Abstract
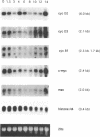
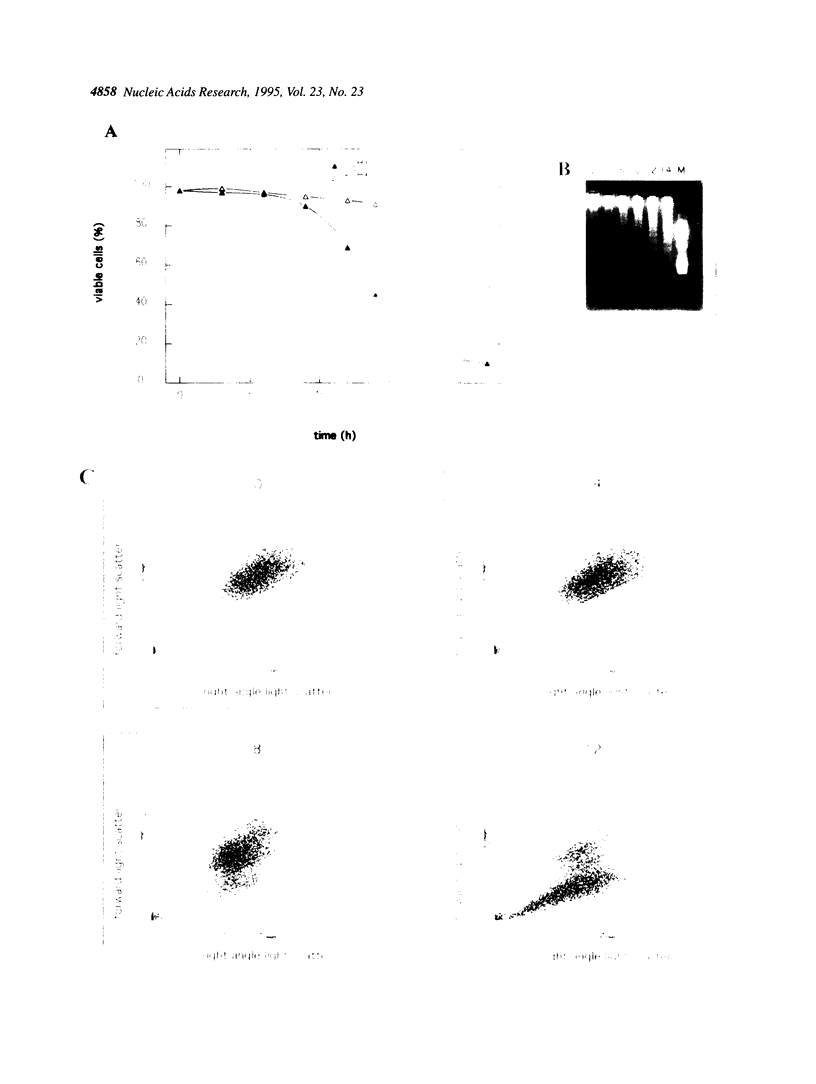
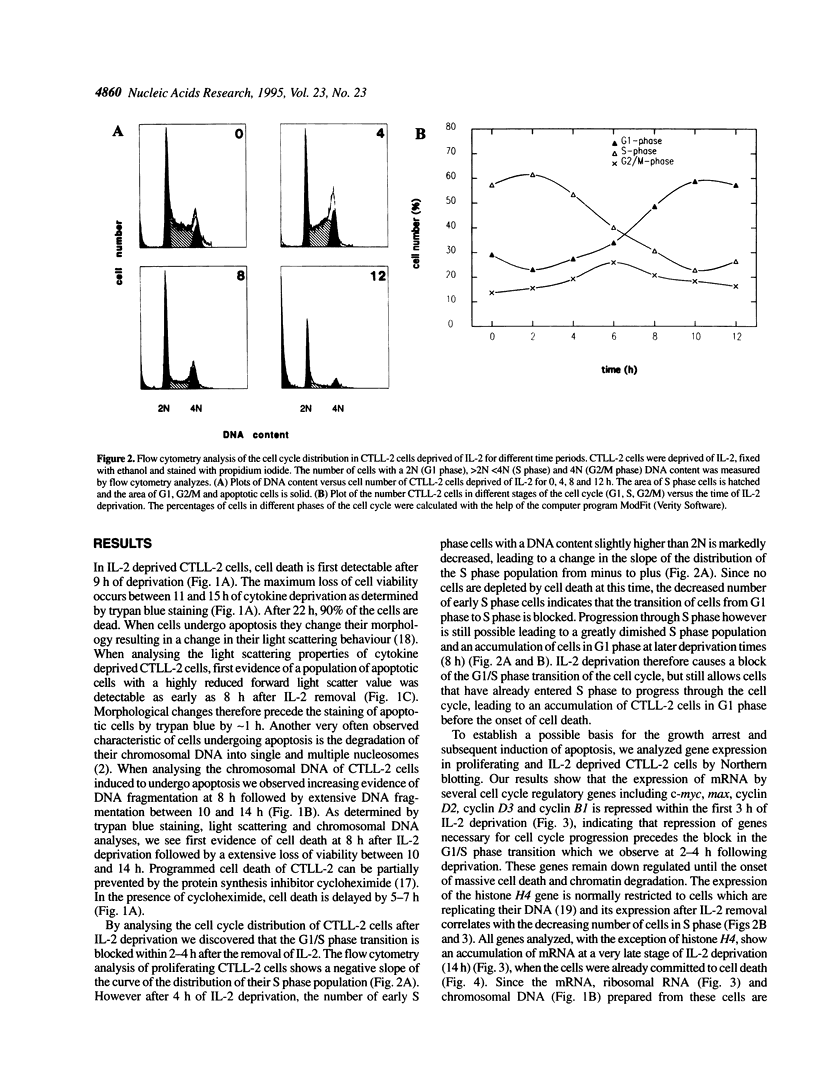
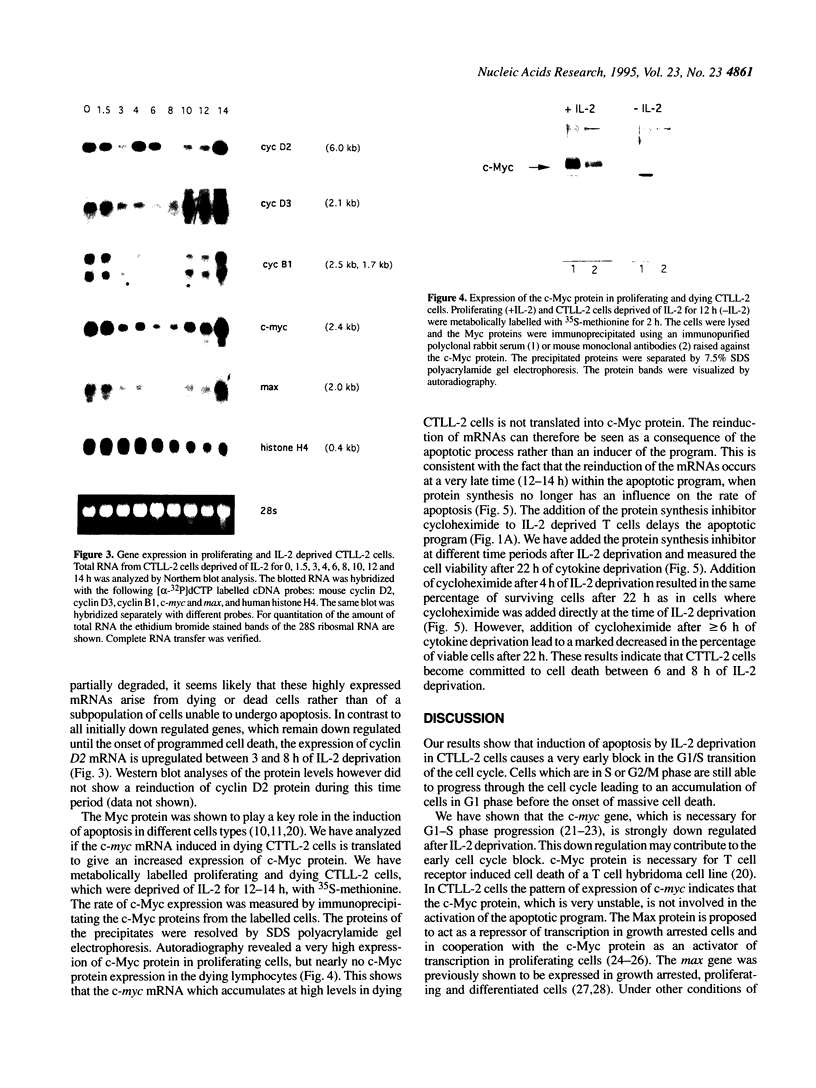
The IL-2 dependent murine cytotoxic T cell line CTLL-2 undergoes programmed cell death when deprived of its specific cytokine. We analyzed the expression of cell cycle related genes after IL-2 deprivation. Here we show that a generalized decrease and re elevation of the levels of mRNA takes place as part of the apoptotic program. The levels of several mRNAs encoding cell cycle functions, including cyclin D2, cyclin D3, cyclin B1, c-myc and max all declined at 1.5-3 h following IL-2 deprivation. Notably, the maxmRNA, which was shown to be expressed in proliferating, growth arrested and differentiated cells, is down regulated with the same kinetics as the other mRNAs. Surprisingly, the mRNAs whose levels declined at 1.5-3 h rose again at 10-14 h, a time which closely followed the time of the first detection of apoptotic DNA degradation, at 8 h, but which precedes actual loss of viability, at 14 h, as measured by trypan blue exclusion. Of all analyzed genes only the expression of the S-phase specific histone H4 gene resists the initial decrease and declines gradually over the course of cell death. Measurement of c-Myc protein synthesis at a late stage of the apoptotic program revealed that the accumulated reinduced mRNA is not translated into protein. Because transcriptional regulation has been shown to be dependent on the chromatin structure, the reinduction may be triggered by relaxation of the chromatin caused by alterations in the chromatin structure of apoptotic cells.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Amati B., Dalton S., Brooks M. W., Littlewood T. D., Evan G. I., Land H. Transcriptional activation by the human c-Myc oncoprotein in yeast requires interaction with Max. Nature. 1992 Oct 1;359(6394):423–426. doi: 10.1038/359423a0. [DOI] [PubMed] [Google Scholar]
