Abstract
Small molecules are important not only as therapeutics to treat disease, but also as chemical tools to probe complex biological processes. The discovery of novel bioactive small molecules has largely been catalyzed by screening diverse chemical libraries for alterations in specific activities in pure proteins assays or in generating cell-based phenotypes. New approaches are needed to close the vast gap between the ability to either study single proteins or whole cellular processes. This review focuses on the growing number of studies aimed at understanding in more detail how small molecules perturb particular signaling pathways and larger networks to yield distinct cellular phenotypes. This type of pathway-level analysis and phenotypic profiling provides valuable insight into mechanistic action of small molecules, can reveal off-target effects, and improve our understanding of how proteins within a pathway regulate signaling.
Introduction
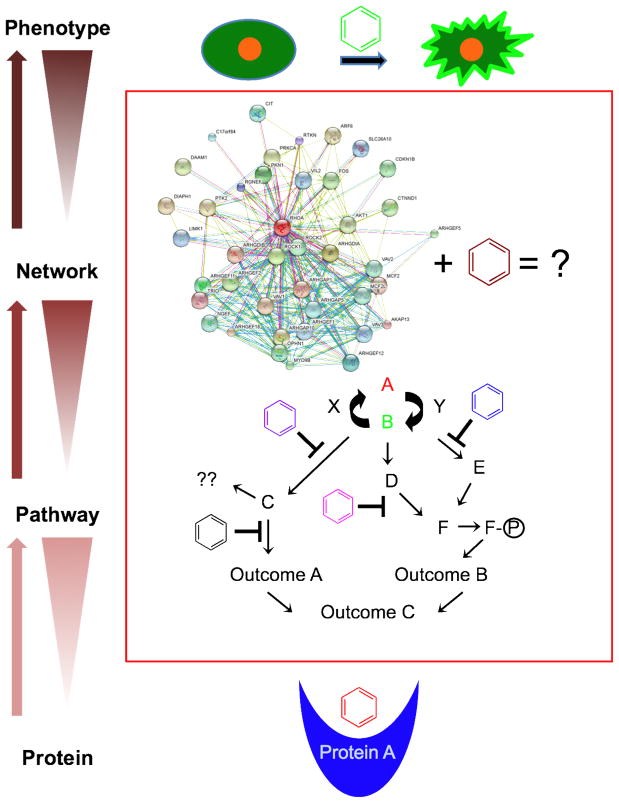
Small molecules are essential as drugs in modern medicine and are valuable as probes of biological mechanism in chemical biology. Significant screening efforts to date have focused on the discovery of small molecules that target specific proteins or that confer interesting cellular phenotypes (Figure 1). Between these two spectra, there is a critical need to expand mechanistic studies into how small molecules modulate signaling pathways and larger networks to generate complex phenotypes (Figure 1). Recent advances in high-content screening methods have greatly enhanced the ability to rapidly acquire large volumes of phenotypic data following the treatment of cells with bioactive small molecules, making it possible to maximize this emerging capability (Figure 1). In this review, we discuss how pathway-level screening and analysis methods have expanded the number of pathways that can be probed with small molecules and that have allowed for a deeper understanding of how compounds alter cellular processes controlled by multiple pathways. This information has provided valuable insight into how small molecules perturb larger signaling networks and has enabled more thorough investigations into mechanisms of action, which is critical for both for drug discovery and chemical probe development.
Figure 1. Small molecule probes of pathways and networks.
Although small molecules can be discovered in, and can target, different levels of complexity, they have been mostly used in “bottom-up” approaches starting at the protein level or in “top-down” approaches starting at the phenotype level. This review focuses on the effects of small molecules on cellular pathways and networks. Pathway/network analyses are important because they offer mechanistic insight into how small molecules perturb proteins in signaling pathways and how disruption of those pathways affects whole networks to yield complex observable phenotypes.
As probes, small molecules are useful and versatile tools to study complex cellular processes requiring integrated inputs from multiple pathways. Small molecules act quickly and often reversibly upon washout, allowing a high degree of spatial and temporal modulation over protein function. Many compounds have been used successfully to probe basic cellular functions. In a historic example, the natural product cytochalasin was shown to disrupt actin filaments and it has long been used to study the role of actin in different processes of the cell including migration, ruffling and division (1, 2). Given their therapeutic potential and possible utility as research tools, it is important to develop methods to characterize in more detail the mechanism of action of existing compounds and discover new bioactive small molecules.
Active small molecules can be discovered by screening chemical libraries for alterations of specific activities in pure protein assays or for desired phenotypes in cell-based assays. Many excellent studies have been directed toward developing biochemical screening assays using purified proteins to identify small molecule inhibitors. While these in vitro assays can be extremely useful in identifying potent and selective compounds, their limits include: 1) They primarily target very specific and measurable functions of a protein or complex of proteins, either particular enzymatic activities or particular functional interactions between defined proteins, 2) potent and selective in vitro activity is not necessarily predictive of actual activity in cells, for example due to poor cell permeability and 3) the scope of off-target cellular effects due to, for example, similarities between related proteins, e.g. kinases, as well as toxicity is not readily discernible.
