Abstract
Despite their name, synonymous mutations have significant consequences for cellular processes in all taxa. As a result, an understanding of codon bias is central to fields as diverse as molecular evolution and biotechnology. Although recent advances in sequencing and synthetic biology have helped resolve longstanding questions about codon bias, they have also uncovered striking patterns that suggest new hypotheses about protein synthesis. Ongoing work to quantify the dynamics of initiation and elongation is as important for understanding natural synonymous variation as it is for designing transgenes in applied contexts.
When the inherent redundancy of the genetic code was discovered, scientists were rightly puzzled by the role of synonymous mutations1. The central dogma of molecular biology suggests that synonymous mutations – those that do not alter the encoded amino acid – will have no effect on the resulting protein sequence and, therefore, no effect on cellular function, organismal fitness, or evolution. Nonetheless, in most sequenced genomes, synonymous codons are not used in equal frequencies. This phenomenon, termed codon usage bias (Figure 1), is now recognized as critical in shaping gene expression and cellular function, through its effects on diverse processes ranging from RNA processing to protein translation and protein folding. Naturally occurring codon biases are pervasive, and they can be extremely strong. Some species, such as Thermus thermophilus, avoid certain codons almost entirely. Synonymous mutations are important in applied settings as well – the use of particular codons can increase the expression of a transgene by over 1,000-fold2.
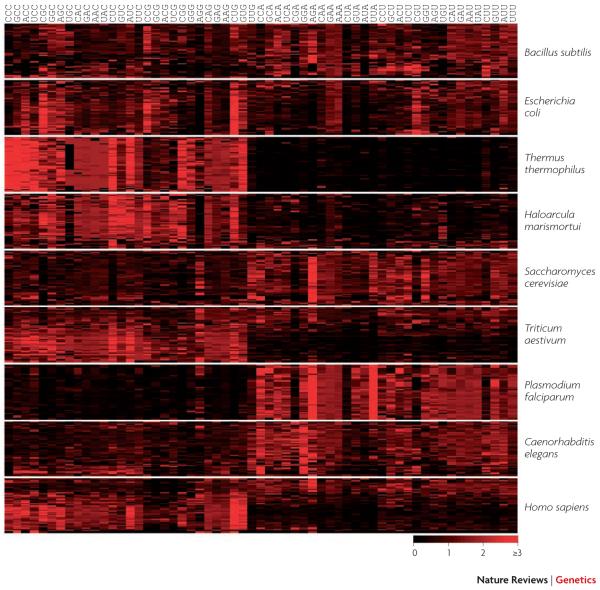
Figure 1. Codon bias within and between genomes.
The Relative Synonymous Codon Usage (RSCU) 127 is plotted for fifty randomly selected genes from each of nine species. RSCU ranges from 0 (when the codon is absent), through 1 (when there is no bias), to 6 (when a single codon is used in a six-codon family). Methionine, tryptophan and stop codons are omitted. Genes are in rows and codons are in columns, with C- and G-ending codons on the left side of each panel. Note the extensive heterogeneity of codon usage among human genes. Other measures of a gene’s codon bias include CAI35 (similarity of codon usage to a reference set of highly expressed genes); FOP28 (the frequency of “optimal” codons), and tAI128 (similarity of codon usage to the relative copy numbers of tRNA genes).
We already enjoy a broad array of often conflicting hypotheses for the mechanisms that induce codon usage biases in nature, and for their effects on protein synthesis and cellular fitness. Until recently we have been unable to systematically interrogate these hypotheses through large-scale experimentation. As a result, despite decades of interest and substantial progress in understanding codon usage biases, there is an over-abundance of plausible explanatory models whose relative, quantitative contributions are seldom compared.
Advances in synthetic biology, mass spectrometry, and sequencing now provide tools for systematically elucidating the molecular and cellular consequences of synonymous nucleotide variation. Such studies have refined our understanding of the relative roles of initiation, elongation, degradation, and mis-folding in determining expression levels of individual genes and the overall fitness of a cell. This information, in turn, is helping researchers to distinguish among the forces that shape naturally occurring patterns of codon usage. Researchers can also leverage high-throughput studies in applied settings that require controlled, heterologous gene expression -- to improve design principles for vaccine development and gene therapy, for example.
Here we review the causes, consequences, and practical utility of codon usage biases. Because we already benefit from several outstanding reviews on naturally occurring codon biases3-7, we focus here on those classical hypotheses that remain unresolved, as well as recent developments arising from high-throughput studies. We begin by summarizing the empirical patterns of codon usage observed across species, across genomes, and across individual genes. We describe the diverse array of mechanistic hypotheses for the causes of such variation, and the sequence signatures that support them. Against this backdrop of hypotheses and sequence analysis, we describe experimental work relating codon usage to endogenous gene expression and cellular fitness. From this, we turn to experimental studies on heterologous gene expression, and their implications both for understanding natural synonymous variation and for engineering novel constructs in applied settings.
Mechanistic hypotheses
Significant deviations from uniform codon choice have been observed in species from all taxa, including bacteria, archaea, yeast, flies, worms, and mammals. The overall codon usage in a genome can differ dramatically between species, although seldom between closely related species6.
Mutational versus selective hypotheses
Explanations for patterns of codon usage, within or between species, fall into two distinct categories associated with two independent forces in molecular evolution: mutation and natural selection3-5.
A mutational explanation posits that codon bias arises from the properties of underlying mutational processes – e.g. biases in nucleotides produced by point mutations8, contextual biases in the point mutation rates, or biases in repair. Mutational explanations are neutral, because they posit no fitness advantage or detriment associated with alternative synonymous codons. Mutational mechanisms are typically invoked to explain inter-specific variation in codon usage, especially among unicellular organisms.
Explanations involving natural selection posit that synonymous mutations somehow influence the fitness of an organism, and they can thus be promoted or repressed throughout evolution. Selective mechanisms are typically invoked to explain variation in codon usage across a genome or across a gene, although some inter-specific variation is also attributable to such mechanisms (see below).
Selective and neutral explanations for codon usage are not mutually exclusive, and both types of mechanisms surely play a role in patterning synonymous variation within and between genomes3, 5, 9. Below we discuss the patterns of codon usage that have been documented, at various levels of biological organization, in light of their mutational or selective causes.
Patterns of codon usage
Patterns across species
The strongest single determinant of codon usage variation across species is genomic GC content. In fact, differences in codon usage between bacterial species can be accurately predicted from the nucleotide content in their non-coding regions3, 10. Genomic GC content is itself typically determined by mutational processes acting genome-wide. As a result, most inter-specific variation in codon usage is attributed to mutational mechanisms3, 10, although the molecular causes of mutation biases are largely unknown10. Contrary to early expectations, the GC content of bacterial genomes or protein-coding genes is not correlated with optimal growth temperature (although, interestingly, structural RNAs show such a correlation)11.
In those species for which the point mutation rate depends strongly on the sequence context of a nucleotide – e.g. in mammals, which experience hypermutable CpG dinucleotides – the mutational model predicts a strong context-dependence of codon usage, which has indeed been observed12. Thus, at the genomic scale, neutral processes that do not discriminate among synonymous mutations remain plausible for explaining interspecific variation in codon usage among higher eukaryotes, and they are well accepted as the primary determinants of inter-specific variation in most other taxa (but see 13).
Aside from mutation biases, adaptation of codon usage to cellular tRNA abundances can also influence synonymous sequence variation across species (see below), since codon usage and tRNA regulation can co-evolve. Finally, some neutral processes responsible for codon bias across taxa are not mutational per se. Even in the absence of selection at synonymous sites, selection at non-synonymous sites can induce differences in nucleotide composition between coding and non-coding regions5, 14-16.
Patterns across a genome
There is often systematic variation in codon usage among the genes in a genome, usually attributed to selection. In organisms including Escherichia coli, Saccharomyes cerevisiae, Caenorhabditis elegans, Arabidopsis thaliana, Drosophila melanogaster and possibly mammals as well (see below), there is a significant positive correlation between a gene’s expression level and the degree of its codon bias, as well a negative correlation between expression level and the rate of synonymous substitutions between divergent species9, 17, 18 – features that are difficult to explain through mutation alone. Although mutational effects could possibly covary with expression levels, because transcription can be mutagenic19, 20, this effect is unlikely to account for the correlations between codon usage and expression levels observed in multiple species5, 19, 21.
The classic explanation for systematic variation across a genome is selectionist: codon bias is more extreme in highly expressed genes in order to match a skew in iso-accepting tRNAs and thereby provide a fitness advantage through increased translation efficiency or accuracy of protein synthesis9, 17, 22-27. There is strong evidence for this hypothesis in several species, mostly in the form of broad correspondences between the “preferred codons” used in highly expressed genes and measures of relative tRNA abundances28-32. As a result, translational selection remains the dominant explanation for systematic variation in codon usage among genes, despite the fact that supporting evidence is sometimes incomplete: direct measurements of tRNA abundances are rare in higher eukaryotes; the correspondence of tRNA abundance with tRNA copy number5 is weak in D. melanogaster and humans33; and 30% of bacterial species show no evidence of translational selection34
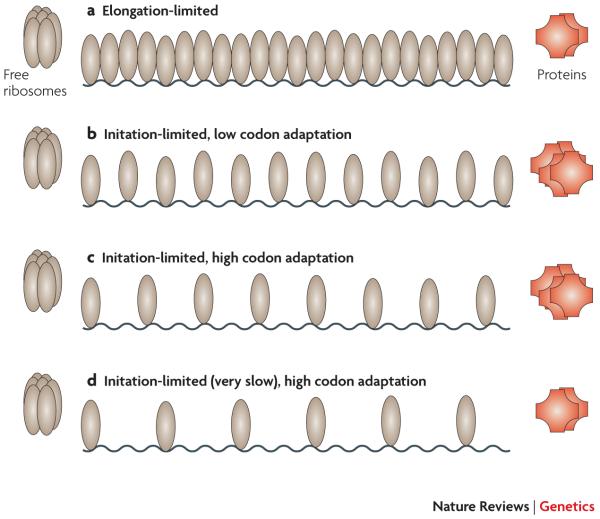
There are two possible directions of causality relating an endogenous gene’s expression level and the degree of its codon adaptation35 to tRNA abundances. Under one view2, 36, 37 high codon adaptation induces strong protein expression, because rapid and/or accurate elongation increases a given protein’s rate of synthesis; under the other view, strong expression selects for high codon adaptation, in order to avoid costs that scale with a gene’s expression level. In the biotechnology literature, the former interpretation is de rigueur whereas the latter interpretation prevails in the literature on molecular evolution3-5. The idea that high codon adaptation induces high protein levels per mRNA molecule does not square well with the notion that initiation is generally rate-limiting for endogenous protein production7, 9, 38, 39 (although it may apply to heterologous genes (see below)). When initiation is limiting, the elongation rate should not influence the amount of protein produced from a given message7, 9 (Figure 2). Moreover, from an evolutionary perspective, if high protein levels are desirable it would seem easier to tune a promoter for increased transcription than to select on hundreds of individual synonymous mutations each of which has only a marginal effect on the overall amount of protein synthesis. Conversely, the use of poorly adapted codons in order to slow the translation of genes expressed at low levels36 would seem wasteful compared to simply reducing transcription or slowing initiation.
