Abstract
S-nitrosylation (SNO) is a reversible protein modification that has the ability to alter the activity of target proteins. However, only a small number of SNO proteins have been found in the myocardium, and even fewer specific sites of SNO have been identified. Therefore, this study aims to characterize potential SNO sites in the myocardium. We utilized a modified version of the SNO-resin-assisted capture technique in tandem with mass spectrometry. In brief, a modified biotin switch was performed using perfused mouse heart homogenates incubated with or without the S-nitrosylating agent S-nitrosoglutathione. Our modified SNO-resin-assisted capture protocol identified 116 unique SNO-modified proteins under basal conditions, and these represent the constitutive SNO proteome. These constitutive SNO proteins are likely to be physiologically relevant targets, since nitric oxide has been shown to play an important role in the regulation of normal cardiovascular physiology. Following S-nitrosoglutathione treatment, we identified 951 unique SNO proteins, many of which contained multiple SNO sites. These proteins show the potential for SNO. This study provides novel information regarding the constitutive SNO proteome of the myocardium, as well as potential myocardial SNO sites, and yields additional information on the SNO sites for many key proteins involved in myocardial contraction, metabolism, and cellular signaling.
Keywords: cardioprotection, ischemic preconditioning
nitric oxide is an essential signaling molecule in the myocardium and is synthesized by three distinct isoforms of nitric oxide synthase (NOS) (37). Neuronal NOS (NOS1) and endothelial NOS (NOS3) are constitutively expressed (2), while inducible NOS (NOS2) is expressed during stress or pathological conditions (1). Nitric oxide can also be produced independent from the NOS isoforms (15, 20, 25). The nonenzymatic reduction of nitrite represents a potential NOS-independent nitric oxide source. Nitric oxide is able to affect end-target proteins through cyclic guanosine monophosphate (cGMP)-dependent and -independent signaling pathways. Nitric oxide can activate guanylate cyclase, which increases cGMP levels and activates the cGMP-dependent protein kinase (36). Nitric oxide also has cGMP-independent effects, which include protein S-nitrosylation (SNO) (10, 12, 21).
SNO is a reversible protein modification in which nitric oxide is covalently bound to a thiol group, leading to the formation of S-nitrosothiols (10, 12, 21). This modification has the ability to alter the activity of target proteins, such as the L-type Ca2+ channel and the sarcoplasmic reticulum Ca2+ release channel (8, 28, 29, 33, 35). In fact, we have shown that treatment with the S-nitrosylating agent S-nitrosoglutathione (GSNO), dose-dependently increased the activity of the sarcoplasmic reticulum Ca2+-ATPase and decreased mitochondrial F1F0-ATPase activity (27). Additional S-nitrosylated proteins that have been identified include aconitate hydratase, aldehyde dehydrogenase, α-ketoglutarate dehydrogenase, creatine kinase, malate dehydrogenase, mitochondrial complex I, and thioredoxin (3, 5, 27). Interestingly, GSNO has been shown to provide cardioprotective effects during ischemia-reperfusion injury (27), and mice lacking GSNO reductase also show cardioprotection (17). This indicates that SNO is likely to play an important role in cardioprotection.
Numerous methodologies have been developed to detect S-nitrosylated proteins (31), such as the biotin switch (8, 14, 28, 34), which includes two-dimensional fluorescence difference gel electrophoresis with DyLight maleimide (2D DyLight Fluor DIGE) (18, 27), and S-nitrosocysteine antibodies (9, 13, 19). However, few studies have identified SNO sites in complex samples (6, 11, 16). Therefore, the goal of this study is to identify both constitutive and GSNO-modifiable SNO sites in the myocardium.
MATERIALS AND METHODS
Animals.
Male C57BL/6 mice were obtained from Jackson Laboratories (Bar Harbor, ME). All animals utilized in this study were between the ages of 12 and 15 wk. This investigation conforms to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH publication no. 85–23, revised 1996) and was approved by the Institutional Laboratory Animal Care and Use Committee.
Solutions and drugs.
Krebs-Henseleit buffer (KHB) consisted of the following (in mmol/l): 120 NaCl, 4.7 KCl, 1.2 NaH2PO4, 25 NaHCO3, 1.2 MgSO4, 10 glucose, and 1.75 CaCl2; pH 7.4. KHB was bubbled with 95% O2/5% CO2. GSNO (Calbiochem, San Diego, CA) was used as an S-nitrosylating agent. Ascorbate (Sigma, St. Louis, MO) was used as an SNO-specific reducing agent. Dithiothreitol (DTT; Pierce, Rockford, IL) was used as a reducing agent. All solutions were made fresh on the day of experimentation.
Whole heart homogenate preparation.
Whole mouse heart homogenates were prepared as described previously (18, 27). Briefly, hearts were Langendorff perfused with KHB in the dark for 60 min. At the end of the perfusion period, hearts were snap frozen in liquid nitrogen. All subsequent procedures were performed in the dark. Each heart was then powdered on liquid nitrogen with a mortar and pestle. Powdered hearts were resuspended in 1.5 ml of homogenization buffer containing the following (in mmol/l): 300 sucrose, 250 HEPES-NaOH (pH 7.7), 1 EDTA, and 0.1 neocuproine. An EDTA-free protease inhibitor tablet (Roche Diagnostics, Indianapolis, IN) was introduced just before use. Samples were then homogenized via Dounce glass homogenization on ice and centrifuged at 1,000 g for 2 min. The supernatant was recovered as total whole heart homogenate. Protein concentration was determined using the Bradford protein assay.
Protein SNO and modified biotin switch.
A modified version of the biotin switch was performed as previously described (see Fig. 1) (14). Briefly, samples [i.e., whole heart homogenate, bovine serum albumin (BSA), glyceraldehyde-3-phosphate dehydrogenase (GAPDH)] were diluted in HEN buffer containing the following (in mmol/l): 250 HEPES-NaOH 7.7, 1 EDTA, and 0.1 neocuproine, with 2.5% SDS (wt/vol) and an EDTA-free protease inhibitor tablet (Roche Diagnostics). Samples were incubated with 1 mmol/l GSNO for 30 min at room temperature; GSNO was removed via cold acetone precipitation (−20°C). Samples were then resuspended in HEN with 2.5% SDS and treated with 50 mmol/l N-ethylmaleimide (NEM; Sigma) for 20 min at 50°C with gentle vortexing every 5 min to block free thiol groups from modification; NEM was removed via cold acetone precipitation (−20°C). Finally, samples were resuspended in HEN with 1% SDS (wt/vol), and treated with ascorbate (Sigma) to reduce SNO cysteine residues, followed by one of the identification strategies listed below.
