Abstract
BACKGROUND
Considering that approximately 2% of Caucasian controls host rare, nonsynonymous variants in the SCN5A-encoded cardiac sodium channel, caution must be exercised when interpreting SCN5A genetic test results for long QT syndrome (LQTS).
OBJECTIVE
The purpose of this study was to determine if A572D-SCN5A is a pathogenic mutation, a possible functional modifier, or background “genetic noise.”
METHODS
The frequency of A572D was compared between 3,741 LQTS referral cases (mostly Caucasian) and 1,437 Caucasian controls. A572D-SCN5A was engineered into SCN5A using the most commonly spliced transcript (Q1077del, hH1c clone) in the setting of either H558 or R558 for heterologous expression/patch clamp studies in HEK293 cells.
RESULTS
A572D-SCN5A was detected in 17 (0.45%) of 3,741 cases compared with 7 (0.49%) of 1,437 controls (P = .82). Among the 17 A572D-positive LQTS referrals, 10 (59%) hosted definite LQTS-causing mutations elsewhere (5 KCNQ1, 3 KCNH2, 2 SCN5A). Functional studies showed no gating kinetic or current density differences compared with wild-type channels in the context of H558 but showed moderate dysfunction when expressed in H558R-SCN5A, with which it is invariably associated.
CONCLUSION
There is sufficient evidence to conclude that A572D-SCN5A is not an independent, LQT3-causative mutation. A572D is present in approximately 0.5% of both cases and controls and has a wild-type phenotype when expressed in HEK293 cells. However, in the context of H558R-SCN5A, persistent late sodium current emerges, indicating that A572D/H558R could be a proarrhythmic factor akin to S1103Y. These findings underscore the scrutiny necessary to distinguish truly pathogenic mutations from functional polymorphisms and otherwise innocuous, rare genetic variants in SCN5A. These results also question how much cellular dysfunction for a mutation is required in vitro to support pathogenicity.
Keywords: Genetic testing, Long QT syndrome, Mutation, SCN5A
Introduction
Long QT syndrome (LQTS), which affecting 1 in 2,500 persons,1 comprises a distinct group of cardiac channelopathies characterized by delayed repolarization of the myocardium, QT prolongation, and increased risk for syncope, seizures, and sudden cardiac death in the setting of a structurally normal heart. Since the sentinel discovery of the pathogenic basis for LQTS in 1995 and the decade of research-based genetic testing from 1995 to 2004, LQTS genetic testing has matured and is recognized as a bona fide clinical test with diagnostic, prognostic, and therapeutic implications. Through genotype–phenotype correlation studies, gene-specific risk stratification algorithms and gene-specific treatments have been developed for LQTS.2–4
Approximately 75% of LQTS is due to mutations in three major genes encoding for ion channel α-subunits that are critically responsible for orchestrating the cardiac action potential: KCNQ1-encoded IKs potassium channel5 (LQT1), KCNH2-encoded IKr potassium channel6 (LQT2), and SCN5A-encoded INa sodium channel7 (LQT3). However, comprehensive open reading frame mutational analysis of these genes in more than 1,300 ostensibly healthy controls has revealed that approximately 3% to 5% of Caucasians and a similar proportion of African Americans, Asians, and Hispanics host rare nonsynonymous variants (<0.5% allelic frequency), with about half of these rare variants residing in SCN5A.8,9
Mutations in the SCN5A-encoded cardiac sodium channel Nav1.5 are responsible for type 3 long QT syndrome (LQT3), type 1 Brugada syndrome (BrS1), and 50% of channelopathic cases of sudden infant death syndrome. One particular SCN5A variant, A572D, has been reported as an LQT3-causing mutation,10,11 recently identified in a postmortem investigation of sudden deaths,12 and functionally characterized in Xenopus oocytes as having a shorter recovery time from inactivation compared to wild-type channels.12 However, A572D resides in what may be considered a “biologically flexible” portion of Nav1.5 (interdomain linker I–II), as evidenced by the plethora of rare SCN5A variants observed in healthy individuals that localize to this specific region of the channel.8,9
Distinguishing authentic pathogenic mutations from otherwise innocuous background variants is of critical importance considering the established and emerging diagnostic, prognostic, and therapeutic implications of a “positive” LQTS genetic test. The presence of a genetic mutation alone cannot supplant clinical evidence, and, even in robust cases of LQTS, a positive genetic test result must be interpreted carefully. As a case in point, we chose to investigate whether A572D-SCN5A is a “pathogenic mutation” responsible for a 1:25,000 condition known as LQT3, a “functional modifier,” or merely “background genetic noise.”
Methods
Case-control study design
From 1997 to 2008, 3,741 unrelated LQTS referral patients were submitted for SCN5A genetic testing to either Mayo Clinic’s Windland Smith Rice Sudden Death Genomics Laboratory (Rochester, MN, USA), the Cardiogenetic Clinic at the Academic Medical Center (Amsterdam, The Netherlands), or PGxHealth (Division of Clinical Data, Inc., New Haven, CT, USA).
The observed frequency of A572D-SCN5A in cases was compared with the frequency in 1,437 unrelated, ostensibly healthy control subjects. The controls consisted of 957 Caucasian healthy volunteers from the United States (US) analyzed previously for SCN5A channel variants by Mayo Clinic and PGxHealth and 480 randomly selected United Kingdom (UK) Caucasian blood donors from the European Collection of Cell Cultures (ECACC), a Health Protection Agency Cell Culture Collection. An ECG showing a normal QT interval was not always a prerequisite for subjects composing this control cohort.
A572D-SCN5A genotyping
The genomic DNA of all cases and controls was analyzed for the presence of A572D-SCN5A using either direct DNA sequencing or a combination of denaturing high-performance liquid chromatography and automated DNA sequencing.
