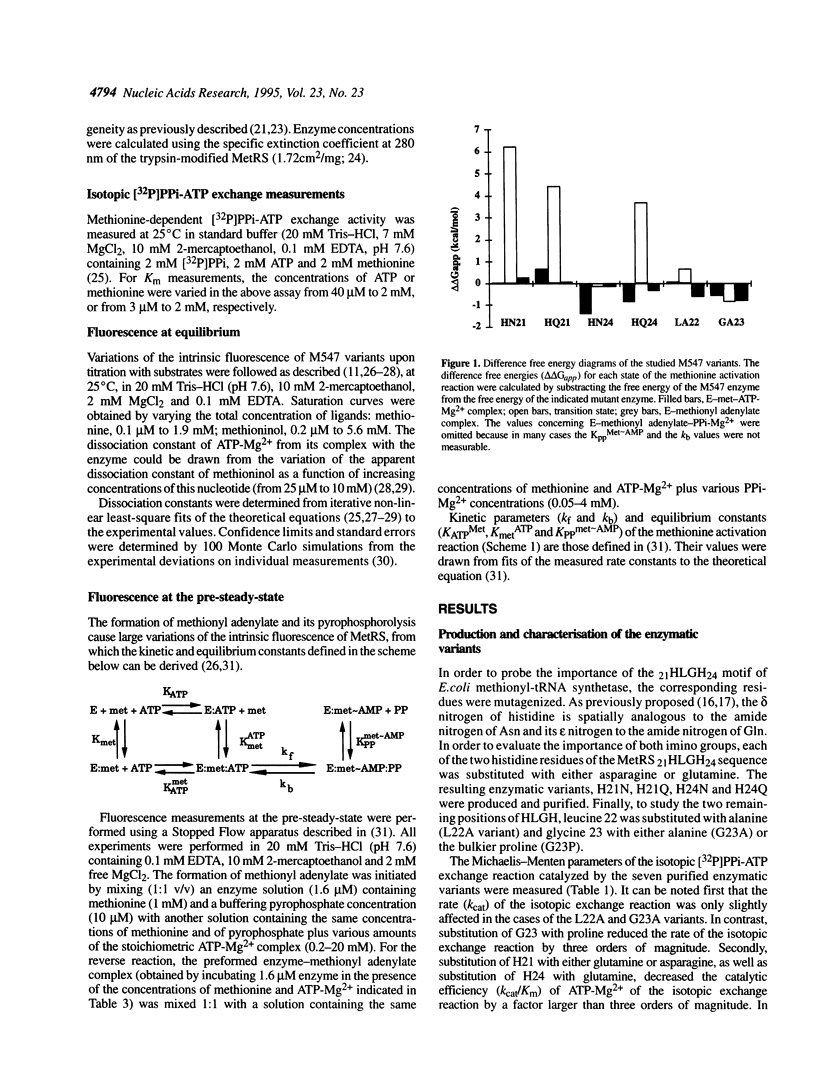
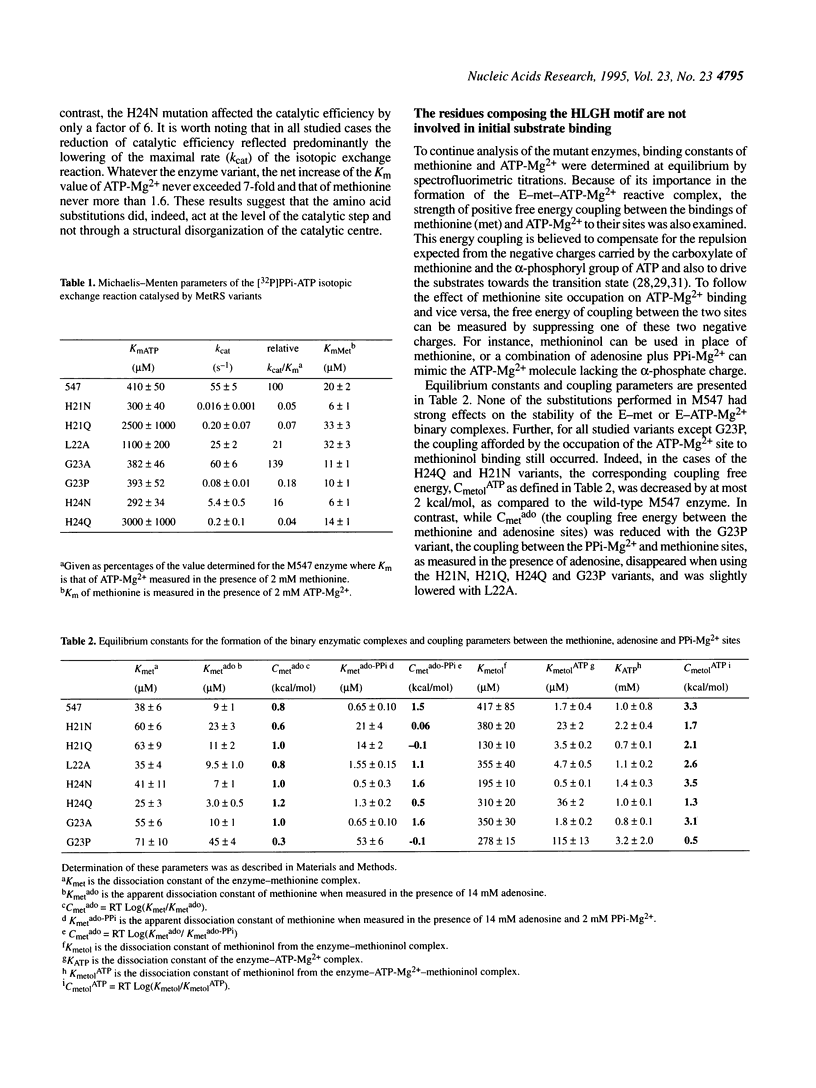
Methionyl-tRNA synthetase belongs to the class I aminoacyl-tRNA synthetase family characterized both by a catalytic center built around a Rossmann Fold and by the presence of the two peptidic marker sequences HIGH and KMSKS. In this study, the role of the 21HLGH24 motif of Escherichia coli methionyl-tRNA synthetase was studied in a systematic fashion by site-directed mutagenesis. It is shown that the two histidine residues play a crucial role in the catalysis of the methionyl adenylate formation by participating in the stabilisation of the ATP phosphate chain during the transition state. Moreover, the results suggest the involvement of the epsilon-imino group of histidine 21 and of the delta-imino group of histidine 24. Notably, the substitution of either the leucine or the glycine residue of the HLGH motif by alanine had no effect on the catalysis. From the data and from other studies with class I aminoacyl-tRNA synthetases, concomitant positive contributions of the HIGH and KMSKS sequences to reach the transition state of aminoacyl adenylate formation can be envisaged.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Barker D. G., Winter G. Conserved cysteine and histidine residues in the structures of the tyrosyl and methionyl-tRNA synthetases. FEBS Lett. 1982 Aug 23;145(2):191–193. doi: 10.1016/0014-5793(82)80165-8. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Fayat G., Waller J. P., Iwatsubo M. The mechanism of reaction of methionyl-tRNA synthetase from Escherichia coli. Interaction of the enzyme with ligands of the amino-acid-activation reaction. Eur J Biochem. 1972 Jan 21;24(3):461–469. doi: 10.1111/j.1432-1033.1972.tb19707.x. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Fayat G., Waller J. P. The amino acid activation reaction catalyzed by methionyl-transfer rna synthetase: evidence for synergistic coupling between the sites for methionine adenosine and pyrophosphate. J Mol Biol. 1975 May 5;94(1):1–15. doi: 10.1016/0022-2836(75)90401-5. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Fayat G., Waller J. P. The mechanism of action of methionyl-tRNA synthetase from Escherichia coli. Mechanism of the amino-acid activation reaction catalyzed by the native and the trypsin-modified enzymes. Eur J Biochem. 1974 May 15;44(2):343–351. doi: 10.1111/j.1432-1033.1974.tb03491.x. [DOI] [PubMed] [Google Scholar]
