Abstract
We show that 1H NMR based metabonomics of serum allows the diagnosis of early stage I/II epithelial ovarian cancer (EOC) required for successful treatment. Because patient specimens are highly precious, we conducted an exploratory study using a micro-flow probe requiring only 20 μL serum. By use of logistic regression on principal components (PCs) of the NMR profiles, we built a 4-variable model for early stage EOC prediction (training set: 69 EOC specimens, 84 healthy controls; test set: 40 EOC, 44 controls) with operating characteristics estimated for the test set at 80% specificity [95% confidence interval (CI): 65% to 90%], 63% sensitivity (95% CI: 46% to 77%), and an area under the Receiver Operator Characteristic Curve (AUC) of 0.796. Independent validation (50 EOC, 50 controls) of the model yielded 95% specificity (95% CI: 86% to 99.5%), 68% sensitivity (95% CI: 53% to 80%) and an AUC of 0.949. A test on cancer type specificity showed that women diseased with renal cell carcinoma were not incorrectly diagnosed with EOC, indicating that metabonomics bears significant potential for cancer type-specific diagnosis. Our model can potentially be applied for women at high risk for EOC, and our study promises to contribute to developing a screening protocol for the general population.
Keywords: Ovarian Cancer, Early Stage Detection, Metabonomics, Cancer-type Specificity, NMR, Micro-flow Probe, Principal Component Analysis, Predictive Statistical Model
Introduction
Epithelial ovarian cancer (EOC) is the leading cause of death from gynecologic malignancies in the United States and Europe. Despite modest improvements in progression-free survival using adjuvant platinum based chemotherapy following cyto-reductive surgery, overall survival rates remain low.1,2 This is because most women are diagnosed at an advanced stage (III/IV) associated with a 5-year survival rate of only 15–20%,3 while the small fraction diagnosed at stage I have 5-year survival rates >90%.4 Hence, early detection is currently by far the most promising approach to reduce EOC morbidity and mortality. The need for such a diagnostic tool is particularly urgent for women of high risk EOC populations with familial or personal history of cancer, and/or abnormalities in cancer predisposition genes such as BRCA1 and BRCA2.
Most investigators agree that a viable strategy for early EOC detection should have a positive predictive value (PPV = true positive/all positive tests) of at least 10% (i.e., no more than nine false positive procedures for each detected case).5 Previously proposed strategies6,7 do not meet this requirement and are not in routine use. To address this challenge, we explored the use of 1H NMR spectroscopy based metabonomics of serum for early EOC detection. Since approaches for non-invasive diagnosis of early stage EOC are not available, serum specimens from women diseased with early stage EOC are highly precious. In fact, they are quite generally obtained only when women are undergoing surgery for other gynecologic malignancies and an EOC tumor is detected by chance. Since the diagnostic potential of NMR-based metabonomics for a given disease can a priori hardly be predicted, it is desirable to be able to explore the diagnostic value with as little serum volume as possible. Hence, we decided to use a NMR micro-flow probe which enables one to record spectra with only 20 μL of serum.
NMR-based metabonomics8 uses NMR spectra as profiles in multivariate data analyses in order to separate classes, i.e. to discriminate diseased and healthy individuals.9–12 Here we show that NMR-based metabonomics of serum allows early stage EOC detection based on a predictive statistical model. Moreover, we discuss preliminary insights into changes of metabolite concentrations which drive class separation in the multi-variate data analyses.
Materials and Methods
Serum Specimens
EOC serum specimens (Supporting Information) were obtained from Gynecologic Oncology Group (GOG) protocol 136 entitled “acquisition of human ovarian and other tissue specimens and serum to be used in studying the causes, diagnosis, prevention and treatment of cancer”. Specimens were collected, processed within 2 hours and frozen (at −80 °C) at the GOG participating institutions, and sent to the GOG Tissue Bank at Children’s Hospital, Columbus, OH, following standard operating procedures (SOP). Specifically, a first set contained 252 specimens (120 from patients with early stage I/II EOC, 132 from age-matched healthy controls). A second set of 100 specimens (50 stage I/II EOC, 50 healthy controls) was solicited from GOG for independent validation after a statistical model for early EOC prediction was obtained with the first set. Renal cell carcinoma (RCC) serum specimens (Supporting Information) were obtained from 30 newly diagnosed female patients at Roswell Park Cancer Institute (RPCI). The samples were collected, processed and stored in liquid nitrogen within 1 hour of the blood draw as previously described.13 15 (50%) of the patients had stage I RCC, and the remainder with stages II–IV. All experimental protocols were approved by Institutional Review Boards at Roswell Park Cancer Institute and the State University of New York at Buffalo.