In contrast, small molecules discovered in phenotypic screens can target any protein, regardless of its activity, and must display some specificity to cause a specific phenotype (3). The potential of phenotypic screens for the discovery of useful compounds is high, but challenges remain because cellular phenotypes that serve as readouts in phenotypic screens often involve intricate biological processes coordinated by multiple signaling pathways. This level of complexity can present significant challenges when performing follow-up secondary assays and target identification, creating a bottleneck, which slows the rate of discovery of useful compounds.
Pathway-directed small molecule screening
To circumvent many of the challenges associated with traditional pure protein or phenotypic screens, several groups have recently developed novel strategies to identify compounds that target specific biological pathways. Pathway-directed screening approaches can simplify target identification and generate a toolbox of small molecules to systematically interrogate a given signaling pathway and identify potential “druggable” targets.
Screening strategies that combine genetic with chemical methods have been successfully implemented in several model systems, including bacteria, yeast, Dictyostelium and Drosophila. We will see below how some progress has been made in screens in different human cells lines, especially in cancer cells. Most of the new developments in pathway screening strategies, however, have taken advantage of model systems. Reasons for this include more straightforward methods for genetic manipulations and less complex, and less redundant, signaling pathways. For example, as discussed below, we performed a screen in Drosophila cells because only one isoform of our target protein/pathway, Rho, exists, while there are three isoforms in humans, and all three need to be depleted to achieve our desired phenotype (4).
Using a screening approach in bacteria that utilized both small molecules and antisense RNA technologies, scientists at Merck discovered the antibiotic platensimycin, a selective inhibitor of FabF, a key enzyme involved in bacterial fatty acid synthesis and thus critical for cell viability (5, 6). The authors screened over 250,000 natural product extracts looking for compounds that selectively inhibited the growth of a bacterial strain genetically engineered to express fabF antisense RNA. The antisense RNA-sensitized strain, because of its already reduced FabF levels, became more susceptible to killing by FabF inhibitors, providing a powerful system to screen for novel antibiotics.
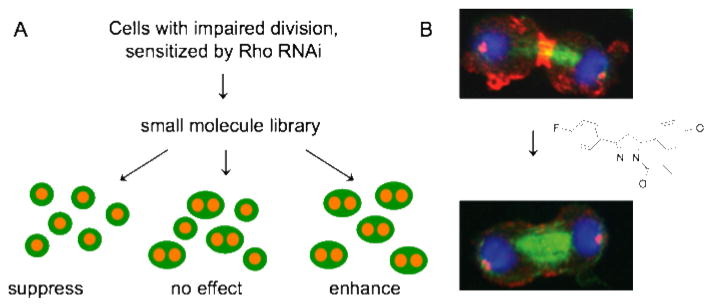
Our lab developed a phenotypic screening approach to discover compounds that specifically target the Rho pathway in cytokinesis (7). Inspired by classical genetic experiments and the Merck study, we developed a screening strategy analogous to a genetic modifier screen, but perturbed cells by small molecules and RNAi instead of genetic mutations. We sensitized Drosophila cells by RNAi treatment, reducing Rho function, and screened for small molecules that modify a Rho-specific RNAi phenotype. We decided to target the pathway rather than Rho GTPase itself because, despite serious efforts by many labs, especially with the oncogenic GTPase Ras, small molecules that inhibit small GTPases have been elusive. GTP affinity in GTPases is much higher than ATP affinity in kinases, making it energetically unfavourable for a small molecule to displace GTP (8). We modestly impaired cytokinesis using partial RNAi depletion of Rho, added small molecules, and identified compounds that suppressed RNAi-induced cytokinesis defects, or aggravated them. We expected to find enhancers and suppressors because the pathway is positively and negatively regulated. An initial analysis resulted in nine compounds that increased the level of binucleated cells in a Rho RNAi background, which we named Rhodblock 1a/b-8. We confirmed that the Rhodblocks truly target the Rho pathway by showing that 8/9 compounds inhibit the phosphorylation of myosin light chain, a key function of the Rho pathway (Figure 2), and identified Rhodblock 6 as an inhibitor of Rho kinase, a key downstream Rho pathway effector. Rhodblocks will be useful tools to dissect Rho signaling and to study cytokinesis regulation.
Figure 2. A pathway screen results in small molecules that target the Rho pathway in cytokinesis.
A. A small molecule/RNAi modifier screen. Cells with two nuclei are a consequence of failed cytokinesis and the readout in the screen. (Whole cells are cartooned in green, DNA in orange). B. Small molecules from the screen inhibit the accumulation of phospho-myosin at the cleavage furrow, a key function of the Rho pathway. Immunofluorescence images show Drosophila Kc167 cells where phospho-myosin (red), tubulin (green) and DNA (blue) have been visualized. Note the decrease of phospho-myosin at the furrow in compound-treated cells. Adapted from (7).
