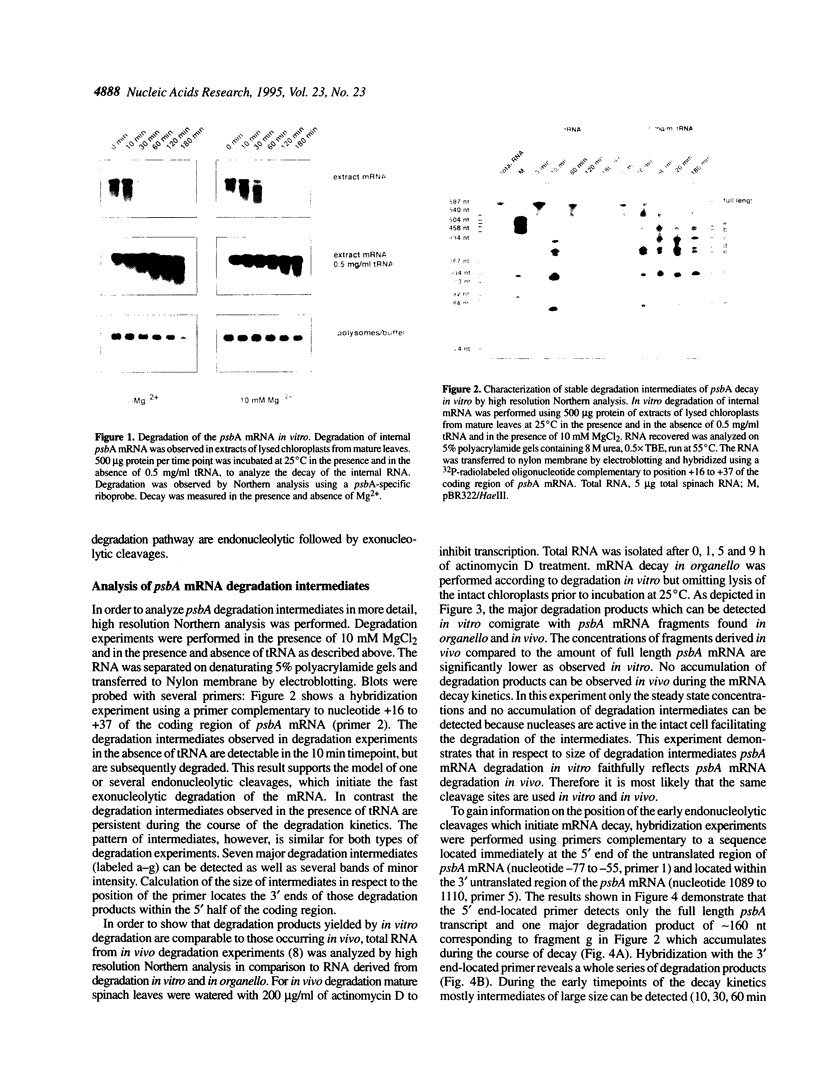
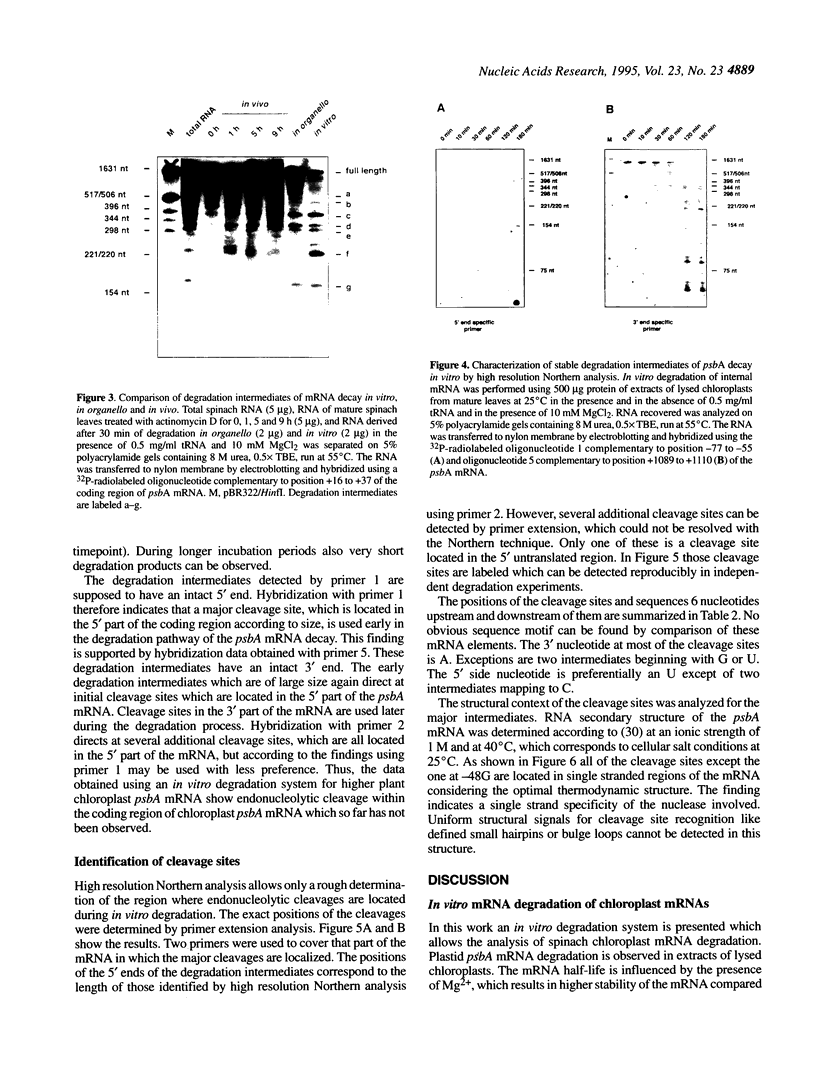
Abstract
The expression of chloroplast genes during leaf development in higher plants is regulated on several levels as transcription, RNA processing and stability, protein stability and turnover. Differential mRNA stability is one major component which contributes to the developmentally controlled accumulation of higher plant chloroplast psbA mRNA, which encodes the D1 protein of photosystem II. To understand the molecular mechanisms of specific mRNA degradation an in vitro mRNA decay system based on lysed chloroplasts from spinach leaves was established. Employing this degradation extract the decay of psbA mRNA was analyzed. Half-life of the psbA mRNA in vitro is dependent on the degradation conditions as the presence of Mg2+, which was found to stabilize the mRNA. Addition of tRNA stabilizes the mRNA and allows the accumulation of distinct degradation intermediates. psbA mRNA derived fragments of the same size were detected in degradation experiments in vitro, in organello and in vivo. 5' ends of the degradation intermediates were identified by primer extension and found to be localized in the 5' part of the coding region. The data indicate a degradation mechanism involving initiation of psbA mRNA decay by specific endonucleolytic cleavage and subsequent exonucleolytic degradation of the fragments. Possible models for cleavage site recognition are discussed.
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Adams C. C., Stern D. B. Control of mRNA stability in chloroplasts by 3' inverted repeats: effects of stem and loop mutations on degradation of psbA mRNA in vitro. Nucleic Acids Res. 1990 Oct 25;18(20):6003–6010. doi: 10.1093/nar/18.20.6003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arraiano C., Yancey S. D., Kushner S. R. Identification of endonucleolytic cleavage sites involved in decay of Escherichia coli trxA mRNA. J Bacteriol. 1993 Feb;175(4):1043–1052. doi: 10.1128/jb.175.4.1043-1052.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bandyopadhyay R., Coutts M., Krowczynska A., Brawerman G. Nuclease activity associated with mammalian mRNA in its native state: possible basis for selectivity in mRNA decay. Mol Cell Biol. 1990 May;10(5):2060–2069. doi: 10.1128/mcb.10.5.2060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barkan A., Miles D., Taylor W. C. Chloroplast gene expression in nuclear, photosynthetic mutants of maize. EMBO J. 1986 Jul;5(7):1421–1427. doi: 10.1002/j.1460-2075.1986.tb04378.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bernstein P. L., Herrick D. J., Prokipcak R. D., Ross J. Control of c-myc mRNA half-life in vitro by a protein capable of binding to a coding region stability determinant. Genes Dev. 1992 Apr;6(4):642–654. doi: 10.1101/gad.6.4.642. [DOI] [PubMed] [Google Scholar]