NMR Sample Preparation
Serum specimens (stored at −80 °C) were thawed at room temperature. Subsequently, NMR samples were prepared by combining 27 μL of serum with 3 μL of a D2O solution required to lock the spectrometer. The D2O solution contained the internal standard formate (27 mM) and NaCl (0.9% w/v). The resulting solution was filtered through a barrier tip (Catalog # 87001-866; VWR International, West Chester, PA, USA) into a 12 × 32 mm glass screw neck vial (Waters Corp., Milford, USA) by centrifugation for 5 minutes at 5 °C.
Operator Certification
Before the start of NMR data acquisition, E.G. was certified as an ‘operator’ for data collection as described.14
NMR Data Collection
After NMR sample (~20 μL volume) preparation, data were acquired following our standard operating procedure14 at 25.0 °C on a Varian INOVA 600 spectrometer equipped with a Protasis microflow probe (Protasis Inc., Marlboro, MA). NMR spectra were acquired for all specimens in a randomized order to minimize potential run-order effects affecting multivariate data analysis. For each sample, one-dimensional (1D) 1H NOESY (100 ms mixing time) and 1H Carr-Purcell-Meiboom-Gill (CPMG; 80 ms spin-lock eliminating the broad resonance lines of high molecular weight compounds in the serum specimens) spectra8 were recorded. For each spectrum, 256 scans were accumulated with 8.5 s relaxation delay and 1.4 s direct acquisition time (other acquisition parameters were similar to those published in ref. 15; Supporting Information) in ~ 45 min. This yielded a total measurement time of 528 hours for all 352 samples. Principal components analyses confirmed the absence of any run order effects. Furthermore, after every 10 serum samples, the entire set-up procedure14 (see also Fig. 1 in ref. 14) was repeated. This included the recording of a 1D NOESY spectrum for a fetal bovine serum test sample. Principal components analyses confirmed that the spectra recorded for the test samples were statistically indistinguishable.
NMR Data Processing and Validation of Spectral Quality
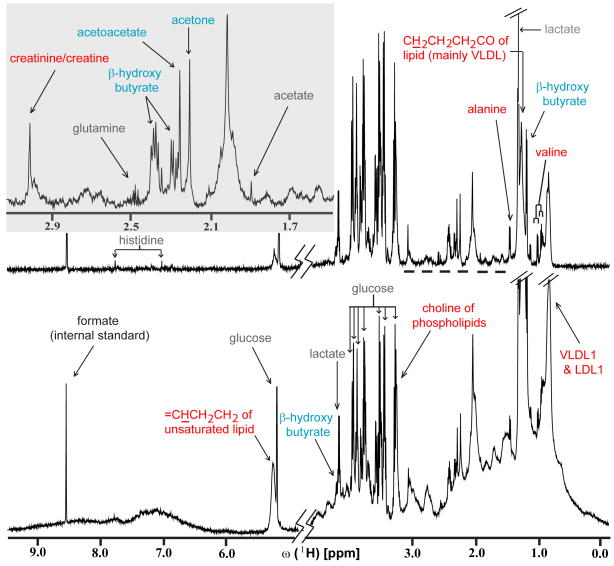
A SOP was defined for NMR data processing and quality validation. Time domain data were zero-filled four-fold to 131,072 points and multiplied by an exponential window function corresponding to a line broadening of 1.2 Hz prior to Fourier transformation. The spectra were phase- and linearly baseline-corrected using VNMRJ (2.1B, Varian), and calibrated to the resonance line of the internal standard formate at 8.444 ppm. Representative NMR spectra are shown in Figure 1 (see also Supporting Information). Prior to statistical analysis, the quality of each frequency domain spectrum was validated by measuring the signal-to-noise (S/N) ratio and line width (at half height and 10% intensity) for the formate signal,14 and inspecting the quality of the ‘water suppression’. Furthermore, figures-of merit (FOM) were calculated to ensure that for each spectrum (i) an unbiased baseline, (ii) a high quality shim and (iii) an accurate phase correction were obtained. For (i), the average and standard deviation were calculated for data points of spectral regions containing only baseline (note that optimal baseline correction yields an average close to zero; for the spectra of the present study, the following cut-offs were chosen: average < 2 × 10−6; standard deviation < 5 × 10−6). For (ii) and (iii), a Lorentzian line was fitted into the resonance line of the internal standard formate and the residual after the fit was integrated (note that poor shims and/or inaccurate phase correction are reflected primarily in non-Lorentzian line-shapes (cut-off: integral of residual < 2% of the total integral of the formate line). For (iii), the symmetry of the formate line was assessed in addition by calculating the difference of the integral obtained for the formate peak segments located, respectively, up- and down-field of the peak maximum (note that inaccurate phase corrections result in peak asymmetry; cut-off: difference < 4% of the total integral).