An important pre-requisite for this screening strategy involves identifying proteins in the candidate pathway that, when perturbed by RNAi or small molecule inhibition, yield robust and measureable phenotypes that can be adapted to high-throughput screening and automated analysis. One can imagine many potential phenotypes amenable to this analysis and therefore our screening strategy should be widely applicable to generate small molecule modulators to dissect other dynamic signaling pathways.
In the previous examples, we describe screening strategies that use amplifications of genetic phenotypes by small molecules as a readout. One can also imagine a reverse approach, i.e. using genetic methods to overcome a small molecule phenotype, which would be especially valuable in small molecule target identification. To study how the mitotic spindle coordinates changes in the cell cortex during cytokinesis in Dictyostelium and to identify potential regulatory pathways, Zhou et al. screened a cDNA library for genetic suppressors of nocodazole-induced growth defects, and identified protein 14–3–3 as a hit in their screen (9). In subsequent follow-up work, the authors dissected the role of 14–3–3 in coordinating the activities of microtubules, RacE and myosin II to modulate cortical remodeling and shape changes during cytokinesis. This work is important because it provides mechanistic insight into how signals emanating from the mitotic spindle initiate cortical changes necessary for cytokinesis.
The work described above largely focuses on how single small molecules in the context of specific genetic perturbations can alter cellular pathways. Combining small molecule treatments to determine synergistic/antagonistic effects can be useful in dissecting connectivity of targets in a given pathway and understanding functional interactions between pathways in disease states (17). For example, Owens et al. systematically combined inhibitors of the sterol biosynthesis pathway and measured effects on Hepatitis C viral replication. The authors determined that sterol biosynthesis inhibitors administered in combination showed synergistic disruptions in HCV viral replication, highlighting the utility/therapeutic potential of combining drug treatments (18).
In addition to using combined genetic and small molecule perturbations to identify pathway-specific small molecules, an alternate approach involves measuring to what extent small molecules disrupt downstream signaling events of a given pathway, such as transcriptional activation or phosphorylation state changes. For example, several groups have used a Wnt/beta-catenin-mediated luciferase reporter assay to screen for modulators of Wnt signaling (19, 20).
The Wnt pathway represents an important therapeutic target because it is misregulated in many cancers. Huang et al. identified a small molecule inhibitor of Wnt signaling (XAV939) that stabilized axin and promoted beta-catenin degradation. The targets of XAV939 were discovered by three-channel iTRAQ quantitative chemical proteomics as the poly-ADP-ribosylating enzymes tankyrase 1 and 2. This study identified tankyrases as new therapeutic targets in Wnt signaling. XAV939 is an important chemical tool to study how tankyrases regulate axin homeostasis and beta-catenin degradation.
Recent work by Hoffman et al. developed a cell-based screening assay to identify small molecule modulators of mTORC1 signaling, which is known to be altered in several disease states including many cancers (21). The authors utilized a novel in cell Western technique to monitor the activation state of the mTORC1 signaling pathway by quantifying the phosphorylation status of a downstream substrate, ribosomal protein S6. The assay is currently being used to screen libraries of small molecules and siRNAs in parallel to enable direct phenotypic comparisons (22), which will provide valuable insights into target protein identification of promising small molecules.
Systems chemical biology and chemical profiling
The work described above largely focused on discovering small molecules that target single pathways. Recently, several groups have taken broader systems-type analyses to determine mechanism of action, where multiple pathways are profiled after small molecule treatment. Using diverse profiling approaches, different types of phenotypic compendia can be created, which are useful both as guides for comparative target identification approaches and to better understand mechanistic context of small molecule perturbations.
Gene expression profiling is commonly used to assess effects of cellular perturbations. Hughes et al. conducted gene expression profiling to functionally characterize 300 genetic mutations and small molecules in yeast (23). Lamb et al. extended this work to mammalian cells and constructed a “Connectivity Map” based on gene-expression profiles and pattern-matching analysis following small molecule treatment. The Connectivity map promises to be a useful resource to group drugs with common mechanisms of action, discover novel mechanisms for uncharacterized compounds, and find small molecules that mimic or suppress a disease state (24).
In yeast, the drug-induced haploinsufficiency profiling method (HIP) has been an important tool for drug discovery and target identification. In HIP, approximately 6,000 heterozygous deletion strains exhibit a partial reduction in gene dosage and can be screened in parallel. Strains that show a decrease in growth/fitness in the presence of a small molecule represent functionally interacting genes (10, 11). Giaever et al. and Baetz et al. used the HIP approach to screen diverse small molecules against the entire genome-wide collection of heterozygous deletions strains to determine which cellular pathways are perturbed by a given small molecule, providing insight into drug mechanism of action (11, 12). The authors validated the HIP approach by identifying previously known targets of several compounds, including methotrexate and statins, while also uncovering new interactions and mechanisms. For example, Baetz et al. tested dihydromotuporamine C, an anti-cancer compound with uncharacterized targets, and discovered that it disrupted key steps in sphingolipid metabolism.