H558R and A572D haplotyping
Because the common SCN5A polymorphism H558R has been shown previously to modify the phenotype of other SCN5A variants, H558R status was determined for all available cases and controls. Further, all available A572D/H558R double heterozygote cases and controls (n = 7) were haplotyped to determine if D572 existed on the same allele as H558 or R558. Following polymerase chain reaction (PCR) amplification of SCN5A exon 12 using the forward (GCCAGTGGCACAAAAGACAGGCT) and reverse primers (GGAACTGCTGATCAGTTTGGGAGA), the PCR products were digested with the restriction enzyme Tsp45I (New England BioLabs, Ipswich, MA, USA), which cleaves only the D572 allele. Digested products are size separated by gel electrophoresis on a 2% agarose gel. A sample heterozygote for A572D will yield bands of 518 bp (representing the noncleaved A572 allele), 275 bp (representing the 5′ end of the D572 allele; containing the nucleotides of codon 558), and 243 bp (representing the 3′ end of the D572 allele) on the agarose gel. The 275-bp fragment is excised from the gel, purified using the Qiaquick gel extraction kit (Qiagen Sciences, Germantown, MD, USA), and DNA sequenced.
A572D site-directed mutagenesis and transfection of cell lines
A572D-SCN5A was engineered into SCN5A with each allele of the common polymorphism H558R using the most commonly spliced transcript13 (Q1077del, Accession No. AY148488) by site-directed mutagenesis (ExSite, Stratagene, La Jolla, CA) using the pcDNA3 vector (Invitrogen, Carlsbad, CA, USA). The SCN5A-wild type constructs (A572/H558 or A572/R558) and the rare variant SCN5A-A572D (referred to as D572/H558 or D572/R588) were confirmed by DNA sequencing analysis (Biotech Center of the University of Wisconsin-Madison) and transiently co-transfected with green fluorescent protein at a 1:5 ratio into HEK293 cells using Fugene (Roche Diagnostics, Indianapolis, IN, USA).
Electrophysiologic characterization
INa was measured from HEK293 cells expressing A572/ H558, D572/H558, A572/R558, or D572/R558. The voltage-clamp protocols are described briefly with the data and have been published previously in detail. The bath (extracellular) solution (pH 7.4 set with NaOH) contained the following (in mM): NaCl 140, KCl 4, CaCl 1.8, MgCl 0.75, and HEPES 5. The pipette (intracellular) solution (pH 7.4 set with CsOH) contained the following (in mM): CsF 120, CsCl 20, EGTA 2, and HEPES 5. Electrodes of resistance 1 to 2 MΩ were made from borosilicate glass using a puller (P-87, Sutter Instrument Co., Novato, CA, USA). Electro-physiologic recordings were carried out at room temperature. Cells were perfused continuously with the bath solution, and voltage-clamp protocols were generated using pClamp software (version 9.0, Axon Instruments, Foster City, CA, USA). The currents were normalized to cell capacity and reported as pA/pF.
Results
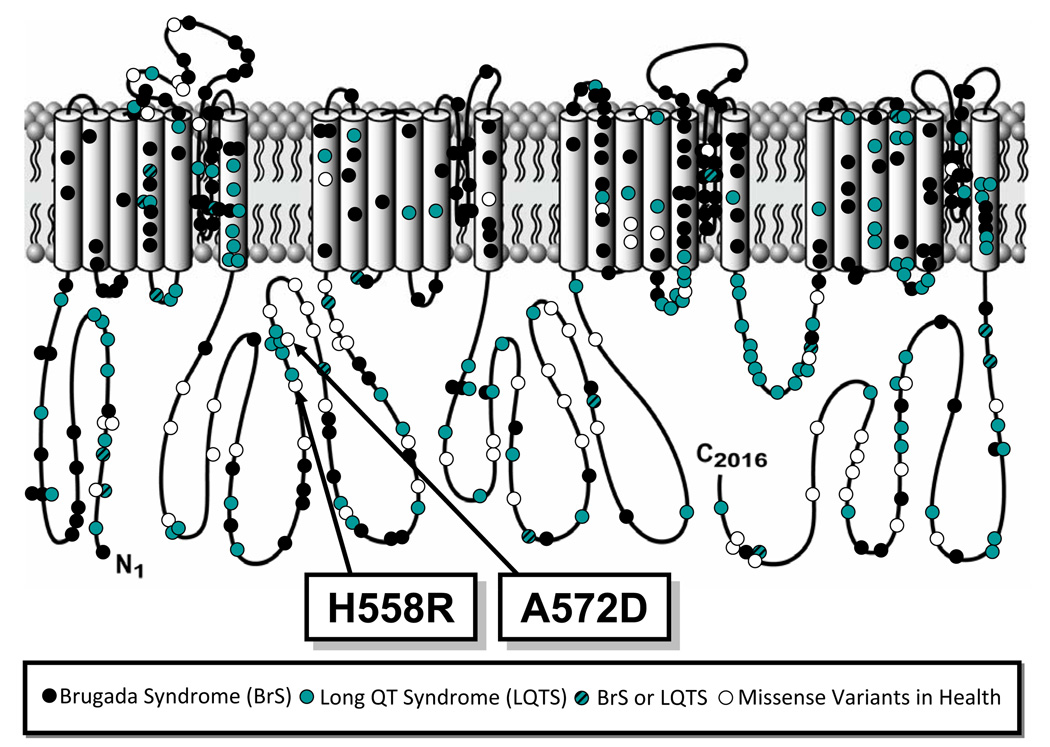
The genetic variant A572D results from a cytosine (C) to alanine (A) substitution at nucleotide position 1715 (1715 c>a), which exchanges a small nonpolar hydrophobic amino acid residue (alanine, A) with a negatively charged acidic amino acid residue (aspartic acid, D) at amino acid position 572. A572 localizes to the DI–DII interdomain linker of Nav1.5 (Figure 1). It is conserved in primates but is not conserved across other species.
Figure 1.
Linear topology of SCN5A-encoded cardiac sodium channel with relative location of A572D along with variants identified in long QT syndrome, Brugada syndrome, and healthy subjects.
