Abstract
Exposure to chronic and acute oxidative stress is correlated with many human diseases, including, but not limited to, cancer, heart disease, diabetes, and obesity. In addition to cellular lipids and proteins, cellular oxidative stress can result in damage to DNA bases, especially in mitochondrial DNA. We previously described the development of spontaneous late-onset obesity, hepatic steatosis, hyperinsulinemia, and hyperleptinemia in mice that are deficient in the DNA glycosylase nei-like 1 (NEIL1), which initiates base excision repair of several oxidatively damaged bases. In the current study, we report that exposure to a chronic oxidative stress in the form of a high-fat diet greatly accelerates the development of obesity in neil1−/− mice. Following a 5-wk high-fat diet challenge, neil1−/− mice gained significantly more body weight than neil1+/+ littermates and had increased body fat accumulation and moderate to severe hepatic steatosis. Analysis of oxygen consumption by indirect calorimetry indicated a modest reduction in total oxygen consumption in neil1−/− mice that was abolished upon correction for lean body mass. Additionally, hepatic expression of several inflammatory genes was significantly upregulated in neil1−/− mice following high-fat diet challenge compared with chow-fed or neil1+/+ counterparts. A long-term high-fat diet also induced glucose intolerance as well as a significant reduction in mitochondrial DNA and protein content in neil1−/− mice. Collectively, these data indicate that NEIL1 deficiency results in an increased susceptibility to obesity and related complications potentially by lowering the threshold for tolerance of cellular oxidative stress in neil1−/− mice.
Keywords: nei-like 1, oxidative stress, base excision repair, metabolic syndrome, mitochondrial DNA
as the collection of diseases comprising the metabolic syndrome continues to threaten public health, there is significant interest in understanding the cellular factors that lead to the development of these pathologies. It is becoming increasingly clear that structural and metabolic alterations within organelles can contribute to total cellular function and consequently to whole body physiology. In this regard, decreased mitochondrial capacity is hypothesized to contribute to a variety of conditions, including obesity, insulin resistance, hypertension, diabetes, and age-related pathologies such as Alzheimer's and Parkinson's disease (reviewed in Refs. 31, 45, and 48). Optimal mitochondrial function is important to several cellular processes, including cell signaling, calcium homeostasis, apoptosis, and fatty acid oxidation, as well as ATP synthesis, to meet cellular energy demands (5, 16, 35). These mitochondrial functions are maintained by mitochondrial proteins, the majority of which are encoded for in the nucleus and then imported into the mitochondria (32). However, mitochondria also express a distinct genome that encodes for multiple tRNAs and rRNAs as well as several subunits of complexes I, III, IV, and V of the electron transport chain (31, 45, 48). Because of the lack of protective features such as histones, intronic regions, and nucleotide excision repair, as well as the close proximity of mitochondrial DNA (mtDNA) to the site of reactive oxygen species (ROS) production, mtDNA is particularly vulnerable to oxidative damage. In response to oxidative stress, mtDNA has been shown to have a 10- to 15-fold higher mutation rate than nuclear DNA, resulting in reduced efficiency of the oxidative phosphorylation system and a decreased capacity of cells to maintain ATP levels (31, 42, 45, 48).
In mammalian cells, oxidative damage to mtDNA is repaired predominantly via the base excision repair (BER) pathway initiated by the activity of several DNA glycosylases, including nei-like (NEIL)1, NEIL2, OGG1, and NTH1 (reviwed in ref. 8). The primary substrates for NEIL1 are the ring-fragmented purines 4,6-diamino-5-formamidopyrimidine (FapyA) and 2,6-diamino-4-hydroxy-5-formamidopyrimidine (FapyG) (9) and the secondary oxidation products of 8-oxoguanine (8-oxoG), guanidinohydantoin, and the two diastereomers of spiroiminodihydantoin (15, 23, 24). In addition, in vitro experiments have revealed that subsets of the ring-saturated pyrimidines, such as both enantiomers of thymine glycol, are substrates for NEIL1 (17, 21, 39).
Previously, we created and characterized mice lacking NEIL1 (neil1−/−) with respect to metabolic disorders, cancer, and immune modulation (4, 34, 49). The majority of the male neil1−/− mice exhibited midlife onset of obesity, with most males weighing ∼50 g at 7–9 mo of age and exhibiting severe fatty liver disease and hyperleptinemia (49). Although these studies were performed in mice of mixed genetic background, they have now been fully backcrossed onto a C57BL/6 background. Neil1+/+ and neil1−/− mice were analyzed for baseline metabolic functions and then challenged with a high-fat diet (HFD). We report here the effects of an acute HFD-induced oxidative stress on organ pathology and energy homeostasis in neil1−/− mice.
MATERIALS AND METHODS
Generation of neil1+/+ and neil1−/− mice.
To generate the mice required to conduct the HFD challenge, neil1+/− males were bred with female neil1+/+ C57BL/6 mice (Charles River Laboratories), and the genotypes of all offspring were determined by PCR, as described previously (49). Subsequent generations were backcrossed in the same manner until 8th-generation backcrossed heterozygotic mice were obtained. A sufficient number of heterozygotic matings were established to yield at least 10 each of the male neil1+/+ and neil1−/− mice. All experiments involving animals were approved by the Institutional Animal Care and Use Committee at Oregon Health and Science University.
Feeding and motor activity measurements.
Animals were individually housed in metabolic cages equipped with a running wheel (Mini-Mitter, Sunriver, OR) for ≥1 wk prior to each experiment being started and habituated for ≥4 days to eating powdered chow. Mice were allowed ad libitum access to the powdered chow that was weighed and replaced daily. Feeding duration and frequency were quantified via a clock recording the number of times and the duration for which an infrared light beam was interrupted when the animal was in the feeding chamber. Voluntary physical activity was measured by recording the magnetic switch closures of a magnet placed on the revolving wheel to quantify wheel revolutions.
Body composition.
Lean body mass and fat mass were determined immediately prior to and at the end of the HFD challenge by dual-energy X-ray absorptiometry (DEXA; PIXImus mouse densitometer; MEC Lunar, Minster, OH) or by low-resolution NMR (EchoMRI 3-in-1 system; Echo Medical Systems, Houston, TX) (44). Animals were fasted overnight prior to DEXA analyses to minimize the effect of ingested food.
Indirect calorimetry.
Oxygen consumption (V̇o2) and carbon dioxide production (V̇co2) were simultaneously determined by indirect calorimetry (Oxymax; Columbus Instruments, Columbus, OH). Animals were individually housed in chambers at 24 ± 1°C and had ad libitum access to chow diet and water. Following a 48-h acclimation period, V̇o2 and V̇co2 were recorded for 24 h, including a 12-h dark and 12-h light phase. Samples were recorded every 2 min, with a room air reference taken every 12 min. Respiratory exchange ratio was calculated as the molar ratio of V̇co2/V̇o2.
HFD feeding and glucose tolerance.
At 23–25 wk of age, neil1+/+ and neil1−/− mice were switched to a HFD (60% fat, 20% protein, and 20% carbohydrate by calories, 5.24 kcal/g metabolizable energy; diet no. D12492; Research Diets, New Brunswick, NJ) for 5 wk. Control animals were maintained on standard rodent chow (Purina 5001, composed of 13% fat, 29% protein, and 58% carbohydrate by calories, 3.04 kcal/g metabolizable energy) for the duration of the experimental period. Body weights were measured prior to and at the indicated intervals during feeding. For the long-term HFD study, 8-wk-old male mice were maintained on chow diet or HFD for 20 wk and were euthanized at 28 wk of age. Glucose tolerance was assessed prior to euthanasia after 16 wk of feeding. Animals were fasted for 4 h and given an intraperitoneal injection of 20% dextrose at a dose of 1 g/kg of body wt. Blood was collected by saphenous vein puncture under manual restraint, and plasma glucose was assessed by the glucose oxidase method at 0, 20, 40, 90, and 180 min postinjection.
Tissue collection and pathology.
Blood was collected from the medial saphenous vein on the inner surface of the hind leg. Prior to tissue collection, mice were euthanized with CO2 overdose. Measurements of circulating insulin, triglycerides, and glucose were carried out as described previously (49). Liver tissues were harvested and processed for histological analyses. Coded samples were sent to IDEXX Reference Laboratories (Westbrook, ME) for sectioning, staining, and interpretation.
Lipid analyses by gas chromatography.