Like HIP, the HOP (homozygous profiling) approach uses homozygous (or haploid) deletion strains (13). Parsons et al. used HOP to generate chemical-genetic fitness profiles to characterize approximately 80 compounds and natural product extracts (14). Through clustering analysis, the authors discovered that the breast cancer drug tamoxifen disrupts calcium homeostasis. Several studies have used multicopy suppression profiling to confirm a drug target (15, 16). Using this approach, overexpression of the target protein should confer resistance to the small molecule inhibitor. Hoon et al. combined the utility of HIP, HOP, and multicopy suppresssion approaches into a systems-level integrated platform to study drug mechanism of action using eight reference compounds and 188 compounds of unknown activity (16). The authors validated the platform by identifying the known molecular targets of the phosphatase inhibitors cantharidin and calyculin A and further characterizing their cellular affects, and also demonstrated its utility in finding novel interactions/potential targets.
To group known compounds of similar mechanisms of action and to suggest targets/mechanisms for new drugs, Perlman et al. developed a high-throughput and quantitative phenotypic profiling method in HeLa cells (25). Using automated fluorescence microscopy and 11 distinct biological probes, the method allowed the extraction of multidimensional phenotypic information at the single-cell level for many compounds (100) over a range of doses (13 threefold dilutions) where a set of 93 descriptors (i.e. measures of size, shape, intensity and ratios of intensities) are measured for nuclear, cytoplasmic and probe regions. Similarly, Tanaka et al. conducted a phenotypic screen of 107 small molecules where changes in cell and organelle morphology were systematically profiled to identify novel bioactive small molecules that generated unique signatures in cancer cell lines (26). From the screen and subsequent affinity-chromatography experiments, the authors identified a hydroxyl-substituted analog of the Src-family kinase inhibitor PP2 as an inhibitor of carbonyl reductase I and show that it can be used to enhance the effectiveness of established anti-cancer drugs. These two studies highlight the utility of using multidimensional phenotypic profiling to discover and characterize interesting new compounds.
While the work discussed above provided useful snapshots of the effects of different small molecule treatments, it would be very helpful to have an opportunity to observe if functional interactions in cells have been perturbed. MacDonald et al. utilized a method to probe the effects of over one hundred compounds on multiple signaling pathways simultaneously in living cells by imaging of protein-fragment complementation assays (PCAs) (27). PCA technology involves fragments of inactive reporter polypetides fused to proteins that are known to interact upon activation of a signaling pathway. When these pathway proteins interact and come into close proximity with each other, the reporter fragments are able to complex into an active reporter to generate a fluorescent signal at the site of interaction. In this study, PCAs served as sensors of specific protein-protein interactions in a given pathway and thus reported on dynamic changes occurring in that pathway in response to a compound. 49 PCAs acted as readouts for pathway activation or inhibition and reported on the status of multiple pathways involved in cell division, apoptosis, inflammation, DNA damage responses, and nuclear hormone receptor signaling. From these data, the authors assembled a multi-pathway signature profile after compound treatment. These profiles were predictive and provided insight into a drug’s mechanism of action and highlighted/identified previously uncharacterized off-target phenotypes. PCAs have also been used to probe more specifically for modulators of Rho family GTPases (Ras) and MAPK signaling pathways (28).
Cancer Cell Profiling
The studies described above highlight the utility of image-based phenotypic and gene-expression profiling to make informed predictions about a drug’s mechanism of action and make previously unidentified connections between groups of compounds. Other profiling approaches have been developed to discover compounds that specifically kill cancer cells.
For example, Dolma et al. performed a high-throughput synthetic lethal screen to look for compounds that kill genetically engineered tumor cell lines (primary fibroblasts expressing hTERT and oncogenic proteins Large T, Small T and HRAS) but not the isogenic parent cell line (29). The readout in the screen utilized calcein AM dye, which is cleaved in living cells by esterases and remains trapped in cells and exhibits green fluorescence. The authors identified nine compounds including five previously known anti-cancer drugs (doxorubicin, daunorubicin, mitoxantrone, camptothecin, and echinomycin). They examined the killing potential of the nine hit compounds in their full complement of engineered tumor cell lines (14 cell lines total) to tease apart the killing mechanism and identified which genetic elements were important for compound activity. They identified three groups of compounds (1) compounds that killed the engineered tumor cells indiscriminately (2) compounds that killed only tumor cells expressing hTERT and inactive RB, and (3) compounds that required oncogenic RAS and ST expression. This is interesting because it enables insight into which compounds might be more effective against specific cancers depending on their particular genetic expression profile.