Figure 2. Relationships between initiation rate, elongation rate, ribosome density, and rate of protein synthesis for endogenous genes.
The steady-state rate of protein synthesis and density of ribosomes bound on an mRNA both depend on the rates of initiation and elongation. When elongation is the rate-limiting step in a gene’s translation (case A), the message will be covered as densely as possible by ribosomes, and faster elongation will tend to increase the rate of protein synthesis. However, most endogenous genes are believed to be initiation-limited (cases B, C, D), so that their transcripts are not completely covered by ribosomes; this is evidenced by extensive variability in ribosome densities across endogenous mRNAs67. For two initiation-limited genes with the same initiation rate, the mRNA with faster elongation (afforded by higher codon adaptation to tRNA pools, say) will exhibit a lower density of translating ribosomes (C versus B) but no greater rate of termination. Thus, when initiation is limiting, high codon adaptation should not be expected to increase the amount of protein produced per mRNA molecule. A lower density of ribosomes can also occur when two initiation-limited genes have the same elongation rate, but one has a slower initiation rate (D versus C). The extent to which variation in ribosome densities67 arises from variation in initiation versus elongation rates remains to be determined. In all cases shown here, like most endogenous genes, the gene’s mRNA does not account for a substantial proportion of total cellular mRNA, so that the rates of initiation and elongation do not substantially alter the pool of free ribosomes (cf Figure 4).
Although evolutionary studies generally agree that high expression selects for high codon adaptation in endogenous genes (as opposed to the converse), the precise nature of fitness gains associated with translationally adapted codons remains a topic of active debate (see Box 1, Selection for Accuracy or Efficiency). Furthermore, even though translational efficiency is energetically beneficial to the cell, efficient translation generally increases the amount of cell-to-cell variation in expression levels40, and this noise is typically deleterious41. Even though translational selection has received the most attention, systematic variation in codon usage across a genome can also be caused by neutral processes in certain species; these processes include horizontal gene transfer42, different nucleotide bias in leading and lagging strands of replication in bacteria43, and isochore structure in mammals (see Mammals Are Different, Box 2).
Box 1 Selection for accuracy or efficiency?
The nature of translational selection remains a topic of active debate. Codons adapted to tRNA pools might be preferentially used in highly expressed genes because such genes experience greater pressure for translational efficiency28, 29, 110, accuracy22-27, or both. Efficient elongation of a transcript might increase its protein yield2, 36, 37, 92, or it may provide a global benefit to the cell by increasing the number of ribosomes available to translate other messages even if it does not increase the yield of the transcript itself3, 7, 9, 55. Accurate elongation, by contrast, benefits the cell by reducing the costs of useless mistranslation products or the toxicity of harmful mistranslation products111. These two models can make different predictions for the fitness costs of maladaptive codons, as a function of transcript level.
There are several lines of sequence-based evidence that discriminate between the efficiency and accuracy hypotheses. Some of the most compelling evidence in favor of accuracy was introduced by Akashi23, 112, who found a greater tendency towards tRNA-adapted codons at residues that are strongly conserved across divergent Drosophila species – suggesting that sites under strong negative selection at the amino acid level also exhibit stronger codon adaptation, presumably to reduce mis-translation. The same finding was later extended to C. elegans113 and unicellular organisms25, 27. A separate line of evidence arises from the correlation between codon adaptation and gene length in E. coli24, reflecting a greater energetic cost of mis-sense and nonsense translation errors in a long protein, especially if they occur near the 3′ end. However, the relationship with gene length does not hold in C. elegans, D.melanogaster or A. thaliana21. Other evidence for the accuracy hypothesis comes from simulations of sequence evolution, protein translation, and protein folding26.
There is also convincing evidence in favor of translational efficiency, especially in prokaryotes. The most compelling observation is a broad correlation between the minimum generation time of a bacterial species and the strength of selection it experiences for codon adaptation in highly expressed genes34, 114. We would expect to see this correlation if preferred codons increase the elongation rate, which is beneficial for rapid growth, but it unclear why we would observe this correlation if preferred codons increase only the accuracy of elongation. Furthermore, Zhang and others have recently shown that codon usage in highly expressed yeast genes is consistent with selection to avoid unnecessary ribosomal sequestration of messages (Zhang et al., in preparation).
The accuracy and efficiency hypotheses are not mutually exclusive, in general. However, in a recent computational study, Shah and Gilchrist115 demonstrated that codons corresponding to more abundant tRNAs are not always expected to produce lower mis-sense error rates, as has been commonly assumed. Moreover, they found that for some amino acids, pressure for elongation speed would result in a different codon choice than pressure for elongation accuracy. Whether patterns of codon bias in evolutionary conserved residues22 occurs only for those amino acids for which efficiency and accuracy selection have the same predicted effect on codon choice remains unresolved and might help to distinguish between these two modes of selection.
Box 2 Mammals are different.
Intra-genomic patterns of codon usage are markedly different in mammals than in other taxa. Selective mechanisms were initially ruled out for humans, on the basis of their small effective population size, which limits the efficacy of selection4. Moreover, the most obvious pattern of gene-to-gene codon usage variation in mammals arises not from selection but from large-scale variation in the GC content -- i.e. the isochores116. Isochores themselves are likely caused by processes primarily related to recombination and repair, such as biased gene conversion117.
Over the past decade, however, researchers have identified several sources of potentially strong selection on synonymous mutations in mammals – a trend that was highlighted by Hurst and others4. Some of these observations fit within the classical model of translational selection – e.g. the presence of a weak but positive relationship between gene expression and codon bias118, 119, especially after accounting for the local GC content120. But studies comparing expression levels to codon adaptation (that is, to tRNA abundances) have been contradictory33, 36, 119. Researchers have also observed significant differences in codon usage between genes specifically expressed in several different tissues121, as well as variation in relative tRNA abundances by tissue type122. But there is little evidence for systematic variation associated with tissue type123; and the quantification of mammalian tRNAs, which contain numerous nucleotide modifications, is still relatively noisy122.
Instead, researchers have identified other mechanistic explanations for codon usage variation in mammals, aside from translational selection. One possibility is selection for the overall stability of mRNA transcripts124, 125, via a skew towards C at fourfold degenerate sites. In mice, computational analyses suggest that such skews have been selected to promote mRNA stability124. Moreover, several diseases arise from mutations that disrupt mRNA structure4, 126, providing a clear target of selection. Another possibility, related to splicing control, is described in the main text.
Patterns across a gene
Codon usage can vary dramatically even within a single gene. Synonymous mutations at specific sites may experience selection because they disrupt motifs recognized by transcription factors or by post-transcriptional regulatory mechanisms – eg, micro-RNAs. Sites that require ribosomal pausing for proper co-translational protein folding or ubiquitin modification44 may experience selection for poorly adapted codons45 or strong mRNA folding 46. Codon choice that promotes proper nucleosome positioning is selectively advantageous in eukaryotes, especially in 5′ regions47. And finally, in mammals, synonymous mutations near an intron–exon boundary can create spurious splice sites or disrupt splicing-control elements4, 48, causing disease4. This phenomenon helps to explain the reduced rate of synonymous substitutions and SNP density near splicing control elements49, 50. Selection for proper splicing also extends to D. melanogaster, and sequence variation suggests it is probably an even stronger force than translational selection in shaping codon usage near intron–exon boundaries51
Although important, the mechanisms of intragenic codon usage variation described above are typically restricted to specific taxa or special classes of sites. Recent studies have argued for three mechanisms that produce systematic variation in codon usage across the sites in a gene, in a diverse range of species.