Fatty acid compositions from blood samples were determined by gas chromatography (GC) as a blinded investigation. Total lipid was extracted by the Folch method (11), and lipid classes (phospholipids, free fatty acids, triglycerides, and cholesterol esters) were separated by one-dimensional thin-layer chromatography. An internal standard was added prior to formation of methyl esters using the procedure of Lepage and Roy (28). Fatty acid methyl esters were recovered, and the extract was evaporated under nitrogen and redissolved in hexane in preparation for GC separations. Aliquots of the extract (1.0 μl) were injected into an Agilent GC 6890 gas chromatograph equipped with an autoinjector, a flame ionization detector (Agilent Technology, Santa Clara, CA), and a fused polar silica capillary column (100 m, 0.25 mm ID, SPE-2560; Supelco, Bellafonte, PA). The GC parameters were as described previously (27). Fatty acid peaks were identified by comparison of their retention times with those of commercial standards, and the amounts were calculated using Chemstation software (Agilent Technology). Precision and accuracy were evaluated using model mixtures of known fatty acid methyl esters and an established in-house control pool.
Gene expression profiling and quantitative real-time PCR.
Gene expression analyses were performed using Affymetrix gene chips (mouse 430 2.0). Each gene on a gene chip is represented by a set of probe pairs (11–20 pairs/gene), and each probe is a 25-mer oligodeoxynucleotide. The probe pair contains one “perfect match” oligodeoxynucleotide whose sequence is derived from the targeted mRNA. A “mismatch” probe, differing only at position 13 in the oligodeoxynucleotide, completes the probe pair and serves as a control for nonspecific hybridization. Reverse transcriptase-directed cDNA synthesis was performed using 10 μg of total RNA and an oligo-dT primer that encodes a bacteriophage T7 RNA polymerase promoter. Second-strand synthesis converts the cDNA into a double-stranded DNA template for use in an in vitro transcription reaction. Biotin-tagged NTPs were used in a T7 RNA polymerase-directed in vitro transcription to produce biotin-labeled cRNA for hybridization to the chip probes. Hybridization was performed at 45°C for 16 h in 0.1 M MES, pH 6.6, 1 M sodium chloride, 0.02 M EDTA, and 0.01% Tween 20. RNAs representing eight prokaryotic genes (dap, thr, lys, and phe from Bacillus subtilis, bio B, bio C, and bio D from Escherichia coli, and cre from bacteriophage P1) were added to each total RNA sample before the initiation of first-strand cDNA synthesis. Each of the prokaryotic genes has been cloned into a transcription vector (bio B, bio C, bio D, and cre into pBluescript-PA; dap, thr, lys, and Phe into pT7PA) that expresses the respective RNA as a polyadenylated transcript. These RNAs are compatible with the biotin-labeling strategy and probe sets since all of these genes are present on all Affymetrix chips. The spiked RNAs were added at different copy number concentrations within the experimental RNA sample, thereby providing a diagnostic for the labeling and hybridization process and serving as a relative standard for evaluating the level of expression for a specific gene in the sample RNA. Gene expression was analyzed using the Affymetrix analysis software (GCOS). An absolute analysis for each chip was used to determine the genes that were expressed in the total RNA sample. Comparative analyses of data sets from two chips were used to determine and quantify changes in gene expression between two RNA populations. Data derived from the neil1+/+ mice fed a chow diet were designated as the baseline for each comparative analysis. Two sets of algorithms were used in determining each gene's change decision (increased, decreased, or no change) and in determining a quantitative estimate of the change in gene expression relative to the baseline sample. This quantitative estimate is termed the signal log ratio and is expressed as a log scale to the base 2. Concordance assays were used to identify genes that showed the same change (increase or decrease) in expression for each RNA sample within an experimental set.
Validations of expression analyses of specific genes were performed using quantitative real-time PCR (QPCR) SYBR Green analysis (Applied Biosystems, Foster City, CA). RNA samples were quantified using a Nanodrop Spectrophotometer (Nanodrop Technologies, Wilmington, DE) and analyzed for quality on a RNA Nano Chip using the Agilent 2100 Bioanalyzer (Agilent Technologies). Synthesis of cDNA was performed using the reagents provided in the Taqman Reverse Transcription Reagents Kit (Applied Biosystems) per specifications. QPCR amplifications were performed in triplicate using SYBR Green PCR Master Mix, 900 mM target-specific primers (Integrated DNA Technologies, Coralville, IA), and 2 μl of cDNA in a 25-μl reaction volume. Specific primer sequences are available on request. All assays were performed as relative QPCR, using 18S as a control. Assays were run on an ABI Prism 7000 Sequence Detection System (Applied Biosystems) using the following protocol: 50°C for 2 min, 95°C for 10 min, and 40 cycles of 95°C for 15 s and 60°C for 60 s.
mtDNA quantitation.
mtDNA content was measured as described previously (14). Briefly, total DNA was isolated from tissues, and three separate mitochondrial DNA targets were amplified by real-time PCR using gene-specific primers. Results are expressed normalized to 18S rRNA for genomic DNA content. Primer sequences are available upon request.
Western blot analyses.
Liver sections were homogenized in modified RIPA buffer, quantified by the Bradford method, and separated by SDS-PAGE. Mitochondrial protein content was determined by blotting for mitochondrial proteins using primary antibodies against complex IV, subunit 1, or porin (Mitosciences, Eugene, OR) followed by blotting with fluorescently tagged secondary antibodies. Blots were quantitatively imaged using the LiCor immunofluorescence detection system. Protein loading was confirmed by blotting for GAPDH (Sigma-Aldrich, St. Louis, MO).
RESULTS
NEIL1 deficiency increases susceptibility to obesity and hyperlipidemia.
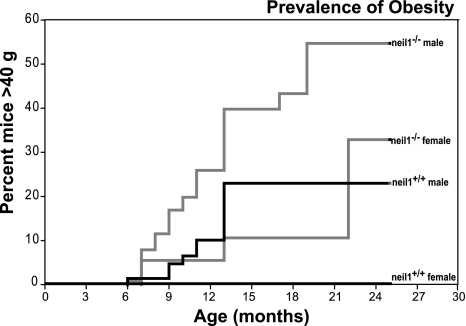
As described previously, deletion of the neil1 gene was frequently associated with visceral obesity and fatty liver disease predominantly in male mice (49). To gain insight into the prevalence of obesity in our neil1−/− colony, we undertook retrospective analyses of body weights in 187 male and female neil1−/− mice relative to neil1+/+ counterparts. The analyses spanned 5 years and included neil1−/− mice at varying stages of backcrossing into a C57BL/6 background. The data shown in Fig. 1 represent the percentage of each of the colonies that achieved a body weight in excess of 40 g at the indicated age, corresponding to a body fat percentage ≥35%. Both male and female neil1−/− mice demonstrated stronger tendencies to be heavier relative to neil1+/+ counterparts (Fig. 1). These data indicate that NEIL1 deficiency increases the susceptibility to weight gain and obesity in mice. It is interesting to note that, even at 24 mo of age, we did not observe 100% penetrance of the obesity phenotype in neil1−/− mice. This is not entirely unexpected, as varying extents of DNA damage in neil1−/− mice, as well as the localization of this damage to specific tissues, may affect the penetrance of obesity in these animals. Therefore, although NEIL1 deficiency predisposes animals to weight gain, it is not an absolute determinant of increased body weight in mice.
Fig. 1.
Rate and frequency of the development of obesity. Weight gain was analyzed in a total of 187 mice, including male and female nei-like 1 (neil1)−/− and neil1+/+ mice. Data shown represent the percentage of each of the colonies (neil1+/+ and neil1−/− male and female mice) that achieved a weight in excess of 40 g (corresponding to ≥35% body fat). Neil1−/− males and females demonstrated a stronger tendency toward weight gain relative to their respective neil1+/+ counterparts.