Cancer stem cells (as well as most other stem cells) have been difficult to screen with small molecules because they exist in low numbers in tumors and are challenging to maintain in culture. Gupta et al. used hTERT-immortalized mammary epithelial cells treated with E-cadherin RNAi to induce a cancer stem cell-like state (30). The authors used their enriched cells to perform a high-throughput screen to look for compounds that selectively kill E-cadherin RNAi-induced cancer stem cells, but not control cells. They identified salinomycin as a compound that selectively killed cells with breast cancer stem cell-like properties. This is important because cancer stem cells drive cancer progression and contribute to relapse following treatment, so targeting these cells to specifically is a promising potential therapy for breast cancer.
To identify new inhibitors of Bcr-abl kinase for Chronic Myeloid Leukemia (CML) treatment, Adrian et al. utilized a cytotoxicity screen to look for compounds that selectively killed Bcr-abl transformed cells, but not non-transformed cells (31). Their screen identified the non-ATP-competitive inhibitor GNF-2. GNF-2 was shown to be effective against imatinib-resistant Bcr-abl mutants. Zhang et al. extended these studies to show that combining GNF-5, a more potent analog of GNF-2, with imatinib had additive inhibitory effects in cell-based assays and suppressed imatinib resistance emergence in vitro and in vivo (32). These studies were important because they demonstrated the therapeutic potential of combining allosteric and ATP-competitive kinase inhibitors to suppress resistance to either compound alone.
Whole organism/IN VIVO chemical screening
As discussed in the above papers, profiling in established cell lines has been very informative in identifying cellular pathways affected/perturbed by small molecules. Many diseases, however, are caused by problems in three-dimensional structures that involve connected cells (e.g. an organ) and can therefore not be studied in a cultured cell model. For this reason, whole organism high content chemical screening is becoming an important resource for expanding drug discovery and target identification efforts.
Since C. elegans was one of the first organisms where RNAi was possible, much insight has been gained into biological signaling networks using this model organism. As we have seen above, combinations of small molecule and RNAi experiments are appealing and therefore C. elegans would have been a natural model for whole organism screening (33). Despite some promising screens having been performed in C. elegans (34, 35), it appears that the general consensus now is that small molecule screens in C. elegans are difficult because small molecules typically do not accumulate in the worms due to significant physical barriers and powerful xenobiotic defenses (36).
A number of chemical screens have been conducted in zebrafish to probe a diverse array of biological processes. Zebrafish are readily adaptable to screening because the embryo, unlike C. elegans, readily absorbs small molecules. This quality makes zebrafish a nice system to study early development, a highly dynamic process involving the coordinate regulation of multiple signaling cascades. Because the embryo is quite transparent, phenotypes can be visualized in detail. For example, Peterson et al. screened for chemical suppressors of a genetic mutation disrupting aortic blood flow in zebrafish (gridlock mutation) (37). The compound GS4012 rescued the gridlock phenotype and restored blood flow by activating the VEGF pathway. Mathew et al. utilized chemical screening in zebrafish to identify modulators of tissue regeneration (38). The authors identified glucocorticoids as inhibitors of caudal fin regeneration.
Many screens involve measuring changes in visual phenotypes in response to small molecule treatment. In an exiting new study, Rihel et al. extended these approaches by monitoring animal behavior as a readout for active compounds (39). The authors profiled dynamic changes in zebrafish locomotion during rest and wake periods. Behavioral profiling allowed the authors to cluster drugs of known mechanism of action with less characterized drugs and allowed them to make meaningful predictions as to the targets and mechanisms of these unknown drugs.
Outlook
In this review, we discussed recent work utilizing pathway-level screening and analysis methods. These approaches have: 1) expanded the number of pathways that can be probed with small molecules, 2) allowed for a more comprehensive understanding of how compounds alter larger networks of pathways by generating phenotypic profiles based on these changes, and 3) enabled compound clustering to shed light on new mechanisms of action, including identifying the target proteins of previously uncharacterized compounds.
Although much progress has been made in the effort to discover and develop small molecules as therapeutics and probes, a host of significant challenges remain: Which model systems are best to utilize for chemical screening? How do we determine which pathways/processes are best to profile? How do we integrate large profile data sets with secondary assays to make informed choices as to which leads/hits to follow-up for detailed studies? As the field of small molecule discovery moves to address these questions, we expect that significant inroads will be made. A major advance will be a better understanding of what constitutes the “druggable genome”.
Acknowledgments
A.B.C. and U.E. were supported by NIH grant R01 GM082834, the Claudia Adams Barr Program and the Dana-Farber Cancer Institute.