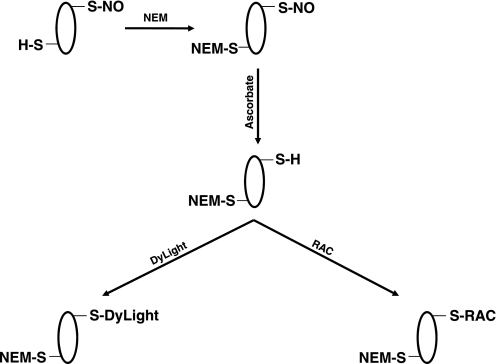
Fig. 1.
Methodology. A modified version of the biotin switch protocol was used to identify S-nitrosylated proteins with DyLight maleimide and resin-assisted capture (RAC). NO, nitric oxide; NEM, N-ethylmaleimide.
SNO protein identification and site determination with DyLight maleimide.
BSA (200 μg) was treated with GSNO and the modified biotin switch, as described above. Following incubation with 1 mmol/l ascorbate (Sigma), BSA was labeled with DyLight488 maleimide (Pierce) for 1 h at room temperature; DyLight 488 was removed via cold acetone precipitation (−20°C). BSA was then subjected to trypsin digestion (sequencing grade modified; Promega, Madison, WI) overnight at 37°C with agitation in buffer containing the following (in mmol/l): 50 NH4HCO3 and 1 EDTA. Trypsin-digested peptides were concentrated via speedvac. Peptides were then resuspended in 0.1% formic acid (vol/vol) and cleaned with a C18 column (ZipTip; Millipore, Billerica, MA). Matrix-assisted laser desorption/ionization-time of flight tandem mass spectrometry (MALDI-TOF/TOF) was then performed using a 4700 Proteomics Discovery System (Applied Biosystems, Foster City, CA). A purified cysteine-containing peptide (H6387: His-Cys-Lys-Phe-Trp-Trp; Sigma) was also subjected to an identical procedure. However, the peptide was not treated with GSNO, blocking reagent, or ascorbate because of the difficulty in removing the various reagents (i.e., GSNO, NEM, etc.). Instead, the peptide was simply incubated with DyLight488 maleimide at a ratio of 1.1 mmol/l H6387 peptide to 40 μmol/l DyLight488 maleimide. In this particular instance, the DyLight488 was bound directly to the free cysteine residue of the peptide, thus negating the need for the biotin switch. Peptides were then cleaned with a C18 column (ZipTip; Millipore). Mass spectrometry (MS) was performed via direct infusion using an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific, San Jose, CA), as described below.
SNO protein identification and site determination with SNO-resin-assisted capture.
A modified version of the SNO-resin-assisted capture (SNO-RAC) protocol was developed to examine SNO (6). Samples (whole heart homogenate; 1 mg) were treated with GSNO and the modified biotin switch, as described above. Samples were then resuspended in HEN with 1% SDS (HENS). All buffers were degassed before use with the SNO-RAC protocol to prevent oxidation of the resin. Thiopropyl sepharose resin (GE Healthcare, Piscataway, NJ) was rehydrated for 25 min in diethyl pyrocarbonate (DEPC) H2O. Following rehydration, 25 μl of the resin slurry were added to a Handee Mini Spin Column (Pierce) and washed with 5 × 0.5 ml DEPC H2O, followed by 10 × 0.5 ml HEN buffer. Blocked samples were then added to the thiopropyl sepharose-containing spin column, along with 20 mmol/l ascorbate, and rotated for 4 h in the dark at room temperature. Resin-bound proteins were then washed with 8 × 0.5 ml HENS buffer, followed by 4 × 0.5 ml HENS buffer diluted 1:10. Samples were then subjected to trypsin digestion (sequencing grade modified; Promega) overnight at 37°C with rotation in buffer containing the following (in mmol/l): 50 NH4HCO3 and 1 EDTA. Resin-bound peptides were then washed with 5 × 0.5 ml HENS buffer diluted 1:10, 5 × 0.5 ml 2 mol/l NaCl, 5 × 0.5 ml 80% acetonitrile (vol/vol)-0.1% trifluoroacetic acid (vol/vol), and 5 × 0.5 ml HEN buffer diluted 1:10. Peptides were eluted for 30 min at room temperature in elution buffer containing the following (in mmol/l): 20 DTT, 10 NH4HCO3, and 50% methanol (vol/vol). The resin was then washed with an additional volume of elution buffer, followed by two volumes of DEPC H2O. All fractions were combined and concentrated via speedvac. Samples were then resuspended in 0.1% formic acid and cleaned with a C18 column (ZipTip; Millipore). Liquid chromatography-tandem MS (LC-MS/MS) was then performed using an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific), as described below. Each of four biological replicates was run in triplicate to increase protein/peptide identifications. The MASCOT database search function was used for protein identification, as described below.
Several control experiments were performed to confirm the specificity of the modified SNO-RAC protocol. As a positive control, we repeated the same procedure with purified GAPDH (Sigma). In a separate set of experiments with whole heart homogenates, DTT was omitted from the elution buffer as a negative control; ascorbate was also omitted from the procedure as a negative control. As an additional negative control, samples were treated with 20 mmol/l ascorbate for 45 min before treatment with NEM in the SNO-RAC protocol.
LC-MS/MS analysis.
LC-MS/MS was performed using either an Eksigent nano-LC 1D plus system (Dublin, CA) coupled to an LTQ Orbitrap XL mass spectrometer (Thermo Fisher Scientific), or an Eksigent nanoLC-Ultra 1D plus system coupled to an LTQ Orbitrap Velos mass spectrometer (Thermo Fisher Scientific). Both systems utilized CID fragmentation. For the LTQ Orbitrap XL, 10 μl of sample (in 50% acetonitrile-0.1% formic acid) was directly infused using a 4-μm-ID Pico Tip (New Objective) with a syringe pump. LTQ Orbitrap XL settings were as follows: spray voltage 1.5 kV, and full MS mass range of mass-to-charge ratio (m/z) of 200–2,000. The LTQ Orbitrap XL was operated in a data-dependent mode [i.e., one MS1 high-resolution (30,000) scan for precursor ions, followed by six data-dependent MS/MS scans for precursor ions above a threshold ion count of 2,000 with collision energy of 35%]. For the LTQ Orbitrap Velos, peptides were first loaded onto a Zorbax 300SB-C18 trap column (Agilent) at a flow rate of 6 μl/min for 6 min, and then separated on a reversed-phase PicoFrit analytical column (New Objective) using a 40-min linear gradient of 5–40% acetonitrile in 0.1% formic acid at a flow rate of 250 nl/min. LTQ Orbitrap Velos settings were as follows: spray voltage 1.5 kV, and full MS mass range of m/z 230–2,000. The LTQ Orbitrap Velos was operated in a data-dependent mode [i.e., one MS1 high-resolution (60,000) scan for precursor ions, followed by six data-dependent MS/MS scans for precursor ions above a threshold ion count of 2,000 with collision energy of 35%].
MASCOT database search.