- Blanquet S., Petrissant G., Waller J. P. The mechanism of action of methionyl-tRNA synthetase. 2. Interaction of the enzyme with specific and unspecific tRNAs. Eur J Biochem. 1973 Jul 2;36(1):227–233. doi: 10.1111/j.1432-1033.1973.tb02904.x. [DOI] [PubMed] [Google Scholar]
- Borgford T. J., Gray T. E., Brand N. J., Fersht A. R. Site-directed mutagenesis reveals transition-state stabilization as a general catalytic mechanism for aminoacyl-tRNA synthetases. Biochemistry. 1987 Nov 17;26(23):7246–7250. doi: 10.1021/bi00397a008. [DOI] [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Cassio D., Waller J. P. Modification of methionyl-tRNA synthetase by proteolytic cleavage and properties of the trypsin-modified enzyme. Eur J Biochem. 1971 May 28;20(2):283–300. doi: 10.1111/j.1432-1033.1971.tb01393.x. [DOI] [PubMed] [Google Scholar]
- Chan K. W., Koeppe R. E., 2nd Role of the TIGN sequence in E. coli tryptophanyl-tRNA synthetase. Biochim Biophys Acta. 1994 Apr 13;1205(2):223–229. doi: 10.1016/0167-4838(94)90237-2. [DOI] [PubMed] [Google Scholar]
- Cusack S., Härtlein M., Leberman R. Sequence, structural and evolutionary relationships between class 2 aminoacyl-tRNA synthetases. Nucleic Acids Res. 1991 Jul 11;19(13):3489–3498. doi: 10.1093/nar/19.13.3489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dardel F. MC-Fit: using Monte-Carlo methods to get accurate confidence limits on enzyme parameters. Comput Appl Biosci. 1994 Jun;10(3):273–275. doi: 10.1093/bioinformatics/10.3.273. [DOI] [PubMed] [Google Scholar]
- Doublié S., Bricogne G., Gilmore C., Carter C. W., Jr Tryptophanyl-tRNA synthetase crystal structure reveals an unexpected homology to tyrosyl-tRNA synthetase. Structure. 1995 Jan 15;3(1):17–31. doi: 10.1016/s0969-2126(01)00132-0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Fayat G., Fromant M., Blanquet S. Couplings between the sites for methionine and adenosine 5'-triphosphate in the amino acid activation reaction catalyzed by trypsin-modified methionyl-transfer RNA synthetase from Escherichia coli. Biochemistry. 1977 May 31;16(11):2570–2579. doi: 10.1021/bi00630a039. [DOI] [PubMed] [Google Scholar]