References
- 1.Peterson JR, Mitchison TJ. Small molecules, big impact: a history of chemical inhibitors and the cytoskeleton. Chem Biol. 2002;9:1275–1285. doi: 10.1016/s1074-5521(02)00284-3. [DOI] [PubMed] [Google Scholar]
- 2.Spudich JA, Lin S. Cytochalasin B, its interaction with actin and actomyosin from muscle (cell movement-microfilaments-rabbit striated muscle) Proc Natl Acad Sci U S A. 1972;69:442–446. doi: 10.1073/pnas.69.2.442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Eggert US, Mitchison TJ. Small molecule screening by imaging. Curr Opin Chem Biol. 2006;10:232–237. doi: 10.1016/j.cbpa.2006.04.010. [DOI] [PubMed] [Google Scholar]
- 4.Kamijo K, Ohara N, Abe M, Uchimura T, Hosoya H, Lee JS, Miki T. Dissecting the role of Rho-mediated signaling in contractile ring formation. Mol Biol Cell. 2006;17:43–55. doi: 10.1091/mbc.E05-06-0569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Young K, Jayasuriya H, Ondeyka JG, Herath K, Zhang C, Kodali S, Galgoci A, Painter R, Brown-Driver V, Yamamoto R, Silver LL, Zheng Y, Ventura JI, Sigmund J, Ha S, Basilio A, Vicente F, Tormo JR, Pelaez F, Youngman P, Cully D, Barrett JF, Schmatz D, Singh SB, Wang J. Discovery of FabH/FabF inhibitors from natural products. Antimicrob Agents Chemother. 2006;50:519–526. doi: 10.1128/AAC.50.2.519-526.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wang J, Soisson SM, Young K, Shoop W, Kodali S, Galgoci A, Painter R, Parthasarathy G, Tang YS, Cummings R, Ha S, Dorso K, Motyl M, Jayasuriya H, Ondeyka J, Herath K, Zhang C, Hernandez L, Allocco J, Basilio A, Tormo JR, Genilloud O, Vicente F, Pelaez F, Colwell L, Lee SH, Michael B, Felcetto T, Gill C, Silver LL, Hermes JD, Bartizal K, Barrett J, Schmatz D, Becker JW, Cully D, Singh SB. Platensimycin is a selective FabF inhibitor with potent antibiotic properties. Nature. 2006;441:358–361. doi: 10.1038/nature04784. [DOI] [PubMed] [Google Scholar]
- 7.Castoreno AB, Smurnyy Y, Torres AD, Vokes MS, Jones TR, Carpenter AE, Eggert US. Small molecules discovered in a pathway screen target the Rho pathway in cytokinesis. Nat Chem Biol. 2010;6:457–463. doi: 10.1038/nchembio.363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sprang SR. G protein mechanisms: insights from structural analysis. Annu Rev Biochem. 1997;66:639–678. doi: 10.1146/annurev.biochem.66.1.639. [DOI] [PubMed] [Google Scholar]
- 9.Zhou Q, Kee YS, Poirier CC, Jelinek C, Osborne J, Divi S, Surcel A, Will ME, Eggert US, Muller-Taubenberger A, Iglesias PA, Cotter RJ, Robinson DN. 14–3–3 Coordinates Microtubules, Rac, and Myosin II to Control Cell Mechanics and Cytokinesis. Curr Biol. 2010 doi: 10.1016/j.cub.2010.09.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lum PY, Armour CD, Stepaniants SB, Cavet G, Wolf MK, Butler JS, Hinshaw JC, Garnier P, Prestwich GD, Leonardson A, Garrett-Engele P, Rush CM, Bard M, Schimmack G, Phillips JW, Roberts CJ, Shoemaker DD. Discovering modes of action for therapeutic compounds using a genome-wide screen of yeast heterozygotes. Cell. 2004;116:121–137. doi: 10.1016/s0092-8674(03)01035-3. [DOI] [PubMed] [Google Scholar]
- 11.Giaever G, Flaherty P, Kumm J, Proctor M, Nislow C, Jaramillo DF, Chu AM, Jordan MI, Arkin AP, Davis RW. Chemogenomic profiling: identifying the functional interactions of small molecules in yeast. Proc Natl Acad Sci U S A. 2004;101:793–798. doi: 10.1073/pnas.0307490100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Baetz K, McHardy L, Gable K, Tarling T, Reberioux D, Bryan J, Andersen RJ, Dunn T, Hieter P, Roberge M. Yeast genome-wide drug-induced haploinsufficiency screen to determine drug mode of action. Proc Natl Acad Sci U S A. 2004;101:4525–4530. doi: 10.1073/pnas.0307122101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lee W, St Onge RP, Proctor M, Flaherty P, Jordan MI, Arkin AP, Davis RW, Nislow C, Giaever G. Genome-wide requirements for resistance to functionally distinct DNA-damaging agents. PLoS Genet. 2005;1:e24. doi: 10.1371/journal.pgen.0010024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons AB, Lopez A, Givoni IE, Williams DE, Gray CA, Porter J, Chua G, Sopko R, Brost RL, Ho CH, Wang J, Ketela T, Brenner C, Brill JA, Fernandez GE, Lorenz TC, Payne GS, Ishihara S, Ohya Y, Andrews B, Hughes TR, Frey BJ, Graham TR, Andersen RJ, Boone C. Exploring the mode-of-action of bioactive compounds by chemical-genetic profiling in yeast. Cell. 2006;126:611–625. doi: 10.1016/j.cell.2006.06.040. [DOI] [PubMed] [Google Scholar]