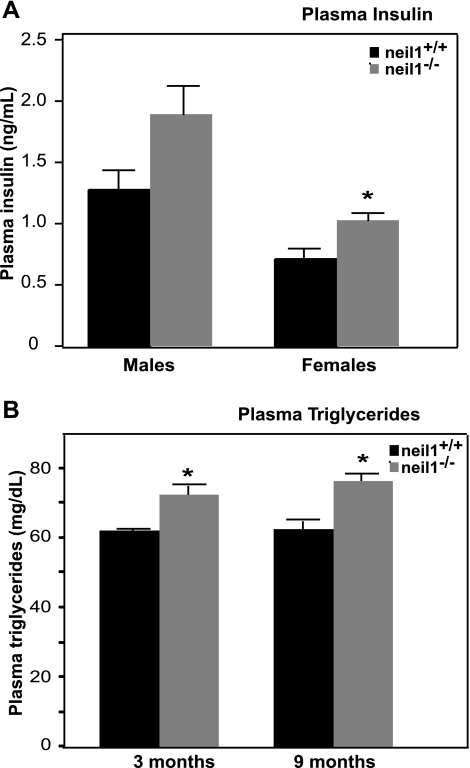
In addition to following the progression of obesity, plasma glucose, insulin, and lipids were also evaluated periodically in these animals between 3 and 12 mo of age. Overnight fasting glucose values showed no significant differences throughout the lifespans of both male and female neil1−/− relative to controls (data not shown). Circulating insulin levels were modestly elevated in both male and female neil1−/− mice, but this difference was statistically significant only in female mice (Fig. 2A). Starting as early as 3 mo of age, prior to the development of overt differences in body weight, plasma triglycerides were significantly elevated in male neil1−/− mice (Fig. 2B). In addition to triglycerides, other circulating lipid species as well as the fatty acyl composition of these lipids are known to influence disease susceptibility (33). Plasma free fatty acids, phospholipids, and cholesterol esters were all slightly elevated in male neil1−/− mice relative to neil1+/+ controls (Table 1). Plasma triglycerides were significantly higher in neil1−/− mice relative to neil1+/+ controls. To determine whether NEIL1 deficiency results in altered fatty acid composition of circulating lipids, fatty acyl chain composition of plasma lipids was determined by GC (Table 1). Rather than a selective increase in particular fatty acids, neil1−/− mice had increased levels of almost all fatty acids in the triglyceride fraction (Table 1). Although neil1−/− mice had elevated levels of these circulating lipids, visualization of aortic tissues from obese (45–55 g) neil1−/− mice showed no significant evidence of atherosclerotic plaque formation (Dr. Scott Ballinger, University of Alabama Birmingham, personal communication).
Fig. 2.
Plasma insulin and triglycerides. The levels of circulating insulin (A) and triglycerides (B) were measured in fasted adult male and female neil1+/+ and neil1−/− mice at 6–9 mo of age (A) and in male neil1−/− mice at 3 and 9 mo of age (B), respectively. Plasma insulin levels were modestly elevated in neil1−/− mice but only reached statistical significance in female mice. Plasma triglycerides were significantly elevated in male neil1−/− mice starting as early as 3 mo of age relative to age-matched neil1+/+ counterparts. Data are expressed as means ± SE of 6–10 animals/group. *P < 0.05 vs. neil1+/+.
Table 1.
Plasma fatty acid composition
| Phospholipid, μg/ml |
Free Fatty Acid, μg/ml |
Cholesterol Ester, μg/ml |
Triglyceride, μg/ml |
|||||
|---|---|---|---|---|---|---|---|---|
| neil1+/+ | neil1−/− | neil1+/+ | neil1−/− | neil1+/+ | neil1−/− | neil1+/+ | neil1−/− | |
| C14:0 | 0.71 ± 0.06 | 0.72 ± 0.06 | 0.43 ± 0.12 | 0.41 ± 0.05 | 1.25 ± 0.17 | 1.03 ± 0.15 | 1.26 ± 0.20 | 3.08 ± 0.33* |
| C16:0 | 208.4 ± 13.1 | 252.2 ± 29.5 | 6.6 ± 1.01 | 10.7 ± 1.4 | 13.7 ± 2.8 | 15.5 ± 1.4 | 39.0 ± 6.0 | 84.2 ± 7.9* |
| C16:1 (n-7) | 6.2 ± 0.6 | 5.1 ± 0.8 | 1.3 ± 0.4 | 2.7 ± 1.0 | 9.0 ± 1.9 | 12.3 ± 2.2 | 7.0 ± 1.4 | 16.9 ± 3.0* |
| C18:0 | 124.6 ± 5.8 | 172.3 ± 12.1* | 2.3 ± 0.14 | 4.0 ± 0.35 | 1.00 ± 0.32 | 0.90 ± 0.17 | 3.5 ± 0.4 | 7.7 ± 0.8* |
| C18:1 (n-9) | 53.2 ± 4.6 | 61.2 ± 6.8 | 5.25 ± 1.34 | 7.8 ± 0.8 | 17.3 ± 1.7 | 28.5 ± 5.1 | 55.2 ± 8.6 | 123.5 ± 8.9* |
| C18:1 (n-7) | 12.8 ± 1.2 | 21.2 ± 3.4* | 0.46 ± 0.10 | 1.2 ± 0.1* | 1.5 ± 0.2 | 3.02 ± 0.53 | 4.4 ± 0.7 | 15.3 ± 1.8* |
| C18:2 (n-6) | 205.1 ± 17.2 | 240.2 ± 26.7 | 5.9 ± 1.4 | 8.0 ± 1.1 | 153.2 ± 27.0 | 203.6 ± 23.3 | 59.7 ± 12.7 | 103.9 ± 9.9* |
| C20:4 (n-6) | 86.0 ± 3.4 | 96.7 ± 4.5 | 0.65 ± 0.13 | 1.27 ± 0.1* | 83.3 ± 8.8 | 124.3 ± 7.1* | 4.2 ± 0.7 | 6.3 ± 0.7 |
| C20:5 (n-3) | 7.7 ± 0.6 | 11.9 ± 1.6* | 0.29 ± 0.04 | 0.62 ± 0.1* | 11.0 ± 1.6 | 20.2 ± 2.8* | 9.7 ± 1.9 | 7.3 ± 1.1 |
| C22:6 (n-3) | 53.8 ± 4.2 | 81.5 ± 9.5* | 0.9 ± 0.18 | 2.0 ± 0.2* | 16.0 ± 2.8 | 29.9 ± 3.7* | 9.60 ± 1.8 | 24.8 ± 3.1* |
| Total | 818.4 ± 46.8 | 1,020.0 ± 92.9 | 26.5 ± 5.2 | 40.8 ± 4.6 | 332.5 ± 41.9 | 455.0 ± 37.2 | 205.2 ± 35.7 | 424.5 ± 31.4* |
Data are expressed as means ± SE for 4–5 neil1+/+ and 5–6 neil1−/− animals. Neil1, nei-like 1. Plasma fatty acids were extracted and separated by thin-layer chromatography prior to gas chromatographic analyses. Neil1−/− mice had a generalized increase in almost all major fatty acids in circulating triglycerides relative to neil1+/+ animals. Levels of major plasma fatty acid species as well as total fatty acids in each fraction are shown.
P < 0.05 vs. neil1+/+.
Neil1−/− mice have no difference in food intake or wheel running but display significantly reduced metabolic rates.
Since previous analyses were carried out on first-generation backcrossed neil1−/− mice that were a genetic hybrid between C57BL/6 and 129/SvEv, it was hypothesized that this genetic heterogeneity could be a potential source of phenotypic variability with regard to symptoms associated with metabolic syndrome. To address this possibility, male heterozygotic mice were bred with female C57BL/6 mice until 8th-generation backcrossed heterozygotic mice were obtained. These mice were mated and offspring genotyped, with expected Mendelian ratios of neil1−/−, neil1+/−, and neil1+/+ observed. A sufficient number of heterozygotic matings to yield at least 10 each of male neil1+/+ and neil1−/− mice were established. All the studies described below were carried out in age-matched male neil1+/+ and neil1−/− mice fully backcrossed onto a C57BL/6 background.
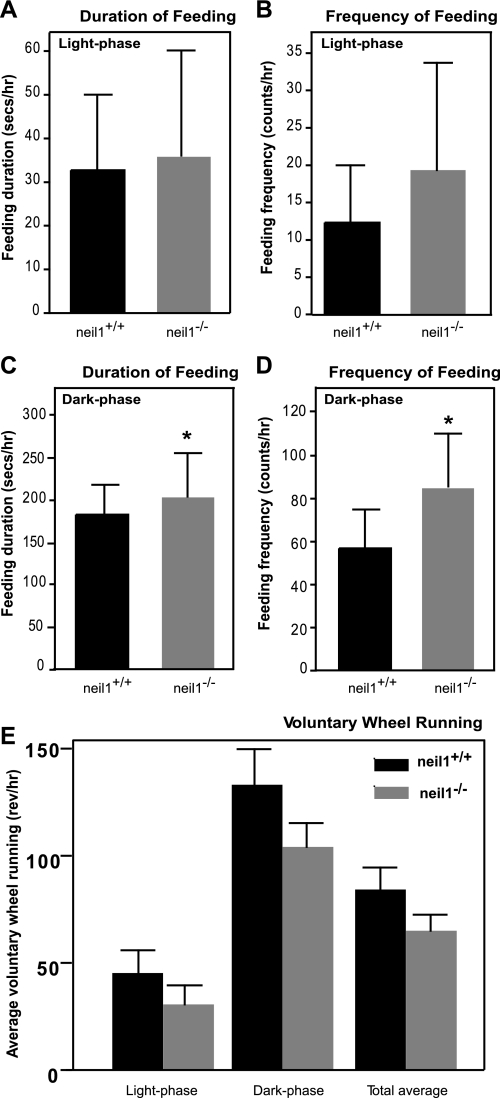
Given the increased susceptibility to weight gain in male neil1−/− mice, we were interested in determining the mechanisms leading to a positive energy balance in these animals. Food intake and voluntary physical activity were measured in individually housed male neil1−/− and neil1+/+ littermates. There were no differences between genotypes in feeding patterns during the light phase (Fig. 3, A and B). However, during the dark phase, neil1−/− mice spent slightly more time feeding and frequenting the hoppers relative to neil1+/+ littermates (Fig. 3, C and D, respectively). Despite these modest differences in feeding behavior, neil1−/− mice did not consume significantly more food than neil1+/+ counterparts (not shown). To determine whether voluntary physical activity was different between neil1+/+ and neil1−/− animals, mice were placed in cages fitted with a running wheel, and wheel running was measured continuously for 10 consecutive 12:12-h dark-light cycles. Neil1−/− mice tended to have a modest reduction in voluntary wheel running relative to neil1+/+ mice across both dark and light cycles, but these differences were not statistically significant (Fig. 3E).