- Carpousis A. J., Van Houwe G., Ehretsmann C., Krisch H. M. Copurification of E. coli RNAase E and PNPase: evidence for a specific association between two enzymes important in RNA processing and degradation. Cell. 1994 Mar 11;76(5):889–900. doi: 10.1016/0092-8674(94)90363-8. [DOI] [PubMed] [Google Scholar]
- Chen H. C., Stern D. B. Specific binding of chloroplast proteins in vitro to the 3' untranslated region of spinach chloroplast petD mRNA. Mol Cell Biol. 1991 Sep;11(9):4380–4388. doi: 10.1128/mcb.11.9.4380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deng X. W., Stern D. B., Tonkyn J. C., Gruissem W. Plastid run-on transcription. Application to determine the transcriptional regulation of spinach plastid genes. J Biol Chem. 1987 Jul 15;262(20):9641–9648. [PubMed] [Google Scholar]
- Gruissem W., Barkan A., Deng X. W., Stern D. Transcriptional and post-transcriptional control of plastid mRNA levels in higher plants. Trends Genet. 1988 Sep;4(9):258–263. doi: 10.1016/0168-9525(88)90033-9. [DOI] [PubMed] [Google Scholar]
- Gruissem W. Chloroplast gene expression: how plants turn their plastids on. Cell. 1989 Jan 27;56(2):161–170. doi: 10.1016/0092-8674(89)90889-1. [DOI] [PubMed] [Google Scholar]
- Gruissem W., Greenberg B. M., Zurawski G., Hallick R. B. Chloroplast gene expression and promoter identification in chloroplast extracts. Methods Enzymol. 1986;118:253–270. doi: 10.1016/0076-6879(86)18077-3. [DOI] [PubMed] [Google Scholar]
- Jackson A. O., Larkins B. A. Influence of Ionic Strength, pH, and Chelation of Divalent Metals on Isolation of Polyribosomes from Tobacco Leaves. Plant Physiol. 1976 Jan;57(1):5–10. doi: 10.1104/pp.57.1.5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Eichacker L. A., Rudiger W., Mullet J. E. Chlorophyll regulates accumulation of the plastid-encoded chlorophyll proteins P700 and D1 by increasing apoprotein stability. Plant Physiol. 1994 Mar;104(3):907–916. doi: 10.1104/pp.104.3.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim J., Klein P. G., Mullet J. E. Ribosomes pause at specific sites during synthesis of membrane-bound chloroplast reaction center protein D1. J Biol Chem. 1991 Aug 15;266(23):14931–14938. [PubMed] [Google Scholar]
- Kim J., Klein P. G., Mullet J. E. Synthesis and turnover of photosystem II reaction center protein D1. Ribosome pausing increases during chloroplast development. J Biol Chem. 1994 Jul 8;269(27):17918–17923. [PubMed] [Google Scholar]