- Arends M. J., Morris R. G., Wyllie A. H. Apoptosis. The role of the endonuclease. Am J Pathol. 1990 Mar;136(3):593–608. [PMC free article] [PubMed] [Google Scholar]
- Askew D. S., Ashmun R. A., Simmons B. C., Cleveland J. L. Constitutive c-myc expression in an IL-3-dependent myeloid cell line suppresses cell cycle arrest and accelerates apoptosis. Oncogene. 1991 Oct;6(10):1915–1922. [PubMed] [Google Scholar]
- Ayer D. E., Eisenman R. N. A switch from Myc:Max to Mad:Max heterocomplexes accompanies monocyte/macrophage differentiation. Genes Dev. 1993 Nov;7(11):2110–2119. doi: 10.1101/gad.7.11.2110. [DOI] [PubMed] [Google Scholar]
- Berberich S., Hyde-DeRuyscher N., Espenshade P., Cole M. max encodes a sequence-specific DNA-binding protein and is not regulated by serum growth factors. Oncogene. 1992 Apr;7(4):775–779. [PubMed] [Google Scholar]
- Chapman D. L., Wolgemuth D. J. Identification of a mouse B-type cyclin which exhibits developmentally regulated expression in the germ line. Mol Reprod Dev. 1992 Nov;33(3):259–269. doi: 10.1002/mrd.1080330305. [DOI] [PubMed] [Google Scholar]
- Chomczynski P., Sacchi N. Single-step method of RNA isolation by acid guanidinium thiocyanate-phenol-chloroform extraction. Anal Biochem. 1987 Apr;162(1):156–159. doi: 10.1006/abio.1987.9999. [DOI] [PubMed] [Google Scholar]
- Clarke A. R., Maandag E. R., van Roon M., van der Lugt N. M., van der Valk M., Hooper M. L., Berns A., te Riele H. Requirement for a functional Rb-1 gene in murine development. Nature. 1992 Sep 24;359(6393):328–330. doi: 10.1038/359328a0. [DOI] [PubMed] [Google Scholar]
- Cohen J. J., Duke R. C., Fadok V. A., Sellins K. S. Apoptosis and programmed cell death in immunity. Annu Rev Immunol. 1992;10:267–293. doi: 10.1146/annurev.iy.10.040192.001411. [DOI] [PubMed] [Google Scholar]
- Croston G. E., Kadonaga J. T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Curr Opin Cell Biol. 1993 Jun;5(3):417–423. doi: 10.1016/0955-0674(93)90006-c. [DOI] [PubMed] [Google Scholar]