Fig. 3.
Feeding behavior and physical activity. The duration (A and C) and frequency (B and D) of access to food hoppers during light and dark phases were measured in male mice. Compared with neil1+/+ animals, neil1−/− mice spent significantly more time feeding and frequenting the hoppers only during the dark phase. Despite this modest difference in feeding behavior, there was no difference in average food intake between neil1+/+ and neil1−/− mice (not shown). E: voluntary wheel running activity was assessed over 10 consecutive dark and light cycles in neil1+/+ and neil1−/− mice. Although neil1−/− mice tended to have reduced voluntary wheel running relative to neil1+/+ counterparts, these differences were not statistically significant. Data are expressed as means ± SE of 5–6 animals/group. *P < 0.05 vs. neil1+/+.
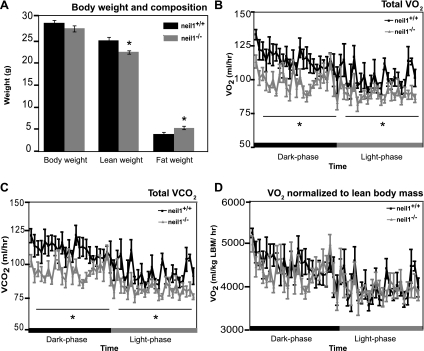
Previously, no significant phenotypic differences were observed in neil1−/− mice relative to their neil1+/+ littermates through their first 6 mo of life, with the exception of increased circulating leptin levels, which, similarly to plasma triglycerides (Fig. 2B), were observed as early as 3 mo of age (49). To determine whether alterations in metabolic rate may also precede changes in total body weight in neil1−/− mice, energy expenditure was measured by indirect calorimetry in 14-wk-old male mice. Immediately prior to the analyses, mice were weighed, and body composition was measured by NMR (Fig. 4A). Although neil1−/− mice were not significantly heavier, they had reduced lean body mass and increased fat mass compared with neil1+/+ counterparts (Fig. 4A). Total O2 consumption (Fig. 4B) as well as CO2 production (Fig. 4C) measured by indirect calorimetry were significantly decreased in neil1−/− mice relative to their neil1+/+ counterparts. This difference was significant across both day and night cycles and remained significant even when normalized to total body weight (P < 0.005; data not shown). Interestingly, when normalized to lean body mass, the more metabolically active component of total body mass (3), the difference between genotypes in O2 consumption (Fig. 4D) and CO2 production (not shown) was ablated, suggesting that alterations in body composition may underlie the reduction in energy expenditure (Fig. 4, B and C) observed in neil1−/− mice. The respiratory exchange ratio, an indicator of substrate oxidation in animals, was not significantly different between neil1+/+ and neil1−/− animals (not shown).
Fig. 4.
Fasted weight loss and energy expenditure by indirect calorimetry. Body composition (A) and 24-h energy expenditures (B–D) were measured in 14-wk-old male mice. Although total body weights were similar (A), lean body mass measured by MRI was reduced, and fat mass was increased in neil1−/− mice. Total O2 consumption (V̇o2; B) and CO2 production (V̇co2; C) were significantly lower in neil1−/− mice across both day and night cycles. When normalized to lean body mass (D), there were no significant differences in V̇o2 between neil1+/+ and neil1−/− animals. Data are expressed as means ± SE of 6 animals/group. *P < 0.05 vs. neil1+/+.
Neil1−/− mice have increased adiposity following a HFD challenge.
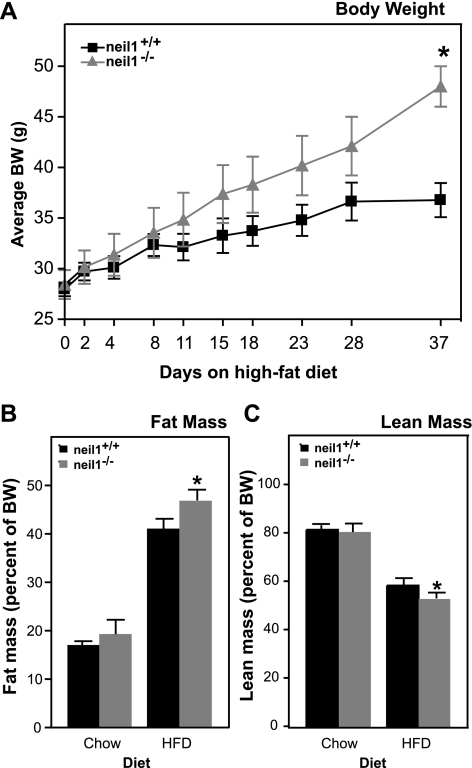
Since NEIL1 catalyzes the repair of oxidatively damaged DNA bases, we hypothesized that the development of metabolic abnormalities may be accelerated in neil1−/− mice upon exposure to oxidative stress. Therefore, neil1+/+ and neil1−/− mice were exposed to an acute oxidative stress in the form of a short-term HFD. Twenty-two-week-old male neil1−/− mice and neil1+/+ counterparts were individually housed and placed on a 60% HFD for 5 wk. Over the feeding period, neil1−/− mice gained significantly more body weight than their neil1+/+ counterparts (Fig. 5A). Body composition evaluated by DEXA analyses was not significantly different between neil1+/+ and neil1−/− mice at this age prior to HFD feeding (Fig. 5, B and C). However, following 5 wk of HFD-feeding, neil1−/− mice had a significantly higher fat mass than neil1+/+ mice (Fig. 5B). Lean body mass as percent of body weight was slightly reduced in neil1−/− mice relative to neil1+/+ mice after HFD feeding (Fig. 5C).
Fig. 5.
High-fat diet (HFD)-induced weight gain and changes in body composition. A: 22-wk-old male neil1+/+ and neil1−/− mice were placed on a HFD containing 60% fat by calories for 5 wk, and body weights were measured at the indicated time points. B and C: additionally, body composition was measured by dual-energy X-ray absorptiometry analysis analysis before and after the HFD challenge. B: %fat mass was increased by HFD feeding in all animals but was significantly higher in neil1−/− mice relative to neil1+/+. C: lean mass, as a percent of body weight, was slightly lower in HFD-fed neil1−/− mice compared with neil1+/+ counterparts. Data are expressed as means ± SE of 5–6 animals/group. *P < 0.05 vs. neil1+/+.
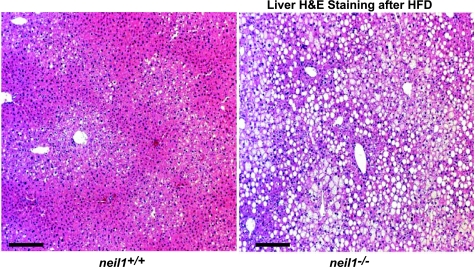
Since increased adiposity is a known risk factor for hepatic lipid accumulation, mice were euthanized and hepatic lipids visualized by hematoxylin and eosin staining of fixed liver sections. Coded tissues were analyzed by IDEXX Reference Laboratories; each IDEXX descriptor was assigned a numeric value corresponding to the level of severity of hepatic steatosis as follows: normal (0), mild steatosis (1), mild to moderate (2), moderate (3), moderate with cytoplasmic microvesiculation (4), moderate to severe (5), moderate to severe with cytoplasmic microvesiculation (6), and severe (7). Whereas livers from HFD-fed neil1+/+ samples showed only mild steatosis with an average score of 1.0 (Fig. 6), neil1−/− livers showed moderate-to-severe steatosis with an average score of 5.6 (Fig. 6).
Fig. 6.