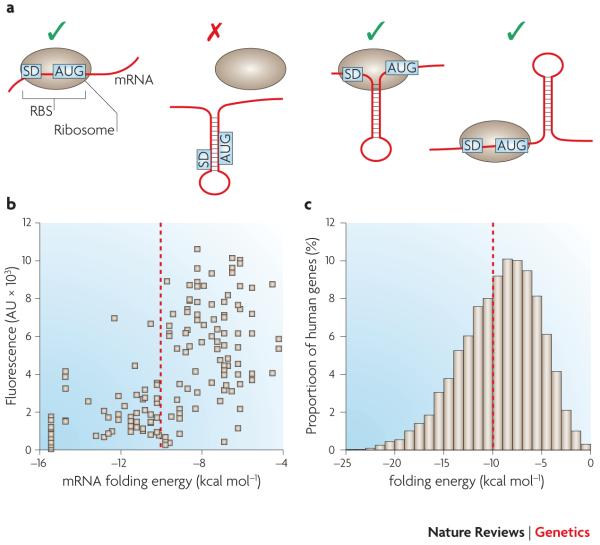
One of these mechanisms is selection against strong 5′ mRNA structure, in order to facilitate translation initiation. mRNA structure near the 5′ end of a coding region is generally disadvantageous9 as it can inhibit ribosomal initiation52, 53 (Figure 3A). Eyre-Walker and Bulmer proposed selection against mRNA structure to explain a trend towards reduced codon adaptation in the 5′ region of E. coli genes, and a corresponding reduced rate of synonymous substitutions across divergent species54. More recently, following similar observations in E. coli55, Gu et al. demonstrated a broad trend in all sequenced prokaryotes and eukaryotes towards reduced mRNA stability near the translationin-itiation sites of genes, especially for GC-rich genes56. This study relied on computational predictions of mRNA structure in short windows; combined with large-scale experimental studies (see below), this work suggests a systematic role for selection on mRNA structure in shaping codon usage in the first 30-60 nucleotides of genes.
Figure 3. Effects of mRNA secondary structure on translation initiation in Bacteria.
A) Structure in the ribosome binding site (RBS) usually inhibits initiation. However, initiation can occur when the structured element is positioned between the Shine-Dalgarno sequence (SD) and start codon (AUG)129, or 15 nucleotides downstream of the start codon130, 131. B) Synonymous mutations in the region from nt −4 to +37 of a GFP gene alter predicting folding energies by up to 12 kcal/mol. 5′ mRNA folding energies below −10 kcal/mol strongly inhibits GFP expression in E. coli55. C) More than 40% of human genes have predicted 5′ folding energies below the −10 kcal/mol threshold, and are therefore expected to express poorly in E. coli without modification.
Tuller et al.57 recently described a second, systematic trend in the pattern of intragenic codon usage: a ‘ramp’ of poorly adapted codons in the first 90-150 nucleotides of genes, which had earlier been observed in bacteria, yeast, and fly58, 59. This pattern has been preserved across divergent species even when tRNA pools (estimated from gene copy numbers) have changed57. A ramp of poorly adapted codons presumably slows elongation at the start of a gene, which may provide several physiological benefits. Slow 5′ elongation is predicted to reduce the frequency of ribosomal traffic jams towards the 3′ end57, 60, thus reducing the cost of wasted ribosomes and of spontaneous or collision-induced abortions. Alternatively, a ramp of slow elongation may facilitate recruitment of chaperone proteins to the emergent peptide61. Other explanations, unrelated to elongation rate, are also plausible -- such as weaker selection for accurate translation near the start of a gene, where mis-sense and non-sense errors would be less costly24, 59. The earliest interpretation of unusual 5′ codon usage posited selection to increase the initiation rate9 – and, interestingly, the 5′ region of poorly-adapted codons identified by Tuller et al. overlaps significantly with the region in which synonymous codon choice systematically reduces mRNA stability54-56, 58. It remains unclear which selective mechanisms are primarily responsible for the unusual and nearly universal pattern of 5′ codon usage. Multiple mechanisms may certainly operate in different genes; however, it is unclear why a single gene should experience selection both to increase its rate of ribosomal initiation9 and to reduce the subsequent rate of its early elongation57.
Cannarozzi et al.62 recently exposed a third, novel pattern of intra-genic codon usage in eukaryotes: the re-use or autocorrelation of codons across a gene sequence, driven, they argue, to improve elongation efficiency through tRNA recycling. If a recently used tRNA molecule is bound to the ribosome, or if it diffuses slowly compared to ribosomal progression and re-acytilation63, then it would be efficient to re-use the same tRNA molecule for subsequent incorporations of the same amino acid. This physical model predicts selection for using the same codon, or, more generally, a codon that is read by the same tRNA species, at nearby sites in a gene that encode the same amino acid. Indeed, Cannarozzi et al. observed significant autocorrelation of codons across gene sequences in most eukaryotes, especially in genes that are rapidly up-regulated in response to stress. Of course, autocorrelation would also be predicted if all sites in a gene independently experience pressure for biased codon usage – e.g. in order to match the global pool of tRNAs. To control for overall codon usage, Cannarozzi et al compared the degree of autocorrelation in actual gene sequences to gene sequences that had been re-shuffled at random, finding more autocorrelation on average in the un-shuffled genes, although only marginally so. More convincing, they observed that autocorrelation is strongest for isoaccepting codons of rare tRNAs in highly expressed genes – which is predicted by the tRNA recycling hypothesis but not by a selective pressure that applies at all sites independently.
Measurements of endogenous expression
Recent developments in mass spectrometry and fluorescence microscopy allow for large-scale measurements of endogenous protein levels64-66. Together with techniques for quantifying ribosomal occupancy67 and measuring elongation dynamics 68, these advances provide a spectacularly detailed accounting of basic cellular processes, with implications for our understanding of codon biases.
Variation in protein/mRNA ratios
Shotgun proteomics have revealed an extensive role for post-transcriptional processes in determining eventual protein levels in bacteria, yeast69, worm70, fly70, and especially mammals65, 66. Whereas the imperfect correlations between protein and mRNA levels (R2 ≈ 47%-77% in E. coli65, 71, 73% in yeast65, and 29% in humans66) may once have been seen as measurement noise, researchers have since attributed much of the variation in protein/mRNA ratios to sequence-derived characteristics of genes. In a recent study in human cells66, the strongest correlates of steady-state protein levels, controlling for mRNA levels, were coding-sequence length (reflecting that longer transcripts are less stable72 or slower to initiate73), amino acid content (reflecting variable costs associated with synthesizing different amino acids, or variable rates of protein degradation), and predicted 5′ mRNA structure (reflecting lower initiation rates when 5′ structure is strong). Importantly, the codon adaptation index35, which correlates strongly with mRNA levels in yeast65 and weakly in human66, shows little or no significant correlation with protein levels per mRNA molecule in either organism65, 66 – suggesting that codon adaptation does not significantly increase the protein yield from a given message, at least among endogenous genes74, 75. It is important to note that steady-state protein levels are influenced by both protein production and protein degradation; and so any variation in degradation rates unrelated to codon usage will further reduce the correlation between codon usage and protein/mRNA ratios.
Ribosomal footprints
Ingolia et al. recently devised a clever application of RNAseq to quantify ribosome-protected RNA fragments in a cell, thereby estimating ‘ribosomal footprints’ across the transcriptome67. This method has provided rich information about translational regulation, and it has uncovered some startling phenomena – such as an abundance of upstream open reading frames with non-AUG start codons. The footprint data in yeast show a greater mean density of ribosomes in the first 100-150 codons of genes, suggesting locally slow elongation; this is consistent with the observation of poorly adapted codons in the 5′ region58, 59. There is also significant negative correlation, genome-wide, between a transcript’s ribosome density and the experimentally measured strength of mRNA structure near its start site 76 – suggesting that strong 5′ mRNA structure retards initiation and reduces the density of translating ribosomes. Remarkably, upon averaging data from all yeast genes, Tuller et al.37 also observed a negative correlation between predicted mRNA folding energy and ribosome density among the first 65 codons – suggesting that strong mRNA structure downstream of start retards elongation. This observation is somewhat surprising, given the helicase activity of translating ribosomes77; however, the correlation between the genome-wide average profiles of mRNA folding and ribosome density does not imply a correlation at the level of individual sites. Ingolia et al. also measured ribosomal footprints under amino-acid starvation, finding one-third of yeast genes with either substantially increased or decreased translational efficiency67. A detailed parsing of the relationship between a gene’s amino-acid content and translational response to starvation may improve design principles for over-expressed heterologous genes, which often induce starvation78, 79 (see below).
Translational efficiency
Notions of translational efficiency differ in the literature on gene expression. Ingolia et al.67 defined the translational efficiency of a gene as the number of bound ribosomes per mRNA molecule; whereas Tuller et al.37, 57 and others defined efficiency as protein yield per mRNA molecule (i.e. the ratio of protein abundance to mRNA abundance). The latter definition is more relevant to issues of total protein synthesis, whereas the former definition may be more relevant to ribosomal availability and overall cellular fitness. These two notions of translational efficiency are only weakly correlated for endogenous genes (R2 < 2.5 % comparing the data by Ingolia et al.67 to ref. 80 – indicating that the density of ribosomes on a given mRNA molecule does not determine the amount of protein produced from it. Similarly, in yeast, a gene’s codon adaptation index35 explains less than 3% of the variance in protein abundances per mRNA67. Both of these observations are consistent with the idea that, for most endogenous genes, initiation is rate-limiting for protein production38, 39 and thus determines the amount of protein produced from each message, regardless of ribosome density or codon adaptation7, 9 (Figure 2); however, this logic may not apply to over-expressed heterologous genes, described below.
Measurements of heterologous expression
Codon bias plays a critical role in heterologous gene expression. However, there is often a disconnect between technological and evolutionary studies of codon bias – a gap that partly reflects genuine differences between endogenous and heterologous situations. In many biotechnological applications, a transgene is massively over-expressed, accounting for up to 30% of the protein mass in cell. As a result, the principles relating heterologous codon usage to protein levels may differ substantially from the endogenous case.