Raw files generated from the LTQ Orbitrap Velos were analyzed using Proteome Discoverer 1.1 (Thermo Fisher Scientific) with the NIH six-processor MASCOT cluster search engine (http://biospec.nih.gov, version 2.3). The following search criteria were used: database, Swiss-Prot (Swiss Institute of Bioinformatics); taxonomy, Mus musculus (mouse); enzyme, trypsin; miscleavages, 3; variable modifications, oxidation (M), NEM (C), deamidation (NQ); MS peptide tolerance 25 ppm; MS/MS tolerance as 0.8 Da. All peptides were assigned an ion score. The ion score is a measure of how well the MS/MS spectra matches the stated peptide; higher scores represent more confident matches. Ion scores were generated as −10 × log10(P), where P represents the probability that the match is random. P is also referred to as the expectation value. A more detailed explanation of the ion score is provided in Ref. 24. For each protein identification, the %coverage (∑coverage) is also reported, and this represents the number of amino acid residues identified compared with the total number in the protein sequence; higher percentages represent more confident protein identifications. However, the SNO-RAC protocol enriches for cysteine-containing peptides, specifically SNO-cysteines, and, therefore, %coverage is not necessarily the best indicator for confidence in protein identifications. With the use of SNO-RAC, the ion score and the expectation value are better measures of confidence. Peptides with ion scores <25 were not accepted. Peptides were filtered at a false discovery rate of 1%, as determined by a targeted decoy database search with a significance threshold of 0.03.
Label-free peptide quantification and analysis.
Relative quantification of SNO was performed using QUOIL (quantification without isotope labeling), an in-house software program designed as a label-free approach to peptide quantification by LC-MS/MS (32). Label-free peptide quantification is a common approach for quantifying peptide intensity when stable-isotope labeling is not utilized (22). This label-free approach relies on the direct comparison of peptide area-under-the-curve peaks from each LC-MS/MS run. More specifically, a peptide's chromatogram peak in each LC-MS/MS run was reconstructed based on its precursor m/z value. Quantitative ratios were then obtained by normalizing the peptide peak areas from GSNO-treated samples against non-GSNO-treated samples. The resulting ratios reflect the relative quantity of a peptide (and hence the corresponding SNO level) in different samples, but the absolute amounts of the protein SNO cannot be determined, since unmodified protein does not bind to the column and was not measured. The ratio maximum was capped at 1,000 for this study.
Statistics.
Statistical significance (P < 0.05) was determined between groups using a Student's t-test for two groups.
RESULTS
Although a number of studies have identified S-nitrosylated proteins (5, 8, 18, 27, 28, 34), very few sites of SNO have been determined. This lack of SNO site identifications has limited the analysis of the role of SNO in myocardial function. Because of the importance of identifying sites of SNO in the myocardium, we attempted several different protocols. These protocols included the use of resin-assisted capture and DyLight maleimide labeling.
DyLight maleimide.
We previously utilized a modified biotin switch method with DyLight maleimide and two-dimensional fluorescence DIGE (2D DyLight Fluor DIGE) to identify S-nitrosylated proteins (Fig. 1). We reasoned that we should be able to determine the sites of SNO by detecting the mass addition of the DyLight maleimide via MS. Therefore, we first examined DyLight488 maleimide alone with MALDI-TOF/TOF, to determine the exact molecular mass of the molecule. MS revealed two different molecular masses for DyLight488 maleimide: 800 and 778 m/z. These mass differences appeared to correspond with the loss of bound Na+ (DyLight488 maleimide can bind up to 3 Na+). We next subjected DyLight488 maleimide-labeled purified BSA (200 μg, ± GSNO) to tryptic digestion and analyzed the peptides via MALDI-TOF/TOF. However, we were only able to detect very low levels of DyLight488-labeled BSA, despite the fact that we used a high level of purified starting material (200 μg). Additionally, we analyzed a DyLight488 maleimide-labeled cysteine-containing peptide (1.1 mmol/l H6387 peptide: 40 μmol/l DyLight488 maleimide) with static nanospray and observed a small peak that corresponded to a mass addition of 777 m/z for DyLight488 maleimide. This is consistent with the molecular mass of DyLight488 maleimide, with the loss of one Na+ and one H+. Finally, we attempted to identify SNO sites from fluorescent spots obtained via 2D DyLight Fluor DIGE, but DyLight maleimide-labeled peptides were not detected. This may suggest that the DyLight maleimide addition may interfere with the ionization of the labeled peptide. Although we have been able to successfully identify SNO proteins utilizing 2D DyLight Fluor DIGE by extracting the fluorescent spots in the gel (18, 27), the MS identification was based largely on peptides that did not contain cysteine residues. Thus this DyLight maleimide-labeling strategy does not appear to be particularly efficacious in the identification of SNO sites.
SNO-RAC.
We next utilized a SNO-RAC protocol (Fig. 1), which has been shown to be effective in the identification of SNO proteins and the sites of SNO formation (6). We developed a modified version of the protocol, which included several alterations to the original technique. First, we substituted NEM for methyl methanethiosulfonate (MMTS) as a blocking agent. This substitution was made because MMTS is susceptible to DTT reduction during the step in which peptides are eluted from the resin. Conversely, NEM is not affected by DTT. This is particularly important with peptides that contain multiple cysteine residues; nonmodified cysteine residues (i.e., not SNO) will remain blocked with NEM and will be detected as such by MS, thus allowing us to distinguish nonmodified cysteine residues from SNO cysteine residues (which will not be labeled with NEM) in the case of peptides that have multiple cysteines. The MMTS would be removed by DTT during the elution step, and nonmodified cysteine residues would not be distinguishable from SNO cysteine residues in peptides with multiple cysteines. Second, we used thiopropyl sepharose rather than sepharose-N-hydroxysuccinimide, because the thiopropyl sepharose resin has a much higher binding capacity. Finally, we added numerous wash steps (i.e., 2 mol/l NaCl, 80% acetonitrile-0.1% trifluoroacetic acid, etc.) to reduce nonspecific protein interaction (i.e., ionic, hydrophobic, etc.) with the thiopropyl sepharose resin. These measures all serve to enhance the specificity of the technique and increase the confidence of SNO site identification.