Hepatic lipid accumulation. Liver sections from HFD-fed neil1+/+ and neil1−/− mice were stained with hematoxylin and eosin (H & E). Neil1−/− animals displayed dramatic accumulation of hepatic lipids, seen as white droplets, following a 5-wk HFD feeding. A blinded assessment of liver damage revealed neil1−/− mice to have moderate to severe hepatic steatosis, whereas neil1+/+ mice displayed only mild steatosis. Images are representative of 5–6 animals/group, and the bar represents 25 μm in both images.
HFD-fed neil1−/− mice have increased hepatic expression of inflammatory genes.
To gain further insight into the metabolic perturbations elicited by HFD feeding in neil1−/− mice, changes in hepatic gene expression patterns were analyzed on Affymetrix 420 2.0 microarray chips. These chips did not include mitochondrially encoded genes. Comparisons of the data sets were filtered for a minimum twofold change, and a concordance analysis requiring the same directional expression for each set of paired samples was performed. Table 2 lists 12 genes (11 increases, 1 decrease) that demonstrated at least a twofold change in each of the neil1+/+ and neil1−/− liver-derived data sets. The changes identified by Affymetrix gene expression analyses were confirmed by real-time PCR, although the absolute fold change did vary between the two types of assays (Table 2). Since the analyses described above were restricted to differential gene expression between neil1+/+ and neil1−/− mice on HFD, it was of additional interest to determine whether the same genes were also differentially expressed in the mice maintained on a chow diet. Therefore, the expression of these genes was analyzed by real-time PCR in livers of chow-fed neil1+/+ vs. neil1−/− mice.
Table 2.
Hepatic gene expression
| HFD (Fold Change) |
|||||
|---|---|---|---|---|---|
| Gene | Gene Symbol | Direction of Change | Microarray | RT-PCR | Chow, RT-PCR (Fold Change) |
| Apolipoprotein A-IV | Apoa4 | ↑ | 5.7 | 12.8 | 5.7 |
| Lipocalin 2 | Lcn2 | ↑ | 5.3 | 7.8 | No change |
| Serum amyloid A2 | Saa2 | ↑ | 3.5 | 6.3 | No change |
| Serum amyloid A1 | Saa1 | ↑ | 3.5 | 6.0 | No change |
| Annexin A2 | Anxa2 | ↑ | 3.2 | 5.6 | 3.7 |
| Complement factor D | Cfd | ↑ | 5.7 | 4.2 | 1.7 |
| Ecto-5′-nucleotidase | Nt5e | ↑ | 2.6 | 4.2 | No change |
| Metallothionein 2 | Mt2 | ↑ | 2.3 | 4.1 | 2.0 |
| β-Galactoside binding protein | Lgals1 | ↑ | 3.5 | 3.8 | 2.8 |
| S100 calcium binding protein A10 | S100a10 | ↑ | 2.3 | 2.6 | No change |
| Regulator of calcineurin 2 | Rcan2 | ↑ | 2.6 | 2.5 | 3.0 |
| Solute carrier organic anion transporter family, member 1a1 | Slco1a1 | ↓ | 0.2 | 0.3 | 0.2 |
HFD, high-fat diet. Expression of hepatic genes was analyzed by DNA microarray (n = 3 for each genotype), and expression of genes that showed a >2-fold directional change was confirmed by quantitative real-time PCR (n = 3 for each genotype). Results are presented as fold change in HFD- or chow-fed neil1−/− livers relative to HFD- or chow-fed neil1+/+ counterparts, respectively.
Of the 12 genes that were differentially expressed between HFD-fed neil1+/+ vs. neil1−/− mice, five genes were not similarly regulated in chow-fed mice, whereas the rest were regulated in a similar manner in neil1+/+ vs. neil1−/− mice regardless of dietary condition. The majority of altered genes were associated with hepatic injury and inflammation. Genes involved in lipid metabolism and energy homeostasis pathways were not among the genes differentially regulated between neil1+/+ and neil1−/− mice. Additionally, to determine whether the integration of the neil1 targeting vector could have disrupted the expression of genes in the vicinity of the neil1 gene, the Affymetrix data were analyzed for changes in expression of four genes 5′ and three genes 3′ to neil1 [snurportin 1; protein tyrosine phosphatase, nonreceptor type 9 location; 2700012I20Rik hypothetical protein; Sin3a, transcriptional regulator, mannosidase, α, class 2C, member 1; COMM domain containing 4; 1700041C23Rik (protein coding, unknown function); taste receptor protein 1]. None of these genes were either up- or downregulated >1.3-fold, leading us to conclude that the phenotypic changes in neil1−/− mice can be attributed to modulation of neil1 expression alone.
Long-term HFD impairs glucose tolerance in neil1−/− mice.
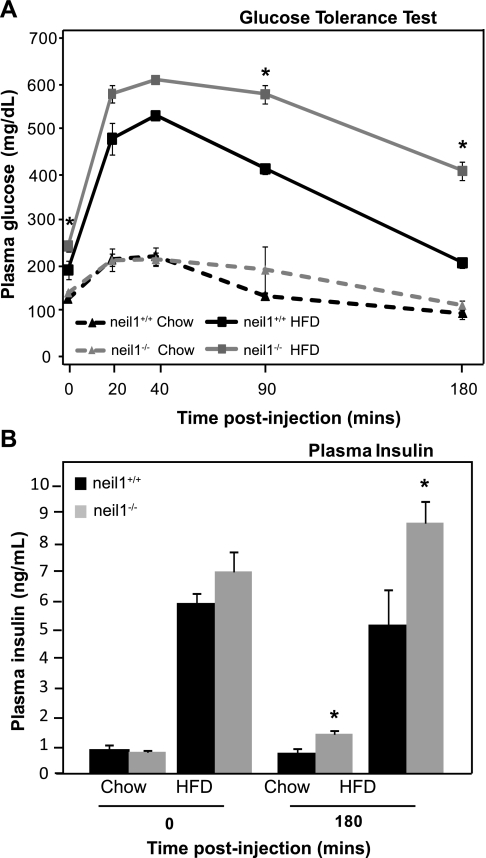
Since neil1−/− mice display several features of the metabolic syndrome, including increased body weight and adiposity and increased hepatic lipid accumulation, we were interested in determining whether a long-term HFD regimen known to impair glucose tolerance would differentially affect neil1−/− mice. Therefore, 8-wk-old male neil1+/+ and neil1−/− animals were maintained on the HFD for 20 wk. Over the feeding period, chow-fed neil1+/+ and neil1−/− mice gained similar amounts of weight (not shown). However, HFD-fed neil1−/− mice gained more body weight than HFD-fed neil1+/+ counterparts (28.1 vs. 25.7 g, respectively, P = 0.02). After 16 wk of feeding, glucose tolerance was measured in both chow-fed and HFD-fed mice. No differences were observed in adipose tissue distribution or histology between neil1+/+ and neil1−/− mice (Supplemental Fig. S1; Supplemental Material is available online at the AJP-Endocrinology and Metabolism web site). On a chow diet, there were no significant differences between neil1+/+ and neil1−/− animals in terms of glucose clearance (Fig. 7A). However, after HFD feeding neil1−/− mice had significantly higher baseline plasma glucose levels than their neil1+/+ counterparts. Additionally, plasma glucose remained elevated above neil1+/+ levels at 90 and 180 min after glucose administration, indicating delayed glucose clearance after HFD feeding in neil1−/− mice. Plasma insulin levels were also measured at 0 and 180 min after glucose injection (Fig. 7B). HFD feeding increased plasma insulin in both neil1+/+ and neil1−/− animals. When measured 180 min after glucose administration, plasma insulin levels were elevated in both chow-fed and HFD-fed neil1−/− mice relative to their respective neil1+/+ counterparts (Fig. 7B). Combined with the sustained hyperglycemia observed in HFD-fed neil1−/− mice (Fig. 7A) at this time point, the elevated plasma insulin levels suggest a state of insulin resistance in neil1−/− mice, especially after HFD feeding.
Fig. 7.
Glucose tolerance and plasma insulin. A: chow-fed and long-term HFD-fed neil1+/+ and neil1−/− animals were injected with 1 g glucose/kg body wt, and plasma glucose was measured at the indicated time points. Glucose clearance was significantly delayed in HFD-fed neil1−/− mice. B: plasma insulin was also measured before and 180 min after glucose injection. Plasma insulin was significantly higher in both chow-fed and HFD-fed neil1−/− mice 180 min after glucose injection relative to their respective neil1+/+ counterparts. Data are expressed as means ± SE of 5–7 animals/group. *P < 0.05 vs. neil1+/+.
HFD-fed neil1−/− mice have reduced hepatic mtDNA content.