Figure 1.
Representative 1D 1H CPMG (top) and NOESY (bottom) spectra recorded for a serum specimen obtained from a patient diseased with early stage EOC. 1H resonance assignments16,17 for metabolites (see also http://www.hmdb.ca) for which significantly lower or higher concentrations were observed in this pilot study when comparing the spectra from early stage EOC and healthy control specimens are indicated in red and blue, respectively. Lower concentrations are observed, for alanine (p-value = 3.48×10−18), the choline moiety of phospholipids (4.44×10−22), creatine/creatinine (<2.0 ×10−9), ‘LDL1’ representing CH3(CH2)n of lipid mainly in LDL (1.13×10−20), CH2CH2CH2CO of lipid mainly in VLDL (5.37×10−4), =CHCH2CH2 of unsaturated lipid (2.09×10−4), valine (6.64×10−9), ‘VLDL1’ representing CH3CH2CH2C= of lipid mainly in VLDL (8.71×10−6). Higher concentrations are observed for acetoacetate (1.16×10−9), acetone (1.69×10−5), and β-hydroxybutyrate (1.07×10−8). Metabolites for which marginal concentration differences were detected are indicated in gray. Future studies have to reveal if those are statistically significant (see ‘Conclusions’).
Resonance Assignment of Metabolites
Resonances observed in 1D CPMG spectra were assigned by combining information on chemical shifts documented in literature16,17 and a database (http://www.hmdb.ca) with Statistical Total Correlation Spectroscopy (STOCSY;18 MATLAB scripts for calculation of co-variations were obtained from J. K. Nicholson and co-workers). Additional broad lines observed in 1D NOESY only were assigned using the same protocol. Subsequently, resonance assignments were confirmed by acquiring 2D NMR spectra8 for a healthy control serum sample on a Varian Inova 600 spectrometer equipped with a cryogenic probe: 2D J-resolved (measurement time = 6.5 hours, relaxation delay between scans = 2.0 s, t1,max = 800 ms, t2,max = 1.365 s, spectral width (sw) 1 = 40 Hz, sw 2 = 12,000 Hz), 2D [1H,1H] TOCSY (mixing time = 50 ms and 80 ms; measurement time for each spectrum = 5.5 hours, relaxation delay between scans = 1.5 s, t1,max = 170 ms, t2,max = 683 ms, sw 1, 2 = 6,000 Hz, spinlock field strength = 8,400 Hz), 2D [13C,1H] HSQC (at natural 13C abundance; measurement time = 16 hours, relaxation delay between scans = 1.5 s, t1,max = 30 ms, t2,max = 683 ms, sw 1 = 27,000 Hz, sw 2 = 6,000 Hz). The assignment of selected resonance lines located in crowded spectral regions were further confirmed by spiking the corresponding metabolites (i.e., acetate, acetoacetate, creatine, creatinine, β-hydroxybutyrate, glutamine and valine) in a healthy control serum.
Statistical Analysis
Well established statistical procedures19 were used (i) to build a predictive model for disease status based on the CPMG and NOESY spectra recorded for the first set of specimens (see above), and (ii) to compare their predictive accuracy. Spectra were normalized to unit integral and binned (0.004 ppm resolution) to reduce effects arising from slight variations of, respectively, total signal and signal positions. The resulting bin intensity arrays contained 3,620 variables and were ‘Pareto-scaled’ (i.e., mean centered and divided by square root of standard deviation8). A principal component (PC) analysis was performed on the training set of spectra to obtain orthogonal linear combinations of bin intensities with maximal variation. The PCs were ordered from largest to smallest variance. A sequence of logistic regression models was then considered: PCs were added in decreasing order of their represented variability into a logistic regression prediction model19 until a new addition was not statistically significant (see Supporting Information). For example, when CPMG and NOESY bin arrays were concatenated to build a ‘joint model’, adding PC1 through PC4 each resulted in a statistically significantly (Wald test19) better model, while adding PC5 did not. For the test and validation sets, the same scaling coefficients, PC rotation, and logistic regression model were used to predict disease status. AUC was computed from the predicted scores, and sensitivity and specificity were computed using a pre-specified cut point of 0.5.
