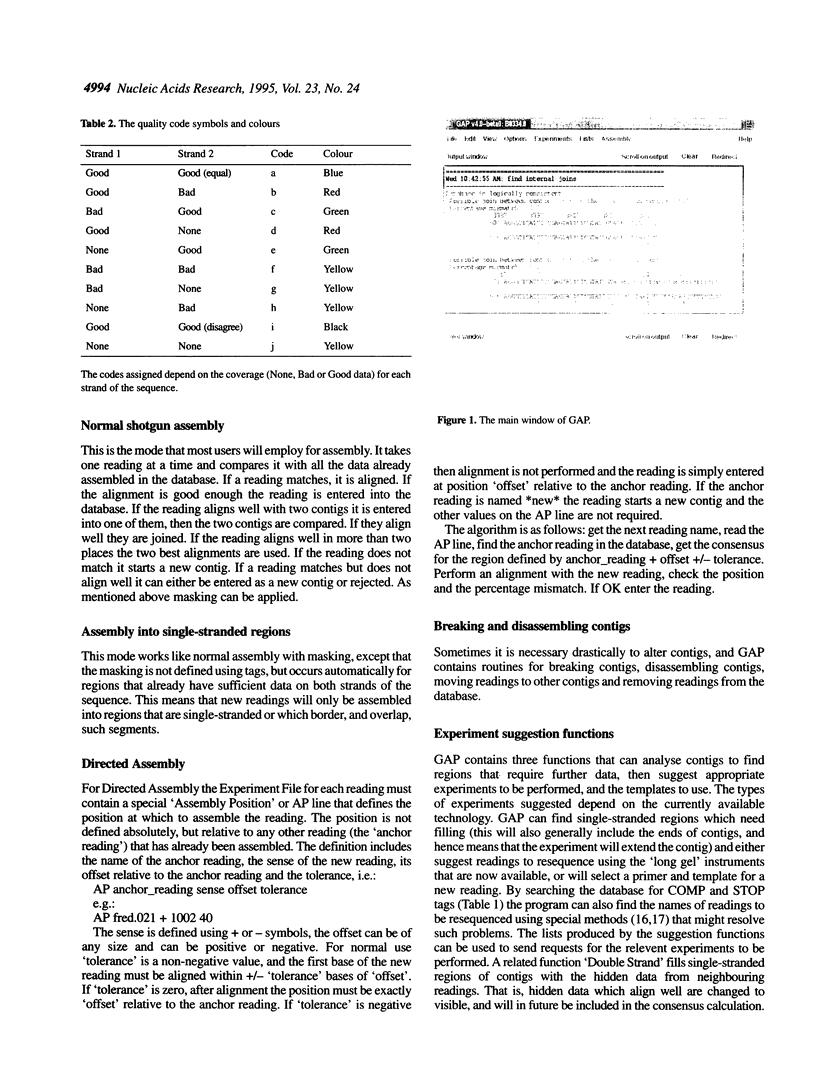
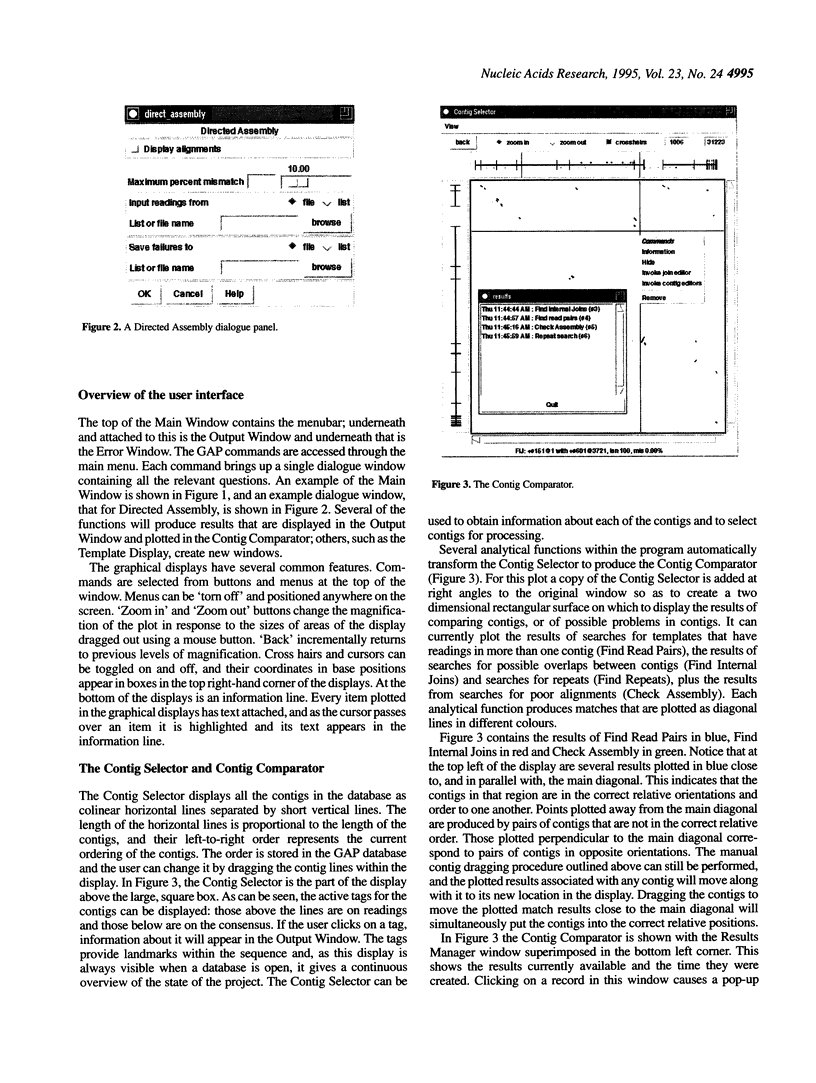
Abstract
We describe the Genome Assembly Program (GAP), a new program for DNA sequence assembly. The program is suitable for large and small projects, a variety of strategies and can handle data from a range of sequencing instruments. It retains the useful components of our previous work, but includes many novel ideas and methods. Many of these methods have been made possible by the program's completely new, and highly interactive, graphical user interface. The program provides many visual clues to the current state of a sequencing project and allows users to interact in intuitive and graphical ways with their data. The program has tools to display and manipulate the various types of data that help to solve and check difficult assemblies, particularly those in repetitive genomes. We have introduced the following new displays: the Contig Selector, the Contig Comparator, the Template Display, the Restriction Enzyme Map and the Stop Codon Map. We have also made it possible to have any number of Contig Editors and Contig Joining Editors running simultaneously even on the same contig. The program also includes a new 'Directed Assembly' algorithm and routines for automatically detecting unfinished segments of sequence, to which it suggests experimental solutions.