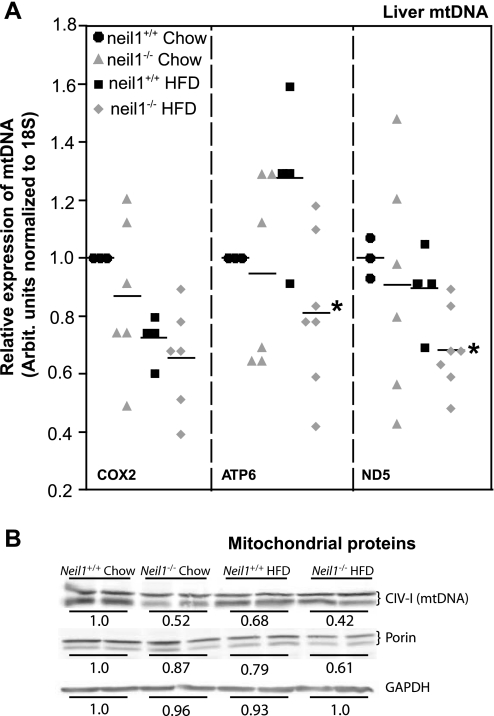
Given the role of NEIL1 in repairing oxidative damage to DNA, we hypothesized that the increased susceptibility to obesity in neil1−/− animals may be a consequence of alterations in mtDNA content, a measure of cellular mitochondrial mass. Therefore, mtDNA levels were determined by real-time PCR in both chow-fed and HFD-fed animals. Despite significant interanimal variability, which is not unexpected given the heteroplasmic nature of mtDNA, Atp6 and Nd5 levels were significantly lower in HFD-fed neil1−/− mice relative to neil1+/+ counterparts (Fig. 8A). Levels of Cox2 were not significantly altered. These data indicate that NEIL1 deficiency causes a measurable reduction in hepatic mtDNA content upon exposure of cells to an oxidative stress such as HFD feeding. Additionally, hepatic content of mitochondrial proteins was also measured by Western blotting. Levels of both a mitochondrially encoded protein (complex IV, subunit 1) as well the mitochondrial marker protein porin were significantly reduced in livers of neil1−/− mice (Fig. 8B). Consistent with the reduction in mtDNA levels (Fig. 8A), the reduction in the mitochondrially encoded protein was particularly substantial (Fig. 8B).
Fig. 8.
Hepatic mitochondrial DNA (mtDNA) and protein content. A: hepatic mtDNA content was measured by real-time PCR and normalized to genomic DNA (18S) content. Three different mtDNA targets were analyzed. HFD-fed neil1−/− mice had significantly reduced levels of ATP synthase, subunit 6 (Atp6), and NADH dehydrogenase 5 (Nd5) relative to neil1+/+ counterparts, indicating reduced mitochondrial content in livers of these mice. Horizontal bar indicates mean value for each group, and data are expressed relative to chow-fed neil1+/+ mice. B: mitochondrial protein content was measured in liver homogenates from chow- and HFD-fed mice. Neil1−/− mice had reduced levels of the mtDNA-encoded subunit of complex IV (CIV-I) as well as the mitochondrial membrane protein porin. Western blotting data are representative of 5–7 animals/group. Quantitation numbers are averages of relative fluorescence units per group expressed relative to chow-fed neil1+/+ mice. *P < 0.05 vs. neil1+/+. Cox2, cytochrome c oxidase subunit 2.
DISCUSSION
Neil1−/− mice display a strong tendency for development of features of the metabolic syndrome, especially when exposed to oxidative stress. It is important to note that the absence of NEIL1 is not an absolute determinant of the development of obesity but rather a predisposing factor. It is possible that subtle variations in environmental conditions and genetic/epigenetic factors significantly influence disease manifestation. Alternatively, it is possible that since neil1 encodes for a DNA repair enzyme, varying extents of accumulation of DNA damage in individual mice and the tissue localization of such damage ultimately influence the development and severity of the metabolic phenotypes in neil1−/− animals. At least two human polymorphic variants of neil1 have been described, resulting in loss of DNA glycosylase activity (39). Although the mechanisms linking DNA repair to metabolic syndrome are yet to be elucidated, it is interesting to note that in addition to the neil1−/− model, male-specific obesity has also been reported in mice lacking OGG1 (ogg1−/−), another DNA glycosylase involved in the repair of oxidatively induced base damage (1). Although this previous study was designed to investigate tumor induction in ogg1−/− mice, male and female ogg1−/− mice were observed to have weight gains nearly identical to neil1−/− mice (1, 49). Further characterization of this obesity phenotype in ogg1−/− mice has not yet been reported.
BER, catalyzed by DNA glycosylases, has often been ascribed a “housekeeping” function in the maintenance of genomic stability. This pathway is almost exclusively responsible for monitoring the genome for endogenous base damage caused by base susceptibility to alkylation, deamination, saturation, and oxidation as well as base loss. BER is also responsible for the repair of the mitochondrial genome that is inherently more susceptible to damage from oxidative species (7, 10, 29). The biological effects of deficiencies in mtDNA repair are not well understood, particularly because knockouts of the coding region of DNA glycosylases inactivate repair in both mitochondria as well as the nucleus. Nevertheless, we reported previously that neil1−/− mice accumulate PCR blocking lesions and deletions in mtDNA at significantly higher frequencies than neil1+/+ mice (49). The reductions in several mtDNA targets in HFD-fed neil1−/− mice (Fig. 8A) suggest accelerated oxidative aging in these animals. Similar reductions in mtDNA have been described in other metabolic conditions, including alcohol-induced hepatic steatosis and a model of adolescent insulin resistance (12, 14, 30, 31). Further investigations are warranted concerning oxidative damage to nucleic acids and cellular proteins in liver and tissues such as skeletal muscle that contribute significantly to organismal energy balance.
Although the issue of mitochondrial damage and mitochondrial biogenesis is still controversial, several lines of evidence suggest that reductions in mtDNA content or quality can result in impairments in mitochondrial function. The most notable example of this is in the case of aged tissue. Aging is associated with reductions in DNA repair capacity and, consequently, increased mtDNA damage and reduced mtDNA content in multiple tissues, including liver and skeletal muscle (2, 26). Therefore, although in the short-term mtDNA damage such as that induced by ROS may trigger a hormetic response by increasing mitochondrial biogenesis (18), in a model of premature oxidative aging it is plausible that reductions in mtDNA repair would in fact result in impairments in mitochondrial number and function. Ongoing studies concerning the effects of NEIL1 deficiency on mitochondrial function should clarify the precise roles of the observed reductions in mtDNA and proteins in neil1−/− mice (Fig. 8).
One of the primary functions of mitochondria is to generate ATP via oxidative phosphorylation, and indeed, several mitochondrial insufficiencies, such as those observed in mice expressing proofreading-deficient DNA polymerase-γ (47), are characterized by reduced energy supplies and consequent weight loss. However, an intact mitochondrial respiratory chain also serves the important function of oxidizing NADH to NAD. This mitochondrially generated NAD is critical to another key mitochondrial function, fatty acid β-oxidation, since the oxidative enzyme 3-hydroxyacyl-CoA dehydrogenase is an NAD-dependent enzyme. Therefore, it is plausible that the reduction in hepatic mitochondrial content in HFD-fed neil1−/− mice could lead to a reduction in fat oxidation in these mice, consistent with their increased adiposity. We have also observed that following a long-term HFD, neil1−/− mice have a markedly different pattern of hepatic lipid accumulation characterized by microvesicular lipid droplets rather than macrovesicular lipid deposition observed in neil1+/+ mice (Sampath H and Lloyd RS, unpublished data). Microvesicular steatosis is a characteristic feature of defects in hepatic fat oxidation and has been described in other metabolic derangements such as alcoholic steatosis (12). Further studies are required to clarify whether the observed reductions in mtDNA and protein content (Fig. 8) result in impairments in mitochondrial function in neil1−/− mice.
Apart from increases in fat mass, it is interesting to note that lean body mass is reduced in neil1−/− animals (Fig. 4A). This is particularly interesting given that the observed reductions in V̇o2 (Fig. 4B) were abolished when the V̇o2 was normalized to lean body mass (Fig. 4D) but not when normalized to total body weight (not shown). These results suggest that NEIL1 function may be important in maintaining lean body mass and that this function of NEIL1 may help protect against accelerated weight gain.