- Darzynkiewicz Z., Bruno S., Del Bino G., Gorczyca W., Hotz M. A., Lassota P., Traganos F. Features of apoptotic cells measured by flow cytometry. Cytometry. 1992;13(8):795–808. doi: 10.1002/cyto.990130802. [DOI] [PubMed] [Google Scholar]
- Drapkin R., Merino A., Reinberg D. Regulation of RNA polymerase II transcription. Curr Opin Cell Biol. 1993 Jun;5(3):469–476. doi: 10.1016/0955-0674(93)90013-g. [DOI] [PubMed] [Google Scholar]
- Duke R. C., Cohen J. J. IL-2 addiction: withdrawal of growth factor activates a suicide program in dependent T cells. Lymphokine Res. 1986 Fall;5(4):289–299. [PubMed] [Google Scholar]
- Evan G. I., Wyllie A. H., Gilbert C. S., Littlewood T. D., Land H., Brooks M., Waters C. M., Penn L. Z., Hancock D. C. Induction of apoptosis in fibroblasts by c-myc protein. Cell. 1992 Apr 3;69(1):119–128. doi: 10.1016/0092-8674(92)90123-t. [DOI] [PubMed] [Google Scholar]
- Feinberg A. P., Vogelstein B. A technique for radiolabeling DNA restriction endonuclease fragments to high specific activity. Anal Biochem. 1983 Jul 1;132(1):6–13. doi: 10.1016/0003-2697(83)90418-9. [DOI] [PubMed] [Google Scholar]
- Ffrench-Constant C. A division too far? Curr Biol. 1992 Nov;2(11):577–579. doi: 10.1016/0960-9822(92)90150-9. [DOI] [PubMed] [Google Scholar]
- Gillis S., Smith K. A. Long term culture of tumour-specific cytotoxic T cells. Nature. 1977 Jul 14;268(5616):154–156. doi: 10.1038/268154a0. [DOI] [PubMed] [Google Scholar]
- Harel-Bellan A., Ferris D. K., Vinocour M., Holt J. T., Farrar W. L. Specific inhibition of c-myc protein biosynthesis using an antisense synthetic deoxy-oligonucleotide in human T lymphocytes. J Immunol. 1988 Apr 1;140(7):2431–2435. [PubMed] [Google Scholar]
- Heikkila R., Schwab G., Wickstrom E., Loke S. L., Pluznik D. H., Watt R., Neckers L. M. A c-myc antisense oligodeoxynucleotide inhibits entry into S phase but not progress from G0 to G1. 1987 Jul 30-Aug 5Nature. 328(6129):445–449. doi: 10.1038/328445a0. [DOI] [PubMed] [Google Scholar]