We first subjected whole heart homogenates (1 mg, ± GSNO) to the modified SNO-RAC procedure and analyzed the tryptic peptides via LC-MS/MS. SNO-RAC detected nearly 200 different constitutive SNO sites from 116 unique proteins (Supplemental Table S1; the online version of this article contains supplemental data) in non-GSNO-treated samples. These SNO sites are modified in the presence of physiological levels of nitric oxide and represent the constitutive SNO proteome. These SNO proteins may also represent key targets that are regulated during normal myocardial function. Following GSNO treatment, MS yielded over 2,000 different SNO sites from 951 unique SNO proteins (Supplemental Table S2). Many of these proteins contain multiple sites for SNO formation (i.e., cysteine and glycine-rich protein 3 contained 14 SNO sites), while other proteins, including A kinase anchoring protein 1, were identified to have only one SNO site. Table 1 shows select protein/peptide identifications for proteins that were previously identified to be SNO with 2D DyLight Fluor DIGE (18, 27). Label-free peptide analysis was performed on the common proteins between GSNO-treated and non-GSNO-treated samples. This label-free analysis revealed that many of the constitutive SNO proteins showed increased SNO, both at the same site and at new sites, on treatment with GSNO. For instance, SNO-RAC identified two constitutive SNO sites for adenine nucleotide translocator 1 (Cys160, Cys247). With GSNO treatment, SNO-RAC detected an increase of nearly 75-fold at those same two sites and also revealed four new SNO sites. The SNO proteins identified in GSNO-treated whole heart homogenates represent potential SNO sites in the myocardium and thus show the potential for endogenous modification. All of the protein/peptide identifications for both GSNO-treated and non-GSNO-treated homogenates can be found in the supplemental tables, which are available online. Please note that, for a small percentage of peptides identified to have multiple SNO sites, some but not all of these cysteine residues may actually be sites of oxidative modification. This possibility exists because oxidative modifications, if present on resin-bound peptides, could be reduced by DTT during the elution step, thus leaving a free cysteine residue. However, resin binding indicates at least one SNO site.
Table 1.
S-nitrosylation sites identified via SNO-RAC proteomic analysis for proteins that were previously determined to be S-nitrosylated with 2D DyLight Fluor DIGE
| Protein Name | Protein ID | Peptide Sequence | SNO Cys | Ion Score | SNO Ratio |
|---|---|---|---|---|---|
| α-Enolase | P17182 | FGANAILGVSLAVCK | 119 | 91 | |
| SCNCLLLK | 337, 339 | 57 | |||
| SGETEDTFIADLVVGLCTGQIK | 379 | 129 | |||
| Aconitate hydratase | Q99KI0 | VAVPSTIHCDHLIEAQVGGEK | 126 | 65 | |
| GGTGAIVEYHGPGVDSISCTGMATICNMGAEIGATTSVFPYNHR | 277, 284 | 107 | |||
| VGLIGSCTNSSYEDMGR | 385 | 117 | 74.7* | ||
| CKSQFTITPGSEQIR | 410 | 86 | |||
| DVGGIVLANACGPCIGQWDR | 448, 451 | 97 | 844.0* | ||
| CTTDHISAAGPWLK | 592 | 89 | 543.5* | ||
| Acyl-CoA dehydrogenase | P45952 | MTEQPMMCAYCVTEPSAGSDVAAIK | 157, 159 | 105 | |
| ELNMGQRCSDTR | 244 | 25 | |||
| ATP synthase-α | Q03265 | LYCIYVAIGQK | 244 | 78 | 336.1* |
| YTIVVSATASDAAPLQYLAPYSGCSMGEYFR | 294 | 63 | |||
| Creatine kinase, M-type | P07310 | GYTLPPHCSR | 146 | 47 | 639.9* |
| FCVGLQK | 254 | 35 | 111.9* | ||
| AGHPFMWNEHLGYVLTCPSNLGTGLR | 283 | 63 | |||
| Electron transfer flavoprotein subunit-α | Q99LC5 | LGGEVSCLVAGTK | 53 | 85 | 71.5* |
| CDKVVQDLCK | 60, 68 | 47 | |||
| QFSYTHICAGASAFGK | 109 | 102 | |||
| TIYAGNALCTVK | 155 | 92 | 61.0* | ||
| Electron transfer flavoprotein subunit-β | Q9DCW4 | HSMNPFCEIAVEEAVR | 42 | 99 | |
| EIIAVSCGPSQCQETIR | 66, 71 | 96 | 109.2* | ||
| QAIDDDCNQTGQMTAGLLDWPQGTFASQVTLEGDK | 131 | 61 | |||
| Heat shock protein 60 kDa | P63038 | CEFQDAYVLLSEK | 237 | 78 | |
| AAVEEGIVLGGGCALLR | 442 | 98 | |||
| CIPALDSLKPANEDQK | 447 | 79 | |||
| Heat shock protein 70 kDa | P63017 | VCNPIITK | 503 | 49 | |
| Malate dehydrogenase, cytoplasmic | P14152 | VIVVGNPANTNCLTASK | 137 | 97 | 149.5* |
| ENFSCLTR | 151 | 48 | 160.4* | ||
| Malate dehydrogenase, mitochondrial | P08249 | GYLGPEQLPDCLK | 89 | 89 | 154.4* |
| GCDVVVIPAGVPR | 93 | 69 | 178.4* | ||
| TIIPLISQCTPK | 212 | 71 | 94.6* | ||
| EGVVECSFVQSK | 275 | 75 | 133.9* | ||
| ETECTYFSTPLLLGK | 285 | 86 | 336.5* | ||
| Myoglobin | P04247 | HGCTVLTALGTILK | 67 | 90 | 1,000* |
| Myosin light chain 3 | P09542 | ITYGQCGDVLR | 85 | 74 | 48.1* |
| LMAGQEDSNGCINYEAFVK | 191 | 120 | 153.7* | ||
| NADH-ubiquinone oxidoreductase, 75-kDa subunit | Q91VD9 | FCYHER | 64 | 31 | |
| VVAACAMPVMK | 92 | 61 | 35.6* | ||
| VDSDNLCTEEIFPTEGAGTDLR | 367 | 90 | |||
| HSFCEVLK | 463 | 53 | |||
| MLFLLGADGGCITR | 554 | 100 | |||
| DCFIVYQGHHGDVGAPMADVILPGAAYTEK | 564 | 69 | |||
| AVTEGAQAVEEPSIC | 727 | 52 | 62.3* |
Peptides were filtered at a false discovery rate of 1%; peptides with ion scores < 25 were not accepted.
M, mitochondrial isoform; C, S-nitrosylation (SNO) cysteine residue; c, N-ethylmaleimide (NEM)-blocked cysteine residue.
Peptide quantitative ratio (SNO ratio) was determined via label-free peptide analysis; ratio maximum was capped at 1,000.
Peptides detected in S-nitrosoglutathione (GSNO)-treated and nontreated samples, P < 0.05 vs. non-GSNO-treated. Peptides not detected under perfusion conditions contain a blank space in the ratio column.
SNO-RAC, SNO-resin-assisted capture; 2D DyLight Fluor DIGE, two-dimensional fluorescence difference gel electrophoresis with DyLight maleimide; CoA, coenzyme A.