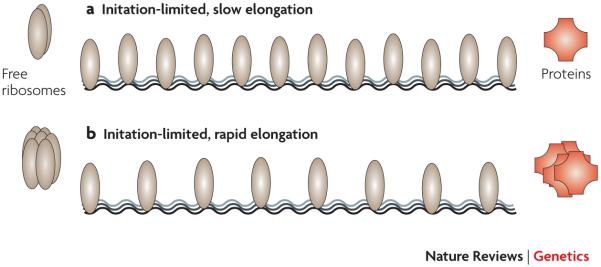
The notion that initiation generally limits translation may not apply to an overexpressed transgene whose mRNA accounts for a very large proportion of total cellular mRNA. In such a case, inefficient use of ribosomes along the over-expressed mRNA may be sufficient to feed back and significantly deplete available ribosomes, thereby reducing initiation rates and retarding further heterologous protein production9 (Figure 4). Thus, we might expect that elongation effects of codon usage will influence protein yields per mRNA molecule for over-expressed genes. Nonetheless, we should not necessarily expect that the codons adapted to efficient elongation for endogenous genes will correspond to the efficient codons for heterologous genes, because over-expression causes amino-acid starvation and concomitant alternations in the abundances of charged tRNAs78, 79, 81. Indeed, there was no significant correlation between codon adaptation35 and expression levels in two large-scale systematic experiments55, 79. In fact, even endogenous genes that are essential during amino-acid starvation, such as amino acid biosynthetic enzymes, preferentially use codons that are poorly adapted to the typical pool of charged tRNAs, but well adapted to starvation-induced tRNA pools78, 79.
Figure 4. The elongation rate may influence the rate of protein synthesis for an over-expressed gene.
Unlike for most endogenous genes, mRNA from an over-expressed transgene may account for a substantial proportion of total cellular mRNA. In this case, slow elongation (caused by poor codon adaptation to charged tRNA pools, say) can increase the density of bound ribosomes and thereby reduce the pool of available ribosomes in the cell. Such a depletion of available ribosomes will feed back to reduce the initiation rate of subsequent translating ribosomes on the message, thereby reducing the rate of protein synthesis. This is illustrated schematically by comparing over-expressed mRNA’s with slow elongation (top) and rapid elongation (bottom), but identical initiation sequences. Thus, the relationship between codon adaptation and the rate of protein synthesis per mRNA molecule may differ for an over-expressed transgene as compared to an endogenous gene (cf Figure 2).
Despite the complications described above, the field of codon optimization has traditionally focused on adjusting codon usage to match cellular tRNA abundances in standard conditions, disregarding other dimensions of bias. However, strategies are now changing. Several recent studies advocate for the role of global nucleotide content82, 83, local mRNA folding55, 84, codon pair bias85, a codon ramp57, or codon correlations62 in optimizing heterologous expression (see Table 1).
Table 1.
Coding-sequence covariates of gene expression, and other sources of codon bias unrelated to gene expression
| Parameter | Species | Relationship with expression |
Type of evidence (0) |
Proposed mechanism | Refs (computation) (1) |
Refs (function) |
|---|---|---|---|---|---|---|
| Codon adaptation | ||||||
| CAI or FOP or tAI | All | Complex (see text) | c | Translation elongation rate/accuracy | CAI35, fop28, tAI128 | 2, 37, 55, 81, 94, 96 |
| Rare codon stretches | Bacteria | − | c | Translation elongation rate/accuracy | 90 | |
| Rare codons between protein domains |
Bacteria | Complex | c | Translation elongation/Protein folding | 91, 132 | |
| Frequency of starvation-resistant codons |
Bacteria | + | c | Translation elongation rate/accuracy | 79, 81 | 81 |
| Frequency of abundant codons | All | Complex | c | Unclear (2) | 133 | 134 |
| mRNA folding | ||||||
| Weak mRNA folding at start codon | Bacteria | + | d | Translation initiation rate | mfold135 | 53, 55, 97 |
| mRNA stem-loop 15 nt downstream of start codon |
Bacteria, mammals | + | c | Translation initiation rate | 130, 131 | 130, 131 |
| mRNA stem-loops further downstream | All | Complex | c | Translation elongation | 135 | 46, 84 |
| Regulatory motifs | ||||||
| Transcription terminators | Bacteria | − | b | Transcription | RNAMotif136 | |
| RNAse E sites | Bacteria | − | b | mRNA stability | 137 | |
| miRNA target sites | Eukaryotes | − | b | Translation, mRNA stability | TargetScan138 | |
| Various mRNA regulatory elements | All | Complex | b | Translation, mRNA stability | TransTerm139 | |
| Nucleotide bias | ||||||
| High GC3 content, low A content | Mammals | + | c | Transcription, mRNA processing, mRNA export |
140, 141 | 82, 83, 101 |
| High CpG content | Mammals | + | c | Transcription | 102 | |
| Others sources of codon bias | ||||||
| Codon pair bias | Bacteria, mammals | + | c | Translation elongation rate | 85, 104 | 85, 104 |
| Codon ramp | All | Complex (3) | b | Translation initiation rate | 57 | |
| Codon correlation | Eukaryotes | + | c | Translation elongation rate | 62 | 62 |
| Unknown | All | Complex | c | Protein folding efficiency | 107 | |
| Unknown | Eukaryotes | Complex | b | Splicing regulation | 4, 142 | |
| Unknown | All | Complex | c | Protein posttranslational modification | 44 | 44 |
| Replication strand nucleotide bias | Bacteria, Mitochondria |
unknown | a | Unknown | ||
| CTAG avoidance | Bacteria | unknown | a | Restriction avoidance | 143 | 143 |
NOTE: This table lists coding-sequence derived parameters that can be changed by synonymous mutations. Other parameters may be important for expression (eg the identity of the N-terminal amino acid or the length of the sequence) but they require nonsynonymous changes.
Type of evidence supporting the influence of the parameter on expression. a: none; b: theoretical; c: anecdotal; d: systematic.
Generic tools to calculate some of these parameters: codonW140 (http://codonw.sourceforge.net/), INCA141, GeneDesigner144.
The frequency of abundant codons is highly correlated with GC content in mammals.
The codon ramp is predicted to decrease the cost of translation, which may indirectly influence expression levels.
Effects of codon adaptation on expression levels
Many studies show strong effects of rare codons on heterologous expression. In E. coli, stretches of rare AGA or AGG codons cause ribosome pausing and co-translational cleavage of mRNA86, ribosomal frameshifting87, or amino-acid mis-incorporation88. Consistent with theoretical expectations, codons read by rare tRNAs can slow elongation several-fold89. And stretches of AGG codons near the ribosome binding site can reduce protein yields by obstructing translation initiation90. Even though such studies are convincing, they usually address the effect of a subset of very rare codons, often in long stretches, in E. coli cells; it is not known how generally these principles apply.
Observations such as the ones above were quickly followed by efforts to adjust the global codon adaptation of transgenes to cellular tRNA abundances. Several approaches have been proposed: ‘CAI maximization’ replaces all codons by the most preferred codons in the target genome, but this could result in unbalanced charged tRNA pools2; ‘codon harmonization’91 puts some nonpreferred codons in positions corresponding to predicted protein domain boundaries; ‘codon sampling’ adjusts the codon usage to reflect the overall usage in the target genome. In the absence of tRNA abundance estimates, codon frequencies in the target genome are sometimes used. It has also been suggested that codon usage should match the profile of charged tRNAs rather than total tRNAs79, 81. The utility of codon adaptation approaches is still unclear, as they have not been systematically compared against each other, and a number of anecdotal studies argue both for (eg. Ref 92) and against (eg Ref 93) their efficiency.
Codon adaptation algorithms typically optimize many sequence properties at once. This makes it difficult to determine which parameter causes observed differences in expression. In two recent multi-gene studies, between 60% and 70% of genes experienced increased expression upon codon optimization94, 95 but whether this was a direct consequence of increased codon adaptation or other sequence properties is unclear. In our study of 154 synonymous variants of GFP, we observed no significant correlation between the codon adaption index35 and expression levels in E. coli55, but a weak positive correlation was later found using non-linear regressions37, 96. In any case, adaptation of codon usage is limited to species with pronounced and well-understood variation in tRNA concentrations, such as bacteria and yeast.
Effects of nucleotide bias on expression levels
Nucleotide biases are pervasive in natural genes and have the potential to alter the interactions of mRNA with DNA, with proteins, and with itself -- thereby influencing RNA production, degradation, and translation rates. Many of these effects are characterized, but this knowledge has yet to find its way into standard codon optimization procedures.
GC-rich mRNAs can form strong secondary structures, and, in bacteria, strong structure near the ribosome binding site prohibits initiation53, 55, 97 (Figure 3B). As a result, more than 40% of human genes would be expected to express poorly when placed in E. coli without modification (Figure 3C). Strong structure near the start codon reduces heterologous expression in yeast as well (G. Kudla, personal communication), consistent with evolutionary analyses56. No such effect has been described in mammals; on the contrary, high GC content generally increases expression levels in mammalian cells (see below). However, a strong mRNA hairpin away from the ribosome binding site (RBS) has been reported to interfere with translation in mammalian cells84 and strong hybrids between RNA and DNA (the R-loops) may interfere with transcription98.
GC-poor mRNAs are unlikely to fold strongly, but they often harbor other sequence elements that limit expression. For example, low GC content is commonly believed to limit the expression of Plasmodium falciparum genes in E. coli, even though the mechanisms are unknown. Such mRNAs may be targets for RNAse E, which cleaves AU-rich sequences with low sequence specificity99. The situation is slightly clearer in mammals, where low GC content (or high A content) has been shown to reduce expression82, 83. This effect is common knowledge in virology, since HIV and HPV genes are very poorly expressed in human cells unless optimized100, 101. The rate-limiting step in these cases may be transcription or nuclear RNA export82, 83, consistent with the efficient expression of GCpoor genes in cytoplasmic transcription systems based on the vaccinia virus101.
Little is known about the functional consequences of replication-strand-related bias or CTAG avoidance that are common in prokaryotes. High CpG content was reported to correlate with high expression in mammalian cells102, possibly by altering the distribution of nucleosomes on DNA.