- First E. A., Fersht A. R. Involvement of threonine 234 in catalysis of tyrosyl adenylate formation by tyrosyl-tRNA synthetase. Biochemistry. 1993 Dec 14;32(49):13644–13650. doi: 10.1021/bi00212a032. [DOI] [PubMed] [Google Scholar]
- First E. A., Fersht A. R. Mutation of lysine 233 to alanine introduces positive cooperativity into tyrosyl-tRNA synthetase. Biochemistry. 1993 Dec 14;32(49):13651–13657. doi: 10.1021/bi00212a033. [DOI] [PubMed] [Google Scholar]
- First E. A., Fersht A. R. Mutational and kinetic analysis of a mobile loop in tyrosyl-tRNA synthetase. Biochemistry. 1993 Dec 14;32(49):13658–13663. doi: 10.1021/bi00212a034. [DOI] [PubMed] [Google Scholar]
- Fourmy D., Meinnel T., Mechulam Y., Blanquet S. Mapping of the zinc binding domain of Escherichia coli methionyl-tRNA synthetase. J Mol Biol. 1993 Jun 20;231(4):1068–1077. doi: 10.1006/jmbi.1993.1352. [DOI] [PubMed] [Google Scholar]
- Hirel P. H., Lévêque F., Mellot P., Dardel F., Panvert M., Mechulam Y., Fayat G. Genetic engineering of methionyl-tRNA synthetase: in vitro regeneration of an active synthetase by proteolytic cleavage of a methionyl-tRNA synthetase--beta-galactosidase chimeric protein. Biochimie. 1988 Jun;70(6):773–782. doi: 10.1016/0300-9084(88)90107-1. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Dessen P., Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986 Sep;68(9):1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- Hyafil F., Jacques Y., Fayat G., Fromant M., Dessen P., Blanquet S. Methionyl-tRNA synthetase from Escherichia coli: active stoichiometry and stopped-flow analysis of methionyl adenylate formaiton. Biochemistry. 1976 Aug 24;15(17):3678–3685. doi: 10.1021/bi00662a006. [DOI] [PubMed] [Google Scholar]
- Leatherbarrow R. J., Fersht A. R. Investigation of transition-state stabilization by residues histidine-45 and threonine-40 in the tyrosyl-tRNA synthetase. Biochemistry. 1987 Dec 29;26(26):8524–8528. doi: 10.1021/bi00400a005. [DOI] [PubMed] [Google Scholar]
- Lowe D. M., Fersht A. R., Wilkinson A. J., Carter P., Winter G. Probing histidine-substrate interactions in tyrosyl-tRNA synthetase using asparagine and glutamine replacements. Biochemistry. 1985 Sep 10;24(19):5106–5109. doi: 10.1021/bi00340a022. [DOI] [PubMed] [Google Scholar]