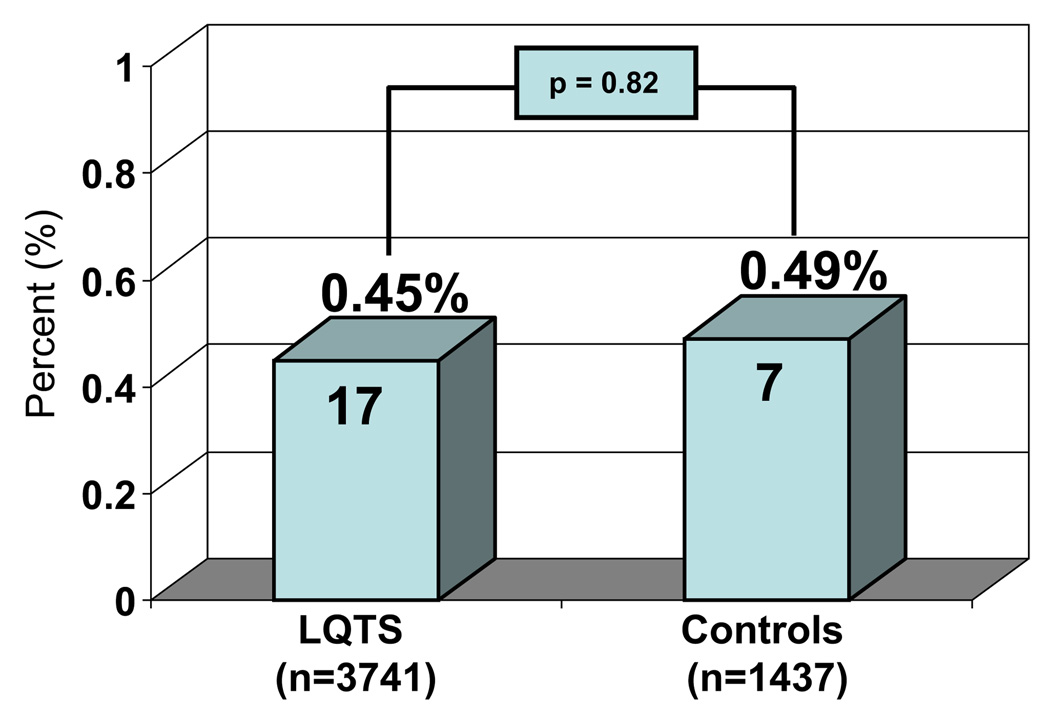
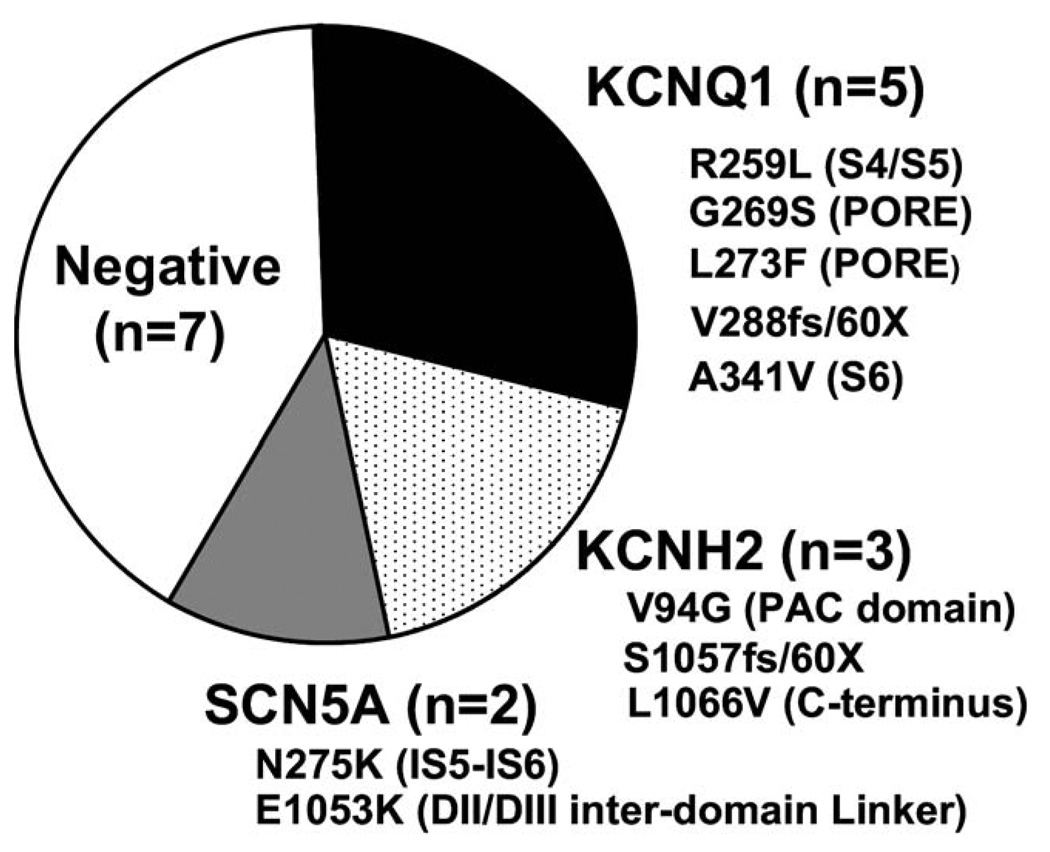
Overall, 17 (0.45%) of 3,741 LQTS referral cases were heterozygous for A572D (Figure 2). In comparison, 7 (0.49%) of 1,437 Caucasian controls hosted A572D (P = .82; Figure 2). Moreover, among the 17 A572D-positive LQTS referrals, 10 (59%) hosted high-probability deleterious, LQTS-causing mutations [5 in KCNQ1 (LQT1): R259L (S4/S5 transmembrane linker), G269S (Pore), L273F (Pore), A287fs/59X (S5/Pore transmembrane frame-shift/premature truncation), and A341V (S6 transmembrane); 3 in KCNH2 (LQT2): V94G (N-terminus PAC domain), S1057fs/60 (c-terminus frameshift/premature truncation) and L1066V (C-terminus); and 2 in SCN5A (LQT3): N275K (DI-S5 transmembrane) and E1053K (DII/DIII inter-domain linker); Figure 3].