We performed several important control experiments to further demonstrate the specificity of our modified SNO-RAC protocol. As a positive control, we repeated the same procedure with purified GAPDH (200 μg, ± GSNO). MS identified two unique peptides (VPTPNVSVVDLTCR, IVSNASCTTNCLAPLAK), which contained three different cysteine residues (Cys150, Cys154, Cys245). These three cysteine residues correspond to the sites of SNO for GAPDH, as previously published (6). Figure 2 shows representative MS/MS spectra for the SNO peptides of purified GAPDH. In a separate set of experiments with whole heart homogenates, DTT was omitted from the elution buffer as a negative control, and no cysteine-containing peptides were identified. When ascorbate was omitted from the procedure, we also failed to detect any cysteine-containing peptides. Additionally, when samples were treated with ascorbate before NEM, only eight cysteine-containing peptides were detected. Identifications from nonspecifically bound peptides (i.e., noncysteine-containing peptides) accounted for <3.6% of all peptide identifications; noncysteine-containing peptides were filtered from the data set. These data demonstrate that the SNO-RAC technique is very specific for the identification of SNO cysteine residues.
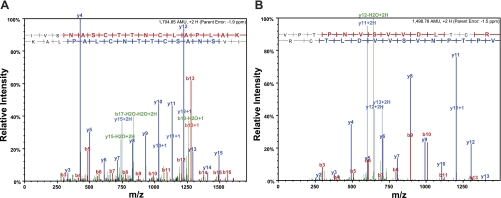
Fig. 2.
Tandem mass spectrometry (MS/MS) spectra for purified GAPDH. Representative MS/MS spectra shows fragmentation of the IVSNASCTTNCLAPLAK peptide (A) and the VPTPNVSVVDLTCR peptide (B) of GAPDH. The peptide sequence above each representative spectrum shows theoretical b ion identifications (red, NH2-terminal fragments) and theoretical y ion identifications (blue, COOH-terminal fragments). Peaks in the spectrum that are marked red correspond to matched b ions, and peaks that are marked blue correspond to matched y ions. The number paired with each ion identification (i.e., b2, y4, etc.) indicates the number of amino acids present on NH2-terminal fragments for b ions and COOH-terminal fragments for y ions. These results were obtained from the positive control experiment conducted with S-nitrosoglutathione (GSNO)-treated GAPDH. m/z, Mass-to-charge ratio.
Gene ontology and pathway analysis.
To identify specific cellular compartments and key signaling pathways that may be targeted by SNO, we performed gene ontology analysis using Scaffold_3_00_3 (Proteome Software, Portland, OR). This analysis showed that SNO targets proteins in many different cellular compartments. The majority of these protein targets were identified to be cytosolic, but mitochondrial proteins also comprised a large group of SNO targets. Consistent with the targeting of mitochondrial proteins by SNO, proteins involved with metabolism comprised a large group of targets related to biological processes. We also performed pathway analysis using Ingenuity IPA 8.7 (Ingenuity Systems, Redwood City, CA). As expected, this analysis identified many different pathways that are targeted by SNO, including myocardial contraction, metabolism, and cellular signaling (Table 2). Interestingly, this analysis also revealed that SNO targets many proteins involved in cell death, and these proteins are shown in Table 3. The targeting of proteins involved in cell death may indicate a potential role for SNO in cardioprotection (27). Interestingly, SNO of metallothionine has been shown to inhibit apoptosis in pulmonary endothelial cells (4, 30).
Table 2.
Biological pathway/function results from Ingenuity IPA 8.7 pathway analysis
| Biological Pathway/Function | P Value | No. of Proteins |
|---|---|---|
| Molecular and cellular functions | ||
| Lipid metabolism | 7.56E-04–4.86E-02 | 57 |
| Cellular assembly and organization | 1.63E-04–4.01E-02 | 51 |
| Molecular transport | 4.67E-04–4.01E-02 | 45 |
| Cell death | 4.12E-06–2.69E-02 | 15 |
| Drug metabolism | 1.80E-04–1.80E-04 | 6 |
| Physiological system development and function | ||
| Skeletal and muscular system development and function | 6.37E-07–4.87E-02 | 43 |
| Cardiovascular system development and function | 1.22E-03–4.94E-02 | 27 |
| Organ morphology | 1.22E-03–4.01E-02 | 21 |
| Tissue morphology | 6.37E-07–9.21E-03 | 18 |
| Diseases and disorders | ||
| Cardiovascular disease | 4.29E-04–4.01E-02 | 37 |
| Immunological disease | 1.24E-06–1.33E-04 | 15 |
| Inflammatory disease | 1.08E-04–1.08E-04 | 13 |
Pathway analysis was performed by searching the Ingenuity Knowledge Base (genes only, species = mouse) for direct and indirect relationships. All SNO site/peptide identifications from GSNO-treated samples (1% false discovery rate) were used for motif analysis.
Table 3.
S-nitrosylation sites identified via SNO-RAC proteomic analysis for proteins that were determined by Ingenuity IPA 8.7 to be involved in cell death
| Protein Name | Protein ID | Peptide Sequence | SNO Cys | Ion Score | SNO Ratio |
|---|---|---|---|---|---|
| Caveolin-3 | P51637 | IIKDIHCK | 19 | 42 | |
| Elongation factor 1-δ | P57776 | SSILLDVKPWDDETDMAQLETCVR | 217 | 66 | |
| Fructose-bisphosphate aldolase A | P05064 | VNPCIGGVILFHETLYQK | 73 | 93 | |
| YASICQQNGIVPIVEPEILPDGDHDLK | 178 | 63 | |||
| CQYVTEK | 202 | 35 | |||
| ALSDHHVYLEGTLLKPNMVTPGHACTQK | 240 | 74 | |||
| ALANSLACQGK | 339 | 80 | 1,000* | ||
| LIM and senescent cell antigen-like-containing domain protein 1 | Q99JW4 | AMNNSWHPECFR | 94 | 38 | |
| CEKPFLGHR | 222 | 37 | |||
| Metallothionein-1 | P02802 | SCCSCCPVGCSK | 33, 34, 36, 37, 41 | 74 | 1,000* |
| GAADKCTCCA | 57, 59, 60 | 36 | 547.8* | ||
| CAQGCVCK | 44, 48, 50 | 43 | |||
| Metallothionein-2 | P02798 | SCCSCCPVGCAK | 33, 34, 36, 37, 41 | 52 | |
| CSQGCICK | 44, 48, 50 | 38 | |||
| NADH dehydrogenase (ubiquinone) flavoprotein 2, mitochondrial | Q9D6J6 | YHIQVCTTTPCMLR | 134, 139 | 46 | |
| FCCEPAGGLTSLTEPPK | 223, 224 | 85 | |||
| Nicotinamide phosphoribosyltransferase | Q99KQ4 | DLLNCSFK | 397 | 38 | |
| Peptidyl-prolyl cis-trans isomerase, mitochondrial | Q99KR7 | VIPAFMCQAGDFTNHNGTGGR | 103 | 118 | |
| HVGPGVLSMANAGPNTNGSQFFICTIK | 156 | 88 | |||
| IVITDCGQLS | 202 | 60 | |||
| Proteasome subunit-β type-1 | O09061 | TVIGCSGFHGDCLTLTK | 81, 88 | 81 | |
| Thioredoxin-dependent peroxide reductase, mitochondrial | P20108 | AFQFVETHGEVCPANWTPESPTIKPSPTASK | 230 | 76 | |
| Trifunctional enzyme subunit-α, mitochondrial | Q8BMS1 | SAVLISSKPGCFVAGADINMLSSCTTPQEATR | 97, 110 | 60 | |
| ALMGLYNGQVLCK | 349 | 73 | 1,000* | ||
| EVESVTPEHCIFASNTSALPINQIAAVSK | 470 | 75 | |||
| CLAPMMSEVMR | 550 | 55 | |||
| YESAYGTQFTPCQLLLDHANNSSK | 747 | 50 | |||
| Ubiquinone biosynthesis monooxygenase | Q8R1S0 | VSSISPGSTTLLSSFGAWDHICNMR | 109 | 64 | |
| Voltage-dependent anion-selective channel protein 1 | Q60932 | EHINLGCDVDFDIAGPSIR | 140 | 102 | |
| YQVDPDACFSAK | 245 | 72 | 1,000* | ||
| Zyxin | Q62523 | CNTCGQPITDR | 336, 339 | 42 |
Peptides were filtered at a false discovery rate of 1%; peptides with ion scores < 25 were not accepted. Peptide quantitative ratio (SNO ratio) was determined via label-free peptide analysis; ratio maximum was capped at 1,000.