In addition to increased weight gain, neil1−/− mice are more susceptible to developing increased circulating triglycerides (Fig. 2B) and HFD-induced hepatic lipid accumulation (Fig. 6). Analyses of hepatic gene expression revealed a significant increase in genes associated with activation of hepatic or systemic inflammatory pathways (Table 2). Upregulation of metallothionein 2 (Mt2) as well as ApoA4 gene transcription has been shown to be important for the regeneration of damaged liver tissue from hepatectomy and injury (20, 38, 46). Mt2 is also known to have a protective role against oxidative stress (25). Thus, the increase in transcript levels of hepatic Mt2 in both chow- and HFD-fed neil1−/− mice is suggestive of increased levels of oxidative stress in the livers of these mice. Apoa4 is also known to be a positive acute-phase protein (22), whereas lipocalin 2 and serum amyloid A1 and A2 are all associated with inflammation and insulin resistance (40, 51, 52). Additionally, ecto5′-nucleotidase contributes to the formation of extracellular adenosine and has been linked to hepatic fibrosis (37). Annexin A2 (Anxa2) and S100 calcium-binding protein A10 are known to exist in a heterotrimeric complex and have also been implicated in the inflammatory response (43). Although the upregulation of inflammatory genes in the fatty livers of neil1−/− mice is not entirely surprising, the increased expression of a subset of these inflammatory genes, including ApoA4 and Anxa2, in chow-fed neil1−/− mice is very intriguing. Additionally, the only gene that was downregulated in neil1−/− livers, solute carrier organic anion transporter (Slco1a1), is also associated with the action of inflammatory cytokines such as TNFα (13). Indeed, chronic activation of inflammatory pathways, especially in organs such as liver and adipose tissue, is triggered by alterations in nutrient availability and often accompanies metabolic dysfunction (6, 19, 41). In fact, a recent study in mice lacking the Toll-like receptor-5 (TLR5) reported metabolic phenotypes generally similar to those observed in neil1−/− mice, including increased body weight, fat mass, and circulating triglycerides, as well as severe hepatic steatosis (50). These metabolic phenotypes in tlr5−/− mice were accompanied by alterations in gut microbiota, leading to increased inflammation in adipose tissue of these mice. Although the effects of NEIL1 deficiency on gut health and energy efficiency are not known, the hepatic induction of inflammatory genes in neil1−/− mice (Table 2) suggests that either systemic or tissue-specific activation of inflammatory cascades in these animals, potentially due to increased signals of oxidative stress, may underlie their predisposition to weight gain.
Based on the results presented here, we propose that NEIL1 deficiency lowers the threshold for tolerance of oxidative damage in animals. A reduced ability to repair oxidative damage is consistent with the increased mtDNA deletions observed previously in neil1−/− mice (49) as well as the reductions in mtDNA and protein content in livers of these mice upon HFD feeding. Increased oxidative stress could also serve as a trigger for the hepatic inflammation observed in neil1−/− mice (Table 2). Additionally, induction of inflammatory pathways has been shown to induce oxidative stress and damage that is, at least in part, repaired by the actions of NEIL1 (36). These changes induced by a reduction in oxidative tolerance in neil1−/− mice could result in the changes in body composition and reduced energy expenditure observed in neil1−/− mice (Fig. 4). Ongoing studies are aimed at understanding the precise nature of mitochondrial dysfunction and hepatic inflammation resulting from inactivation of DNA glycosylases and the mechanistic contribution of DNA repair pathways to the maintenance of lean body mass, substrate oxidation, and energy homeostasis.
GRANTS
This work was supported by National Institute of Diabetes and Digestive and Kidney Diseases Grant RO1-DK-075974 (to R. S. Lloyd).
DISCLOSURES
The authors declare that there are no conflicts of interest.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of the Lloyd and McCullough laboratories for numerous discussions and analyses of these data.
REFERENCES
- 1. Arai T, Kelly VP, Minowa O, Noda T, Nishimura S. The study using wild-type and Ogg1 knockout mice exposed to potassium bromate shows no tumor induction despite an extensive accumulation of 8-hydroxyguanine in kidney DNA. Toxicology 221: 179–186, 2006 [DOI] [PubMed] [Google Scholar]
- 2. Barazzoni R, Short KR, Nair KS. Effects of aging on mitochondrial DNA copy number and cytochrome c oxidase gene expression in rat skeletal muscle, liver, and heart. J Biol Chem 275: 3343–3347, 2000 [DOI] [PubMed] [Google Scholar]
- 3. Butler AA, Kozak LP. A recurring problem with the analysis of energy expenditure in genetic models expressing lean and obese phenotypes. Diabetes 59: 323–329, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Chan MK, Ocampo-Hafalla MT, Vartanian V, Jaruga P, Kirkali G, Koenig KL, Brown S, Lloyd RS, Dizdaroglu M, Teebor GW. Targeted deletion of the genes encoding NTH1 and NEIL1 DNA N-glycosylases reveals the existence of novel carcinogenic oxidative damage to DNA. DNA Repair (Amst) 8: 786–794, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Contreras L, Drago I, Zampese E, Pozzan T. Mitochondria: the calcium connection. Biochim Biophys Acta 1797: 607–618, 2010 [DOI] [PubMed] [Google Scholar]
- 6. Dandona P, Aljada A, Bandyopadhyay A. Inflammation: the link between insulin resistance, obesity and diabetes. Trends Immunol 25: 4–7, 2004 [DOI] [PubMed] [Google Scholar]
- 7. de Souza-Pinto NC, Bohr VA. The mitochondrial theory of aging: involvement of mitochondrial DNA damage and repair. Int Rev Neurobiol 53: 519–534, 2002 [DOI] [PubMed] [Google Scholar]
- 8. Dizdaroglu M. Base-excision repair of oxidative DNA damage by DNA glycosylases. Mutat Res 591: 45–59, 2005 [DOI] [PubMed] [Google Scholar]
- 9. Dizdaroglu M, Kirkali G, Jaruga P. Formamidopyrimidines in DNA: mechanisms of formation, repair, and biological effects. Free Radic Biol Med 45: 1610–1621, 2008 [DOI] [PubMed] [Google Scholar]
- 10. Druzhyna NM, Wilson GL, LeDoux SP. Mitochondrial DNA repair in aging and disease. Mech Ageing Dev 129: 383–390, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Folch J, Lees M, Sloane Stanley GH. A simple method for the isolation and purification of total lipides from animal tissues. J Biol Chem 226: 497–509, 1957 [PubMed] [Google Scholar]
- 12. Fromenty B, Grimbert S, Mansouri A, Beaugrand M, Erlinger S, Rotig A, Pessayre D. Hepatic mitochondrial DNA deletion in alcoholics: association with microvesicular steatosis. Gastroenterology 108: 193–200, 1995 [DOI] [PubMed] [Google Scholar]
- 13. Geier A, Dietrich CG, Voigt S, Kim SK, Gerloff T, Kullak-Ublick GA, Lorenzen J, Matern S, Gartung C. Effects of proinflammatory cytokines on rat organic anion transporters during toxic liver injury and cholestasis. Hepatology 38: 345–354, 2003 [DOI] [PubMed] [Google Scholar]
- 14. Gianotti TF, Sookoian S, Dieuzeide G, Garcia SI, Gemma C, Gonzalez CD, Pirola CJ. A decreased mitochondrial DNA content is related to insulin resistance in adolescents. Obesity (Silver Spring) 16: 1591–1595, 2008 [DOI] [PubMed] [Google Scholar]
- 15. Hailer MK, Slade PG, Martin BD, Rosenquist TA, Sugden KD. Recognition of the oxidized lesions spiroiminodihydantoin and guanidinohydantoin in DNA by the mammalian base excision repair glycosylases NEIL1 and NEIL2. DNA Repair (Amst) 4: 41–50, 2005 [DOI] [PubMed] [Google Scholar]
- 16. Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem 54: 1015–1069, 1985 [DOI] [PubMed] [Google Scholar]