- Kim M., Christopher D. A., Mullet J. E. Direct evidence for selective modulation of psbA, rpoA, rbcL and 16S RNA stability during barley chloroplast development. Plant Mol Biol. 1993 Jun;22(3):447–463. doi: 10.1007/BF00015975. [DOI] [PubMed] [Google Scholar]
- Klaff P., Gruissem W. Changes in Chloroplast mRNA Stability during Leaf Development. Plant Cell. 1991 May;3(5):517–529. doi: 10.1105/tpc.3.5.517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melefors O., von Gabain A. Site-specific endonucleolytic cleavages and the regulation of stability of E. coli ompA mRNA. Cell. 1988 Mar 25;52(6):893–901. doi: 10.1016/0092-8674(88)90431-x. [DOI] [PubMed] [Google Scholar]
- Mullet J. E. Dynamic regulation of chloroplast transcription. Plant Physiol. 1993 Oct;103(2):309–313. doi: 10.1104/pp.103.2.309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet J. E., Klein R. R. Transcription and RNA stability are important determinants of higher plant chloroplast RNA levels. EMBO J. 1987 Jun;6(6):1571–1579. doi: 10.1002/j.1460-2075.1987.tb02402.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nickelsen J., van Dillewijn J., Rahire M., Rochaix J. D. Determinants for stability of the chloroplast psbD RNA are located within its short leader region in Chlamydomonas reinhardtii. EMBO J. 1994 Jul 1;13(13):3182–3191. doi: 10.1002/j.1460-2075.1994.tb06617.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parker R., Jacobson A. Translation and a 42-nucleotide segment within the coding region of the mRNA encoded by the MAT alpha 1 gene are involved in promoting rapid mRNA decay in yeast. Proc Natl Acad Sci U S A. 1990 Apr;87(7):2780–2784. doi: 10.1073/pnas.87.7.2780. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ross J., Kobs G. H4 histone messenger RNA decay in cell-free extracts initiates at or near the 3' terminus and proceeds 3' to 5'. J Mol Biol. 1986 Apr 20;188(4):579–593. doi: 10.1016/s0022-2836(86)80008-0. [DOI] [PubMed] [Google Scholar]
- Sakamoto W., Kindle K. L., Stern D. B. In vivo analysis of Chlamydomonas chloroplast petD gene expression using stable transformation of beta-glucuronidase translational fusions. Proc Natl Acad Sci U S A. 1993 Jan 15;90(2):497–501. doi: 10.1073/pnas.90.2.497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salvador M. L., Klein U., Bogorad L. 5' sequences are important positive and negative determinants of the longevity of Chlamydomonas chloroplast gene transcripts. Proc Natl Acad Sci U S A. 1993 Feb 15;90(4):1556–1560. doi: 10.1073/pnas.90.4.1556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmitz M., Steger G. Base-pair probability profiles of RNA secondary structures. Comput Appl Biosci. 