- Heintz N. Cell death and the cell cycle: a relationship between transformation and neurodegeneration? Trends Biochem Sci. 1993 May;18(5):157–159. doi: 10.1016/0968-0004(93)90103-t. [DOI] [PubMed] [Google Scholar]
- Heintz N., Sive H. L., Roeder R. G. Regulation of human histone gene expression: kinetics of accumulation and changes in the rate of synthesis and in the half-lives of individual histone mRNAs during the HeLa cell cycle. Mol Cell Biol. 1983 Apr;3(4):539–550. doi: 10.1128/mcb.3.4.539. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holt J. T., Redner R. L., Nienhuis A. W. An oligomer complementary to c-myc mRNA inhibits proliferation of HL-60 promyelocytic cells and induces differentiation. Mol Cell Biol. 1988 Feb;8(2):963–973. doi: 10.1128/mcb.8.2.963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacks T., Fazeli A., Schmitt E. M., Bronson R. T., Goodell M. A., Weinberg R. A. Effects of an Rb mutation in the mouse. Nature. 1992 Sep 24;359(6393):295–300. doi: 10.1038/359295a0. [DOI] [PubMed] [Google Scholar]
- Kamakaka R. T., Kadonaga J. T. Biochemical analysis of the role of chromatin structure in the regulation of transcription by RNA polymerase II. Cold Spring Harb Symp Quant Biol. 1993;58:205–212. doi: 10.1101/sqb.1993.058.01.025. [DOI] [PubMed] [Google Scholar]
- Kelekar A., Cole M. D. Tumorigenicity of fibroblast lines expressing the adenovirus E1a, cellular p53, or normal c-myc genes. Mol Cell Biol. 1986 Jan;6(1):7–14. doi: 10.1128/mcb.6.1.7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kerr J. F. Shrinkage necrosis: a distinct mode of cellular death. J Pathol. 1971 Sep;105(1):13–20. doi: 10.1002/path.1711050103. [DOI] [PubMed] [Google Scholar]
- Kerr J. F., Wyllie A. H., Currie A. R. Apoptosis: a basic biological phenomenon with wide-ranging implications in tissue kinetics. Br J Cancer. 1972 Aug;26(4):239–257. doi: 10.1038/bjc.1972.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretzner L., Blackwood E. M., Eisenman R. N. Myc and Max proteins possess distinct transcriptional activities. Nature. 1992 Oct 1;359(6394):426–429. doi: 10.1038/359426a0. [DOI] [PubMed] [Google Scholar]