Figure 2.
Relative frequency of A572D among 3,741 long QT syndrome patients and 1,437 ostensibly healthy controls. Numbers in the bars represent the number of unrelated cases/controls heterozygous for A572D.
Figure 3.
Spectrum and prevalence of additional LQTS-causing mutations among A572D-SCN5A positive index cases. Distribution of additional putative pathogenic mutations identified among the 17 A572D-positive LQTS referral cases is shown. Listed under the gene name are the unique mutations with their topologic channel location shown in parentheses.
Because the common SCN5A polymorphism H558R can have a significant modifying effect on the sodium channel, we assessed the H558R polymorphism status for each A572D-positive case and control. Interestingly, all 17 cases and all 7 controls with A572D were identified as having at least one R558 allele (H558R heterozygotes: 10 cases and 6 controls; R558 homozygotes: 7 cases and 1 control). For those cases and controls that were heterozygous for H558R and for which DNA was available (n = 7, 1 case and 6 controls), we determined whether D572 and R558 were on the same (“cis”) or opposite (“trans”) alleles and found that in all cases and controls tested, D572 and R558 were in fact in “cis” strongly suggesting that D572 is invariably linked to R558. This tight linkage disequilibrium is not surprising given their close physical genetic spacing. Although it appears that D572 always occurs on the same allele as R558, we still functionally characterized D572 in both H558- and R558-containing constructs.
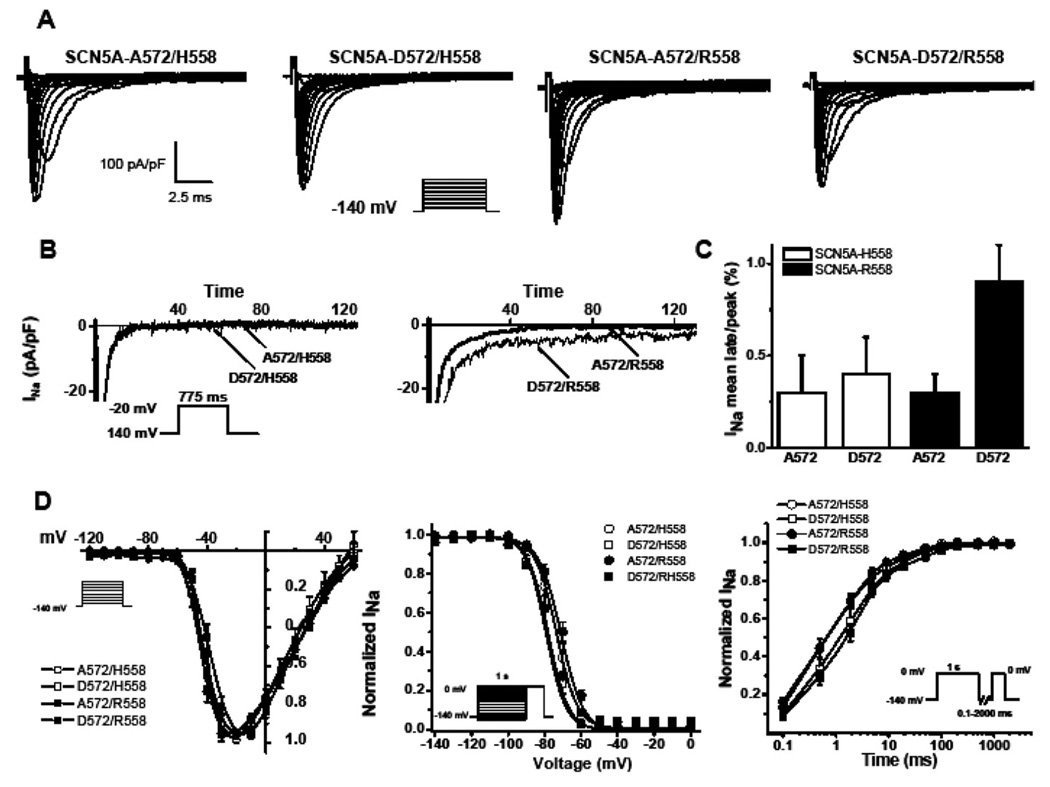
SCN5A-WT splice variant Q1077 del, containing either a histidine (H) at amino acid residue 558 or an arginine (R) at 558, was used to engineer the A572D (D572) variant and expressed in HEK293 cells. Examples of INa traces in response to different step voltages from a holding potential of −140 mV are shown in Figure 4A, and summary data are given in Table 1. When D572 was studied in the presence of H558, no significant differences from wild type were observed. However, when D572 was studied in the presence of R558, significant differences in kinetics were found compared with either A572/R558 or A572/H558 (P <.05). Peak INa was not statistically different among the different backgrounds; however, D572/R558 tends to express about 30% to 40% less INa than D572/H558 channels (Figure 4A and Table 1). Although D572/H558 behaved like wild type, D572/R558 precipitated a threefold increase in late/persistent sodium current compared to A572/R558 (P <.05; Table 1 and Figures 4B and 4C). We observed that D572 negatively shifted the steady-state inactivation, reaching significance for D572/R558 (P <.05; Figure 4D and Table 1) without affecting the activation parameters. Also, slower time constants of recovery from inactivation were observed for D572/R558 (Figure 4D and Table 1). These results suggest that even though D572 in the background of H558 functions normally, the presence of R558 in “cis” with D572 produced channels with altered, moderate LQT3-like gain-of-function properties in vitro.
Figure 4.