Results and Discussion
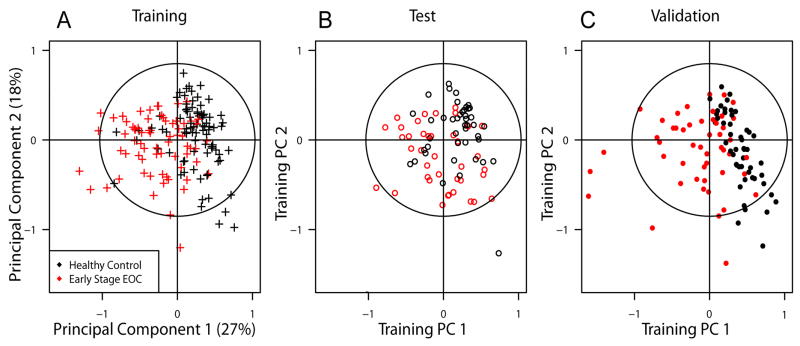
In order to build a predictive statistical model for diagnosis of early stage EOC, two thirds of the first set of specimens (i.e., 80 of 120 early stage EOC and 88 of 132 healthy controls) were randomly selected as the training set, and the remaining specimens formed the test set (Figures 2A,B). Out of the 168 training samples, the spectra of 11 EOC and 4 healthy controls exhibited 1H lines which are generally not observed in serum spectra and were therefore deemed outliers (our SOP14 includes an outlier identification procedure). Thus, those were not considered for the training set used to build a predictive statistical model. Subsequently, three models were built with (a) CPMG or (b) NOESY bin intensity arrays, and (c) both types of bin arrays being concatenated (‘joint model’). Their accuracy for the test set was quite similar (i.e., predictions based on CPMG and NOESY bin arrays were consistent in nearly all cases), but the joint model was slightly superior for differentiating classes (Table S1; see also Figure 4A). For the joint model, four PCs were selected for prediction based on the training set (Figure 3A) yielding a 4-variable logistic regression model with operating characteristics estimated for the test set (no outliers were excluded; Figure 2B) at 80% specificity [95% confidence interval (CI): 65 % to 90 %], 63% sensitivity (95% CI: 46 % to 77 %), and an area under the Receiver Operator Characteristic Curve (AUC) of 0.796 (Figure 4A). Importantly, the predictive model together with an a priori probability of EOC (‘prevalence’ in a population) can be used in a clinical setting to calculate the a posterior probability, p-EOC, of early stage EOC based on the NMR profile (Figure 3). To independently validate the model, we acquired spectra for the second set of 100 samples which we obtained after the predictive model was successfully built. We found that (i) serum samples from early stage EOC patients were well separated from healthy controls in PCA (Figure 2C) and (ii) early stage EOC patients exhibited higher p-EOC values than healthy controls when employing our model (Figure 3C). To confirm statistical robustness, potential outliers identified by our SOP14 among the spectra for the 100 specimens were not excluded for the independent validation (see above). The operating characteristics were estimated at 95% specificity (95% CI: 86% to 99.5%), 68% sensitivity (95% CI: 53% to 80%) and an AUC of 0.949 (Figure 4B).
Figure 2.
(A) Score plot of first and second principal components obtained from Training Set. Using the same components, the data from (B) Test Set, and (C) Validation Set are displayed to show separation of early stage EOC patients and healthy women (see text). EOC patients (red) and healthy controls (black) are also separated in the third and fourth components (not shown). The predictive model was built with logistic regression from Training Set and accuracy was assessed with the Test and Validation Set (see Supporting Information).
Figure 4.
Receiver Operator Characteristic (ROC) Curves for the three logistic regression models built with CPMG bin arrays (‘CPMG’ model), NOESY bin arrays (‘NOESY’ model), and concatenated CPMG and NOESY bin arrays (‘joint’) as obtained for the Validation Set. Area under the ROC Curve (AUC) values measure the quality of predictive models.19 Those values (Table S1) are similar for the three predictive models, with the joint model being slightly superior for both the Test Set and Validation Set.
Figure 3.