1992 Aug;8(4):389–399. doi: 10.1093/bioinformatics/8.4.389. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Gruissem W. Control of plastid gene expression: 3' inverted repeats act as mRNA processing and stabilizing elements, but do not terminate transcription. Cell. 1987 Dec 24;51(6):1145–1157. doi: 10.1016/0092-8674(87)90600-3. [DOI] [PubMed] [Google Scholar]
- Stern D. B., Radwanski E. R., Kindle K. L. A 3' stem/loop structure of the Chlamydomonas chloroplast atpB gene regulates mRNA accumulation in vivo. Plant Cell. 1991 Mar;3(3):285–297. doi: 10.1105/tpc.3.3.285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern S., Moazed D., Noller H. F. Structural analysis of RNA using chemical and enzymatic probing monitored by primer extension. Methods Enzymol. 1988;164:481–489. doi: 10.1016/s0076-6879(88)64064-x. [DOI] [PubMed] [Google Scholar]
- Tanzer M. M., Meagher R. B. Faithful degradation of soybean rbcS mRNA in vitro. Mol Cell Biol. 1994 Apr;14(4):2640–2650. doi: 10.1128/mcb.14.4.2640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P., Buddelmeijer N., Raué H. A. A cell-free extract from yeast cells for studying mRNA turnover. Nucleic Acids Res. 1992 May 25;20(10):2503–2510. doi: 10.1093/nar/20.10.2503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vreken P., Raué H. A. The rate-limiting step in yeast PGK1 mRNA degradation is an endonucleolytic cleavage in the 3'-terminal part of the coding region. Mol Cell Biol. 1992 Jul;12(7):2986–2996. doi: 10.1128/mcb.12.7.2986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Cohen S. N. ard-1: a human gene that reverses the effects of temperature-sensitive and deletion mutations in the Escherichia coli rne gene and encodes an activity producing RNase E-like cleavages. Proc Natl Acad Sci U S A. 1994 Oct 25;91(22):10591–10595. doi: 10.1073/pnas.91.22.10591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wennborg A., Sohlberg B., Angerer D., Klein G., von Gabain A. A human RNase E-like activity that cleaves RNA sequences involved in mRNA stability control. Proc Natl Acad Sci U S A. 1995 Aug 1;92(16):7322–7326. doi: 10.1073/pnas.92.16.7322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zurawski G., Bohnert H. J., Whitfeld P. R., Bottomley W. Nucleotide sequence of the gene for the M(r) 32,000 thylakoid membrane protein from Spinacia oleracea and Nicotiana debneyi predicts a totally conserved primary translation product of M(r) 38,950. Proc Natl Acad Sci U S A. 1982 Dec;79(24):7699–7703. doi: 10.1073/pnas.79.24.7699. [DOI] [PMC free article] [PubMed] [Google Scholar]