A572D-SCN5A functional data in HEK293 cells. A: Whole-cell Na current (INa) from representative experiments for D572 with or without the common polymorphism R558. Activation of sodium channels was measured at different voltage steps −120 mV to +50 mV with a 10-mV step at holding potential of −140 mV. B: Late INa after leak subtraction was measured as the mean current between 600 and 700 ms elicited by depolarization from −140 mV to −20 mV. C: Summary data showing the ratio of late INa to peak INa as a percentage from several cells. D: Protocols used to address the kinetics of INa represented as inset in each plot. Data obtained from the activation protocol to establish peak INa were fitted to a Boltzmann equation to obtain the midpoint of activation. Inactivation was measured by a two-step protocol, with a holding potential of −140 mV and 1-second conditioning pulse from −150 to 0 mV with a 10-mV step and fitted to a Boltzmann equation to obtain the midpoint of inactivation. Recovery from inactivation was measured using a two-pulse protocol where a conditioning step of 1 second to 0 mV inactivated INa, followed by a test pulse to 0 mV after a different recovery period at a recovery potential of −140 mV. Data were normalized to peak and fitted to an exponential to obtain fast and slow time constants (τf and τs) and the proportion of the slow component (As) (Table 1). *P <0.5 vs A572/H558, D572/H558, A572/R558.
Table 1.
Kinetics of a rare variant A572D with or without the common polymorphism H558R
| Samples (variant Q1077 del) |
Peak INa | Activation | Inactivation | Recovery | INaL | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| pA/pF | n | V1/2 (mV) | n | V1/2 (mV) | n | τf (ms) | τs (ms) | As (%) | n | % | n | |
| SCN5A-A572/H558 (WT) | −309 ± 54 | 5 | −42 ± 2 | 4 | −72 ± 1 | 5 | 0.9 ± 0.1 | 23 ± 4 | 25± 3 | 5 | 0.4 ± 0.03 | 4 |
| SCN5A-D572/H558 | −296 ± 52 | 13 | −42 ± 2 | 8 | −75 ± 1 | 10 | 1.7 ± 0.3 | 30 ± 7 | 24 ± 3 | 10 | 0.4 ± 0.02 | 7 |
| SCN5A-A572/R558 (WT) | −347 ± 51 | 10 | −43 ± 2 | 7 | −70 ± 1† | 10 | 1.1 ± 0.1 | 22 ± 25 | 21 ± 5 | 9 | 0.3 ± 0.1 | 8 |
| SCN5A-D572/R558 | −207 ± 55 | 9 | −39 ± 2 | 8 | −76 ± 2* | 8 | 1.7 ± 0.2* | 52 ± 14* | 20 ± 1 | 8 | 0.9 ± 0.3* | 7 |
τf, τs = fast and slow time constants, respectively; As = slow component; WT = wild type.
P < .05 vs – SCN5A-A572/R558.
P < .05 vs – SCN5A-D572/H558 or D572/R558.
Discussion
The cardiac sodium channel missense mutation A572D originally was designated as a probable LQT3-causing mutation largely based solely on its absence in ostensibly healthy controls. First reported by Paulussen et al10 in 2003, A572D was identified in a 10-year-old boy who had experienced multiple syncopal events, QTc of 483 ms, and positive paternal family history of sudden unexplained death in an uncle at age 8 years and multiple childhood syncopal events and prolonged QTc in a grandmother. Besides A572D-SCN5A, however, the investigators also identified an LQT1-associated missense mutation, V254M-KCNQ1, in the index case. Both variants were absent in 90 ethnic matched control subjects; therefore, both were considered most likely pathogenic. However, whereas the LQT1 mutation V254M co-segregated properly with the paternal family history of cardiac events, A572D was determined instead to be maternally derived and present in clinically normal individuals.
In 2005, we reported A572D as a putative pathogenic mutation due to its presence in 3 (0.5%) of 541 unrelated Caucasian LQTS referral cases and its absence in 295 Caucasian, 319 African American, 112 Asian, and 103 Hispanic ostensibly healthy controls.11 Following these sentinel reports, mounting epidemiologic, molecular, and functional evidence now suggests that A572D can not be viewed as a standalone LQT3-causative mutation but instead is invariably linked to the common polymorphism H558R, where it may confer proarrhythmic susceptibility secondary to its accentuated late/persistent sodium current properties, thereby joining S1103Y as a possible “functional polymorphism” in SCN5A. Unlike S1103Y, however, no epidemiologic data to date suggest that A572D has any S1103Y-like, proarrhythmia susceptibility for which an evidence-based recommendation of QT drug avoidance can be given.
We report here that of the 3,741 cases referred for LQTS genetic testing, overall 17 (0.45%) hosted A572D-SCN5A, with 10 (59%) of the 17 having high-probability deleterious channel mutations elsewhere compared to 7 (0.49%) of 1,437 controls. Interestingly, there appears to be a wide disparity in regional frequencies of A572D among the control populations. For example, the frequency of A572D heterozygotes among reported North American controls is 0.09% compared to 2.3% (4/184) in Norwegian controls14 and 6% among more than 5,000 Finnish controls.15 This observation is true even among the controls analyzed here, where none of the 957 US controls hosted A572D compared with 7 (1.5%) of the 480 UK control subjects (P <.0005).
Considering the recent estimated incidence of LQTS in the general population at 1:2,500 (0.04%)1 Caucasian individuals and recognizing that less than 10% of LQTS cases are secondary to pathogenic mutations in SCN5A (LQT3), this implies that approximately 1:25,000 Caucasians have LQTS secondary to a rare, pathogenic mutation in SCN5A. Put another way, if A572D were pathogenic, its penetrance would have to be near zero, meaning it almost never causes LQTS. Accordingly, this rare SCN5A variant, A572D, present in 1:200 of a mixed Caucasian population sample and up to 1:17 Finnish persons, simply cannot be an independent, LQT3-causing pathogenic mutation.
Previous reports on the compendium of rare genetic variants identified in more than 1,300 ostensibly healthy individuals for ion channel genes associated with LQTS have indicated a “background genetic noise rate” of 4% among Caucasians and up to 8% among non-Caucasians, with the majority of variants occurring in SCN5A and in a single control.8 This indicates that rarity alone does not necessarily equal pathogenicity and that careful interpretation of SCN5A genetic variants is crucial to the clinical management of patients. The observed preponderance of unique and rare SCN5A missense variants, over 50% of which localize to these interdomain linkers, identified in healthy controls suggests that some of the missense mutations identified in cases within the DI–DII and DII–DIII linker domains may in fact represent “false-positives,” and our data suggest that A572D is one of them.