- Laybourn P. J., Kadonaga J. T. Role of nucleosomal cores and histone H1 in regulation of transcription by RNA polymerase II. Science. 1991 Oct 11;254(5029):238–245. doi: 10.1126/science.254.5029.238. [DOI] [PubMed] [Google Scholar]
- Lee E. Y., Chang C. Y., Hu N., Wang Y. C., Lai C. C., Herrup K., Lee W. H., Bradley A. Mice deficient for Rb are nonviable and show defects in neurogenesis and haematopoiesis. Nature. 1992 Sep 24;359(6393):288–294. doi: 10.1038/359288a0. [DOI] [PubMed] [Google Scholar]
- Marzluff W. F. Histone 3' ends: essential and regulatory functions. Gene Expr. 1992;2(2):93–97. [PMC free article] [PubMed] [Google Scholar]
- Matsushime H., Roussel M. F., Ashmun R. A., Sherr C. J. Colony-stimulating factor 1 regulates novel cyclins during the G1 phase of the cell cycle. Cell. 1991 May 17;65(4):701–713. doi: 10.1016/0092-8674(91)90101-4. [DOI] [PubMed] [Google Scholar]
- Norris D., Dunn B., Osley M. A. The effect of histone gene deletions on chromatin structure in Saccharomyces cerevisiae. Science. 1988 Nov 4;242(4879):759–761. doi: 10.1126/science.2847314. [DOI] [PubMed] [Google Scholar]
- Oberhammer F., Wilson J. W., Dive C., Morris I. D., Hickman J. A., Wakeling A. E., Walker P. R., Sikorska M. Apoptotic death in epithelial cells: cleavage of DNA to 300 and/or 50 kb fragments prior to or in the absence of internucleosomal fragmentation. EMBO J. 1993 Sep;12(9):3679–3684. doi: 10.1002/j.1460-2075.1993.tb06042.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paranjape S. M., Kamakaka R. T., Kadonaga J. T. Role of chromatin structure in the regulation of transcription by RNA polymerase II. Annu Rev Biochem. 1994;63:265–297. doi: 10.1146/annurev.bi.63.070194.001405. [DOI] [PubMed] [Google Scholar]
- Prendergast G. C., Hopewell R., Gorham B. J., Ziff E. B. Biphasic effect of Max on Myc cotransformation activity and dependence on amino- and carboxy-terminal Max functions. Genes Dev. 1992 Dec;6(12A):2429–2439. doi: 10.1101/gad.6.12a.2429. [DOI] [PubMed] [Google Scholar]