Other effects of synonymous mutations on expression levels
Other examples of synonymous mutations influencing expression have been described as primarily anecdotal observations. In E. coli, over-represented codon pairs103 were proposed to decrease translation elongation rates104, although this conclusion was later disputed105. In an attempt to produce attenuated strains, Coleman et al.85 partially de-optimized codon pairs in the poliovirus genome and observed several-fold reduction in protein yield in mammalian cells, as well as a thousand-fold reduction in viral infectivity. A version of GFP with auto-correlated codon usage exhibited 30% lower ribosome density in yeast, suggesting faster elongation, than a version with anti-correlated codon usage62. And a synonymous mutation in the human MDR1 gene was proposed to influence mRNA stability106 or protein folding and substrate specificity107. These observations are all intriguing, and they form important avenues for future systematic studies to determine their molecular basis.
Conclusions
Recent years have begun to see a convergence of experimental work on endogenous and heterologous gene expression, as both types of studies take advantage of high-throughput, quantitative techniques. Heterologous studies based on large libraries of random or unbiased synonymous sequence variation55, 81, 97 are especially important for uncovering and comparing general rules to optimize expression. By contrast, relatively small-scale studies based on pre-conceived notions of “optimized” codon usage do not provide sufficient power to distinguish among alternative mechanisms, nor do they allow us to discover any new mechanisms that increase expression. Heterologous studies will be complemented by endogenous measurements of initiation and elongation dynamics, and their effects on protein synthesis as a function of a gene’s amino acid content and transcript level.
In the short-term, there will be a tradeoff between gaining predictive power for transgene optimization and deducing the underlying mechanisms that link codon usage and gene expression. High-dimensional, statistical regressions applied to large libraries of synonymous genes81, 96 provide a principled, effective means of increasing heterologous expression. Such techniques are increasingly valuable in applied contexts where high expression is required – such as viral-delivered gene therapies108, 109 – but they do not generally identify molecular mechanisms. Our hope, over the long-term, is that cross-fertilization between biotechnological and molecular biological studies will elucidate effective strategies for designing transgenes, as well as the mechanistic principles that underlie their expression.
Online Summary.
Codon usage varies widely between species, between genes in a genome, and between sites in a gene.
Explanations for natural variation in codon usage fall into two categories: mutational and selective.
Mutational mechanisms are responsible for most codon usage variation between species; whereas selection for translation efficiency accounts for much of the systematic variation across a genome (except in mammals).
Translationally efficient codons may increase elongation rate, accuracy, or both.
Rapid elongation should not be expected to influence protein yield per mRNA molecule for an endogenous gene, but it may be relevant for an over-expressed transgene.
The codons that provide efficient translation of an over-expressed transgene may differ from the efficient codons for an endogenous gene.
High-throughput measurements of endogenous mRNA levels, protein levels, and ribosomal occupancies provide a detailed description of translation processes.
Libraries of randomized genes can elucidate design principles for efficient transgene expression, even without uncovering underlying mechanisms.
Acknowledgements
We thank Laurence Hurst for helpful discussions. We apologize to those whose work we were unable to cite because of space constraints. G.K. acknowledges funding from the Wellcome Trust. J.B.P. acknowledges support from the Burroughs Wellcome Fund, the David and Lucile Packard Foundation, the James S. McDonnell Foundation, the Alfred P. Sloan Foundation, the Defense Advanced Research Projects Agency (HR0011-05-1-0057), and the US National Institute of Allergy and Infectious Diseases (2U54AI057168).
Glossary
| effective population size | Number of individuals in a population who produce viable offspring. |
| biased gene conversion | Recombination event in which one variant of genomic sequence is preferentially “copied/pasted” onto another one. |
| fourfold degenerate sites | Positions within the coding sequence of a gene at which all four nucleotides encode the same amino acid. |
| codon adaptation index | Measure of similarity between the codon usage of a gene and the average codon usage of highly expressed genes in a species. |
| iso-accepting tRNAs | Subset of tRNAs that carry the same amino acid. |
| horizontal gene transfer | Transfer of genetic material from one species into another. |
| isochore | Large fragment of a chromosome characterized by homogeneous GC content. |
| negative selection | A form of natural selection that suppresses alternative genetic variants in favor of the wildtype. |
| ribosomal pausing | A temporary arrest of the ribosome during translation elongation. |
| shotgun proteomics | Methods of quantifying protein levels in a complex sample, typically using mass spectrometry. |
| RNAseq | Quantitative analysis of RNA in a complex sample by high-throughput sequencing. |
| upstream open reading frames | Open reading frames located 5′ from the primary open reading frame; considered to inhibit translation of the primary ORF. |
| ribosomal footprints | Fragments of mRNA that were protected by ribosomes from nuclease digestion in a ribosomal profiling experiment. |
Footnotes
Reference highlights (eleven)
Andersson and Kurland 1990
A classic paper that has framed the field of codon usage adaptation. A must-read.
de Smit and van Duin 1990
A very thorough study of how mRNA folding affects translation initiation, supported by a convincing theoretical model.
Akashi 1994
By quantifying rates of synonymous substitutions at conserved and non-conserved positions in proteins, Akashi shows that such mutations influence translational accuracy in Drosophila.
Bulmer 1991
A foundational study of both the population genetics underlying codon bias and the biophysics of translation. Emphasizes that elongation speed will not generally influence protein yield per mRNA, for endogenous genes.
Lu et al. Nature Biotechnology 2006
A quantitative proteomics approach that promises new insights into coding-sequence determinants of protein levels. See also Vogel et al. Mol. Sys. Biol. (2010)
Ingolia et al. Science 2009
A clever method to map the positions of ribosomes on messages with unprecedented accuracy; an essential tool for the study of translation kinetics.
Elf et al. Science 2003
A detailed theoretical model of what happens to tRNA when cells go hungry.
Chamary, J.V. et al Nat Rev Genet 2006
An excellent review of the many surprisingly strong effects of synonymous mutations on splicing.
Drummond & Wilke. Nat Rev Genet 2009
The various types of errors in protein synthesis, many of them directly related to codon usage, are summarized in this very readable review.
Tuller et al. Cell 2010
Rare codons at the beginning of genes could help prevent “ribosomal traffic jams”. See also Bulmer (1988), Qin et al. (2004).
Kudla et al. 2009
Libraries of synthetic genes isolate the effects of synonymous mutations on gene expression. See also Welch et al. (2009), Voges et al. (2004).
References
- 1.Zuckerkandl E, Pauling L. Molecules as documents of evolutionary history. J Theor Biol. 1965;8:357–66. doi: 10.1016/0022-5193(65)90083-4. [DOI] [PubMed] [Google Scholar]
- 2.Gustafsson C, Govindarajan S, Minshull J. Codon bias and heterologous protein expression. Trends Biotechnol. 2004;22:346–53. doi: 10.1016/j.tibtech.2004.04.006. [DOI] [PubMed] [Google Scholar]
- 3.Hershberg R, Petrov DA. Selection on codon bias. Annu Rev Genet. 2008;42:287–99. doi: 10.1146/annurev.genet.42.110807.091442. [DOI] [PubMed] [Google Scholar]
- 4.Chamary JV, Parmley JL, Hurst LD. Hearing silence: non-neutral evolution at synonymous sites in mammals. Nat Rev Genet. 2006;7:98–108. doi: 10.1038/nrg1770. [DOI] [PubMed] [Google Scholar]
- 5.Duret L. Evolution of synonymous codon usage in metazoans. Curr Opin Genet Dev. 2002;12:640–9. doi: 10.1016/s0959-437x(02)00353-2. [DOI] [PubMed] [Google Scholar]
- 6.Sharp PM, Averof M, Lloyd AT, Matassi G, Peden JF. DNA sequence evolution: the sounds of silence. Philos Trans R Soc Lond B Biol Sci. 1995;349:241–7. doi: 10.1098/rstb.1995.0108. [DOI] [PubMed] [Google Scholar]
- 7.Andersson SG, Kurland CG. Codon preferences in free-living microorganisms. Microbiol Rev. 1990;54:198–210. doi: 10.1128/mr.54.2.198-210.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kimura M. A simple method for estimating evolutionary rates of base substitutions through comparative studies of nucleotide sequences. J Mol Evol. 1980;16:111–20. doi: 10.1007/BF01731581. [DOI] [PubMed] [Google Scholar]