The variant A572D, a seemingly radical amino acid substitution of a small nonpolar hydrophobic residue (alanine) for a larger amino acid with a hydrophilic negatively charged acidic side chain (aspartic acid), is relatively non-conserved across species and resides in what may be considered a “biologically flexible” portion of Nav1.5’s inter-domain linker I–II, as evidenced by the number of rare SCN5A variants that reside in this area of the channel observed in healthy individuals. Unfortunately, this variable region of SCN5A has not been studied in depth functionally, and the prevalence of rare variants observed in controls creates a conundrum for mutation calling efforts. Simply put, one cannot reliably determine if the next novel DI–DII and DII–DIII linker domain variant identified in a case should be considered pathogenic or benign. Although it would be a major undertaking, a systematic functional analysis of all DI–DII and DII–DIII linker domain variants observed in cases or controls would be most helpful in establishing benchmark criteria for the assignment of pathogenicity based on functional data derived from heterologous expression systems.
The presence of a genetic mutation alone cannot supersede clinical evidence, and, even in phenotypically strong cases of LQTS, a positive genetic test result must be interpreted carefully. Ethnicity, mutation type, and mutation location, particularly channel structure–function domains, are critical determinants of a particular mutation’s probability of pathogenicity in the three major LQTS-causing genes.9 Distinguishing rare, pathogenic mutations from similarly rare but otherwise innocuous variants of uncertain functional/clinical significance is of the utmost importance in the age of clinical diagnostic genetic testing for arrhythmia syndromes such as LQTS, considering the established and emerging diagnostic, prognostic, and therapeutic implications of a “positive” LQTS genetic test.9 Overinterpretation of a “positive” LQTS test may lead to unnecessary and invasive therapies and/or the introduction of improper gene-directed therapies. A case in point: several family members, interpreted elsewhere to have LQT3 based in part on a “positive” genetic test result for A572D-SCN5A, already received an implantable cardioverter-defibrillator prior to their second opinion evaluation at the Mayo Clinic.
Various nonsynonymous polymorphisms of the cardiac sodium channel have been described previously as having either a modifying role on the disease or as conferring increased risk for potentially lethal arrhythmias in the setting of exogenous or endogenous factors. For example, the common cardiac sodium channel polymorphism S1103Y, which is present in approximately 13% of African Americans, produces sodium channels that have a wild-type electrophysiologic phenotype under normal conditions but has a persistent late sodium current (LQT3-like) phenotype under conditions mimicking cellular acidosis and has been shown to be overrepresented in African American arrhythmia cases, adolescent and adult cases of autopsy-negative sudden unexplained death, and sudden infant death syndrome.16–18
A572D/H558R could conceivably confer proarrhythmic susceptibility as a functionally significant rare polymorphism seen in less than 1% of Caucasians akin to this far more common African American SCN5A polymorphism, S1103Y. Recently, two studies examining the role of ion channel genetic variants in either the development of life-threatening torsades de pointes dysrhythmia or their association with sudden cardiac death identified A572D in their cohorts. In a study by Mank-Seymour et al,19 A572D seemed overrepresented in proarrhythmic cases, being identified in approximately 3% (1/34) of European torsades de pointes–positive congestive heart failure or myocardial infarction cases compared with approximately 0.7% (4/555) of European controls. Similarly, Albert et al12 identified A572D in 1.6% (1/60) of female sudden cardiac death victims compared with 0.27% (2/733) of controls. However, the findings of both studies did not achieve statistical significance. Marjamaa et al15 recently reported a study on QT-interval duration and common LQTS gene variant association in 5,043 Finnish individuals. For the 6% of the Finnish population hosting A572D, there was no significant difference in QT-interval duration among heterozygote or homozygote carriers compared to noncarriers, suggesting that A572D was unlikely to modify the resting QT interval. However, extrinsic stresses such as exercise, hypokalemia, ischemic or structural heart disease, or drug exposure were not tested.
Because the common SCN5A polymorphism H558R (heterozygous frequency of 30% in healthy Caucasians) can have a profound functional modifying effect on other sodium channel mutations, we assessed H558R status in both A572D-positive cases and controls where DNA was available. All of the 17 A572D-positive subjects in this study also harbored at least one allele containing the common SCN5A R558 polymorphism, with 40% being homozygous for R558. For those cases and controls that were heterozygous for H558R and for which DNA was available, we confirmed that D572 and R558 were in fact in “cis” (i.e., residing on the same chromosome), as the observational data suggested. Furthermore, among a population of 6,200 Finnish subjects, all 7 D572 homozygotes identified were also found to be homozygous for R558, and all 394 subjects heterozygous for A572D had at least one R558 allele (either H558R or homozygous for R558; Dr. Annukka Marjamaa, Personal Communication, March 2010.). This pattern is consistent with the D572 variant having originated once as a point mutation on a chromosome containing R558. Because the DNA alterations resulting in R558 and D572 are so close (within 42 bp in the same exon), these two variants have not been dissociated by recombination between them. The D572 variant has drifted subsequently to a detectable frequency in general population samples, reaching as high as 6% in the Finnish.
Albert et al12 had functionally characterized A572D in Xenopus oocytes as having a shorter recovery time from inactivation compared to wild-type channels. However, their study was performed in the background of the hH1 SCN5A clone, which encodes a histidine (H) at position 558, a glutamine (Q) at 1027, and a glutamine (Q) at both positions 1076 and 1077 as a result of alternative splicing. We previously demonstrated that the most common SCN5A transcript in humans (Q1077del) lacks glutamine at position 1077 due to preferred alternate splicing and contains a histidine at position 558 and an arginine at position 1027, rather than glutamine (Q) 1027 as in hH1.13 In fact, Q1027 may simply represent an artifact or, at best, an extremely rare variant of SCN5A, as we have not observed glutamine at position 1027 in more than 500 LQTS patients or 1,300 control subjects.8,11 Expressing genetic variants in the proper genetic background is paramount when trying to decipher the variant’s true biologic/functional role in health and disease, as varying backgrounds potentially can lead to widely discrepant and erroneous functional results.