- Quelle D. E., Ashmun R. A., Shurtleff S. A., Kato J. Y., Bar-Sagi D., Roussel M. F., Sherr C. J. Overexpression of mouse D-type cyclins accelerates G1 phase in rodent fibroblasts. Genes Dev. 1993 Aug;7(8):1559–1571. doi: 10.1101/gad.7.8.1559. [DOI] [PubMed] [Google Scholar]
- Roeder R. G. The complexities of eukaryotic transcription initiation: regulation of preinitiation complex assembly. Trends Biochem Sci. 1991 Nov;16(11):402–408. doi: 10.1016/0968-0004(91)90164-q. [DOI] [PubMed] [Google Scholar]
- Sachs A. B. Messenger RNA degradation in eukaryotes. Cell. 1993 Aug 13;74(3):413–421. doi: 10.1016/0092-8674(93)80043-e. [DOI] [PubMed] [Google Scholar]
- Shi Y., Glynn J. M., Guilbert L. J., Cotter T. G., Bissonnette R. P., Green D. R. Role for c-myc in activation-induced apoptotic cell death in T cell hybridomas. Science. 1992 Jul 10;257(5067):212–214. doi: 10.1126/science.1378649. [DOI] [PubMed] [Google Scholar]
- White E., Cipriani R., Sabbatini P., Denton A. Adenovirus E1B 19-kilodalton protein overcomes the cytotoxicity of E1A proteins. J Virol. 1991 Jun;65(6):2968–2978. doi: 10.1128/jvi.65.6.2968-2978.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White R. J., Jackson S. P. The TATA-binding protein: a central role in transcription by RNA polymerases I, II and III. Trends Genet. 1992 Aug;8(8):284–288. doi: 10.1016/0168-9525(92)90255-3. [DOI] [PubMed] [Google Scholar]
- Wyllie A. H. Glucocorticoid-induced thymocyte apoptosis is associated with endogenous endonuclease activation. Nature. 1980 Apr 10;284(5756):555–556. doi: 10.1038/284555a0. [DOI] [PubMed] [Google Scholar]
- Yonish-Rouach E., Resnitzky D., Lotem J., Sachs L., Kimchi A., Oren M. Wild-type p53 induces apoptosis of myeloid leukaemic cells that is inhibited by interleukin-6. Nature. 1991 Jul 25;352(6333):345–347. doi: 10.1038/352345a0. [DOI] [PubMed] [Google Scholar]