Probability of early stage Epithelial Ovarian Cancer (p-EOC) calculated for each spectrum in (A) Training, (B) Test, and (C) Validation Set. The predictive model was constructed by logistic regression using four Principal Components of the joint CPMG and NOESY bin arrays based on the Training Set (Figure 2A). For all three sets, early stage EOC patients (red) exhibit significantly higher p-EOC values than their healthy controls (black) (see Supporting Information).
To test the specificity of the model on cancer type, the model was applied to spectra recorded with identical experimental protocols for 30 serum specimens (obtained from RPCI) from women newly diagnosed with renal cancer carcinoma (RCC). Eight of the 30 (23%) RCC specimens tested positive for EOC, which is not significantly different (p = 0.12; Supporting Information) than for the EOC specimens (11% for combined test and validation sets). Hence, RCC NMR profiles were not incorrectly diagnosed as early stage EOC (see Supporting Information for details).
Metabolites were identified for which significant (p-value < 0.02) changes in concentrations are observed when comparing the averaged spectra from early stage EOC and healthy control specimens (see legend of Figure 1). Inspection of the loading plots of the principal components used to build the predictive model confirmed that the signals arising from these metabolites contribute significantly to class separation. Upon onset of early stage EOC, decreased concentrations are registered, for alanine (resonance lines contribute to PC 1 of the predictive model), CH3CH2CH2C= of lipid (mainly in very-low density lipoproteins, VLDL) (PC2), CH3(CH2)n of lipid (mainly in low-density lipoproteins, LDL) (PC2), valine (PC2), creatine/creatinine (PC2), choline of phospholipids (PC1), CH2CH2CH2CO of lipid (mainly in VLDL) (PC2) and =CHCH2CH2 of unsaturated lipid (PC2). On the other hand, higher concentrations are registered for β-hydroxybutyrate (PC1, 3, and 4), acetone (PC1, 3, and 4), and acetoacetate (PC1, 3, and 4). These preliminary findings can be qualitatively compared with concentration profile changes that were described for NMR-based metabonomic studies of serum specimens from patients with other types of cancer. As for early stage EOC, (i) lower VLDL and LDL serum concentrations were associated with human hepatocellular carcinoma and liver cirrhosis,20 (ii) lower alanine, valine and creatine serum concentrations were observed for oral cancer,21 and (iii) increased acetoacetate and β-hydroxybutyrate serum concentrations were associated with colorectal cancer.22 It has been suggested that increased ketone body concentrations in serum can be linked to lypolysis as an alternative route for energy production by tumor cells.22 It is evident that only a quantitative comparison can reveal to which extent which types of cancer are detected as false positives when a predictive model for a given type of cancer is employed. Remarkably, our current model for early stage EOC diagnosis did not identify patients with RCC as false positives, which is consistent with the fact that qualitatively different metabolite concentration changes were associated with RCC23 when compared with early stage EOC (e.g., the acetoacetate serum concentration was found to be lower than in healthy controls).
Conclusions
The detection of the early, asymptomatic invasive stage I/II of EOC has a profound impact on clinical outcome. While there are currently no screening strategies with proven efficacy for early stage EOC detection available, several ovarian cancer screening trials are on-going. Those are based on transvaginal ultrasound, or serum concentration of CA125 combined with transvaginal ultrasound as part of a multimodal screening strategy.24 Although the search for a single biomarker continues, it is more likely that either a panel of several biomarkers and/or a “fingerprint” of easily accessible biofluids will ultimately prove useful for early stage EOC detection.25 For example, the combination of six markers (leptin, prolactin, osteopontin, insulin-like growth factor 2, macrophage inhibitory factor and CA125) exhibited significantly better discrimination compared with CA125 alone.26
In a previous study, we demonstrated that 1H NMR-based metabonomics of serum allows accurate discrimination of advanced stage III/IV EOC and healthy women.27 This finding motivated us to initiate the current study focusing on early stage EOC. Considering that patients with early and advanced stage EOC likely exhibit quite different metabolic phenotypes, it is not surprising that the results of multivariate analyses of the NMR profiles differ (e.g., in terms of sensitivity and specificity of the statistical model). Furthermore, since the previous study focused on exploring the feasibility of profiling27 advanced stage EOC with little effort at metabolite identification, a comparison of changes in serum metabolite concentrations is rather limited. Nevertheless, the observation that β-hydroxybutyrate concentrations were found to be increased in both studies suggests that this change occurs early during development of ovarian cancer, and persists in patients with advanced disease.