Peptides detected in GSNO-treated and nontreated samples, P < 0.05 vs. non-GSNO-treated. Peptides not detected under perfusion conditions contain a blank space in the ratio column.
Consensus SNO motif.
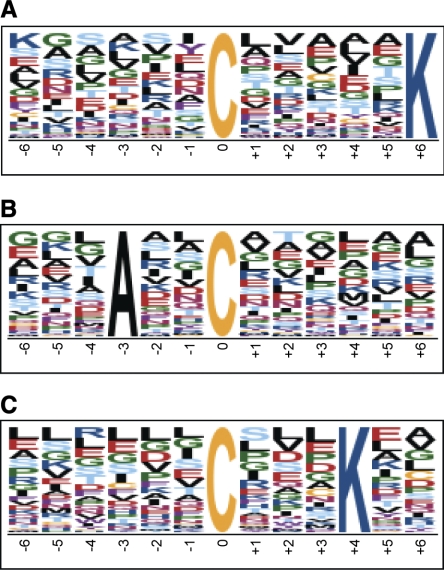
We next examined the large number of SNO site/peptide identifications from GSNO-treated samples in an attempt to identify a potential consensus SNO motif. Motif-X (http://motif-x.med.harvard.edu) was used for motif analysis (26). The search returned three potential SNO motifs, which are as follows: (......C.....K), (...A..C......), and (......C...K..). These motifs are shown in Fig. 3.
Fig. 3.
Consensus S-nitrosylation (SNO) motif. Motif-X search results for a consensus SNO motif are shown. All SNO site/peptide identifications from GSNO-treated samples (1% false discovery rate) were used for motif analysis. Search parameters were as follows: central character, C; width, 13; occurrences, 20; significance, 0.000001; background, international protein index (IPC) mouse proteome. Motif score for A is 8.80, B is 7.35, and C is 6.28. The size of each amino acid character represents the probability of occurrence; larger characters are more likely to be present in the SNO motif.
DISCUSSION
To test whether SNO is a biologically relevant posttranslational modification, it is essential to identify specific sites. Therefore, our goal in this study was to identify constitutive SNO sites. We also used GSNO treatment to identify additional potential SNO sites. Our approach in this study was to be inclusive and identify as many potential sites as possible. Thus we have included all peptides identified from each of the four hearts used in this study. To increase identifications, samples were also run in triplicate. Additionally, we performed these studies using an Orbitrap Velos mass spectrometer, which is a highly sensitive instrument. The identification of potential SNO sites in the myocardium is particularly important, considering that we and others have shown that SNO can modify the activity of target proteins (5, 27–29, 35), and SNO plays a critical role in cardioprotection (27). These results also compare favorably to studies showing protein SNO in the myocardium (5, 8, 18, 27, 28, 34), especially considering that very few studies have successfully identified sites of SNO. Here we confirm the modification of many previously reported S-nitrosylated proteins (18, 23, 27) and provide specific SNO sites. More importantly, we provide the identification and modification sites for many novel S-nitrosylated proteins in the myocardium.
SNO and DyLight maleimide labeling.
In previous studies, we used 2D DyLight Fluor DIGE to examine differences in protein SNO with cardioprotection (18, 27). After picking the fluorescent spots, we were able to successfully identify many different S-nitrosylated proteins, including ATP synthase-α, creatine kinase, malate dehydrogenase, and electron transfer flavoprotein-α. This technique is extremely useful in identifying S-nitrosylated proteins and quantifying SNO levels, but, unfortunately, DyLight maleimide labeling does not appear to be a very effective strategy for the identification of S-nitrosothiol sites. When we examined purified BSA, we were able to detect peptides with the mass addition of DyLight maleimide, but at very low levels. This low-level detection occurred, despite the fact that we used a very high level of starting material and that the BSA was noticeably fluorescent. We also had difficulty detecting a DyLight maleimide-labeled, cysteine-containing peptide, even though we again used a high level of starting material. Finally, we failed to detect any DyLight maleimide-labeled peptides from 2D DyLight Fluor DIGE. This strategy is likely ineffective for the identification of S-nitrosylated cysteine residues in complex samples for several reasons: 1) poor ionization efficiency of DyLight maleimide-labeled peptides; 2) inefficient trypsin digestion of DyLight maleimide-labeled proteins; and/or 3) large mass addition of DyLight maleimide. This approach also requires a large amount of labeled sample. This result is not entirely surprising, given that a similar approach with CyDye maleimide was also shown to be ineffective in the identification of oxidation sites (7).
Identification of potential SNO sites in the myocardium.
Our modified version of the SNO-RAC protocol identified nearly 200 sites from 116 proteins that showed constitutive SNO in the absence of GSNO treatment, thus indicating that these modifications occurred at physiological levels of nitric oxide. Nitric oxide has been shown to play an important role in normal cardiovascular physiology (37), thus indicating that these constitutively S-nitrosylated proteins may represent key targets that are regulated during normal myocardial function. With GSNO treatment, over 2,000 potential SNO sites from 951 unique proteins were identified, and many of these proteins contained multiple SNO sites (Table 1). The proteins identified via SNO-RAC included cytosolic, membrane, and mitochondrial proteins, and many of these proteins are involved with cellular metabolism and signaling (Table 2). Pathway analysis revealed that SNO targets many key pathways like myocardial contraction, and even cell death (Table 3). The targeting of proteins involved in cell death may lend critical insight into a potential role for SNO in cardioprotection, as SNO has been shown to inhibit apoptosis (4, 30). Taken together, these SNO-RAC identifications represent potential SNO sites in the myocardium and show the potential for endogenous modification and regulation. Additionally, many of the constitutively modified proteins showed an increase in S-nitrosothiol formation with GSNO treatment, further supporting a role for S-nitrosothiol formation in the regulation of protein function.