- 17. Hazra TK, Izumi T, Boldogh I, Imhoff B, Kow YW, Jaruga P, Dizdaroglu M, Mitra S. Identification and characterization of a human DNA glycosylase for repair of modified bases in oxidatively damaged DNA. Proc Natl Acad Sci USA 99: 3523–3528, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Hori A, Yoshida M, Shibata T, Ling F. Reactive oxygen species regulate DNA copy number in isolated yeast mitochondria by triggering recombination-mediated replication. Nucleic Acids Res 37: 749–761, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Hotamisligil GS. Inflammation and metabolic disorders. Nature 444: 860–867, 2006 [DOI] [PubMed] [Google Scholar]
- 20. Hsieh HC, Chen YT, Li JM, Chou TY, Chang MF, Huang SC, Tseng TL, Liu CC, Chen SF. Protein profilings in mouse liver regeneration after partial hepatectomy using iTRAQ technology. J Proteome Res 8: 1004–1013, 2009 [DOI] [PubMed] [Google Scholar]
- 21. Jaruga P, Birincioglu M, Rosenquist TA, Dizdaroglu M. Mouse NEIL1 protein is specific for excision of 2,6-diamino-4-hydroxy-5-formamidopyrimidine and 4,6-diamino-5-formamidopyrimidine from oxidatively damaged DNA. Biochemistry 43: 15909–15914, 2004 [DOI] [PubMed] [Google Scholar]
- 22. Khovidhunkit W, Duchateau PN, Medzihradszky KF, Moser AH, Naya-Vigne J, Shigenaga JK, Kane JP, Grunfeld C, Feingold KR. Apolipoproteins A-IV and A-V are acute-phase proteins in mouse HDL. Atherosclerosis 176: 37–44, 2004 [DOI] [PubMed] [Google Scholar]
- 23. Krishnamurthy N, Zhao X, Burrows CJ, David SS. Superior removal of hydantoin lesions relative to other oxidized bases by the human DNA glycosylase hNEIL1. Biochemistry 47: 7137–7146, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Krishnan KJ, Reeve AK, Samuels DC, Chinnery PF, Blackwood JK, Taylor RW, Wanrooij S, Spelbrink JN, Lightowlers RN, Turnbull DM. What causes mitochondrial DNA deletions in human cells? Nat Genet 40: 275–279, 2008 [DOI] [PubMed] [Google Scholar]
- 25. Kumari MV, Hiramatsu M, Ebadi M. Free radical scavenging actions of metallothionein isoforms I and II. Free Radic Res 29: 93–101, 1998 [DOI] [PubMed] [Google Scholar]
- 26. Lee HY, Choi CS, Birkenfeld AL, Alves TC, Jornayvaz FR, Jurczak MJ, Zhang D, Woo DK, Shadel GS, Ladiges W, Rabinovitch PS, Santos JH, Petersen KF, Samuel VT, Shulman GI. Targeted expression of catalase to mitochondria prevents age-associated reductions in mitochondrial function and insulin resistance. Cell Metab 12: 668–674, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Lemaitre RN, King IB, Patterson RE, Psaty BM, Kestin M, Heckbert SR. Assessment of trans-fatty acid intake with a food frequency questionnaire and validation with adipose tissue levels of trans-fatty acids. Am J Epidemiol 148: 1085–1093, 1998 [DOI] [PubMed] [Google Scholar]
- 28. Lepage G, Roy CC. Direct transesterification of all classes of lipids in a one-step reaction. J Lipid Res 27: 114–120, 1986 [PubMed] [Google Scholar]
- 29. Mandavilli BS, Santos JH, Van Houten B. Mitochondrial DNA repair and aging. Mutat Res 509: 127–151, 2002 [DOI] [PubMed] [Google Scholar]
- 30. Mansouri A, Fromenty B, Berson A, Robin MA, Grimbert S, Beaugrand M, Erlinger S, Pessayre D. Multiple hepatic mitochondrial DNA deletions suggest premature oxidative aging in alcoholic patients. J Hepatol 27: 96–102, 1997 [DOI] [PubMed] [Google Scholar]
- 31. Mantena SK, King AL, Andringa KK, Eccleston HB, Bailey SM. Mitochondrial dysfunction and oxidative stress in the pathogenesis of alcohol- and obesity-induced fatty liver diseases. Free Radic Biol Med 44: 1259–1272, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Mokranjac D, Neupert W. Thirty years of protein translocation into mitochondria: unexpectedly complex and still puzzling. Biochim Biophys Acta 1793: 33–41, 2009 [DOI] [PubMed] [Google Scholar]
- 33. Moller DE, Kaufman KD. Metabolic syndrome: a clinical and molecular perspective. Annu Rev Med 56: 45–62, 2005 [DOI] [PubMed] [Google Scholar]
- 34. Mori H, Ouchida R, Hijikata A, Kitamura H, Ohara O, Li Y, Gao X, Yasui A, Lloyd RS, Wang JY. Deficiency of the oxidative damage-specific DNA glycosylase NEIL1 leads to reduced germinal center B cell expansion. DNA Repair (Amst) 8: 1328–1332, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Newmeyer DD, Ferguson-Miller S. Mitochondria: releasing power for life and unleashing the machineries of death. Cell 112: 481–490, 2003 [DOI] [PubMed] [Google Scholar]
- 36. Pal S, Polyak SJ, Bano N, Qiu WC, Carithers RL, Shuhart M, Gretch DR, Das A. Hepatitis C virus induces oxidative stress, DNA damage and modulates the DNA repair enzyme NEIL1. J Gastroenterol Hepatol 25: 627–634, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Peng Z, Fernandez P, Wilder T, Yee H, Chiriboga L, Chan ES, Cronstein BN. Ecto-5′-nucleotidase (CD73)-mediated extracellular adenosine production plays a critical role in hepatic fibrosis. FASEB J 22: 2263–2272, 2008 [DOI] [PubMed] [Google Scholar]
- 38. Richards VE, Chau B, White MR, McQueen CA. Hepatic gene expression and lipid homeostasis in C57BL/6 mice exposed to hydrazine or acetylhydrazine. Toxicol Sci 82: 318–332, 2004 [DOI] [PubMed] [Google Scholar]
- 39. Roy LM, Jaruga P, Wood TG, McCullough AK, Dizdaroglu M, Lloyd RS. Human polymorphic variants of the NEIL1 DNA glycosylase. J Biol Chem 282: 15790–15798, 2007 [DOI] [PubMed] [Google Scholar]
- 40. Scheja L, Heese B, Zitzer H, Michael MD, Siesky AM, Pospisil H, Beisiegel U, Seedorf K. Acute-phase serum amyloid A as a marker of insulin resistance in mice. Exp Diabetes Res 2008: 230837, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Shoelson SE, Lee J, Goldfine AB. Inflammation and insulin resistance. J Clin Invest 116: 1793–1801, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Starkov AA. The role of mitochondria in reactive oxygen species metabolism and signaling. Ann NY Acad Sci 1147: 37–52, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Swisher JF, Khatri U, Feldman GM. Annexin A2 is a soluble mediator of macrophage activation. J Leukoc Biol 82: 1174–1184, 2007 [DOI] [PubMed] [Google Scholar]
- 44. Taicher GZ, Tinsley FC, Reiderman A, Heiman ML. Quantitative magnetic resonance (QMR) method for bone and whole-body-composition analysis. Anal Bioanal Chem 377: 990–1002, 2003 [DOI] [PubMed] [Google Scholar]
- 45. Taylor RW, Turnbull DM. Mitochondrial DNA mutations in human disease. Nat Rev Genet 6: 389–402, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Theocharis SE, Margeli AP, Skaltsas SD, Spiliopoulou CA, Koutselinis AS. Induction of metallothionein in the liver of carbon tetrachloride intoxicated rats: an immunohistochemical study. Toxicology 161: 129–138, 2001 [DOI] [PubMed] [Google Scholar]
- 47. Trifunovic A, Wredenberg A, Falkenberg M, Spelbrink JN, Rovio AT, Bruder CE, Bohlooly YM, Gidlof S, Oldfors A, Wibom R, Tornell J, Jacobs HT, Larsson NG. Premature ageing in mice expressing defective mitochondrial DNA polymerase. Nature 429: 417–423, 2004 [DOI] [PubMed] [Google Scholar]
- 48. Tuppen HA, Blakely EL, Turnbull DM, Taylor RW. Mitochondrial DNA mutations and human disease. Biochim Biophys Acta 1797: 113–128, 2010 [DOI] [PubMed] [Google Scholar]
- 49. Vartanian V, Lowell B, Minko IG, Wood TG, Ceci JD, George S, Ballinger SW, Corless CL, McCullough AK, Lloyd RS. The metabolic syndrome resulting from a knockout of the NEIL1 DNA glycosylase. Proc Natl Acad Sci USA 103: 1864–1869, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Vijay-Kumar M, Aitken JD, Carvalho FA, Cullender TC, Mwangi S, Srinivasan S, Sitaraman SV, Knight R, Ley RE, Gewirtz AT. Metabolic syndrome and altered gut microbiota in mice lacking Toll-like receptor 5. Science 328: 228–231, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Yan QW, Yang Q, Mody N, Graham TE, Hsu CH, Xu Z, Houstis NE, Kahn BB, Rosen ED. The adipokine lipocalin 2 is regulated by obesity and promotes insulin resistance. Diabetes 56: 2533–2540, 2007 [DOI] [PubMed] [Google Scholar]
- 52. Yang RZ, Lee MJ, Hu H, Pollin TI, Ryan AS, Nicklas BJ, Snitker S, Horenstein RB, Hull K, Goldberg NH, Goldberg AP, Shuldiner AR, Fried SK, Gong DW. Acute-phase serum amyloid A: an inflammatory adipokine and potential link between obesity and its metabolic complications. PLoS Med 3: e287, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.