- 9.Bulmer M. The selection-mutation-drift theory of synonymous codon usage. Genetics. 1991;129:897–907. doi: 10.1093/genetics/129.3.897. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chen SL, Lee W, Hottes AK, Shapiro L, McAdams HH. Codon usage between genomes is constrained by genome-wide mutational processes. Proc Natl Acad Sci U S A. 2004;101:3480–5. doi: 10.1073/pnas.0307827100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hurst LD, Merchant AR. High guanine-cytosine content is not an adaptation to high temperature: a comparative analysis amongst prokaryotes. Proc R Soc Lond B Biol Sci. 2001;268:493–7. doi: 10.1098/rspb.2000.1397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fedorov A, Saxonov S, Gilbert W. Regularities of context-dependent codon bias in eukaryotic genes. Nucleic Acids Res. 2002;30:1192–7. doi: 10.1093/nar/30.5.1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hildebrand F, Meyer A, Eyre-Walker A. Evidence of selection upon genomic GC-content in bacteria. PLoS Genet. 2010;6:e1001107. doi: 10.1371/journal.pgen.1001107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Morton BR. Selection at the amino acid level can influence synonymous codon usage: implications for the study of codon adaptation in plastid genes. Genetics. 2001;159:347–58. doi: 10.1093/genetics/159.1.347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cambray G, Mazel D. Synonymous genes explore different evolutionary landscapes. PLoS Genet. 2008;4:e1000256. doi: 10.1371/journal.pgen.1000256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Plotkin JB, Dushoff J. Codon bias and frequency-dependent selection on the hemagglutinin epitopes of influenza A virus. Proc Natl Acad Sci U S A. 2003;100:7152–7. doi: 10.1073/pnas.1132114100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sharp PM, Li WH. The rate of synonymous substitution in enterobacterial genes is inversely related to codon usage bias. Mol Biol Evol. 1987;4:222–30. doi: 10.1093/oxfordjournals.molbev.a040443. [DOI] [PubMed] [Google Scholar]
- 18.Eyre-Walker A, Bulmer M. Synonymous substitution rates in enterobacteria. Genetics. 1995;140:1407–12. doi: 10.1093/genetics/140.4.1407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Francino MP, Ochman H. Deamination as the basis of strand-asymmetric evolution in transcribed Escherichia coli sequences. Mol Biol Evol. 2001;18:1147–50. doi: 10.1093/oxfordjournals.molbev.a003888. [DOI] [PubMed] [Google Scholar]
- 20.Majewski J. Dependence of mutational asymmetry on gene-expression levels in the human genome. Am J Hum Genet. 2003;73:688–92. doi: 10.1086/378134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Duret L, Mouchiroud D. Expression pattern and, surprisingly, gene length shape codon usage in Caenorhabditis, Drosophila, and Arabidopsis. Proc Natl Acad Sci U S A. 1999;96:4482–7. doi: 10.1073/pnas.96.8.4482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Akashi H. Synonymous codon usage in Drosophila melanogaster: natural selection and translational accuracy. Genetics. 1994;136:927–35. doi: 10.1093/genetics/136.3.927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Akashi H, Schaeffer SW. Natural selection and the frequency distributions of “silent” DNA polymorphism in Drosophila. Genetics. 1997;146:295–307. doi: 10.1093/genetics/146.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Eyre-Walker A. Synonymous codon bias is related to gene length in Escherichia coli: selection for translational accuracy? Mol Biol Evol. 1996;13:864–72. doi: 10.1093/oxfordjournals.molbev.a025646. [DOI] [PubMed] [Google Scholar]
- 25.Stoletzki N, Eyre-Walker A. Synonymous codon usage in Escherichia coli: selection for translational accuracy. Mol Biol Evol. 2007;24:374–81. doi: 10.1093/molbev/msl166. [DOI] [PubMed] [Google Scholar]
- 26.Drummond DA, Wilke CO. Mistranslation-induced protein misfolding as a dominant constraint on coding-sequence evolution. Cell. 2008;134:341–52. doi: 10.1016/j.cell.2008.05.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhou T, Weems M, Wilke CO. Translationally optimal codons associate with structurally sensitive sites in proteins. Mol Biol Evol. 2009;26:1571–80. doi: 10.1093/molbev/msp070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ikemura T. Correlation between the abundance of Escherichia coli transfer RNAs and the occurrence of the respective codons in its protein genes: a proposal for a synonymous codon choice that is optimal for the E. coli translational system. J Mol Biol. 1981;151:389–409. doi: 10.1016/0022-2836(81)90003-6. [DOI] [PubMed] [Google Scholar]
- 29.Ikemura T. Correlation between the abundance of yeast transfer RNAs and the occurrence of the respective codons in protein genes. Differences in synonymous codon choice patterns of yeast and Escherichia coli with reference to the abundance of isoaccepting transfer RNAs. J Mol Biol. 1982;158:573–97. doi: 10.1016/0022-2836(82)90250-9. [DOI] [PubMed] [Google Scholar]
- 30.Post LE, Nomura M. Nucleotide sequence of the intercistronic region preceding the gene for RNA polymerase subunit alpha in Escherichia coli. J Biol Chem. 1979;254:10604–6. [PubMed] [Google Scholar]
- 31.Moriyama EN, Powell JR. Codon usage bias and tRNA abundance in Drosophila. J Mol Evol. 1997;45:514–23. doi: 10.1007/pl00006256. [DOI] [PubMed] [Google Scholar]
- 32.Duret L. tRNA gene number and codon usage in the C. elegans genome are co-adapted for optimal translation of highly expressed genes. Trends Genet. 2000;16:287–9. doi: 10.1016/s0168-9525(00)02041-2. [DOI] [PubMed] [Google Scholar]
- 33.Kanaya S, Yamada Y, Kinouchi M, Kudo Y, Ikemura T. Codon usage and tRNA genes in eukaryotes: correlation of codon usage diversity with translation efficiency and with CG-dinucleotide usage as assessed by multivariate analysis. J Mol Evol. 2001;53:290–8. doi: 10.1007/s002390010219. [DOI] [PubMed] [Google Scholar]
- 34.Sharp PM, Bailes E, Grocock RJ, Peden JF, Sockett RE. Variation in the strength of selected codon usage bias among bacteria. Nucleic Acids Res. 2005;33:1141–53. doi: 10.1093/nar/gki242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Sharp PM, Li WH. The codon Adaptation Index--a measure of directional synonymous codon usage bias, and its potential applications. Nucleic Acids Res. 1987;15:1281–95. doi: 10.1093/nar/15.3.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lavner Y, Kotlar D. Codon bias as a factor in regulating expression via translation rate in the human genome. Gene. 2005;345:127–38. doi: 10.1016/j.gene.2004.11.035. [DOI] [PubMed] [Google Scholar]
- 37.Tuller T, Waldman YY, Kupiec M, Ruppin E. Translation efficiency is determined by both codon bias and folding energy. Proc Natl Acad Sci U S A. 2010;107:3645–50. doi: 10.1073/pnas.0909910107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Bergmann JE, Lodish HF. A kinetic model of protein synthesis. Application to hemoglobin synthesis and translational control. J Biol Chem. 1979;254:11927–37. [PubMed] [Google Scholar]
- 39.Mathews MB, Sonenberg N, Hershey JWB, editors. Translational Control in Biology and Medicine. 2007. Mathews, M.B., Sonenberg, N. & Hershey, J.W.B. [Google Scholar]
- 40.Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nat Genet. 2002;31:69–73. doi: 10.1038/ng869. [DOI] [PubMed] [Google Scholar]
- 41.Fraser HB, Hirsh AE, Giaever G, Kumm J, Eisen MB. Noise minimization in eukaryotic gene expression. PLoS Biol. 2004;2:e137. doi: 10.1371/journal.pbio.0020137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lawrence JG, Ochman H. Molecular archaeology of the Escherichia coli genome. Proc Natl Acad Sci U S A. 1998;95:9413–7. doi: 10.1073/pnas.95.16.9413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Karlin S, Campbell AM, Mrazek J. Comparative DNA analysis across diverse genomes. Annu Rev Genet. 1998;32:185–225. doi: 10.1146/annurev.genet.32.1.185. [DOI] [PubMed] [Google Scholar]
- 44.Zhang F, Saha S, Shabalina SA, Kashina A. Differential arginylation of actin isoforms is regulated by coding sequence-dependent degradation. Science. 2010;329:1534–7. doi: 10.1126/science.1191701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Thanaraj TA, Argos P. Ribosome-mediated translational pause and protein domain organization. Protein Sci. 1996;5:1594–612. doi: 10.1002/pro.5560050814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Watts JM, et al. Architecture and secondary structure of an entire HIV-1 RNA genome. Nature. 2009;460:711–6. doi: 10.1038/nature08237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Warnecke T, Batada NN, Hurst LD. The impact of the nucleosome code on protein-coding sequence evolution in yeast. PLoS Genet. 2008;4:e1000250. doi: 10.1371/journal.pgen.1000250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Eskesen ST, Eskesen FN, Ruvinsky A. Natural selection affects frequencies of AG and GT dinucleotides at the 5′ and 3′ ends of exons. Genetics. 2004;167:543–50. doi: 10.1534/genetics.167.1.543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Chamary JV, Hurst LD. Biased codon usage near intron-exon junctions: selection on splicing enhancers, splice-site recognition or something else? Trends Genet. 2005;21:256–9. doi: 10.1016/j.tig.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 50.Orban TI, Olah E. Purifying selection on silent sites -- a constraint from splicing regulation? Trends Genet. 2001;17:252–3. doi: 10.1016/s0168-9525(01)02281-8. [DOI] [PubMed] [Google Scholar]
- 51.Warnecke T, Hurst LD. Evidence for a trade-off between translational efficiency and splicing regulation in determining synonymous codon usage in Drosophila melanogaster. Mol Biol Evol. 2007;24:2755–62. doi: 10.1093/molbev/msm210. [DOI] [PubMed] [Google Scholar]
- 52.Bettany AJ, et al. 5′-secondary structure formation, in contrast to a short string of non-preferred codons, inhibits the translation of the pyruvate kinase mRNA in yeast. Yeast. 1989;5:187–98. doi: 10.1002/yea.320050308. [DOI] [PubMed] [Google Scholar]