As a positive control, purified GAPDH was subjected to SNO-RAC analysis (Fig. 2). Three distinct S-nitrosothiol formation sites were identified, and these are consistent with the sites previously published by Forrester et al. (6). This technique is highly selective for S-nitrosylated proteins, as illustrated by the negative control experiments (no cysteine-containing peptides present when DTT or ascorbate was omitted, few cysteine containing peptides upon ascorbate pretreatment) and the low number of nonspecific peptide identifications (<3.6%). Therefore, our modified SNO-RAC protocol is very effective at identifying S-nitrosylated proteins and the specific sites of formation, especially compared with previous methodologies (31), even in complex samples.
Study limitations.
The methods used in the present study will tend to favor high-abundance protein identifications, which likely show higher levels of SNO. Consequently, lower abundance proteins and/or proteins with low levels of SNO may not be detected. Additionally, certain peptides just do not ionize well for one reason or another and, therefore, will not be detected by the mass spectrometer. As a result, the possibility exists for highly abundant/highly SNO peptides to be missed because of the limitations of MS. The use of a single concentration of one particular nitric oxide donor (GSNO) may also be a limiting factor in the number of SNO site identifications. In a previous study, our laboratory showed that GSNO has a dose-dependent effect on the activity of many different enzymes [i.e., sarco(endo)plasmic reticulum Ca2+-ATPase 2a, F1F0-ATPase, etc.], indicating that a dose dependency is likely to exist with GSNO-induced protein SNO (27). In the present study, we used a high concentration of GSNO (1 mmol/l) to identify all potential SNO sites. The use of other agents (i.e., S-nitroso-N-acetyl penicillamine, peroxynitrite, etc.) may also yield slightly different SNO profiles, resulting, in part, from differences in the target reactivity. The possibility also exists for the identification of S-nitrosylated proteins in the present study, which may or may not be S-nitrosylated under more physiological conditions. Furthermore, we demonstrated that Langendorff perfusion with GSNO increased SNO of many different proteins, as determined via two-dimensional gel electrophoresis (27). These same proteins were identified to be S-nitrosylated using the SNO-RAC protocol in our present study.
Conclusions.
This study provides novel information regarding the constitutive S-nitrosothiol proteome of the murine myocardium and also provides a large number of potential SNO sites. The modified version of the SNO-RAC protocol was very successful in the identification of nearly 1,000 unique S-nitrosylated proteins, ranging from aconitate hydratase to the sarcoplasmic reticulum Ca2+-ATPase. The diverse array of S-nitrosylated proteins identified indicates that S-nitrosothiol formation likely plays a critical role in the regulation of many different cellular functions within the myocardium, including myocardial contraction, metabolism, and cellular signaling. S-nitrosothiol formation is also likely to play an important role in cardioprotection, given that GSNO is a cardioprotective agent and appears to target many different proteins that are involved in cell death. Furthermore, this modified protocol can be applied to any organ or cell type and can also be used to examine SNO during disease states and with various drug treatments, thus yielding a valuable research tool for the identification of S-nitrosothiols.
GRANTS
This work was supported by NHLBI Grants 1F32HL096142 (M. J. Kohr) and 5R01HL039752 (C. Steenbergen), and the NHLBI/NIH Intramural Program (A. M. Aponte, E. Murphy, G. Wang, J. Sun, and M. Gucek).
DISCLOSURES
No conflicts of interest, financial or otherwise, are declared by the author(s).
Supplementary Material
ACKNOWLEDGMENTS
We thank the Proteomics Core Facility of the National Heart, Lung, and Blood Institute (NHLBI) for help with LC-MS/MS and MALDI-TOF/TOF peptide mass fingerprinting and label-free peptide quantification and analysis.
REFERENCES
- 1. Balligand JL, Ungureanu-Longrois D, Simmons WW, Pimental D, Malinski TA, Kapturczak M, Taha Z, Lowenstein CJ, Davidoff AJ, Kelly RA, et al. Cytokine-inducible nitric oxide synthase (iNOS) expression in cardiac myocytes. Characterization and regulation of iNOS expression and detection of iNOS activity in single cardiac myocytes in vitro. J Biol Chem 269: 27580– 27588, 1994 [PubMed] [Google Scholar]
- 2. Barouch LA, Harrison RW, Skaf MW, Rosas GO, Cappola TP, Kobeissi ZA, Hobai IA, Lemmon CA, Burnett AL, O'Rourke B, Rodriguez ER, Huang PL, Lima JA, Berkowitz DE, Hare JM. Nitric oxide regulates the heart by spatial confinement of nitric oxide synthase isoforms. Nature 416: 337– 339, 2002 [DOI] [PubMed] [Google Scholar]
- 3. Burwell LS, Nadtochiy SM, Tompkins AJ, Young S, Brookes PS. Direct evidence for S-nitrosation of mitochondrial complex I. Biochem J 394: 627– 634, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Ceneviva GD, Tzeng E, Hoyt DG, Yee E, Gallagher A, Engelhardt JF, Kim YM, Billiar TR, Watkins SA, Pitt BR. Nitric oxide inhibits lipopolysaccharide-induced apoptosis in pulmonary artery endothelial cells. Am J Physiol Lung Cell Mol Physiol 275: L717– L728, 1998 [DOI] [PubMed] [Google Scholar]
- 5. Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RA, Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J 430: 49– 59, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Forrester MT, Thompson JW, Foster MW, Nogueira L, Moseley MA, Stamler JS. Proteomic analysis of S-nitrosylation and denitrosylation by resin-assisted capture. Nat Biotechnol 27: 557– 559, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Fu C, Hu J, Liu T, Ago T, Sadoshima J, Li H. Quantitative analysis of redox-sensitive proteome with DIGE and ICAT. J Proteome Res 7: 3789– 3802, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Gonzalez DR, Beigi F, Treuer AV, Hare JM. Deficient ryanodine receptor S-nitrosylation increases sarcoplasmic reticulum calcium leak and arrhythmogenesis in cardiomyocytes. Proc Natl Acad Sci U S A 104: 20612– 20617, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Gow AJ, Chen Q, Hess DT, Day BJ, Ischiropoulos H, Stamler JS. Basal and stimulated protein S-nitrosylation in multiple cell types and tissues. J Biol Chem 277: 9637– 9640, 2002 [DOI] [PubMed] [Google Scholar]