Here, we expressed in HEK293 cells the A572D-SCN5A variant in the most common SCN5A transcript (Q1077del). Although it appears that D572 is invariably linked with R558 on the same chromosome, we nevertheless functionally characterized D572 in both constructs containing either a histidine at position 558 (H558) or an arginine at 558 (R558). Interestingly, although D572 in the setting of H558 behaved like wild type (A572/H558), D572 in its invariably linked context of R558 produced sodium channels with a 6-mV negative shift in steady-state inactivation, slower time constants of recovery from inactivation, and a threefold increase in late/persistent sodium current. Whether this degree of channel dysfunction is enough to provide a mechanistic basis for proarrhythmia susceptibility in the less than 1% of Caucasians who host A572D/H558R on the same allele is debatable because no epidemiologic data have yet reached statistical significance. Furthermore, it brings into question how much channel dysfunction is required in an in vitro heterologous expression system before we can conclude that a particular channel variant/mutation is truly a pathogenic one.
Other common DI–DII and DII–DIII linker domain sodium channel variants observed in health, such as R481W, R1193Q, and P1090L) have shown similar degrees of biophysical dysfunction in vitro, suggesting that this level of impairment (6-mV voltage shift and threefold increase in late sodium current) may not always represent sufficient evidence to conclude that a particular variant is an independent, LQT3-causative mutation.20 However, investigators, including ourselves, have previously used this level of dysfunction to support a causal link with disease for mutations that are absent in controls.21–23 Based solely on heterologous expression system data using the current loosely held in vitro criteria for invoking pathogenicity, A572D would indeed be wrongly assigned the label of a pathogenic, LQT3-causative mutation.
Instead, the epidemiologic evidence argues strongly for a conclusion of nonpathogenicity rather than A572D-SCN5A being an independent genetic cause for LQT3. Further, the epidemiologic evidence for A572D strongly suggests that, despite the recorded level of in vitro dysfunction, the heterologous expression data are insufficient to elevate A572D to an LQT3-causative mutation. Instead, akin to the potentially proarrhythmic, functional common polymorphism status assigned to S1103Y, A572D in the setting of R558 may similarly confer S1103Y-like susceptibility.
Although it would be a major undertaking, a systematic functional analysis of all DI–DII and DII–DIII linker domain variants observed in cases or controls would be most helpful in establishing benchmark criteria for the assignment of pathogenicity based on functional data derived from heterologous expression systems. Data derived from a systematic functional analysis of unequivocal LQT3-causative SCN5A mutations, based on proper familial co-segregation with disease and their absence in controls, compared with data derived from a simultaneous functional analysis of SCN5A variants previously identified in ostensibly healthy control panels may permit such in vitro benchmarking. Until such a study is performed, we can only speculate on the degree of channel dysfunction necessary to make this call based on heterologous expression systems involving SCN5A. Furthermore, this type of study may yield sobering data that could dampen enthusiasm for the continued reliance on heterologous expression systems in general for making claims of pathogenicity. Such an observation could accelerate the need for, and use of, novel techniques, such as channel expression studies in the patient’s own skin biopsy–generated, induced pluripotent stem cell– derived cardiomyocytes, where the variant/mutation of interest would be characterized in its true native environment, including the host patient’s entire proteome.
Conclusion
There is sufficient evidence to conclude that A572D-SCN5A is not an independent, LQT3-causative mutation. Rather, epidemiologic, molecular, and functional evidence presented here suggests that A572D, invariably linked with H558R, is functionally aberrant but nonpathogenic. Functionally, R558/D572-containing sodium channels exhibit a moderate LQT3-like dysfunction that might contribute to a heightened risk for arrhythmias in some individuals, perhaps under certain exogenous factor(s). However, this heightened risk, if it exists, must be small as no epidemiologic sign of it is found. Furthermore, these findings underscore the scrutiny necessary to distinguish true LQT3-causative mutations from both functional modifiers and otherwise innocuous, rare genetic variants in SCN5A.
Acknowledgments
The analyses were performed in Dr. Ackerman’s research program with support from the Mayo Clinic Windland Smith Rice Comprehensive Sudden Cardiac Death Program. Dr. Wilde’s research program is supported by Interuniversity Cardiology Institute the Netherlands (ICIN) project 27 and a Leducq programm grant “Alliance against sudden cardiac death.” Dr. Carole Harris-Kerr and Dr. Benjamin A. Salisbury are employees of PGxHealth, which offers the FAMILION LQTS Test, and are stockholders of the parent company, Clinical Data. Dr. Wilde is on PGxHealth’s Scientific Advisory Board. Dr. Ackerman is a consultant for PGxHealth and chairs their FAMILION Medical/Scientific Advisory Board (approved by Mayo Clinic’s Medical-Industry Relations Office and Conflict of Interests Review Board). “Cardiac channel gene screen” and “know-how relating to long QT genetic testing” license agreements, resulting in consideration and royalty payments, were established between Genaissance Pharmaceuticals (now PGxHealth) and Mayo Medical Ventures (now Mayo Clinic Health Solutions) in 2004.