- Mechulam Y., Dardel F., Le Corre D., Blanquet S., Fayat G. Lysine 335, part of the KMSKS signature sequence, plays a crucial role in the amino acid activation catalysed by the methionyl-tRNA synthetase from Escherichia coli. J Mol Biol. 1991 Feb 5;217(3):465–475. doi: 10.1016/0022-2836(91)90750-z. [DOI] [PubMed] [Google Scholar]
- Meinnel T., Mechulam Y., Le Corre D., Panvert M., Blanquet S., Fayat G. Selection of suppressor methionyl-tRNA synthetases: mapping the tRNA anticodon binding site. Proc Natl Acad Sci U S A. 1991 Jan 1;88(1):291–295. doi: 10.1073/pnas.88.1.291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mellot P., Mechulam Y., Le Corre D., Blanquet S., Fayat G. Identification of an amino acid region supporting specific methionyl-tRNA synthetase: tRNA recognition. J Mol Biol. 1989 Aug 5;208(3):429–443. doi: 10.1016/0022-2836(89)90507-x. [DOI] [PubMed] [Google Scholar]
- Nureki O., Vassylyev D. G., Katayanagi K., Shimizu T., Sekine S., Kigawa T., Miyazawa T., Yokoyama S., Morikawa K. Architectures of class-defining and specific domains of glutamyl-tRNA synthetase. Science. 1995 Mar 31;267(5206):1958–1965. doi: 10.1126/science.7701318. [DOI] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993 Aug 31;32(34):8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- RAVEL J. M., WANG S. F., HEINEMEYER C., SHIVE W. GLUTAMYL AND GLUTAMINYL RIBONUCLEIC ACID SYNTHETASES OF ESCHERICHIA COLI W. SEPARATION, PROPERTIES, AND STIMULATION OF ADENOSINE TRIPHOSPHATE-PYROPHOSPHATE EXCHANGE BY ACCEPTOR RIBONUCLEIC ACID. J Biol Chem. 1965 Jan;240:432–438. [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Schmitt E., Meinnel T., Blanquet S., Mechulam Y. Methionyl-tRNA synthetase needs an intact and mobile 332KMSKS336 motif in catalysis of methionyl adenylate formation. J Mol Biol. 1994 Sep 30;242(4):566–576. doi: 10.1006/jmbi.1994.1601. [DOI] [PubMed] [Google Scholar]
- Webster T., Tsai H., Kula M., Mackie G. A., Schimmel P. Specific sequence homology and three-dimensional structure of an aminoacyl transfer RNA synthetase. Science. 1984 Dec 14;226(4680):1315–1317. doi: 10.1126/science.6390679. [DOI] [PubMed] [Google Scholar]