- 10. Handy DE, Loscalzo J. Nitric oxide and posttranslational modification of the vascular proteome: S-nitrosation of reactive thiols. Arterioscler Thromb Vasc Biol 26: 1207– 1214, 2006 [DOI] [PubMed] [Google Scholar]
- 11. Hao G, Derakhshan B, Shi L, Campagne F, Gross SS. SNOSID, a proteomic method for identification of cysteine S-nitrosylation sites in complex protein mixtures. Proc Natl Acad Sci U S A 103: 1012– 1017, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Hess DT, Matsumoto A, Kim SO, Marshall HE, Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150– 166, 2005 [DOI] [PubMed] [Google Scholar]
- 13. Iwakiri Y, Satoh A, Chatterjee S, Toomre DK, Chalouni CM, Fulton D, Groszmann RJ, Shah VH, Sessa WC. Nitric oxide synthase generates nitric oxide locally to regulate compartmentalized protein S-nitrosylation and protein trafficking. Proc Natl Acad Sci U S A 103: 19777– 19782, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Jaffrey SR, Snyder SH. The biotin switch method for the detection of S-nitrosylated proteins. Sci STKE 2001: PL1, 2001 [DOI] [PubMed] [Google Scholar]
- 15. Kirsch M, Buscher AM, Aker S, Schulz R, de Groot H. New insights into the S-nitrosothiol-ascorbate reaction. The formation of nitroxyl. Org Biomol Chem 7: 1954– 1962, 2009 [DOI] [PubMed] [Google Scholar]
- 16. Lee JH, Yang ES, Park JW. Inactivation of NADP+-dependent isocitrate dehydrogenase by peroxynitrite. Implications for cytotoxicity and alcohol-induced liver injury. J Biol Chem 278: 51360– 51371, 2003 [DOI] [PubMed] [Google Scholar]
- 17. Lima B, Lam GK, Xie L, Diesen DL, Villamizar N, Nienaber J, Messina E, Bowles D, Kontos CD, Hare JM, Stamler JS, Rockman HA. Endogenous S-nitrosothiols protect against myocardial injury. Proc Natl Acad Sci U S A 106: 6297– 6302, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Lin J, Steenbergen C, Murphy E, Sun J. Estrogen receptor-beta activation results in S-nitrosylation of proteins involved in cardioprotection. Circulation 120: 245– 254, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Maejima Y, Adachi S, Morikawa K, Ito H, Isobe M. Nitric oxide inhibits myocardial apoptosis by preventing caspase-3 activity via S-nitrosylation. J Mol Cell Cardiol 38: 163– 174, 2005 [DOI] [PubMed] [Google Scholar]
- 20. Martin C, Schulz R, Post H, Boengler K, Kelm M, Kleinbongard P, Gres P, Skyschally A, Konietzka I, Heusch G. Microdialysis-based analysis of interstitial NO in situ: NO synthase-independent NO formation during myocardial ischemia. Cardiovasc Res 74: 46– 55, 2007 [DOI] [PubMed] [Google Scholar]
- 21. Martinez-Ruiz A, Lamas S. S-nitrosylation: a potential new paradigm in signal transduction. Cardiovasc Res 62: 43– 52, 2004 [DOI] [PubMed] [Google Scholar]
- 22. Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res 7: 51– 61, 2008 [DOI] [PubMed] [Google Scholar]
- 23. Murray CI, Kane LA, Uhrigshardt H, Wang SB, Van Eyk JE. Site-mapping of in vitro S-nitrosation in the cardiac mitochondrial: implications for cardioprotection. Mol Cell Proteomics. In press [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Perkins DN, Pappin DJ, Creasy DM, Cottrell JS. Probability-based protein identification by searching sequence databases using mass spectrometry data. Electrophoresis 20: 3551– 3567, 1999 [DOI] [PubMed] [Google Scholar]
- 25. Schulz R, Rassaf T, Massion PB, Kelm M, Balligand JL. Recent advances in the understanding of the role of nitric oxide in cardiovascular homeostasis. Pharmacol Ther 108: 225– 256, 2005 [DOI] [PubMed] [Google Scholar]
- 26. Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol 23: 1391– 1398, 2005 [DOI] [PubMed] [Google Scholar]
- 27. Sun J, Morgan M, Shen RF, Steenbergen C, Murphy E. Preconditioning results in S-nitrosylation of proteins involved in regulation of mitochondrial energetics and calcium transport. Circ Res 101: 1155– 1163, 2007 [DOI] [PubMed] [Google Scholar]
- 28. Sun J, Picht E, Ginsburg KS, Bers DM, Steenbergen C, Murphy E. Hypercontractile female hearts exhibit increased S-nitrosylation of the L-type Ca2+ channel alpha1 subunit and reduced ischemia/reperfusion injury. Circ Res 98: 403– 411, 2006 [DOI] [PubMed] [Google Scholar]
- 29. Sun J, Xin C, Eu JP, Stamler JS, Meissner G. Cysteine-3635 is responsible for skeletal muscle ryanodine receptor modulation by NO. Proc Natl Acad Sci U S A 98: 11158– 11162, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Tang ZL, Wasserloos KJ, Liu X, Stitt MS, Reynolds IJ, Pitt BR, St, Croix CM. Nitric oxide decreases the sensitivity of pulmonary endothelial cells to LPS-induced apoptosis in a zinc-dependent fashion. Mol Cell Biochem 234–235: 211– 217, 2002 [PubMed] [Google Scholar]
- 31. Torta F, Usuelli V, Malgaroli A, Bachi A. Proteomic analysis of protein S-nitrosylation. Proteomics 8: 4484– 4494, 2008 [DOI] [PubMed] [Google Scholar]
- 32. Wang G, Wu WW, Zeng W, Chou CL, Shen RF. Label-free protein quantification using LC-coupled ion trap or FT mass spectrometry: reproducibility, linearity, and application with complex proteomes. J Proteome Res 5: 1214– 1223, 2006 [DOI] [PubMed] [Google Scholar]
- 33. Wang H, Viatchenko-Karpinski S, Sun J, Gyorke I, Benkusky NA, Kohr MJ, Valdivia HH, Murphy E, Gyorke S, Ziolo M. Regulation of myocyte contraction via neuronal nitric oxide synthase: role of ryanodine receptor S-nitrosylation. J Physiol 588: 2905– 2917, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Wang W, Zhu W, Wang S, Yang D, Crow MT, Xiao RP, Cheng H. Sustained beta1-adrenergic stimulation modulates cardiac contractility by Ca2+/calmodulin kinase signaling pathway. Circ Res 95: 798– 806, 2004 [DOI] [PubMed] [Google Scholar]
- 35. Xu L, Eu JP, Meissner G, Stamler JS. Activation of the cardiac calcium release channel (ryanodine receptor) by poly-S-nitrosylation. Science 279: 234– 237, 1998 [DOI] [PubMed] [Google Scholar]
- 36. Ziolo MT. The fork in the nitric oxide road: cyclic GMP or nitrosylation? Nitric Oxide 18: 153– 156, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Ziolo MT, Kohr MJ, Wang H. Nitric oxide signaling and the regulation of myocardial function. J Mol Cell Cardiol 45: 625– 632, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.