ABBREVIATIONS
- LQTS
long QT syndrome
- PCR
polymerase chain reaction
References
- 1.Schwartz PJ, Stramba-Badiale M, Crotti L, et al. Prevalence of the congenital long-QT syndrome. Circulation. 2009;120:1761–1767. doi: 10.1161/CIRCULATIONAHA.109.863209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Priori SG, Schwartz PJ, Napolitano C, et al. Risk stratification in the long-QT syndrome. N Engl J Med. 2003;348:1866–1874. doi: 10.1056/NEJMoa022147. [DOI] [PubMed] [Google Scholar]
- 3.Zareba W, Moss AJ, Locati EH, et al. Modulating effects of age and gender on the clinical course of long QT syndrome by genotype. J Am Coll Cardiol. 2003;42:103–109. doi: 10.1016/s0735-1097(03)00554-0. [DOI] [PubMed] [Google Scholar]
- 4.Ruan Y, Liu N, Napolitano C, et al. Therapeutic strategies for long-QT syndrome: does the molecular substrate matter? Circ Arrhythm Electrophysiol. 2008;1:290–297. doi: 10.1161/CIRCEP.108.795617. [DOI] [PubMed] [Google Scholar]
- 5.Splawski I, Shen J, Timothy K, et al. Genomic structure of three long QT syndrome genes: KVLQT1, HERG, and KCNE1. Genomics. 1998;51:86–97. doi: 10.1006/geno.1998.5361. [DOI] [PubMed] [Google Scholar]
- 6.Curran ME, Splawski I, Timothy KW, et al. A molecular basis for cardiac arrhythmia: HERG mutations cause long QT syndrome. Cell. 1995;80:795–803. doi: 10.1016/0092-8674(95)90358-5. [DOI] [PubMed] [Google Scholar]
- 7.Wang Q, Shen J, Splawski I, et al. SCN5A mutations associated with an inherited cardiac arrhythmia, long QT syndrome. Cell. 1995;80:805–811. doi: 10.1016/0092-8674(95)90359-3. [DOI] [PubMed] [Google Scholar]
- 8.Ackerman MJ, Splawski I, Makielski JC, et al. Spectrum and prevalence of cardiac sodium channel variants among Black, White, Asian, and Hispanic individuals: implications for arrhythmogenic susceptibility and Brugada/Long QT Syndrome genetic testing. Heart Rhythm. 2004;1:600–607. doi: 10.1016/j.hrthm.2004.07.013. [DOI] [PubMed] [Google Scholar]
- 9.Kapa S, Tester DJ, Salisbury BA, et al. Genetic testing for long-QT syndrome distinguishing pathogenic mutations from benign variants. Circulation. 2009;120:1752–1760. doi: 10.1161/CIRCULATIONAHA.109.863076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Paulussen A, Matthijs G, Gewillig M, et al. Mutation analysis in congenital Long QT Syndrome—a case with missense mutations in KCNQ1 and SCN5A. Genet Test. 2003;7:57–61. doi: 10.1089/109065703321560958. [DOI] [PubMed] [Google Scholar]
- 11.Tester DJ, Will ML, Haglund CM, et al. Compendium of cardiac channel mutations in 541 consecutive unrelated patients referred for long QT syndrome genetic testing. Heart Rhythm. 2005;2:507–517. doi: 10.1016/j.hrthm.2005.01.020. [DOI] [PubMed] [Google Scholar]
- 12.Albert CM, Nam EG, Rimm EB, et al. Cardiac sodium channel gene variants and sudden cardiac death in women. Circulation. 2008;117:16–23. doi: 10.1161/CIRCULATIONAHA.107.736330. [DOI] [PubMed] [Google Scholar]
- 13.Makielski JC, Ye B, Valdivia CR, et al. A ubiquitous splice variant and a common polymorphism affect heterologous expression of recombinant human SCN5A heart sodium channels. Circ Res. 2003;93:821–828. doi: 10.1161/01.RES.0000096652.14509.96. [DOI] [PubMed] [Google Scholar]
- 14.Arnestad M, Crotti L, Rognum TO, et al. Prevalence of long-QT syndrome gene variants in sudden infant death syndrome. Circulation. 2007;115:361–367. doi: 10.1161/CIRCULATIONAHA.106.658021. [DOI] [PubMed] [Google Scholar]
- 15.Marjamaa A, Newton-Cheh C, Porthan K, et al. Common candidate gene variants are associated with QT interval duration in the general population. J Intern Med. 2009;265:448–458. doi: 10.1111/j.1365-2796.2008.02026.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Burke A, Creighton W, Mont E, et al. Role of SCN5A Y1102 polymorphism in sudden cardiac death in blacks. Circulation. 2005;112:798–802. doi: 10.1161/CIRCULATIONAHA.104.482760. [DOI] [PubMed] [Google Scholar]
- 17.Plant LD, Bowers PN, Liu QY, et al. A common cardiac sodium channel variant associated with sudden infant death in African Americans, SCN5A S1103Y. J Clin Invest. 2006;116:430–435. doi: 10.1172/JCI25618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Van Norstrand DW, Tester DJ, Ackerman MJ. Overrepresentation of the proarrhythmic, sudden death predisposing sodium channel polymorphism S1103Y in a population-based cohort of African-American sudden infant death syndrome. Heart Rhythm. 2008;5:712–715. doi: 10.1016/j.hrthm.2008.02.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mank-Seymour AR, Richmond JL, Wood LS, et al. Association of torsades de pointes with novel and known single nucleotide polymorphisms in long QT syndrome genes. Am Heart J. 2006;152:1116–1122. doi: 10.1016/j.ahj.2006.08.020. [DOI] [PubMed] [Google Scholar]
- 20.Tan B-H, Valdivia CR, Rok BA, et al. Common human SCN5A polymorphisms have altered electrophysiology when expressed in Q1077 splice variants. Heart Rhythm. 2005;2:741–747. doi: 10.1016/j.hrthm.2005.04.021. [DOI] [PubMed] [Google Scholar]
- 21.Vatta M, Ackerman MJ, Ye B, et al. Mutant caveolin-3 induces persistent late sodium current and is associated with long-QT syndrome. Circulation. 2006;114:2104–2112. doi: 10.1161/CIRCULATIONAHA.106.635268. [DOI] [PubMed] [Google Scholar]
- 22.Ueda K, Valdivia C, Medeiros-Domingo A, et al. Syntrophin mutation associated with long QT syndrome through activation of the nNOS-SCN5A macromolecular complex. Proc Natl Acad Sci U S A. 2008;105:9355–9360. doi: 10.1073/pnas.0801294105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wehrens XHT, Rossenbacker T, Jongbloed RJ, et al. A novel mutation L619F in the cardiac Na+ channel SCN5A associated with long-QT syndrome (LQT3): a role for the I–II linker in inactivation gating. Hum Mutat. 2003;21:552. doi: 10.1002/humu.9136. [DOI] [PubMed] [Google Scholar]