Since only an early stage diagnosis can significantly impact on the clinical outcome of EOC patients, it is an important finding that 1H NMR-based metabonomics of serum also allows discrimination of stage I/II EOC and healthy women. For example, carriers of mutations in BRCA1 and BRCA2 genes are at high risk of EOC and develop ovarian cancer by age 70 with 18%–54% probability.28 Assuming a corresponding EOC prevalence of ~3%, we then obtain for the PPV lower bounds of 6%, 18% and 11%, respectively, for Test Set, Validation Set, and Combined Test/Validation Set (the corresponding sensitivities and specificities are provided along with their 95% CIs in Table S1). Given the fact that the combined set comprises the largest number of specimens and that even the corresponding lower bound of the PPV is around 10%, we conclude that our current predictive model might be suitable for populations at high risk of EOC. The potential of NMR-based metabonomics to discriminate body fluids from cancer patients and healthy individuals has by now been proven for several types of cancer. However, our finding that the false positive rate for EOC diagnosis does not significantly increase for women diseased with RCC indicates, to the best of our knowledge for the first time, that NMR-based metabonomics also bears great promise for cancer-type specific diagnosis.
Taken together, our pilot study paves the way for future research focusing on (i) increasing predictive accuracy to obtain a model suited for the general population, which may result from increased spectrometer sensitivity and/or recording other types of NMR experiments,8 (ii) the calculation of composite predictive models relying on both NMR profile and other EOC biomarker concentrations such as CA125, HE-4 and/or mesothelin,29 (iii) testing and refinement of models in large prospective cohort studies,7 and (iv) obtaining new insights into EOC associated changes of the metabolic phenotype, which may potentially open new avenues to target tumor metabolism for EOC treatment.30
Supplementary Material
Acknowledgments
This work was supported by the John Oshei and Roswell Park Alliance Foundations (to TS and KO), and the U.S. National Institutes of Health (R21CA106949-02 to KO). We are indebted to Drs. Jeremy K. Nicholson and Hector Keun, Imperial College, UK, for helpful discussions.
Footnotes
Supporting Information Available: clinical characteristics of EOC/RCC patients, additional plots of NMR spectra, additional information on multivariate data analysis, and Tables S1 and S2. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Armstrong DK, Bundy B, Wenzel L, Huang HQ, Baergen R, Lele S, Copeland LJ, Walker JL, Burger RA. Intraperitoneal cisplatin and paclitaxel in ovarian cancer. N Engl J Med. 2006;354:34–43. doi: 10.1056/NEJMoa052985. [DOI] [PubMed] [Google Scholar]
- 2.Bookman MA, Brady MF, McGuire WP, Harper PG, Alberts DS, Friedlander M, Colombo N, Fowler JM, Argenta PA, De Geest K, Mutch DG, Burger RA, Swart AM, Trimble EL, Accario-Winslow C, Roth LM. Evaluation of new platinum-based treatment regimens in advanced-stage ovarian cancer: a phase III trial of the Gynecologic Cancer Intergroup. J Clin Oncol. 2009;27:1419–1425. doi: 10.1200/JCO.2008.19.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Greenlee RT, Hill-Harmon MB, Murray T, Thun M. Cancer statistics, 2001. CA Cancer J Clin. 2001;51:15–36. doi: 10.3322/canjclin.51.1.15. [DOI] [PubMed] [Google Scholar]
- 4.Young RC, Walton LA, Ellenberg SS, Homesley HD, Wilbanks GD, Decker DG, Miller A, Park R, Major F., Jr Adjuvant therapy in stage I and stage II epithelial ovarian cancer. Results of two prospective randomized trials. N Engl J Med. 1990;322:1021–1027. doi: 10.1056/NEJM199004123221501. [DOI] [PubMed] [Google Scholar]