- 15.Butcher RA, Bhullar BS, Perlstein EO, Marsischky G, LaBaer J, Schreiber SL. Microarray-based method for monitoring yeast overexpression strains reveals small-molecule targets in TOR pathway. Nat Chem Biol. 2006;2:103–109. doi: 10.1038/nchembio762. [DOI] [PubMed] [Google Scholar]
- 16.Hoon S, Smith AM, Wallace IM, Suresh S, Miranda M, Fung E, Proctor M, Shokat KM, Zhang C, Davis RW, Giaever G, St Onge RP, Nislow C. An integrated platform of genomic assays reveals small-molecule bioactivities. Nat Chem Biol. 2008;4:498–506. doi: 10.1038/nchembio.100. [DOI] [PubMed] [Google Scholar]
- 17.Lehar J, Stockwell BR, Giaever G, Nislow C. Combination chemical genetics. Nat Chem Biol. 2008;4:674–681. doi: 10.1038/nchembio.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Owens CM, Mawhinney C, Grenier JM, Altmeyer R, Lee MS, Borisy AA, Lehar J, Johansen LM. Chemical combinations elucidate pathway interactions and regulation relevant to Hepatitis C replication. Mol Syst Biol. 2010;6:375. doi: 10.1038/msb.2010.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang SM, Mishina YM, Liu S, Cheung A, Stegmeier F, Michaud GA, Charlat O, Wiellette E, Zhang Y, Wiessner S, Hild M, Shi X, Wilson CJ, Mickanin C, Myer V, Fazal A, Tomlinson R, Serluca F, Shao W, Cheng H, Shultz M, Rau C, Schirle M, Schlegl J, Ghidelli S, Fawell S, Lu C, Curtis D, Kirschner MW, Lengauer C, Finan PM, Tallarico JA, Bouwmeester T, Porter JA, Bauer A, Cong F. Tankyrase inhibition stabilizes axin and antagonizes Wnt signalling. Nature. 2009;461:614–620. doi: 10.1038/nature08356. [DOI] [PubMed] [Google Scholar]
- 20.Ewan K, Pajak B, Stubbs M, Todd H, Barbeau O, Quevedo C, Botfield H, Young R, Ruddle R, Samuel L, Battersby A, Raynaud F, Allen N, Wilson S, Latinkic B, Workman P, McDonald E, Blagg J, Aherne W, Dale T. A useful approach to identify novel small-molecule inhibitors of Wnt-dependent transcription. Cancer Res. 2010;70:5963–5973. doi: 10.1158/0008-5472.CAN-10-1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hoffman GR, Moerke NJ, Hsia M, Shamu CE, Blenis J. A high-throughput, cell-based screening method for siRNA and small molecule inhibitors of mTORC1 signaling using the In Cell Western technique. Assay Drug Dev Technol. 2010;8:186–199. doi: 10.1089/adt.2009.0213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Eggert US, Kiger AA, Richter C, Perlman ZE, Perrimon N, Mitchison TJ, Field CM. Parallel chemical genetic and genome-wide RNAi screens identify cytokinesis inhibitors and targets. PLoS Biol. 2004;2:e379. doi: 10.1371/journal.pbio.0020379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hughes TR, Marton MJ, Jones AR, Roberts CJ, Stoughton R, Armour CD, Bennett HA, Coffey E, Dai H, He YD, Kidd MJ, King AM, Meyer MR, Slade D, Lum PY, Stepaniants SB, Shoemaker DD, Gachotte D, Chakraburtty K, Simon J, Bard M, Friend SH. Functional discovery via a compendium of expression profiles. Cell. 2000;102:109–126. doi: 10.1016/s0092-8674(00)00015-5. [DOI] [PubMed] [Google Scholar]
- 24.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, Lerner J, Brunet JP, Subramanian A, Ross KN, Reich M, Hieronymus H, Wei G, Armstrong SA, Haggarty SJ, Clemons PA, Wei R, Carr SA, Lander ES, Golub TR. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006;313:1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 25.Perlman ZE, Slack MD, Feng Y, Mitchison TJ, Wu LF, Altschuler SJ. Multidimensional drug profiling by automated microscopy. Science. 2004;306:1194–1198. doi: 10.1126/science.1100709. [DOI] [PubMed] [Google Scholar]