- 53.de Smit MH, van Duin J. Secondary structure of the ribosome binding site determines translational efficiency: a quantitative analysis. Proc Natl Acad Sci U S A. 1990;87:7668–72. doi: 10.1073/pnas.87.19.7668. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Eyre-Walker A, Bulmer M. Reduced synonymous substitution rate at the start of enterobacterial genes. Nucleic Acids Res. 1993;21:4599–603. doi: 10.1093/nar/21.19.4599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Kudla G, Murray AW, Tollervey D, Plotkin JB. Coding-sequence determinants of gene expression in Escherichia coli. Science. 2009;324:255–8. doi: 10.1126/science.1170160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gu W, Zhou T, Wilke CO. A universal trend of reduced mRNA stability near the translation-initiation site in prokaryotes and eukaryotes. PLoS Comput Biol. 2010;6:e1000664. doi: 10.1371/journal.pcbi.1000664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Tuller T, et al. An evolutionarily conserved mechanism for controlling the efficiency of protein translation. Cell. 2010;141:344–54. doi: 10.1016/j.cell.2010.03.031. [DOI] [PubMed] [Google Scholar]
- 58.Bulmer M. Codon usage and intragenic position. J Theor Biol. 1988;133:67–71. doi: 10.1016/s0022-5193(88)80024-9. [DOI] [PubMed] [Google Scholar]
- 59.Qin H, Wu WB, Comeron JM, Kreitman M, Li WH. Intragenic spatial patterns of codon usage bias in prokaryotic and eukaryotic genomes. Genetics. 2004;168:2245–60. doi: 10.1534/genetics.104.030866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang S, Goldman E, Zubay G. Clustering of low usage codons and ribosome movement. J Theor Biol. 1994;170:339–54. doi: 10.1006/jtbi.1994.1196. [DOI] [PubMed] [Google Scholar]
- 61.Fredrick K, Ibba M. How the sequence of a gene can tune its translation. Cell. 2010;141:227–9. doi: 10.1016/j.cell.2010.03.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Cannarozzi G, et al. A role for codon order in translation dynamics. Cell. 2010;141:355–67. doi: 10.1016/j.cell.2010.02.036. [DOI] [PubMed] [Google Scholar]
- 63.Zouridis H, Hatzimanikatis V. Effects of codon distributions and tRNA competition on protein translation. Biophys J. 2008;95:1018–33. doi: 10.1529/biophysj.107.126128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Huh WK, et al. Global analysis of protein localization in budding yeast. Nature. 2003;425:686–91. doi: 10.1038/nature02026. [DOI] [PubMed] [Google Scholar]
- 65.Lu P, Vogel C, Wang R, Yao X, Marcotte EM. Absolute protein expression profiling estimates the relative contributions of transcriptional and translational regulation. Nat Biotechnol. 2007;25:117–24. doi: 10.1038/nbt1270. [DOI] [PubMed] [Google Scholar]
- 66.Vogel C, et al. Sequence signatures and mRNA concentration can explain two-thirds of protein abundance variation in a human cell line. Mol Syst Biol. 2010;6:400. doi: 10.1038/msb.2010.59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Ingolia NT, Ghaemmaghami S, Newman JR, Weissman JS. Genome-wide analysis in vivo of translation with nucleotide resolution using ribosome profiling. Science. 2009;324:218–23. doi: 10.1126/science.1168978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uemura S, et al. Real-time tRNA transit on single translating ribosomes at codon resolution. Nature. 2010;464:1012–7. doi: 10.1038/nature08925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Futcher B, Latter GI, Monardo P, McLaughlin CS, Garrels JI. A sampling of the yeast proteome. Mol Cell Biol. 1999;19:7357–68. doi: 10.1128/mcb.19.11.7357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Schrimpf SP, et al. Comparative functional analysis of the Caenorhabditis elegans and Drosophila melanogaster proteomes. PLoS Biol. 2009;7:e48. doi: 10.1371/journal.pbio.1000048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Taniguchi Y, et al. Quantifying E. coli proteome and transcriptome with single-molecule sensitivity in single cells. Science. 329:533–8. doi: 10.1126/science.1188308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Feng L, Niu DK. Relationship between mRNA stability and length: an old question with a new twist. Biochem Genet. 2007;45:131–7. doi: 10.1007/s10528-006-9059-5. [DOI] [PubMed] [Google Scholar]
- 73.Arava Y, Boas FE, Brown PO, Herschlag D. Dissecting eukaryotic translation and its control by ribosome density mapping. Nucleic Acids Res. 2005;33:2421–32. doi: 10.1093/nar/gki331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Welch M, Villalobos A, Gustafsson C, Minshull J. You’re one in a googol: optimizing genes for protein expression. J R Soc Interface. 2009;6(Suppl 4):S467–76. doi: 10.1098/rsif.2008.0520.focus. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Gouy M, Gautier C. Codon usage in bacteria: correlation with gene expressivity. Nucleic Acids Res. 1982;10:7055–74. doi: 10.1093/nar/10.22.7055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Kertesz M, et al. Genome-wide measurement of RNA secondary structure in yeast. Nature. 2010;467:103–7. doi: 10.1038/nature09322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Takyar S, Hickerson RP, Noller HF. mRNA helicase activity of the ribosome. Cell. 2005;120:49–58. doi: 10.1016/j.cell.2004.11.042. [DOI] [PubMed] [Google Scholar]
- 78.Dittmar KA, Sorensen MA, Elf J, Ehrenberg M, Pan T. Selective charging of tRNA isoacceptors induced by amino-acid starvation. EMBO Rep. 2005;6:151–7. doi: 10.1038/sj.embor.7400341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Elf J, Nilsson D, Tenson T, Ehrenberg M. Selective charging of tRNA isoacceptors explains patterns of codon usage. Science. 2003;300:1718–22. doi: 10.1126/science.1083811. [DOI] [PubMed] [Google Scholar]
- 80.Ghaemmaghami S, et al. Global analysis of protein expression in yeast. Nature. 2003;425:737–41. doi: 10.1038/nature02046. [DOI] [PubMed] [Google Scholar]
- 81.Welch M, et al. Design parameters to control synthetic gene expression in Escherichia coli. PLoS One. 2009;4:e7002. doi: 10.1371/journal.pone.0007002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kudla G, Lipinski L, Caffin F, Helwak A, Zylicz M. High guanine and cytosine content increases mRNA levels in mammalian cells. PLoS Biol. 2006;4:e180. doi: 10.1371/journal.pbio.0040180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Han JS, Szak ST, Boeke JD. Transcriptional disruption by the L1 retrotransposon and implications for mammalian transcriptomes. Nature. 2004;429:268–74. doi: 10.1038/nature02536. [DOI] [PubMed] [Google Scholar]
- 84.Nackley AG, et al. Human catechol-O-methyltransferase haplotypes modulate protein expression by altering mRNA secondary structure. Science. 2006;314:1930–3. doi: 10.1126/science.1131262. [DOI] [PubMed] [Google Scholar]
- 85.Coleman JR, et al. Virus attenuation by genome-scale changes in codon pair bias. Science. 2008;320:1784–7. doi: 10.1126/science.1155761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Hayes CS, Bose B, Sauer RT. Stop codons preceded by rare arginine codons are efficient determinants of SsrA tagging in Escherichia coli. Proc Natl Acad Sci U S A. 2002;99:3440–5. doi: 10.1073/pnas.052707199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Spanjaard RA, van Duin J. Translation of the sequence AGG-AGG yields 50% ribosomal frameshift. Proc Natl Acad Sci U S A. 1988;85:7967–71. doi: 10.1073/pnas.85.21.7967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Kramer EB, Farabaugh PJ. The frequency of translational misreading errors in E. coli is largely determined by tRNA competition. RNA. 2007;13:87–96. doi: 10.1261/rna.294907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Sorensen MA, Kurland CG, Pedersen S. Codon usage determines translation rate in Escherichia coli. J Mol Biol. 1989;207:365–77. doi: 10.1016/0022-2836(89)90260-x. [DOI] [PubMed] [Google Scholar]
- 90.Chen GF, Inouye M. Suppression of the negative effect of minor arginine codons on gene expression; preferential usage of minor codons within the first 25 codons of the Escherichia coli genes. Nucleic Acids Res. 1990;18:1465–73. doi: 10.1093/nar/18.6.1465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Angov E, Hillier CJ, Kincaid RL, Lyon JA. Heterologous protein expression is enhanced by harmonizing the codon usage frequencies of the target gene with those of the expression host. PLoS One. 2008;3:e2189. doi: 10.1371/journal.pone.0002189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Rosenberg AH, Goldman E, Dunn JJ, Studier FW, Zubay G. Effects of consecutive AGG codons on translation in Escherichia coli, demonstrated with a versatile codon test system. J Bacteriol. 1993;175:716–22. doi: 10.1128/jb.175.3.716-722.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Gursky YG, Beabealashvilli R. The increase in gene expression induced by introduction of rare codons into the C terminus of the template. Gene. 1994;148:15–21. doi: 10.1016/0378-1119(94)90228-3. [DOI] [PubMed] [Google Scholar]
- 94.Burgess-Brown NA, et al. Codon optimization can improve expression of human genes in Escherichia coli: A multi-gene study. Protein Expr Purif. 2008;59:94–102. doi: 10.1016/j.pep.2008.01.008. [DOI] [PubMed] [Google Scholar]
- 95.Maertens B, et al. Gene optimization mechanisms: a multi-gene study reveals a high success rate of full-length human proteins expressed in Escherichia coli. Protein Sci. 2010;19:1312–26. doi: 10.1002/pro.408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Supek F, Smuc T. On relevance of codon usage to expression of synthetic and natural genes in Escherichia coli. Genetics. 2010;185:1129–34. doi: 10.1534/genetics.110.115477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Voges D, Watzele M, Nemetz C, Wizemann S, Buchberger B. Analyzing and enhancing mRNA translational efficiency in an Escherichia coli in vitro expression system. Biochem Biophys Res Commun. 2004;318:601–14. doi: 10.1016/j.bbrc.2004.04.064. [DOI] [PubMed] [Google Scholar]
- 98.El Hage A, French SL, Beyer AL, Tollervey D. Loss of Topoisomerase I leads to R-loop-mediated transcriptional blocks during ribosomal RNA synthesis. Genes Dev. 2010;24:1546–58. doi: 10.1101/gad.573310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.McDowall KJ, Lin-Chao S, Cohen SN. A+U content rather than a particular nucleotide order determines the specificity of RNase E cleavage. J Biol Chem. 1994;269:10790–6. [PubMed] [Google Scholar]