- 5.Jacobs I. Overview-progress in screening for ovarian cancer. In: Sharp F, Blackett A, Berek J, Bast R, editors. Ovarian Cancer. Vol. 5. Oxford: Isis Medical Media Ltd; 1998. pp. 173–185. [Google Scholar]
- 6.Buys SS, Partridge E, Greene MH, Prorok PC, Reding D, Riley TL, Hartge P, Fagerstrom RM, Ragard LR, Chia D, Izmirlian G, Fouad M, Johnson CC, Gohagan JK. Ovarian cancer screening in the Prostate, Lung, Colorectal and Ovarian (PLCO) cancer screening trial: findings from the initial screen of a randomized trial. Am J Obstet Gynecol. 2005;193:1630–1639. doi: 10.1016/j.ajog.2005.05.005. [DOI] [PubMed] [Google Scholar]
- 7.Partridge E, Kreimer AR, Greenlee RT, Williams C, Xu JL, Church TR, Kessel B, Johnson CC, Weissfeld JL, Isaacs C, Andriole GL, Ogden S, Ragard LR, Buys SS. Results from four rounds of ovarian cancer screening in a randomized trial. Obstet Gynecol. 2009;113:775–782. doi: 10.1097/AOG.0b013e31819cda77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lindon JC, Nicholson JK, Holmes E. The Handbook of Metabonomics and Metabolomics. Amsterdam: Elsevier; 2007. [Google Scholar]
- 9.Nicholson JK, Lindon JC, Holmes E. ‘Metabonomics’: understanding the metabolic responses of living systems to pathophysiological stimuli via multivariate statistical analysis of biological NMR spectroscopic data. Xenobiotica. 1999;29:1181–1189. doi: 10.1080/004982599238047. [DOI] [PubMed] [Google Scholar]
- 10.Lindon JC, Nicholson JK, Everett JR. NMR spectroscopy of biofluids. Annu Rep NMR Spectrosc. 1999;38:1–88. [Google Scholar]
- 11.Lindon JC, Nicholson JK, Holmes E, Everett JR. Metabonomics: metabolic processes studied by NMR spectroscopy of biofluids. Concepts Magn Reson. 2000;12:289–320. [Google Scholar]
- 12.Nicholson JK, Connelly J, Lindon JC, Holmes E. Metabonomics: a platform for studying drug toxicity and gene function. Nat Rev Drug Discov. 2002;1:153–161. doi: 10.1038/nrd728. [DOI] [PubMed] [Google Scholar]
- 13.Ambrosone CB, Nesline MK, Davis W. Establishing a cancer center data bank and biorepository for multidisciplinary research. Cancer Epidemiol Biomarkers Prev. 2006;15:1575–7. doi: 10.1158/1055-9965.EPI-06-0628. [DOI] [PubMed] [Google Scholar]
- 14.Sukumaran D, Garcia E, Hua J, Tabaczynski W, Odunsi K, Andrews C, Szyperski T. Standard operating procedure for metabonomics studies of blood serum and plasma samples using a 1H-NMR micro-flow probe. Magn Reson Chem. 2009;47 (Suppl 1):S81–85. doi: 10.1002/mrc.2469. [DOI] [PubMed] [Google Scholar]
- 15.Beckonert O, Keun HC, Ebbels TM, Bundy J, Holmes E, Lindon JC, Nicholson JK. Metabolic profiling, metabolomic and metabonomic procedures for NMR spectroscopy of urine, plasma, serum and tissue extracts. Nat Protoc. 2007;2:2692–2703. doi: 10.1038/nprot.2007.376. [DOI] [PubMed] [Google Scholar]
- 16.Nicholson JK, Foxall PJD, Spraul M, Farrant RD, Lindon JC. 750 MHz 1H and 1H-13C NMR spectroscopy of human blood plasma. Anal Chem. 1995;67:793–811. doi: 10.1021/ac00101a004. [DOI] [PubMed] [Google Scholar]
- 17.Lindon JC, Nicholson JK, Everette JR. NMR spectroscopy of biofluids. Ann Rep NMR Spectrosc. 1999;38:1–88. [Google Scholar]
- 18.Cloarec O, Dumas M, Craig A, Barton RH, Trygg J, Hudson J, Blancher C, Gauguier D, Lindon JC, Holmes E, Nicholson J. Statistical Total Correlation Spectroscopy: An Exploratory Approach for Latent Biomarker Identification from Metabolic 1H NMR Data Sets. Anal Chem. 2005;77:1282–1289. doi: 10.1021/ac048630x. [DOI] [PubMed] [Google Scholar]
- 19.Steyerberg EW. Clinical Prediction Models: A Practical Approach the Development, Validation, and Updating. New York: Springer; 2009. [Google Scholar]