- 26.Tanaka M, Bateman R, Rauh D, Vaisberg E, Ramachandani S, Zhang C, Hansen KC, Burlingame AL, Trautman JK, Shokat KM, Adams CL. An unbiased cell morphology-based screen for new, biologically active small molecules. PLoS Biol. 2005;3:e128. doi: 10.1371/journal.pbio.0030128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.MacDonald ML, Lamerdin J, Owens S, Keon BH, Bilter GK, Shang Z, Huang Z, Yu H, Dias J, Minami T, Michnick SW, Westwick JK. Identifying off-target effects and hidden phenotypes of drugs in human cells. Nat Chem Biol. 2006;2:329–337. doi: 10.1038/nchembio790. [DOI] [PubMed] [Google Scholar]
- 28.Westwick JK, Michnick SW. Protein-fragment complementation assays (PCA) in small GTPase research and drug discovery. Methods Enzymol. 2006;407:388–401. doi: 10.1016/S0076-6879(05)07032-1. [DOI] [PubMed] [Google Scholar]
- 29.Dolma S, Lessnick SL, Hahn WC, Stockwell BR. Identification of genotype-selective antitumor agents using synthetic lethal chemical screening in engineered human tumor cells. Cancer Cell. 2003;3:285–296. doi: 10.1016/s1535-6108(03)00050-3. [DOI] [PubMed] [Google Scholar]
- 30.Gupta PB, Onder TT, Jiang G, Tao K, Kuperwasser C, Weinberg RA, Lander ES. Identification of selective inhibitors of cancer stem cells by high-throughput screening. Cell. 2009;138:645–659. doi: 10.1016/j.cell.2009.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Adrian FJ, Ding Q, Sim T, Velentza A, Sloan C, Liu Y, Zhang G, Hur W, Ding S, Manley P, Mestan J, Fabbro D, Gray NS. Allosteric inhibitors of Bcr-abl-dependent cell proliferation. Nat Chem Biol. 2006;2:95–102. doi: 10.1038/nchembio760. [DOI] [PubMed] [Google Scholar]
- 32.Zhang J, Adrian FJ, Jahnke W, Cowan-Jacob SW, Li AG, Iacob RE, Sim T, Powers J, Dierks C, Sun F, Guo GR, Ding Q, Okram B, Choi Y, Wojciechowski A, Deng X, Liu G, Fendrich G, Strauss A, Vajpai N, Grzesiek S, Tuntland T, Liu Y, Bursulaya B, Azam M, Manley PW, Engen JR, Daley GQ, Warmuth M, Gray NS. Targeting Bcr-Abl by combining allosteric with ATP-binding-site inhibitors. Nature. 2010;463:501–506. doi: 10.1038/nature08675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Eggert US, Field CM, Mitchison TJ. Small molecules in an RNAi world. Mol BioSyst. 2006;2:93–96. doi: 10.1039/b515335b. [DOI] [PubMed] [Google Scholar]
- 34.Kwok TC, Ricker N, Fraser R, Chan AW, Burns A, Stanley EF, McCourt P, Cutler SR, Roy PJ. A small-molecule screen in C. elegans yields a new calcium channel antagonist. Nature. 2006;441:91–95. doi: 10.1038/nature04657. [DOI] [PubMed] [Google Scholar]
- 35.Petrascheck M, Ye X, Buck LB. An antidepressant that extends lifespan in adult Caenorhabditis elegans. Nature. 2007;450:553–556. doi: 10.1038/nature05991. [DOI] [PubMed] [Google Scholar]
- 36.Burns AR, Wallace IM, Wildenhain J, Tyers M, Giaever G, Bader GD, Nislow C, Cutler SR, Roy PJ. A predictive model for drug bioaccumulation and bioactivity in Caenorhabditis elegans. Nat Chem Biol. 2010;6:549–557. doi: 10.1038/nchembio.380. [DOI] [PubMed] [Google Scholar]
- 37.Peterson RT, Shaw SY, Peterson TA, Milan DJ, Zhong TP, Schreiber SL, MacRae CA, Fishman MC. Chemical suppression of a genetic mutation in a zebrafish model of aortic coarctation. Nat Biotechnol. 2004;22:595–599. doi: 10.1038/nbt963. [DOI] [PubMed] [Google Scholar]
- 38.Mathew LK, Sengupta S, Kawakami A, Andreasen EA, Lohr CV, Loynes CA, Renshaw SA, Peterson RT, Tanguay RL. Unraveling tissue regeneration pathways using chemical genetics. J Biol Chem. 2007;282:35202–35210. doi: 10.1074/jbc.M706640200. [DOI] [PubMed] [Google Scholar]
- 39.Rihel J, Prober DA, Arvanites A, Lam K, Zimmerman S, Jang S, Haggarty SJ, Kokel D, Rubin LL, Peterson RT, Schier AF. Zebrafish behavioral profiling links drugs to biological targets and rest/wake regulation. Science. 2010;327:348–351. doi: 10.1126/science.1183090. [DOI] [PMC free article] [PubMed] [Google Scholar]