Full text
PDF





Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bonfield J. K., Staden R. The application of numerical estimates of base calling accuracy to DNA sequencing projects. Nucleic Acids Res. 1995 Apr 25;23(8):1406–1410. doi: 10.1093/nar/23.8.1406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear S., Staden R. A sequence assembly and editing program for efficient management of large projects. Nucleic Acids Res. 1991 Jul 25;19(14):3907–3911. doi: 10.1093/nar/19.14.3907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dear S., Staden R. A standard file format for data from DNA sequencing instruments. DNA Seq. 1992;3(2):107–110. doi: 10.3109/10425179209034003. [DOI] [PubMed] [Google Scholar]
- Gleizes A., Hénaut A. A global approach for contig construction. Comput Appl Biosci. 1994 Jul;10(4):401–408. doi: 10.1093/bioinformatics/10.4.401. [DOI] [PubMed] [Google Scholar]
- Hillier L., Green P. OSP: a computer program for choosing PCR and DNA sequencing primers. PCR Methods Appl. 1991 Nov;1(2):124–128. doi: 10.1101/gr.1.2.124. [DOI] [PubMed] [Google Scholar]
- Huang X. A contig assembly program based on sensitive detection of fragment overlaps. Genomics. 1992 Sep;14(1):18–25. doi: 10.1016/s0888-7543(05)80277-0. [DOI] [PubMed] [Google Scholar]
- Huang X. On global sequence alignment. Comput Appl Biosci. 1994 Jun;10(3):227–235. doi: 10.1093/bioinformatics/10.3.227. [DOI] [PubMed] [Google Scholar]
- Lawrence C. B., Honda S., Parrott N. W., Flood T. C., Gu L., Zhang L., Jain M., Larson S., Myers E. W. The genome reconstruction manager: a software environment for supporting high-throughput DNA sequencing. Genomics. 1994 Sep 1;23(1):192–201. doi: 10.1006/geno.1994.1477. [DOI] [PubMed] [Google Scholar]
- Lee L. G., Connell C. R., Woo S. L., Cheng R. D., McArdle B. F., Fuller C. W., Halloran N. D., Wilson R. K. DNA sequencing with dye-labeled terminators and T7 DNA polymerase: effect of dyes and dNTPs on incorporation of dye-terminators and probability analysis of termination fragments. Nucleic Acids Res. 1992 May 25;20(10):2471–2483. doi: 10.1093/nar/20.10.2471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers E. W., Miller W. Optimal alignments in linear space. Comput Appl Biosci. 1988 Mar;4(1):11–17. doi: 10.1093/bioinformatics/4.1.11. [DOI] [PubMed] [Google Scholar]
- Pearson W. R. Using the FASTA program to search protein and DNA sequence databases. Methods Mol Biol. 1994;25:365–389. doi: 10.1385/0-89603-276-0:365. [DOI] [PubMed] [Google Scholar]
- Peltola H., Söderlund H., Ukkonen E. SEQAID: a DNA sequence assembling program based on a mathematical model. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 1):307–321. doi: 10.1093/nar/12.1part1.307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prober J. M., Trainor G. L., Dam R. J., Hobbs F. W., Robertson C. W., Zagursky R. J., Cocuzza A. J., Jensen M. A., Baumeister K. A system for rapid DNA sequencing with fluorescent chain-terminating dideoxynucleotides. Science. 1987 Oct 16;238(4825):336–341. doi: 10.1126/science.2443975. [DOI] [PubMed] [Google Scholar]
- Smith S., Welch W., Jakimcius A., Dahlberg T., Preston E., Van Dyke D. High throughput DNA sequencing using an automated electrophoresis analysis system and a novel sequence assembly program. Biotechniques. 1993 Jun;14(6):1014–1018. [PubMed] [Google Scholar]
- Staden R. A computer program to enter DNA gel reading data into a computer. Nucleic Acids Res. 1984 Jan 11;12(1 Pt 2):499–503. doi: 10.1093/nar/12.1part2.499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staden R. Automation of the computer handling of gel reading data produced by the shotgun method of DNA sequencing. Nucleic Acids Res. 1982 Aug 11;10(15):4731–4751. doi: 10.1093/nar/10.15.4731. [DOI] [PMC free article] [PubMed] [Google Scholar]