- 100.Nguyen KL, et al. Codon optimization of the HIV-1 vpu and vif genes stabilizes their mRNA and allows for highly efficient Rev-independent expression. Virology. 2004;319:163–75. doi: 10.1016/j.virol.2003.11.021. [DOI] [PubMed] [Google Scholar]
- 101.Sokolowski M, Tan W, Jellne M, Schwartz S. mRNA instability elements in the human papillomavirus type 16 L2 coding region. J Virol. 1998;72:1504–15. doi: 10.1128/jvi.72.2.1504-1515.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Bauer AP, et al. The impact of intragenic CpG content on gene expression. Nucleic Acids Res. 2010;38:3891–908. doi: 10.1093/nar/gkq115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Gutman GA, Hatfield GW. Nonrandom utilization of codon pairs in Escherichia coli. Proc Natl Acad Sci U S A. 1989;86:3699–703. doi: 10.1073/pnas.86.10.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Irwin B, Heck JD, Hatfield GW. Codon pair utilization biases influence translational elongation step times. J Biol Chem. 1995;270:22801–6. doi: 10.1074/jbc.270.39.22801. [DOI] [PubMed] [Google Scholar]
- 105.Cheng L, Goldman E. Absence of effect of varying Thr-Leu codon pairs on protein synthesis in a T7 system. Biochemistry. 2001;40:6102–6. doi: 10.1021/bi010236v. [DOI] [PubMed] [Google Scholar]
- 106.Wang D, Johnson AD, Papp AC, Kroetz DL, Sadee W. Multidrug resistance polypeptide 1 (MDR1, ABCB1) variant 3435C>T affects mRNA stability. Pharmacogenet Genomics. 2005;15:693–704. [PubMed] [Google Scholar]
- 107.Kimchi-Sarfaty C, et al. A “silent” polymorphism in the MDR1 gene changes substrate specificity. Science. 2007;315:525–8. doi: 10.1126/science.1135308. [DOI] [PubMed] [Google Scholar]
- 108.Foster H, et al. Codon and mRNA sequence optimization of microdystrophin transgenes improves expression and physiological outcome in dystrophic mdx mice following AAV2/8 gene transfer. Mol Ther. 2008;16:1825–32. doi: 10.1038/mt.2008.186. [DOI] [PubMed] [Google Scholar]
- 109.Arruda VR, et al. Peripheral transvenular delivery of adeno-associated viral vectors to skeletal muscle as a novel therapy for hemophilia B. Blood. 115:4678–88. doi: 10.1182/blood-2009-12-261156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Fuglsang A. The relationship between palindrome avoidance and intragenic codon usage variations: a Monte Carlo study. Biochem Biophys Res Commun. 2004;316:755–62. doi: 10.1016/j.bbrc.2004.02.117. [DOI] [PubMed] [Google Scholar]
- 111.Drummond DA, Wilke CO. The evolutionary consequences of erroneous protein synthesis. Nat Rev Genet. 2009;10:715–24. doi: 10.1038/nrg2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Akashi H, Kliman RM, Eyre-Walker A. Mutation pressure, natural selection, and the evolution of base composition in Drosophila. Genetica. 1998;102-103:49–60. [PubMed] [Google Scholar]
- 113.Marais G, Duret L. Synonymous codon usage, accuracy of translation, and gene length in Caenorhabditis elegans. J Mol Evol. 2001;52:275–80. doi: 10.1007/s002390010155. [DOI] [PubMed] [Google Scholar]
- 114.Higgs PG, Ran W. Coevolution of codon usage and tRNA genes leads to alternative stable states of biased codon usage. Mol Biol Evol. 2008;25:2279–91. doi: 10.1093/molbev/msn173. [DOI] [PubMed] [Google Scholar]
- 115.Shah P, Gilchrist M. Effect of Correlated tRNA Abundances on Translation Errors and Evolution of Codon Usage Bias. PLoS Genet. 2010;6:e1001128. doi: 10.1371/journal.pgen.1001128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Bernardi G, et al. The mosaic genome of warm-blooded vertebrates. Science. 1985;228:953–8. doi: 10.1126/science.4001930. [DOI] [PubMed] [Google Scholar]
- 117.Galtier N, Piganeau G, Mouchiroud D, Duret L. GC-content evolution in mammalian genomes: the biased gene conversion hypothesis. Genetics. 2001;159:907–11. doi: 10.1093/genetics/159.2.907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Urrutia AO, Hurst LD. The signature of selection mediated by expression on human genes. Genome Res. 2003;13:2260–4. doi: 10.1101/gr.641103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Comeron JM. Selective and mutational patterns associated with gene expression in humans: influences on synonymous composition and intron presence. Genetics. 2004;167:1293–304. doi: 10.1534/genetics.104.026351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Karlin S, Mrazek J. What drives codon choices in human genes? J Mol Biol. 1996;262:459–72. doi: 10.1006/jmbi.1996.0528. [DOI] [PubMed] [Google Scholar]
- 121.Plotkin JB, Robins H, Levine AJ. Tissue-specific codon usage and the expression of human genes. Proc Natl Acad Sci U S A. 2004;101:12588–91. doi: 10.1073/pnas.0404957101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Dittmar KA, Goodenbour JM, Pan T. Tissue-specific differences in human transfer RNA expression. PLoS Genet. 2006;2:e221. doi: 10.1371/journal.pgen.0020221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Semon M, Lobry JR, Duret L. No Evidence for Tissue-Specific Adaptation of Synonymous Codon Usage in Human. Mol Biol Evol. 2005 doi: 10.1093/molbev/msj053. [DOI] [PubMed] [Google Scholar]
- 124.Chamary JV, Hurst LD. Evidence for selection on synonymous mutations affecting stability of mRNA secondary structure in mammals. Genome Biol. 2005;6:R75. doi: 10.1186/gb-2005-6-9-r75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Seffens W, Digby D. mRNAs have greater negative folding free energies than shuffled or codon choice randomized sequences. Nucleic Acids Res. 1999;27:1578–84. doi: 10.1093/nar/27.7.1578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Duan J, et al. Synonymous mutations in the human dopamine receptor D2 (DRD2) affect mRNA stability and synthesis of the receptor. Hum Mol Genet. 2003;12:205–16. doi: 10.1093/hmg/ddg055. [DOI] [PubMed] [Google Scholar]
- 127.Sharp PM, Tuohy TM, Mosurski KR. Codon usage in yeast: cluster analysis clearly differentiates highly and lowly expressed genes. Nucleic Acids Res. 1986;14:5125–43. doi: 10.1093/nar/14.13.5125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.dos Reis M, Savva R, Wernisch L. Solving the riddle of codon usage preferences: a test for translational selection. Nucleic Acids Res. 2004;32:5036–44. doi: 10.1093/nar/gkh834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Nivinskas R, Malys N, Klausa V, Vaiskunaite R, Gineikiene E. Posttranscriptional control of bacteriophage T4 gene 25 expression: mRNA secondary structure that enhances translational initiation. J Mol Biol. 1999;288:291–304. doi: 10.1006/jmbi.1999.2695. [DOI] [PubMed] [Google Scholar]
- 130.Kozak M. Downstream secondary structure facilitates recognition of initiator codons by eukaryotic ribosomes. Proc Natl Acad Sci U S A. 1990;87:8301–5. doi: 10.1073/pnas.87.21.8301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Paulus M, Haslbeck M, Watzele M. RNA stem-loop enhanced expression of previously non-expressible genes. Nucleic Acids Res. 2004;32:e78. doi: 10.1093/nar/gnh076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Zhang G, Hubalewska M, Ignatova Z. Transient ribosomal attenuation coordinates protein synthesis and co-translational folding. Nat Struct Mol Biol. 2009;16:274–80. doi: 10.1038/nsmb.1554. [DOI] [PubMed] [Google Scholar]
- 133.Nakamura Y, Gojobori T, Ikemura T. Codon usage tabulated from international DNA sequence databases: status for the year 2000. Nucleic Acids Res. 2000;28:292. doi: 10.1093/nar/28.1.292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Zolotukhin S, Potter M, Hauswirth WW, Guy J, Muzyczka N. A “humanized” green fluorescent protein cDNA adapted for high-level expression in mammalian cells. J Virol. 1996;70:4646–54. doi: 10.1128/jvi.70.7.4646-4654.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Markham NR, Zuker M. DINAMelt web server for nucleic acid melting prediction. Nucleic Acids Res. 2005;33:W577–81. doi: 10.1093/nar/gki591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Lesnik EA, et al. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–94. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137.Bernstein JA, Khodursky AB, Lin PH, Lin-Chao S, Cohen SN. Global analysis of mRNA decay and abundance in Escherichia coli at single-gene resolution using two-color fluorescent DNA microarrays. Proc Natl Acad Sci U S A. 2002;99:9697–702. doi: 10.1073/pnas.112318199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Lewis BP, Shih IH, Jones-Rhoades MW, Bartel DP, Burge CB. Prediction of mammalian microRNA targets. Cell. 2003;115:787–98. doi: 10.1016/s0092-8674(03)01018-3. [DOI] [PubMed] [Google Scholar]
- 139.Jacobs GH, et al. Transterm: a database to aid the analysis of regulatory sequences in mRNAs. Nucleic Acids Res. 2009;37:D72–6. doi: 10.1093/nar/gkn763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Peden JF. Department of Genetics. University of Nottingham; Nottingham: 1999. p. 226. [Google Scholar]
- 141.Supek F, Vlahovicek K. INCA: synonymous codon usage analysis and clustering by means of self-organizing map. Bioinformatics. 2004;20:2329–30. doi: 10.1093/bioinformatics/bth238. [DOI] [PubMed] [Google Scholar]
- 142.Pagani F, Raponi M, Baralle FE. Synonymous mutations in CFTR exon 12 affect splicing and are not neutral in evolution. Proc Natl Acad Sci U S A. 2005;102:6368–72. doi: 10.1073/pnas.0502288102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Burge C, Campbell AM, Karlin S. Over- and under-representation of short oligonucleotides in DNA sequences. Proc Natl Acad Sci U S A. 1992;89:1358–62. doi: 10.1073/pnas.89.4.1358. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Villalobos A, Ness JE, Gustafsson C, Minshull J, Govindarajan S. Gene Designer: a synthetic biology tool for constructing artificial DNA segments. BMC Bioinformatics. 2006;7:285. doi: 10.1186/1471-2105-7-285. [DOI] [PMC free article] [PubMed] [Google Scholar]