- 20.Gao H, Lu Q, Liu X, Cong H, Zhao L, Wang H, Lin D. Application of 1H NMR-based metabonomics in the study of metabolic profiling of human hepatocellular carcinoma and liver cirrhosis. Cancer Sci. 2009;100:782–785. doi: 10.1111/j.1349-7006.2009.01086.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tiziani S, Lopes V, Günther UL. Early stage diagnosis of oral cancer using 1H NMR-based metabolomics. Neoplasia. 2009;11:269–276. doi: 10.1593/neo.81396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ludwig C, Ward DG, Martin A, Viant MR, Ismail T, Johnson PJ, Wakelam MJO, Günther UL. Fast targeted multidimensional NMR metabolomics of colorectal cancer. Magn Reson Chem. 2009;47:S68–S73. doi: 10.1002/mrc.2519. [DOI] [PubMed] [Google Scholar]
- 23.Zira AN, Theocharis SE, Mitropoulos D, Migdalis V, Mikros E. 1H NMR metabonomics analysis in renal cell carcinoma: a possible diagnostic tool. J Proteome Res. 2010;9:4038–4044. doi: 10.1021/pr100226m. [DOI] [PubMed] [Google Scholar]
- 24.Menon U, Gentry-Maharaj A, Hallett R, Ryan A, Burnell M, Sharma A, Lewis S, Davies S, Philpott S, Lopes A, Godfrey K, Oram D, Herod J, Williamson K, Seif MW, Scott I, Mould T, Woolas R, Murdoch J, Dobbs S, Amso NN, Leeson S, Cruickshank D, Mcguire A, Campbell S, Fallowfield L, Singh N, Dawnay A, Skates SJ, Parmar M, Jacobs I. Sensitivity and specificity of multimodal and ultrasound screening for ovarian cancer, and stage distribution of detected cancers: results of the prevalence screen of the UK Collaborative Trial of Ovarian Cancer Screening (UKCTOCS) Lancet Oncol. 2009;10:327–340. doi: 10.1016/S1470-2045(09)70026-9. [DOI] [PubMed] [Google Scholar]
- 25.Kulasingam V, Pavlou MP, Diamandis EP. Integrating high-throughput technologies in the quest for effective biomarkers for ovarian cancer. Nat Rev Cancer. 2010;10:371–378. doi: 10.1038/nrc2831. [DOI] [PubMed] [Google Scholar]
- 26.Visintin I, Feng Z, Longton G, Ward DC, Alvero AB, Lai Y, Tenthorey J, Leiser A, Flores-Saaib R, Yu H, Azori M, Rutherford T, Schwartz PE, Mor G. Diagnostic markers for early detection of ovarian cancer. Clin Cancer Res. 2008;14:1065–1072. doi: 10.1158/1078-0432.CCR-07-1569. [DOI] [PubMed] [Google Scholar]
- 27.Odunsi K, Wollman RM, Ambrosone CB, Hutson A, McCann SE, Tammela J, Geisler JP, Miller G, Sellers T, Cliby W, Qian F, Keitz B, Intengan M, Lele S, Alderfer JL. Detection of epithelial ovarian cancer using 1H-NMR-based metabonomics. Int J Cancer. 2005;113:782–8. doi: 10.1002/ijc.20651. [DOI] [PubMed] [Google Scholar]
- 28.Antoniou A, Pharoah PD, Narod S, Risch HA, Eyfjord JE, Hopper JL, Loman N, Olsson H, Johannsson O, Borg A, Pasini B, Radice P, Manoukian S, Eccles DM, Tang N, Olah E, Anton-Culver H, Warner E, Lubinski J, Gronwald J, Gorski B, Tulinius H, Thorlacius S, Eerola H, Nevanlinna H, Syrjäkoski K, Kallioniemi OP, Thompson D, Evans C, Peto J, Lalloo F, Evans DG, Easton DF. Average risks of breast and ovarian cancer associated with BRCA1 and BRCA2 mutations detected in case series unselected for family history: a combined analysis of 22 studies. Am J Hum Genet. 2003;72:1117–1130. doi: 10.1086/375033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anderson GL, McIntosh M, Wu L, Barnett M, Goodman G, Thorpe JD, Bergan L, Thornquist MD, Scholler N, Kim N, O’Briant K, Drescher C, Urban N. Assessing lead time of selected ovarian cancer biomarkers: a nested case-control study. J Natl Cancer Inst. 2010;102:26–38. doi: 10.1093/jnci/djp438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Merida I, Avila-Flores A. Tumor metabolism: new opportunities for cancer treatment. Clin Transl Ocol. 2006;8:711–716. doi: 10.1007/s12094-006-0117-6. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.